Introduction
Colorectal cancer (CRC) is one of the most commonly
diagnosed malignancies, and in recent years, the incidence rate of
CRC has shown an increasing tendency; it was estimated that there
were >1.2 million newly diagnosed patients with CRC and ~600,000
patients died due to CRC in 2019, worldwide (1). However, the pathogenesis of CRC remains
unclear, and current standard treatments for CRC remain a
combination of surgery, chemotherapy and radiotherapy; in numerous
cases, none of the current treatment methods achieve the desired
therapeutic effects, and the prognosis of patients with CRC remains
poor (1–3). Therefore, there is an urgent need to
identify novel therapeutic targets and develop new methods for the
treatment of CRC with improved therapeutic efficacy.
In recent years, the roles of microRNA (miRNAs or
miRs) in carcinogenesis have been investigated in numerous studies
(4,5). miRNAs are endogenous non-coding mRNAs
with a length of 18–25 nucleotides, which can bind to the
3′-untranslated region (UTR) of mRNAs and suppress the expression
of their target proteins at the post-transcriptional level
(6). In the field of CRC, miRNAs
have been reported to participate in the occurrence and development
of the disease, either as tumor suppressors or promoters (7,8). For
example, it has been observed that miR-503 can promote the
migration and invasion of CRC cells via targeting programmed cell
death 4 (PDCD4) (9). By contrast,
miR-185 has been reported to enhance the radiosensitivity of CRC
cells (10). These results suggest
that miRNAs are promising therapeutic targets for CRC. miR-429 is a
member of the miR-200 family, and we previously reported the tumor
suppressive role of miR-429 in colon cancer cell lines (11). High mobility group box 3 (HMGB3) has
been reported to be an oncogene (12–15).
However, to the best of our knowledge, the molecular mechanisms
underlying the association between miR-429 and HMGB3 in CRC are yet
to be elucidated.
The present study aimed to identify the association
between miR-429 and HMGB3 in CRC and the underlying mechanism.
Materials and methods
Patients and samples
A total of 65 patients with primary CRC who
underwent curative resection at Heping Hospital (Changzhi, China)
were recruited between May 2016 and July 2017. Patients who did not
receive preoperative neoadjuvant therapy were included in the
study. The main characteristics of these 65 patients are presented
in Table I. Fresh frozen tumor
tissue and adjacent normal tissue samples (>5 cm from the margin
of the tumors) were collected from each patient. The experiments
were performed with the understanding and written informed consent
of each patient, and the investigation was performed in accordance
with the Declaration of Helsinki. The present study was approved by
the Medical Ethics Committee of Heping Hospital, Changzhi Medical
College.
 | Table I.Main characteristics of the patients
included in the study (n=65). |
Table I.
Main characteristics of the patients
included in the study (n=65).
| Characteristic | Number of
patients |
|---|
| Han nationality |
|
| Yes | 65 |
| No | 0 |
| Family history |
|
| Yes | 0 |
| No | 65 |
| Smoking |
|
| Yes | 0 |
| No | 65 |
| Drinking |
|
| Yes | 0 |
| No | 65 |
| Sex |
|
| Male | 38 |
|
Female | 27 |
| Age, years |
|
|
>50 | 47 |
| ≤50 | 18 |
| Age range, years | 45–71 |
| Median age,
years | 58 |
| Tumor site |
|
|
Colon | 39 |
|
Rectum | 26 |
| Tumor size, cm |
|
|
<3 | 34 |
| ≥3 | 31 |
| Number of
lesions |
|
| 1 | 15 |
|
>1 | 31 |
| Not
available | 19 |
| TNM stage |
|
| I–II | 22 |
|
III–IV | 43 |
| Differentiation |
|
|
Well/moderate | 39 |
| Poor | 26 |
| Lymphatic
metastasis |
|
|
Positive | 27 |
|
Negative | 38 |
| Preoperative
neoadjuvant therapy |
|
|
Yes | 0 |
| No | 65 |
Cell culture and transfection
The human CRC LOVO cell line was obtained from the
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. LOVO cells were cultured in high-glucose Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (Cytiva) and 1%
penicillin/streptomycin (Thermo Fisher Scientific, Inc.), and
maintained in a humidified incubator at 37°C with 5%
CO2. DNA constructs, including miR-429 mimics
(5′-UGCCAAAAUGGUCUGUCAUAAU-3′), miR-429 mimic negative control (NC;
scrambled sequence; 5′-UUCUCCGAACGUGUCACGUTT-3′) and HMGB3
overexpressing plasmid (empty vector was used as NC), were designed
by Shanghai GenePharma Co., Ltd. miR-429 mimics and miR-429 mimic
NC sequences were ligated into a pGCMV vector. The cells were
transfected with Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), using plasmid DNA
(1–2 µg). The cells were transfected with miR-429 mimics or miR-429
mimics in combination with HMGB3 overexpressing plasmids. The
transfected cells were incubated at 37°C with 5% CO2,
and the subsequent experiments were performed 48 h after
transfection. The successful transfections of miR-429 mimics and
HMGB3 overexpressing constructs in LOVO cells are shown in Fig. S1. Researchers performing the
aforementioned assays were blinded to sample information, and all
experiments were repeated with at least three independent culture
preparations.
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8) cell proliferation
assay (Sigma-Aldrich; Merck KGaA) was performed to detect the
viability of cells at 12, 24 and 48 h post-transfection. LOVO cells
were seeded onto 96-well plates at 1×103 cells/well and
incubated with 100 µl DMEM containing 10 µl CCK-8 solution at 37°C
for 2 h at each time point. The optical density value at 450 nm was
then examined to determine the proliferative ability of the
cells.
Apoptosis assay
The apoptosis of LOVO cells was determined using an
Annexin V apoptosis detection kit (Nanjing KeyGen Biotech Co.,
Ltd.). Briefly, cells were stained with Annexin V-FITC and
propidium iodide (PI) for 10–15 min at room temperature, and the
apoptosis of cells was examined using a BD FACSCalibur flow
cytometer (BD Biosciences). The experimental results were analyzed
using FlowJo version 10.4 (FlowJo LLC). The apoptosis between
different groups was compared and illustrated in results at 48 h
post-transfection.
Bioinformatics analysis
TargetScan (http://www.targetScan.org/vert_72) predicts biological
targets of miRNAs by searching for the presence of conserved 8-mer,
7-mer and 6-mer sites that match the seed region of each miRNA. The
human HMGB3 gene was searched on the website, with miR-429 as the
microRNA.
Dual-luciferase reporter assay
293 cells (Cell Bank of Type Culture Collection of
the Chinese Academy of Sciences) were used for the dual-luciferase
reporter assay. The cells were cultured in DMEM supplemented with
10% fetal calf serum (Cytiva). pGL3 plasmids (Promega Corporation)
containing the wild-type (WT) 3′-UTR of HMGB3 or mutated (MT)
3′-UTR of HMGB3 were constructed. WT or MT plasmids were
co-transfected with miR-429 mimics or NC using Lipofectamine 3000
reagent. Cells were harvested 24 h later, and luciferase activity,
which was compared with Renilla luciferase activity and was
determined using a dual luciferase assay system (Promega
Corporation) according to the manufacturer's protocol.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissue and cell samples
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. RT was
performed using PrimeScript™ RT Master mix (Perfect Real Time) kit
(Takara Bio, Inc.) according to the manufacturer's protocol. qPCR
was subsequently performed using the Mir-X™ miRNA qRT-PCR
SYBR® kit (Takara Bio, Inc.) for miR-429 mRNA expression
(with U6 used as the internal reference gene) and SYBR Green qPCR
Master mix (Takara Bio, Inc.) for HMGB3 mRNA expression (with GAPDH
used as the internal reference gene). The following thermocycling
conditions were used: Initial denaturation at 95°C for 15 sec
followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. All
reactions were performed in duplicates, and the comparative
2−ΔΔCq method was used for comparisons between samples
unless otherwise stated (16).
Primers are listed in Table SI.
Western blot analysis
Tissues and cell samples were harvested using
radioimmunoprecipitation lysis buffer (Beyotime Institute of
Biotechnology), and the protein concentration was determined using
a BCA Protein assay kit (Beyotime Institute of Biotechnology).
Total protein was then separated using 10% SDS-PAGE, and
transferred to a PVDF membrane. Subsequently the membrane was
blocked with 5% skimmed milk in PBS for 4 h. Next, the membrane was
incubated with specific primary antibodies for 3 h at room
temperature. After being washed with PBS three times, for 5 min
each time, the membrane was incubated with the appropriate
secondary antibody for 2 h at room temperature. After washing with
PBS three times, for 5 min each time, an enhanced chemiluminescence
western blotting kit (Cytiva) was used to detect the immune
complexes on the membrane. The following antibodies were used:
Anti-HMGB3 (1:1,000; cat. no. PA5-68709; Thermo Fisher Scientific,
Inc.), anti-GAPDH (1:2,000; cat. no. ab8227; Abcam) and HRP goat
anti-rabbit (1:2,000; cat. no. ab7090; Abcam).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism (version 6.01; GraphPad Software, Inc.). Data are presented
as the mean ± standard deviation. The differences between two
groups were compared by paired or unpaired t-test, and the
differences among multiple groups were analyzed by one-way analysis
of variance with Turkey's post-hoc test. Pearson's correlation
coefficient was used to determine the correlation between miR-429
and HMGB3 expression in tumor samples. P<0.05 was considered to
indicate a statistically significant difference.
Results
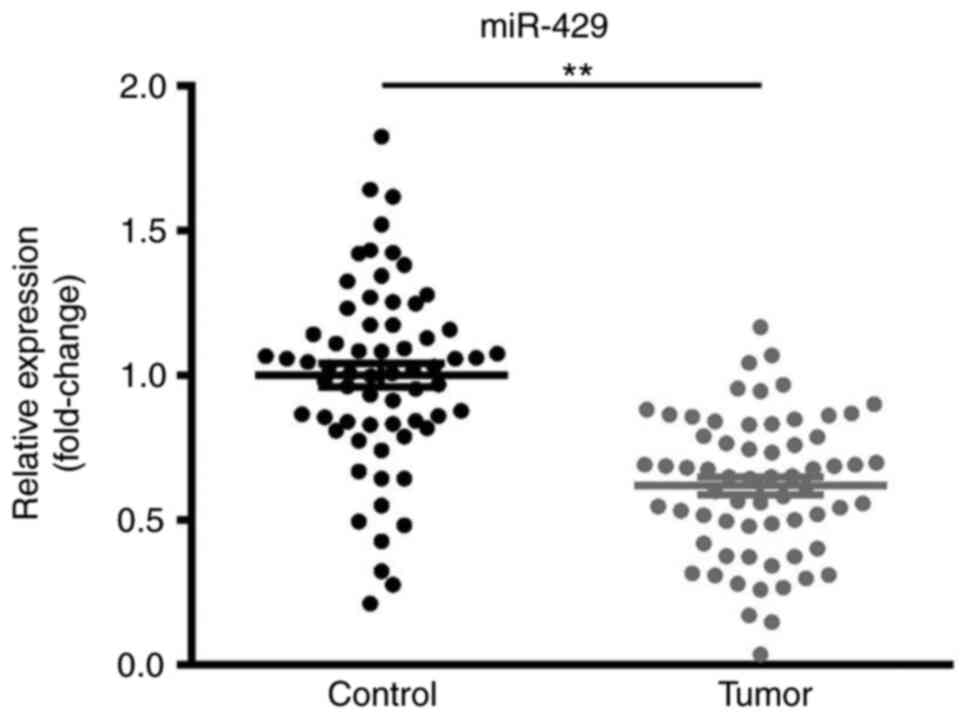
Downregulation of miR-429 in CRC
tissue samples
The expression levels of miR-429 in CRC and matched
adjacent non-cancer tissues were examined by RT-qPCR. As presented
in Fig. 1, miR-429 expression was
significantly decreased in CRC tissues compared with in the
corresponding non-cancer colorectal tissues.
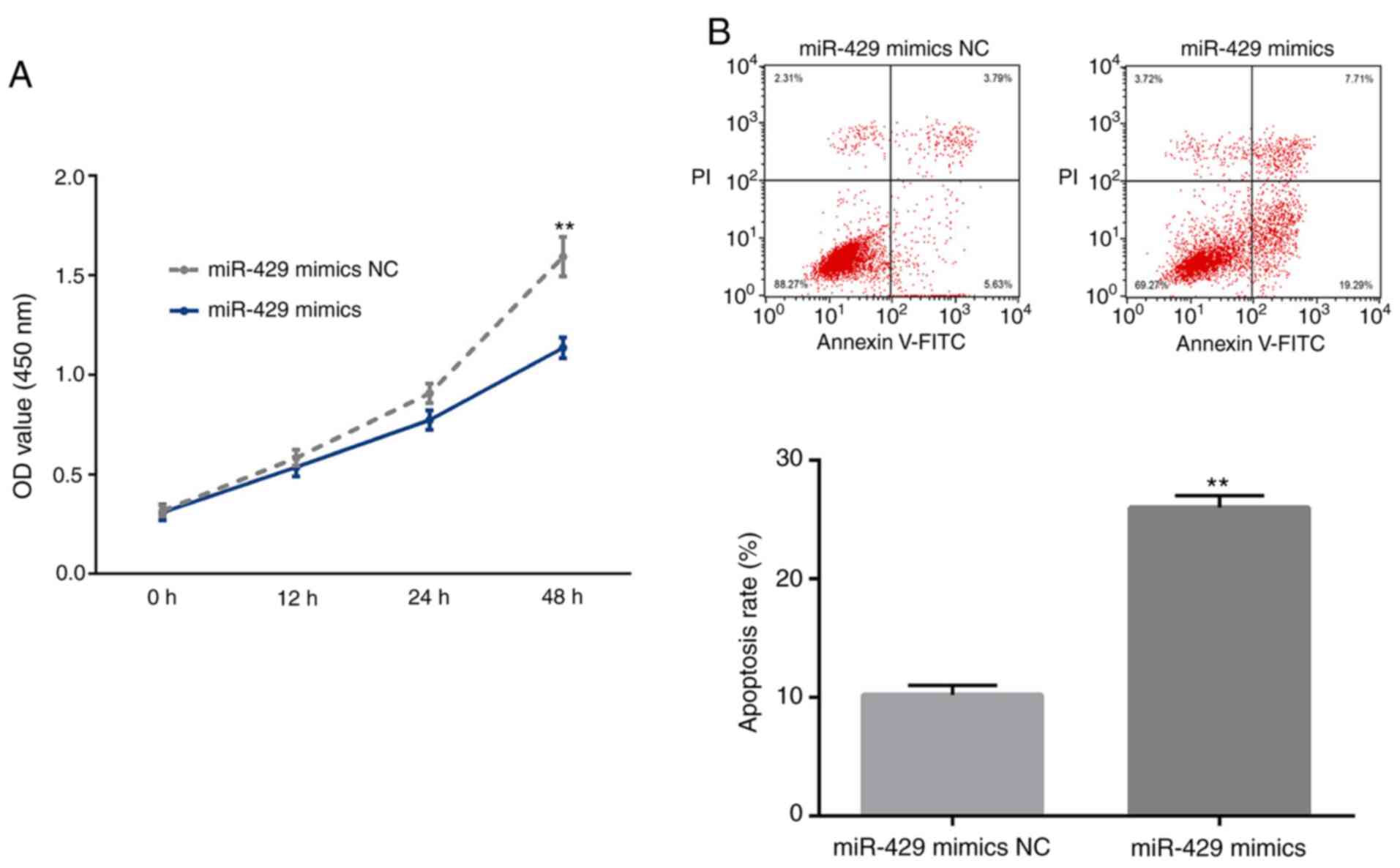
Tumor suppressive function of miR-429
in LOVO cells
The tumor suppressive role of miR-429 in CRC has
been reported previously (11). The
present study further validated these previous findings by
investigating the effects of miR-429 on the proliferation and
apoptosis of LOVO cells. As a result, transient overexpression of
miR-429 induced a significant decrease in the proliferation and a
significant increase in the apoptosis of LOVO cells (Fig. 2A and B).
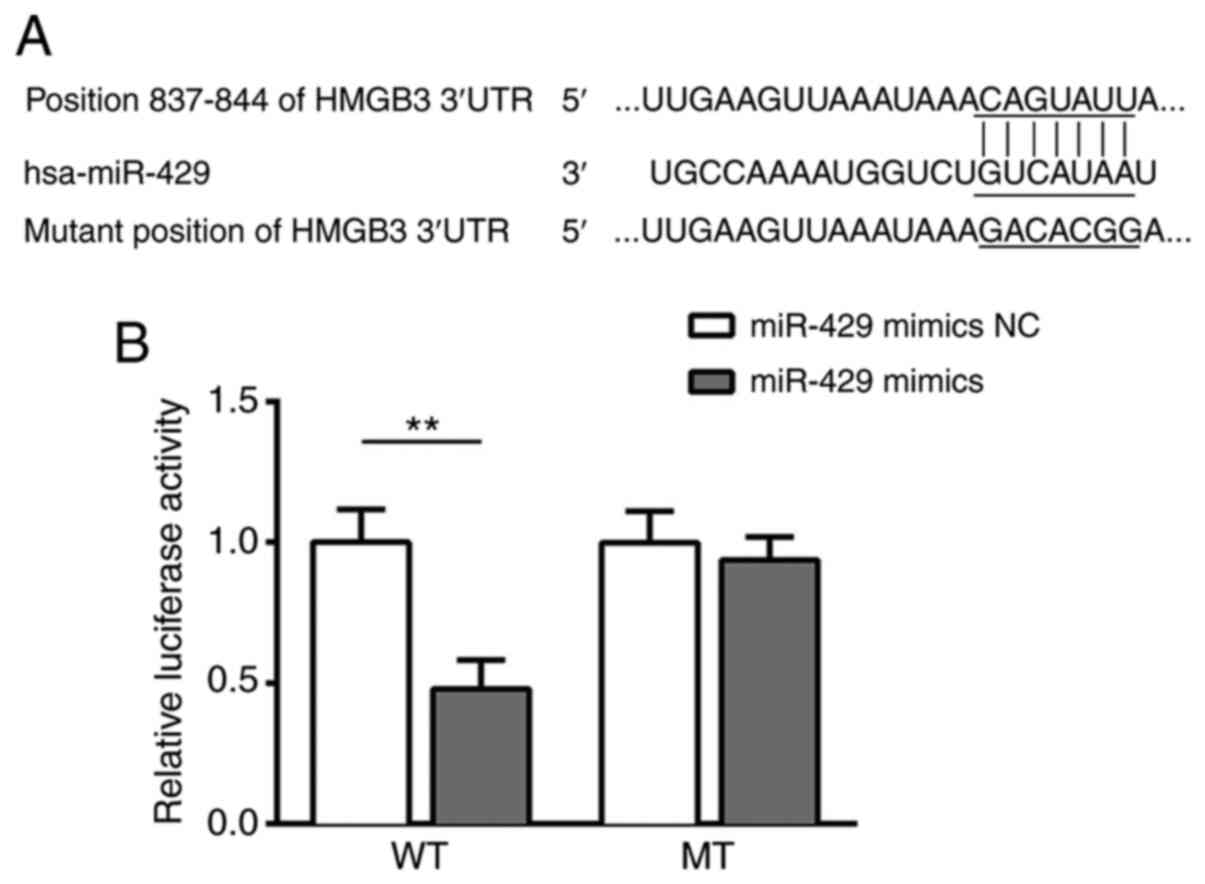
HMGB3 is a direct target of
miR-429
Using the online bioinformatics tool Targetscan,
HMGB3 was predicted as a direct target of miR-429 (Fig. 3A). To confirm the association between
miR-429 and HMGB3, a dual-luciferase reporter assay was conducted.
Notably, luciferase activity was significantly decreased in 293
cells co-transfected with the reporter containing the WT 3′-UTR of
HMGB3 and miR-429 mimics, while co-transfection of the reporter
containing MUT 3′-UTR of HMGB3 and miR-429 mimics did not affect
luciferase activity (Fig. 3B). These
results provided direct evidence that HMGB3 was a target of
miR-429.
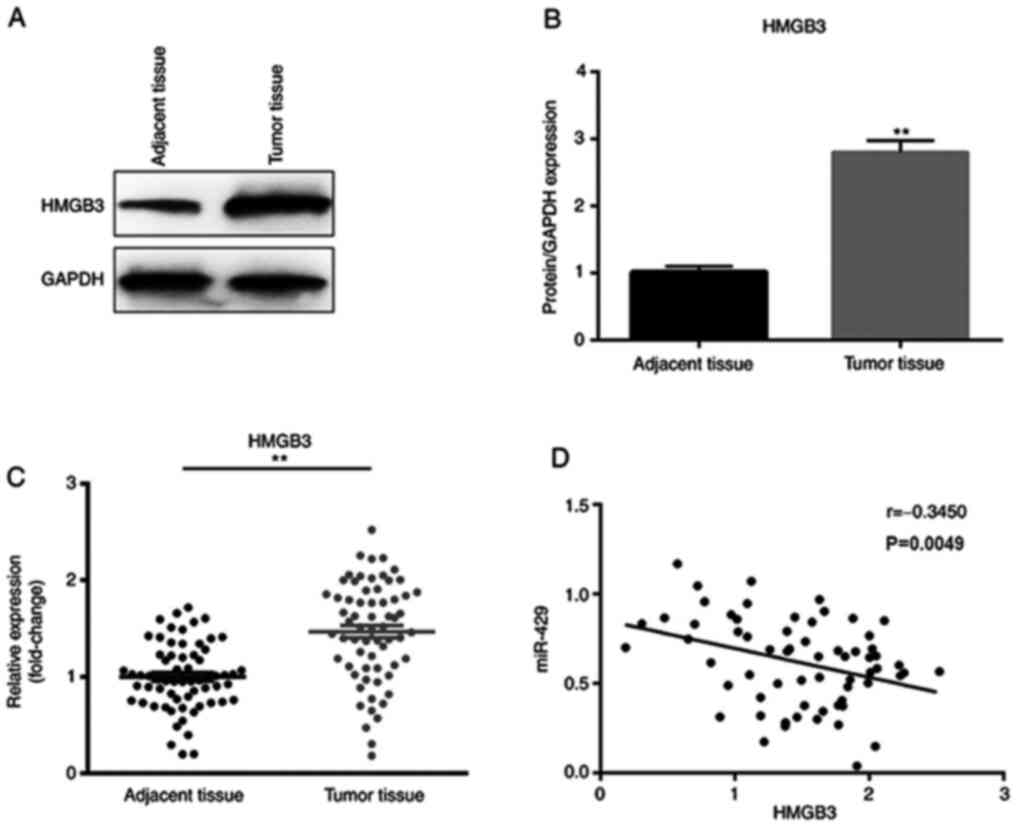
Expression levels of miR-429 and HMGB3
are negatively correlated in CRC tissue samples
To further investigate the association between
miR-429 and HMGB3 in CRC, the expression levels of HMGB3 in CRC
tissues and matched adjacent non-cancer tissues were examined. It
was observed that HMGB3 was significantly upregulated in CRC tumor
tissues compared with in the adjacent tissues at both the protein
and mRNA levels (Fig. 4A-C).
Furthermore, the expression levels of HMGB3 were negatively
correlated with those of miR-429 in CRC tissue samples (Fig. 4D).
HMGB3 is involved in the antitumor
effect of miR-429
To further identify whether HMGB3 is involved in the
antitumor effect of miR-429, LOVO cells were divided into the
following three groups according to the transfection performed: i)
Control group; ii) miR-429 mimics group; and iii) miR-429 mimics
and HMGB3 overexpression plasmid group. The protein expression
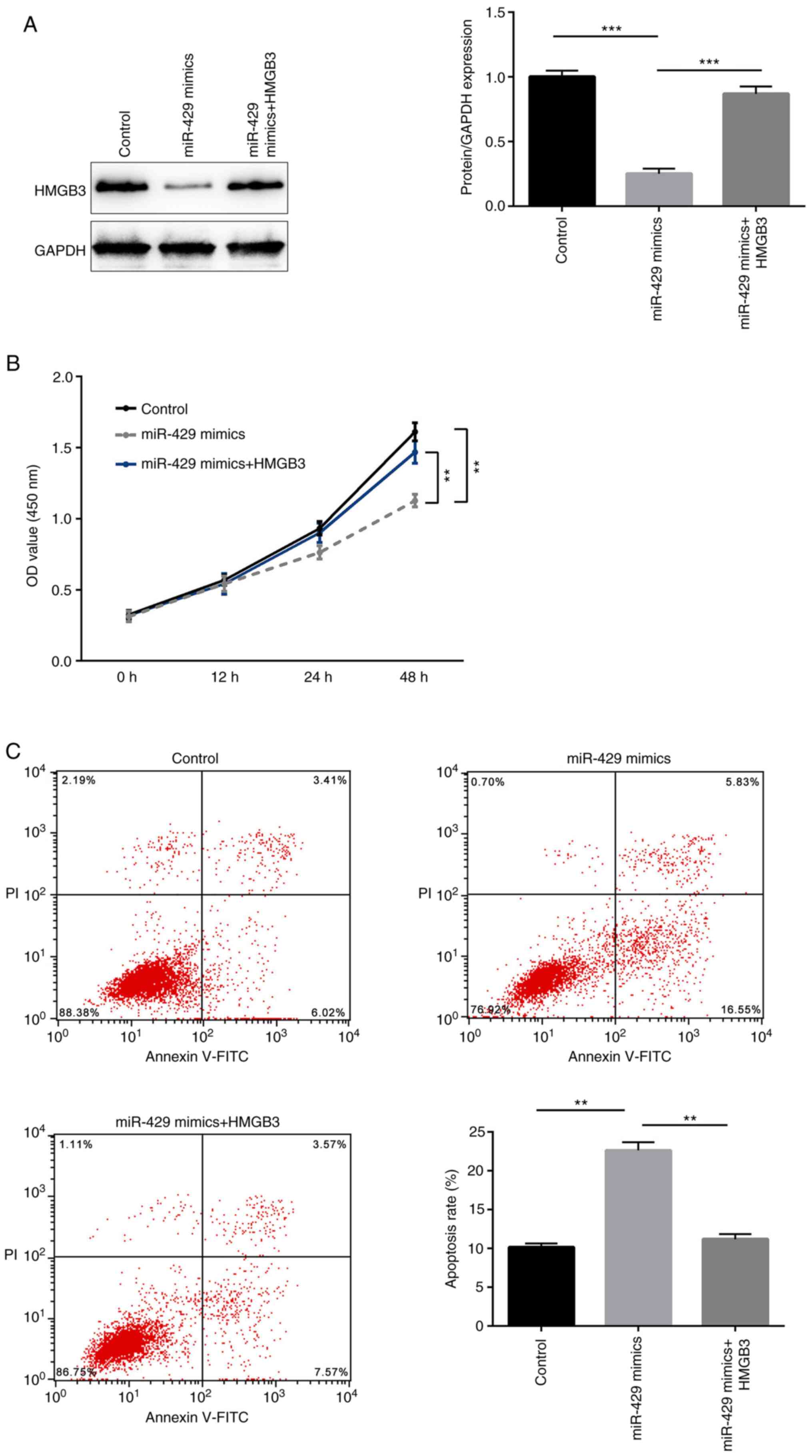
levels of HMGB3 in each group were determined by western blotting.
It was demonstrated that transfection with miR-429 mimics
significantly decreased the protein expression levels of HMGB3,
which were then restored by HMGB3 overexpression (Fig. 5A). Furthermore, transient
overexpression of miR-429 mimics induced a significant decrease in
the proliferation and a significant increase in the apoptosis of
LOVO cells, while concomitant overexpression of HMGB3 significantly
rescued these tumor suppressive effects of miR-429 (Fig. 5B and C). Thus, it was demonstrated
for the first time that miR-429 functioned as a tumor suppressor in
CRC via targeting HMGB3.
Discussion
CRC is one of the most common and lethal
malignancies worldwide. Therefore, there is an urgent requirement
to improve the understanding of the pathological mechanisms of CRC
and explore more effective methods for CRC diagnosis and treatment.
miR-429 exerts a tumor suppressive function in CRC (11); however, the molecular mechanism
remains unclear. The present study investigated miR-429 and its
target gene HMGB3, and their underlying mechanism in the
progression of CRC.
The role of miR-429 in CRC is not clearly defined.
Han et al (17) observed that
miR-429 expression was upregulated in CRC tissue compared with that
in adjacent non-tumor tissue, and increased the proliferation and
migration of CRC cells by targeting homeobox A5, a transcription
factor of HOX families. Comparatively, Sun et al (18) suggested that miR-429 inhibits cell
proliferation and invasion, and regulates epithelial-mesenchymal
transition-associated marker genes by targeting Onecut2 in CRC. In
addition, we previously reported that miR-429 suppresses the
migration and invasion of CRC cells by targeting p21-activated
kinase 6/cofilin signaling (11).
These differences may be a result of the limited sample size. The
present study identified that miR-429 expression was downregulated
in CRC tissues compared with that in adjacent tissues, which was
consistent with the results of Sun et al (18) and our previous study (11). Furthermore, the present study
demonstrated that transient overexpression of miR-429 significantly
decreased the proliferation and promoted the apoptosis of CRC cells
in vitro. Overall, the present results indicated the tumor
suppressive function of miR-429 in CRC.
It is well-known that miRNAs exert their function
through suppressing the expression of their target genes. HMGB3
belongs to the HMGB family, which consists of HMGB1, HMGB2, HMGB3
and HMGB4 (19). Increasing evidence
has demonstrated that HMGB3 may act as an oncogene in different
types of cancer, including non-small cell lung cancer (20), breast cancer (21), esophageal squamous cell cancer
(22), gastric cancer (23) and CRC (24). Notably, the current study confirmed
that HMGB3 was a target gene of miR-429 using a dual-luciferase
reporter assay. Furthermore, it was identified that HMGB3
expression was upregulated in CRC tissues compared with in adjacent
tissues, which was consistent with the findings of Zhang et
al (24). Notably, the present
study first reported the negative correlation between the
expression levels of miR-429 and HMGB3 in CRC tissue samples. In
addition, it was identified that transfection of LOVO cells with
miR-429 mimics significantly decreased HMGB3 expression, and
transient overexpression of HMGB3 blocked the antitumor effects of
miR-429. Thus, miR-429 may act as a tumor suppressor in the
progression of CRC via suppressing HMGB3 expression. Further
studies are required to explore the upstream regulatory molecules
of miR-429 in CRC progression and verify whether the miR-429/HMGB3
signaling pathway is associated with patient outcome.
Overall, the present study investigated the function
of miR-429 in CRC progression. Using correlation analysis and a
dual-luciferase reporter assay, it was identified that HMGB3 acted
as a direct target of miR-429. In addition, it was observed that
miR-429 regulated the proliferation and apoptosis of CRC cells via
targeting HMGB3. How precisely the regulation occurs and/or whether
miR-429 interacts with other known signaling pathways regulating
CRC cell proliferation and apoptosis remains to be determined in
future studies. Meanwhile, other colon cancer cell lines should be
used to further verify the tumor suppressive role of the
miR-429/HMGB3 signaling pathway in the future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Supporting Fund
for Scientific Research of Changzhi Medical College (grant no.
QDZ201610).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JY and SW conceived the study and designed the
experiments. XT performed the experiments and analyzed the data. HL
conceived the study, analyzed the data and wrote the manuscript. JC
and NZ collected samples and analyzed the clinical data. All
authors read and approved the final manuscript. JY and SW confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
Medical Ethics Committee of Heping Hospital (Changzhi, China), and
with the 1964 Declaration of Helsinki and its later amendments or
comparable ethical standards. Written informed consent was obtained
from all individual participants that were included in the present
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gormley JA, Hegarty SM, O'Grady A,
Stevenson MR, Burden RE, Barrett HL, Scott CJ, Johnston JA, Wilson
RH, Kay EW, et al: The role of cathepsin S as a marker of prognosis
and predictor of chemotherapy benefit in adjuvant CRC: A pilot
study. Br J Cancer. 105:1487–1494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nikolouzakis TK, Vassilopoulou L,
Fragkiadaki P, Mariolis Sapsakos T, Papadakis GZ, Spandidos DA,
Tsatsakis AM and Tsiaoussis J: Improving diagnosis, prognosis and
prediction by using biomarkers in CRC patients (Review). Oncol Rep.
39:2455–2472. 2018.PubMed/NCBI
|
|
3
|
Wu J, Wang F, Liu X, Zhang T, Liu F, Ge X,
Mao Y and Hua D: Correlation of IDH1 and B7H3 expression with
prognosis of CRC patients. Eur J Surg Oncol. 44:1254–1260. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Costa DF and Torchilin VP: Micelle-like
nanoparticles as siRNA and miRNA carriers for cancer therapy.
Biomed Microdevices. 20:592018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu B, Liu J, Xiang X, Liu S, Zhong P, Xie
F, Mou T and Lai L: Expression of miRNA-143 in pancreatic cancer
and its clinical significance. Cancer Biother Radiopharm.
33:373–379. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen Y, Zhao J, Luo Y, Wang Y and Jiang Y:
Downregulated expression of miRNA-149 promotes apoptosis in side
population cells sorted from the TSU prostate cancer cell line.
Oncol Rep. 36:2587–2600. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ju H, Tan JY, Cao B, Song MQ and Tian ZB:
Effects of miR-223 on colorectal cancer cell proliferation and
apoptosis through regulating FoxO3a/BIM. Eur Rev Med Pharmacol Sci.
22:3771–3778. 2018.PubMed/NCBI
|
|
8
|
Su M, Qin B, Liu F, Chen Y and Zhang R:
miR-885-5p upregulation promotes colorectal cancer cell
proliferation and migration by targeting suppressor of cytokine
signaling. Oncol Lett. 16:65–72. 2018.PubMed/NCBI
|
|
9
|
Li L, Zhang X, Yi Z, Liang X, Yin W and Li
S: MiR-503 promotes the migration and invasion of colorectal cancer
cells by regulating PDCD4. J BUON. 23:579–586. 2018.PubMed/NCBI
|
|
10
|
Afshar S, Najafi R, Sedighi Pashaki A,
Sharifi M, Nikzad S, Gholami MH, Khoshghadam A, Amini R, Karimi J
and Saidijam M: MiR-185 enhances radiosensitivity of colorectal
cancer cells by targeting IGF1R and IGF2. Biomed Pharmacother.
106:763–769. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tian X, Wei Z, Wang J, Liu P, Qin Y and
Zhong M: MicroRNA-429 inhibits the migration and invasion of colon
cancer cells by targeting PAK6/cofilin signaling. Oncol Rep.
34:707–714. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Petit A, Ragu C, Della-Valle V,
Mozziconacci MJ, Lafage-Pochitaloff M, Soler G, Schluth C, Radford
I, Ottolenghi C, Bernard OA, et al: NUP98-HMGB3: A novel oncogenic
fusion. Leukemia. 24:654–658. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo S, Wang Y, Gao Y, Zhang Y, Chen M, Xu
M, Hu L, Jing Y, Jing F, Li C, et al: Knockdown of high mobility
group-box 3 (HMGB3) expression inhibits proliferation, reduces
migration, and affects chemosensitivity in gastric cancer cells.
Med Sci Monit. 22:3951–3960. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li M, Cai Y, Zhao H, Xu Z, Sun Q, Luo M,
Gu L, Meng M, Han X and Sun H: Overexpression of HMGB3 protein
promotes cell proliferation, migration and is associated with poor
prognosis in urinary bladder cancer patients. Tumour Biol.
36:4785–4792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamada Y, Nishikawa R, Kato M, Okato A,
Arai T, Kojima S, Yamazaki K, Naya Y, Ichikawa T and Seki N:
Regulation of HMGB3 by antitumor miR-205-5p inhibits cancer cell
aggressiveness and is involved in prostate cancer pathogenesis. J
Hum Genet. 63:195–205. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han Y, Zhao Q, Zhou J and Shi R: miR-429
mediates tumor growth and metastasis in colorectal cancer. Am J
Cancer Res. 7:218–233. 2017.PubMed/NCBI
|
|
18
|
Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu
Z, Li X and Wu M: MiR-429 inhibits cells growth and invasion and
regulates EMT-related marker genes by targeting Onecut2 in
colorectal carcinoma. Mol Cell Biochem. 390:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li L, Huang J, Ju Z, Li Q, Wang C, Qi C,
Zhang Y, Hou Q, Hang S and Zhong J: Multiple promoters and targeted
microRNAs direct the expressions of HMGB3 gene transcripts in dairy
cattle. Anim Genet. 44:241–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song N, Liu B, Wu JL, Zhang RF, Duan L, He
WS and Zhang CM: Prognostic value of HMGB3 expression in patients
with non-small cell lung cancer. Tumour Biol. 34:2599–2603. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Elgamal OA, Park JK, Gusev Y,
Azevedo-Pouly AC, Jiang J, Roopra A and Schmittgen TD: Tumor
suppressive function of mir-205 in breast cancer is linked to HMGB3
regulation. PLoS One. 8:e764022013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao J, Zou Z, Gao J, Zhang H, Lin Z, Zhang
Y, Luo X, Liu C, Xie J and Cai C: Increased expression of HMGB3: A
novel independent prognostic marker of worse outcome in patients
with esophageal squamous cell carcinoma. Int J Clin Exp Pathol.
8:345–352. 2015.PubMed/NCBI
|
|
23
|
Gong Y, Cao Y, Song L, Zhou J, Wang C and
Wu B: HMGB3 characterization in gastric cancer. Genet Mol Res.
12:6032–6039. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Z, Chang Y, Zhang J, Lu Y, Zheng L,
Hu Y, Zhang F and Li X, Zhang W and Li X: HMGB3 promotes growth and
migration in colorectal cancer by regulating WNT/β-catenin pathway.
PLoS One. 12:e01797412017. View Article : Google Scholar : PubMed/NCBI
|