Non-small cell lung cancer (NSCLC) is one of the
most prevalent malignant tumors and accounts for ~85% of the lung
cancer related deaths globally (1).
As reported in 2017, lung cancer related deaths in Europe were the
leading cause of cancer deaths in both sexes, accounting for 24%
male deaths and 15% female deaths (2,3). Data
from 2017 predicted a 10.7% fall in 5 years for males
(corresponding to 33.3 deaths per one lakh residents), individuals
for females a 5.1% increment (accounting for 14.6 deaths per one
lakh individuals) (1–3). Unfortunately, current therapies against
NSCLCs are ineffective due to the advanced stage tumor progression
at diagnosis and post therapy relapses (4,5). Within
the USA and Europe, the 5-year overall survival rate of patients
with NSCLC is only 13–17% (6).
Numerous studies have indicated that sex disparities
exist in the development and complications of lung cancer (7–16). For
instance, Jemal et al (7)
reported higher lung cancer susceptibility in young females
compared with males in the USA. Possible reasons for this disparity
include sex distinctions in genetics and epigenetics (8,9), sex
hormone levels (10,11), sex hormone receptors levels (12), post-menopausal hormone replacement
therapy (13,14) and smoking history (15,16).
Racial and ethnic differences also contribute to the development
and complications of lung cancer (17,18).
In non-smokers with NSCLC, biomarkers including
epidermal growth factor receptor (EGFR), ELK (Ets like
transcription factor-1; highly expressed in NSCLC, irrespective of
patient's age, sex, smoking status and histology) and KRAS
mutations are more frequently observed in women compared with men
(21,22). These mutations mostly occur in
adenocarcinoma (23). Notably, women
exhibit greater benefit compared with men when treated with EGFR
inhibitors (24). In contrast, women
have less benefit from anti programmed death 1 inhibitors compared
with men (25). There is no sex
distinction in response to ALK (anaplastic lymphoma kinase)
inhibitors (26). Of note, ALK
inhibitors are anticancer drugs which act on tumours with ALK
varied expressions (27). ALK
inhibitors are tyrosine kinase inhibitors and act by inhibiting the
proteins responsible for abnormal tumour cell growth (28). The higher response rate to anti-EGFR
in women may be due to a greater intrinsic EGFR expression
(9,29). Notably, female smokers exhibit a
higher likelihood of developing lung cancer compared with males
(15). The higher female
susceptibility to tobacco carcinogens could be due to an enhanced
expression of the cytochrome P450 (CYP) enzyme CYP1A, which is
responsible for polycyclic aromatic hydrocarbon activation in human
lungs (16). Also, female smokers
have a higher frequency of TP53 gene mutations compared to
non-smoking females or males (30–32).
p53, the protein product of TP53, is a potent tumor
suppressor (33). Women are also
more likely to have mutations in the GSTM1 (Glutathione
S-transferase Mu 1) gene, which normally inactivates toxic
metabolites and has been linked to lung cancer development in
smokers (34). Additional studies
are needed in both smokers and non-smokers to fully understand the
genetic and epigenetic factors contributing to increased lung
cancer incidence in women compared with men.
Physiologically, mammalian lungs are continuously
exposed to estrogens by the blood circulation (10). Females produce higher levels of
estrogens compared with males, owing to higher aromatase (the
enzyme involved in conversion of androgen/testosterone to
estrogens) synthesis in gonadal tissues (35–37).
Besides major synthesis in the gonads such as ovary, aromatases are
locally expressed in non-gonadal tissues including the lungs,
brain, liver, bone, intestines, skin, blood vessels and spleen
(38,39). Hence, estrogens are synthesized
within the lungs normally (40) as
well as during various pathologic states including NSCLC (41). Estrogen receptors (ERs: ERα and ERβ)
are also detected in lung tissues in the normal physiological state
as well as in lung cancers (42,43).
While estrogens are normally involved in lung development (44,45),
pathophysiologically these hormones serve an important role in lung
carcinogenesis and its complications (46–48). At
present, a number of clinical trials are ongoing to assess the
efficacy of antiestrogen/antiER therapies against NSCLC development
and complications (49,50). This approach has been summarized in
multiple comprehensive reviews and is therefore not discussed in
the present review.
The estrogen related receptors (ERRs) were initially
identified from a cDNA library screen by Giguere et al
(51). Using rat and human tissue
samples, the investigators identified unique clones in kidney and
heart cDNA libraries that encoded previously unknown proteins with
conserved features of nuclear steroid hormone receptors,
particularly ERs (51). The clones
were designated as estrogen-related receptor α (ERRα) and
estrogen-related receptor β (ERRβ) (51). A third isoform of ERR, ERR-γ (ERRγ)
was subsequently identified by Eudy et al (52) through its linkage to the Usher's
Syndrome locus. Hong et al (53) using yeast two-hybrid screening and
the nuclear receptor co-activator glutamate receptor-interacting
protein 1 as bait also identified ERRγ.
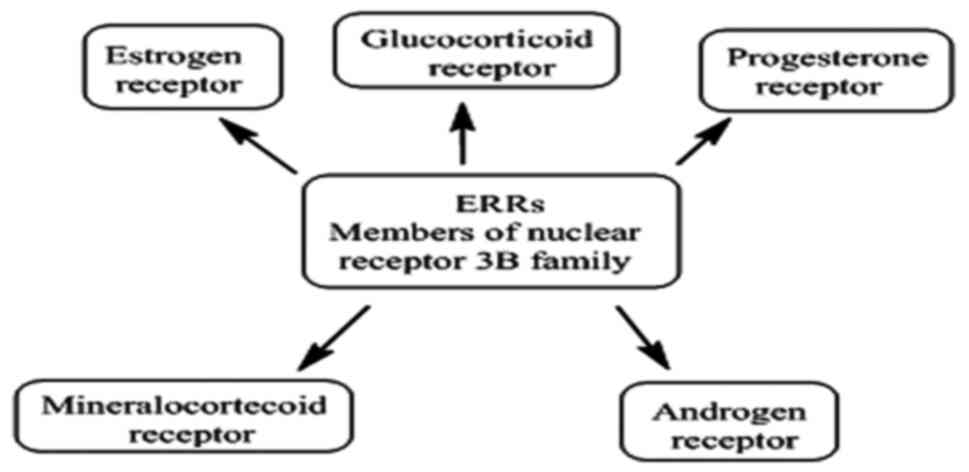
ERRs do not bind endogenous estrogens or their
derivatives and are therefore recognized as orphan nuclear
receptors, exhibiting considerable structural and functional
homology with ERs (Fig. 1) (51). The ERRs involvement in ER-dependent
signaling is associated with breast cancer cell proliferation
(54). ERRs pathological
significance is additionally noted by resistance to tamoxifen, a
competitive ER inhibitor used for breast cancer treatment (55) and activity in highly metastatic
triple negative (ER−, PR−, HER−)
(estrogen, progesterone and human Epidermal growth factor receptor
2 negative) (56). Hence, ERRs
appear to serve important pathological roles in both explicitly ER
positive and negative−breast cancers.
Numerous studies have indicated that ERRs serve
pathological roles in other estrogen dependent and independent
cancers, including ovarian (57),
endometrial (58), prostate
(59) colon/colorectal (60) and lung (61). Compounds that modulate ERRα
activity may serve critical roles in disease progression as well as
homeostasis (62). No endogenous
ligand for ERRα has been identified, although several
synthetic antagonists have been reported (63–65).
Recently, dietary products, such as genistein, apigenin,
resveratrol, rutacarpine, piceatanol, daidzein, flavone and
cholesterol have been reported as potential ERRα agonists
(66–68). The primary aim of the present review
is to highlight the emerging role of ERRs in NSCLCs.
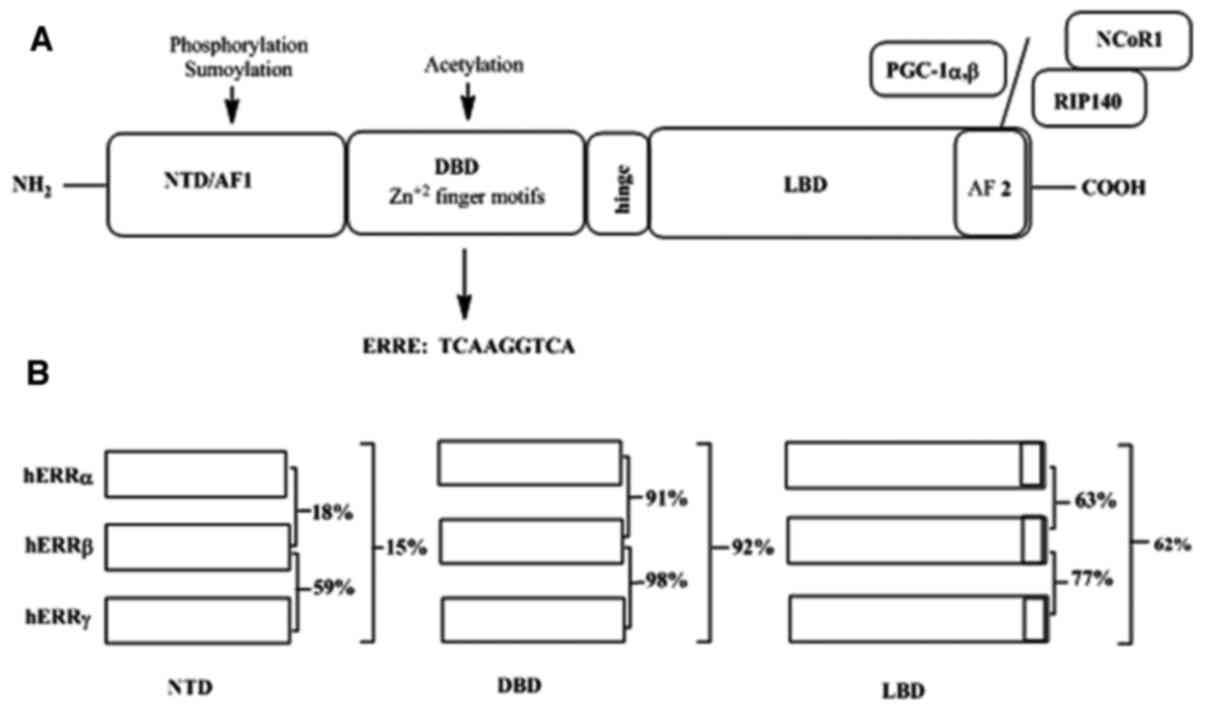
The C-terminal LBDs of ERRs have a conserved AF-2
helix motif essential for cofactor interactions (73). A distinctive aspect of ERRs unlike
other conventional NRs is their ability to activate transcription
without need for exogenous ligands, because the LBD conformation in
the absence of ligand supports the involvement of NR co-activators,
which are necessary for ERR regulated transcriptional activation
(82,83). Inspection of the ERRα and ERRγ LBD
conformations reveals the importance of amino acids that have bulky
side chains occupying the ligand binding pocket, hence mimicking a
ligand bound conformation that facilitates cofactor binding
(73). As one example, the ERRα LBD
crystal structure revealed a significant Phe328 hold of the ligand
binding pocket that confers an agonist conformation to the LBD,
which further binds the PPARγ co-activator-1α peptide (84). Of note, PPARγ is a type II proton
regulating protein encoded by PPARG gene in humans, substantially
prevalent in adipose tissue, colon and macrophages (64). While transcriptional activity of ERRs
is mostly independent of agonists, structural studies have revealed
an open ligand binding pocket of ~220 cubic Ȧ in ERRγ and of ~100
cubic Ȧ in ERRα, allowing transcriptional intervention by synthetic
molecules (85–89).
ERs (ERα and ERβ) are members of the steroid/nuclear
receptor superfamily and are activated via ligand binding (90). Mammalian ERs function both as signal
transducers and transcription factors to modulate target gene
expression (91). In response to
ligand binding, ERs undergo conformational changes and
‘activation’, accompanied by heat shock protein hsp90, hsp70 or
other proteins dissociations (92),
forming a ligand-occupied ER dimer (93). Stimulation of target gene expression
in response to 17β-estradiol (E2), or other agonists, is thought to
be mediated either via ‘direct binding’ to DNA specific genes, such
as vitellogenin A2 and oxytocin or through ‘indirect binding’ by
transcription factors, such as NF-κB, specificity protein-1 (SP-1)
and activator protein-1 (AP-1) (94). In the former, E2-liganded ER dimer
(E2-ER-ER) binds directly to a specific estrogen responsive gene
sequence, called an estrogen response element (ERE) before
interacting with co-activator proteins and RNA polymerase II
transcription initiation complex components resulting in enhanced
transcription (95). The EREs are
permutations of the 5′-GGT CAn nnTG ACC-3′ DNA palindrome, wherein
‘n’ denotes a nonspecific 3 nucleotide spacer located at varying
distances from the transcription start site and/or within a gene
locus (96). The regulation of gene
expression by the E2-ER-ER binding to EREs is referred to as the
ER-dependent signaling pathway (97,98). A
second mechanism of regulation is the transcriptional modulation of
target genes through E2-ER-ER and transcription factors
interactions, referred to as ‘tethering’ (99). The prominent transcription factors
involved in this interaction include SP1 (100,101),
AP1 (102–104), and a number of other proteins
(105). In a comprehensive review,
Klinge (106) described the
molecular mechanism by which ligand bound ER dimers modulate ERE
dependent and independent transcription, i.e. transcription factor
dependent transcription of various estrogen regulated genes, such
as cytochrome c, insulin like growth factor binding protein 4,
early estrogen-induced gene 1 and 4, heat shock 70 kDa protein 8,
keratin 8 and nuclease sensitive element binding protein 1.
Like ERα, ERRα binds to the classical ERE of
estrogen responsive genes, characterized by 5′-AGGTCANNNTGACCT−3′
sequence (N denoting a typical nucleotide) (106). ERRα also has binding sites for an
extended half of palindromic ERE as ERR response elements (ERRE),
having 5′-TNAAGGTCA-3′ sequence (91,107,108).
Hence, ERRα can affect ERα transcriptional activity. Although ERR
dimers can bind to the ERE, ERα dimers (not those of ERβ) also can
recognize a functional ERRE, hence demonstrating a nearly identical
binding specificity (109).
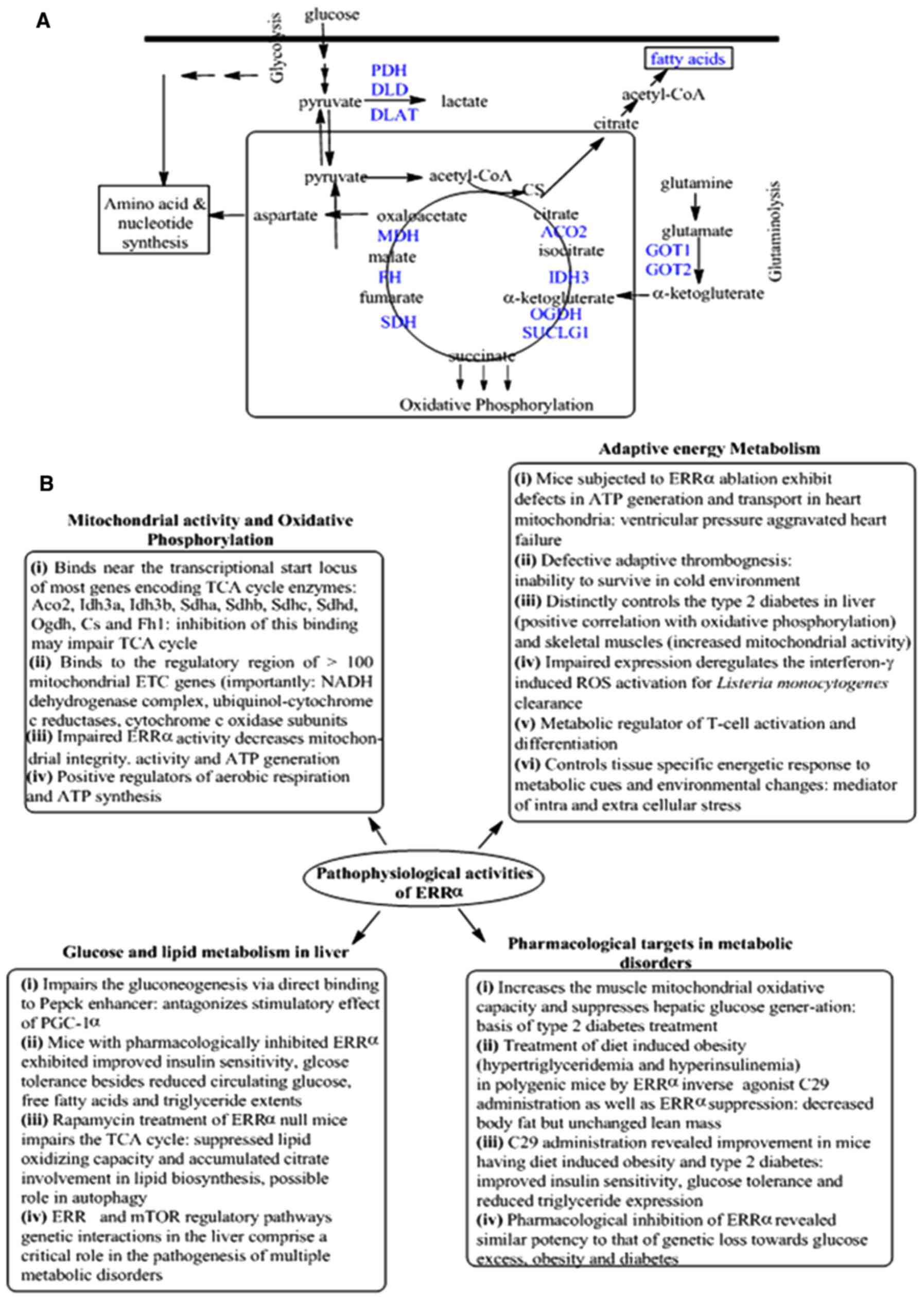
Basic physiological functions of ERRs include a
central role in regulating cellular metabolism by modulating genes
involved in glycolysis, the TCA cycle and mitochondrial oxidative
phosphorylation (Fig. 3) (110). Normally, an association of
proliferator activated receptor γ co-activator 1 (PGC-1) with the
ERR transcriptional axis controls mitochondrial biogenesis
(111). Besides a role in normal
physiology, roles of other PGC-1/ERR pathways are observed in
cancers, which depend on tissue specific and environmental stimuli
(112–116). For instance, the PGC-1/ERR axis has
been identified as necessary for tumor cell motility and metastasis
driven malignant transformation in breast and melanoma cancer
progression, whereas in prostate cancer the same pathway suppresses
tumor progression and metastasis (Table
I) (111,112,117–122).
ERRα is present in tissues actively engaged in high
glucose and lipid metabolism including heart, kidney, intestinal
tract, skeletal muscles and brown adipose tissues (Fig. 3A) (111,120,122–125).
Compared to ERRα, ERRβ and ERRγ expression is much more restricted,
with heart and kidney being the major sites (125,126).
Expression of both ERRα and ERRγ are increased in preadipocytes and
pluripotent mesenchymal cells under adipogenic conditions
indicating regulation by lipid accumulation (127,128).
In the central nervous system and spinal cord, ERRβ and ERRγ are
expressed during early embryonic development (129–131).
Specific roles for each ERR were demonstrated using
ERR specific knockout (KO) mice (132–134).
ERRα KO mice are viable, but exhibit a phenotype characterized by
reduced body weight, peripheral fat deposition and resistance to
high-fat diet-induced obesity (132). ERRα KO mice also exhibit cardiac
defects in bioenergetics and functional adaptation to pressure
overload, but their development and function under normal,
unstressed conditions is unaffected (133). ERRα KO mice also exhibit a loss of
normal mitochondrial biogenesis (134). In contrast, ERRβ KO mice are lethal
due to impaired placenta formation (130). ERRγ KO mice exhibit impaired
oxidative phosphorylation of perinatal heart mitochondria resulting
in 100% mortality within 48 h of birth (135). In summary, ERRs are essential for
maintaining normal physiological functions. While ERRα is detected
in the lung, the exact physiologic role of ERRα in the lung is not
known. ERRβ and ERRγ, have not yet been detected in lung tissues
(136).
In recent years, several studies have reported a
close association between ERRα expression and progression of
estrogen-dependent tumors including breast, ovarian, endometrial,
prostate and lung cancers as well as non-estrogen-dependent tumors
such as gastric, colon and colorectal cancers (47,49–51).
This suggests the involvement of ERRα both in estrogen dependent
and independent processes for a wide range of tumors (111,137).
Initial studies of various rat and human tissues
indicated that high level ERRα expression was a hallmark of
metabolically active organs, such as the heart, liver and brain
(128,132,134).
Low ERRα expression was detected in several other organs including
lung (51). Subsequently, using
embryonic and adult mouse tissues, low ERRα levels were
demonstrated in bone and skin (138). ERRα has been detected in human
NSCLC samples (59). In rats, ERRβ
is detected at low levels in kidney, heart, testis, brain and
prostate (49), whereas in mouse, it
is weakly expressed in adult kidney and heart (139). ERRγ is detected in embryonic lung
tissues including humans, but is not detected in adult lungs
(71). To the best of our knowledge
no study to date, has demonstrated ERRβ and ERRγ expression in
adult human lungs.
Regarding the role of ERRα in NSCLC, a number of
studies demonstrated elevated ERRα expression in NSCLC cells,
xenograft NSCLC mouse models and clinical NSCLC samples, indicating
possible diagnostic or post-therapeutic prognostic roles of ERRα in
NSCLC (91,138–140).
One study elucidated a cell-specific ERRα transactivator
functioning though SFRE sequence, wherein ERRα contributed in
transcriptional activation in rat osteosarcoma cell line (ROS
17.2/8) and HeLa, NB-E and FREJ4 cells, but not in COS1 and HepG2
cells (138). The investigators
reasoned such distinctions in ERRα functioning were due to the
osteopontin gene promoter as a transcription regulating target for
ERRα (138). Pettersson et
al (139) obbserved the
expression of nuclear receptors in embryonal carcinoma stem cells.
This study found that adequate homodimerization and DNA binding of
mERRβ was exclusively dependent on interaction with heat shock
protein 90, a molecular chaperone known to interact exclusively
with steroid hormone receptor subgroup of nuclear receptors
(140). In summary, the mouse
orphan receptor mERRβ exhibited the potential to control the
coinciding gene networks with the estrogen receptor, simultaneously
participating in signal transduction pathways during a limited time
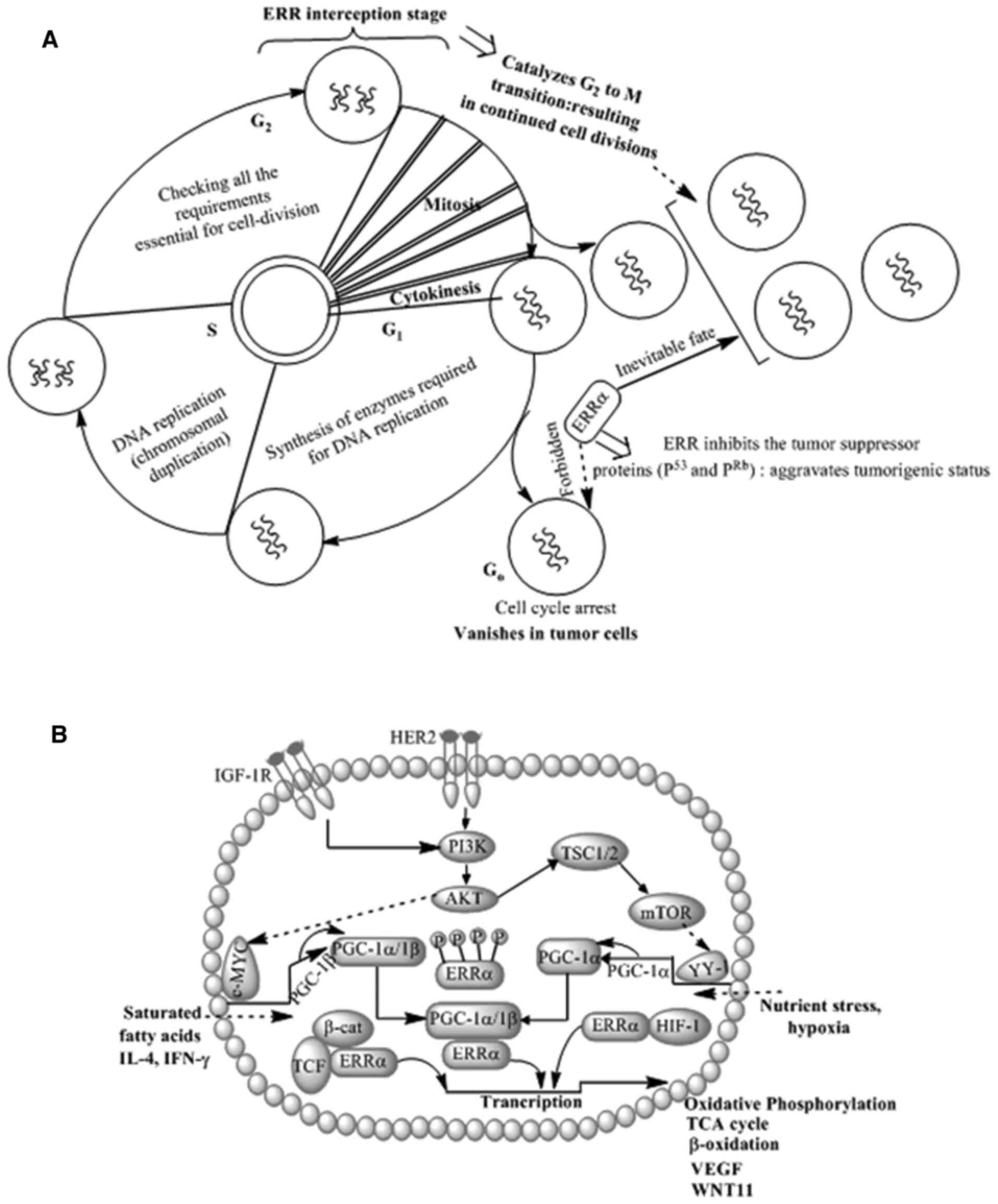
span analogous to chorion formation (138). Wang et al (140) demonstrated the tumorigenic
potential of ERRα via studying the effect of administered XCT-790,
an ERRα specific inverse agonist in A549 NSCLC cells. The findings
of the aforementioned study revealed reduced mitochondrial mass and
enhanced ROS generation through interception of TCA cycle. These
changes manifested in elevated mitochondrial membrane potential and
suppressed superoxide dismutase expression (140). It was also noticed that XCT-790
modulated the p53 and pRB signaling pathways (via ROS involvement)
and consequently suppressed cell replication (140). These observations led to the
generalization that disrupting ERRα regulated cell cycle mechanisms
could modulate tumour suppressor activities and arrest the cell
cycle (140). The specific role of
ERRα in NSCLCs has not been determined, but studies have
demonstrated its involvement in regulating the cell cycle and
cell-extracellular matrix interactions. These observations infer
likely ERRα involvement in regulating cell proliferation as well as
subsequent invasion/migration (metastasis). The mechanism by which
ERRα regulates NSCLC cell division and migration is discussed in
the following sections.
NSCLC cell culture-based investigations demonstrated
ERRα specific inverse agonists/small interfering (si)RNA/shRNA
effect cell cycle regulation (144). In one such investigation using
NSCLC A549 cells, Wang et al (140) noticed significant alterations in
mitochondrial mass, mitochondrial membrane potential and
mitochondrial reactive oxygen species (ROS) generation following
the administration of the ERRα inverse agonist XCT-790. The ROS
produced by XCT-790 activated the tumor suppressor proteins p53 and
pRB, which arrested the cell cycle in NSCLC cells (140). These in vitro cell
culture-based observations suggest that in A549 NSCLC cells, ERRα
decreases the tumor suppressor proteins p53 and pRB expression by
effecting mitochondrial physiology and quenching ROS generation
resulting in unopposed cell-cycle progression (Figs. 3A and 4A). Modulation of multiple signaling
pathways by ERRα presents implicit cell-division acceleration
strategies, which collectively result in tumor progression
(Fig. 4B).
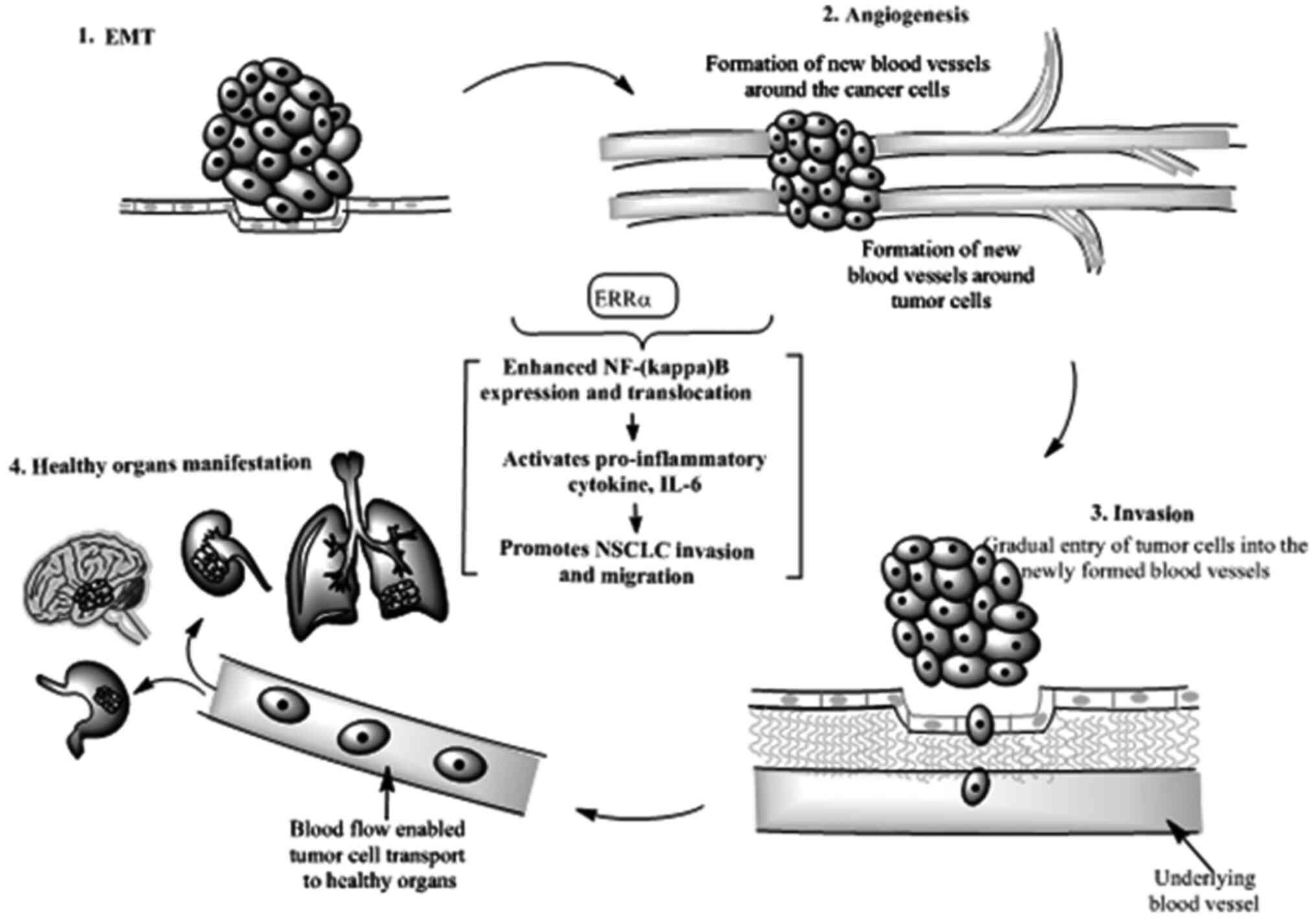
Capacity for invasion and migration remains a
hallmark of cancer cell metastasis to distant organs (145). Epithelial to mesenchymal transition
(EMT) is an important early step in invasion and metastasis
(141,145). In course of acquiring mesenchymal
phenotypes, tumor cells progressively develop enhanced motility and
the ability to invade through the tumor vasculature (Fig. 5). Acquiring mesenchymal status is an
important feature of tumor progression, drug resistance and
metastasis (146,147). Several transcription factors are
involved in EMT including Snail, Slug, Twist and Zeb (146,148).
Notable markers of EMT initiation and progression involve
activation of multiple cellular signalling pathways including MAPK,
PI3K and pro-inflammatory transcription factors, such as NF-κB
(146,149).
In lung cancers, circulating tumour cells
expressing epithelial cell adhesion molecules have much lower
expression compared with other solid tumours, indicating a loss of
epithelial markers (150). The EMT
phenotype in NSCLC is associated with EGFR mutations, drug
resistance (151–153) and formation of cancer stem cells
(154). A number of studies have
indicated that EMT related to NSCLC requires immune evasion
(155,156). In lung adenocarcinoma, intratumoral
CD8+ Tc (T cytotoxic) cell suppression is mediated
through ZEB1, which activates EMT and represses micro RNA-200, an
EMT and programmed death ligand-1 suppressor (157).
While the role of ERs in NSCLC is established, that
of ERRs in NSCLC is only beginning to be elucidated. A body of
literature has recently developed that suggests an important role
of ERRs in the development and progression of various cancers
including NSCLCs. In particular, ERRα expression by cancer cells
has emerged as an important prognostic indicator associated with
poor survival in several cancers including NSCLC (129,130,132).
In contrast, the role of ERRβ and ERRγ in NSCLC remains unknown,
due to undetectable low level or null expression of these molecules
in adult mammalian lungs (133). A
number of antiERRα molecules have been developed, including
diethylstilbestrol (DES), that bind to ERRα and inhibit its
activity (83). At present, most of
the studies of the effects of ERRα modulation in NSCLC are based on
in vitro cell culture experiments (129–131,162–164).
It is now imperative that the molecular mechanisms by which ERRα
promotes NSCLC development and progression be examined using in
vivo models (137,162–164).
The implicit involvement of ERRα in NSCLCs could be screened using
ERRα antagonists or activating ERRα dependent signaling pathways
using specific agonists. In this age of individualized medicine,
the effects of antiERR molecules alone or in combination with
aromatase inhibitors (e.g. anastrazole), selective estrogen
receptor modulators (SERMs e.g. tamoxifen) or selective estrogen
receptor down regulators (SERDs e.g. fulvestrant) should be
evaluated in specific NSCLC types.
Not applicable.
The present study was supported by a grant from the
Renzetti Presidential Endowed Chair, Department of Internal
Medicine, University of Utah, USA.
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
TKM conceptualized the basic theme of the
manuscript, prepared the first draft of the manuscript and
suggested the contents of figures and tables. PM compiled the
information about the physiological functions of estrogen related
receptors (ERRs) and the specific role of ERRα in cell cycle
regulation, NSCLC proliferation and the role of ERRα in NSCLC and
migration after discussion and gaining inputs from TKM. TKM and PM
addressed the reviewer's comments. JRH participated in manuscript
development, particularly in the writing of the lung cancer
statistics in males vs. females and the role of epithelial to
mesenchymal transition in lung cancer metastasis. JRH also
scrutinized, corrected and approved the finalized manuscript in the
initial and revised versions. All authors read and approved the
final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
International Agency for Research on
Cancer (IARC): GLOBOCAN 2012, . Estimated Cancer Incidence.
Mortality and Prevalence Worldwide. 2013.
|
|
2
|
Malvezzi M, Carioli G, Bertuccio P,
Boffetta P, Levi F, La Vecchia C and Negri E: European cancer
mortality predictions for the year 2017, with focus on lung cancer.
Ann Oncol. 28:1117–1123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Ward EM, Johnson CJ, Cronin KA,
Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, et al:
Annual report to the nation on the status of cancer, 1975–2014,
featuring survival. J Natl Cancer Inst. 109:djx0302017. View Article : Google Scholar
|
|
4
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morgensztern D, Ng SH, Gao F and Govindan
R: Trends in stage distribution for patients with non-small cell
lung cancer: A National cancer database survey. J Thorac Oncol.
5:29–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jemal A, Miller KD, Ma J, Siegel RL,
Fedewa SA, Farhad I, Devesa SS and Thun MJ: Higher lung cancer
incidence in young women than young men in the United States. N
Engl J Med. 378:1999–2009. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ragavan MV and Patel MI: Understanding sex
disparities in lung cancer incidence: Ar women more at risk? Lung
Cancer Manag. 9:LMY342020. View Article : Google Scholar
|
|
9
|
Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao
H, Li J, Mills GB, Shu Y, Li L and Liang H: Comprehensive
characterization of molecular differences in cancer between male
and female patients. Cancer Cell. 29:711–722. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia B, Feldman R, Cozen W, Kang I, Raez
LE, Borghaei H, Kim C, Nagasaka M, Mamdani H, Vanderwalde AM, et
al: Sex disparities in hormone positive lung cancer. J Clin Oncol.
38 (15_suppl):e215522020. View Article : Google Scholar
|
|
11
|
Fidler-Benaoudia MM, Torre LA, Bray F,
Ferlay J and Jemal A: Lung cancer incidence in young women vs.
young men: A systematic analysis in 40 countries. Int J Cancer.
147:811–819. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Skov BG, Fischer BM and Pappot H:
Oestrogen receptor beta over expression in males with non-small
cell lung cancer is associated with better survival. Lung Cancer.
59:88–94. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Greiser CM, Greiser EM and Dören M:
Menopausal hormone therapy and risk of lung cancer: Systematic
review and meta-analysis. Maturitas. 65:198–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adami HO, Persson I, Hoover R, Schairer C
and Bergkvist L: Risk of cancer in women receiving hormone
replacement therapy. Int J Cancer. 44:833–839. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Risch HA, Howe GR, Jain M, Burch JD,
Holowaty EJ and Miller AB: Are female smokers at higher risk for
lung cancer than male smokers? A case-control analysis by
histologic type. Am J Epidemiol. 138:281–293. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mollerup S, Berge G, Baera R, Skaug V,
Hewer A, Phillips DH, Stangeland L and Haugen A: Sex differences in
risk of lung cancer: Expression of genes in the PAH bioactivation
pathway in relation to smoking and bulky DNA adducts. Int J Cancer.
119:741–744. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Klugman M, Xue X and Hosgood HD III:
Race/ethnicity and lung cancer survival in the United States: A
meta-analysis. Cancer Causes Control. 30:1231–1241. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schabath MB, Cress D and Muñoz-Antonia T:
Racial and ethnic differences in the epidemiology and genomics of
lung cancer. Cancer Control. 23:338–346. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Izbicka E, Streeper RT, Michalek JE,
Louden CL, Diaz A III and Campos DR: Plasma biomarkers distinguish
non-small cell lung cancer from asthma and differ in men and women.
Cancer Genomics Proteomics. 9:27–35. 2012.PubMed/NCBI
|
|
20
|
Hastings RH, Laux AM, Casillas A, Xu R,
Lukas Z, Ernstrom K and Deftos LJ: Sex-specific survival advantage
with parathyroid hormone-related protein in non-small cell lung
carcinoma patients. Clin Cancer Res. 12:499–506. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang YL, Yuan JQ, Wang KF, Fu XH, Han XR,
Threapleton D, Yang ZY, Mao C and Tang JL: The prevalence of EGFR
mutation in patients with non-small cell lung cancer: A systematic
review and meta-analysis. Oncotarget. 7:78985–78993. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim HR, Shim HS, Chung JH, Lee YJ, Hong
YK, Rha SY, Kim SH, Ha SJ, Kim SK, Chung KY, et al: Distinct
clinical features and outcomes in never-smokers with nonsmall cell
lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement.
Cancer. 118:729–739. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Patel MI, McKinley M, Cheng I, Haile R,
Wakelee H and Gomez SL: Lung cancer incidence trends in California
by race/ethnicity, histology, sex and neighbourhood socioeconomic
status: An analysis spanning 28 years. Lung Cancer. 108:140–149.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang CH, Lee CH, Ho CC, Wang JY and Yu
CJ: Sex-based impact of epidermal growth factor receptor mutation
in patients with non-small cell lung cancer and previous
tuberculosis. Medicine (Baltimore). 94:e4442015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang C, Qiao W, Jiang Y, Zhu M, Shao J,
Ren P, Liu D and Li W: Effect of sex on the efficacy of patients
receiving immune checkpoint inhibitors in advanced non-small cell
lung cancer. Cancer Med. 8:4023–4031. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pinto JA, Vallejos CS, Raez LE, Mas LA,
Ruiz R, Torres-Roman JS, Morante Z, Araujo JM, Gomez HL, Aguilar A,
et al: Sex and outcomes in non-small cell lung cancer: An old
prognostic variable comes back for targeted therapy and
immunotherapy? ESMO Open. 3:e0003442018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nelson R: ALK inhibitors: Possible new
treatment for lung cancer. Medscape Medical News AACR-IASLC Joint
Conference on Molecular Origins of Lung Cancer. Jan 15–2010.
|
|
28
|
Wang WC, Shiao HY, Lee CC, Fung KS and
Hsieh HP: Anaplastic lymphnoma kinase (ALK) inhibitors: A review of
deign and discovery. Med Chem Comm. 5:1266–1279. 2014. View Article : Google Scholar
|
|
29
|
Shepherd FA, Rodrigues Pereira J, Ciuleanu
T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S,
Smylie M, Martins R, et al: Erlotinib in previously treated
non-small-cell lung cancer. N Engl J Med. 353:123–132. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yim SH and Chung YJ: Molecular
epidemiology of female lung cancer. Cancers (Basel). 3:1861–1876.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guinee DG Jr, Travis WD, Trivers GE, De
Benedetti VM, Cawley H, Welsh JA, Bennett WP, Jett J, Colby TV,
Tazelaar H, et al: Sex comparisons in human lung cancer: Analysis
of p53 mutations, anti-p53 serum antibodies and C-erbB-2
expression. Carcinogenesis. 16:993–1002. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kure EH, Ryberg D, Hewer A, Phillips DH,
Skaug V, Baera R and Haugen A: p53 mutations in lung tumours:
Relationship to sex and lung DNA adduct levels. Carcinogenesis.
17:2201–2205. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rivlin N, Brosh R, Oren M and Rotter V:
Mutations in the p53 tumor suppressor gene. Genes Cancers.
2:466–474. 2011. View Article : Google Scholar
|
|
34
|
Rivera MP: Lung cancer in women:
Differences in epidemiology, biology, histology and treatment
outcomes. Semin Respir. Crit Care Med. 34:792–801. 2013.
|
|
35
|
Stocco C: Tissue physiology and pathology
of aromatase. Steroids. 77:27–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Barakat R, Oakley O, Kim H, Jin J and Ko
CJ: Extra-gonadal sites of estrogen biosynthesis and function. BMV
Rep. 49:488–496. 2016.
|
|
37
|
Hammes SR and Levin ER: Impact of
estrogens in males and androgens in females. J Clin Invest.
129:1818–1826. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Reckelhoff JF: Sex differences in the
regulation of blood pressure. Hypertension. 37:1199–1208. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Blenck CL, Harvey PA, Reckelhoff JF and
Leinwand LA: The importance of biological sex and estrogen in
rodent models of cardiovascular health and disease. Circ Res.
118:1294–1312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Simpson ER, Clyne C, Rubin G, Boon WC,
Robertson K, Britt K, Speed C and Jones M: Aromatase-A brief
Overview. Ann Rev Physiol. 64:93–127. 2002. View Article : Google Scholar
|
|
41
|
Weinberg OK, Marquez-Garban DC, Fishbein
MJ, Goodglick L, Garban HJ, Dubinett SM and Pietras RJ: Aromatase
inhibitors in human lung cancer therapy. Cancer Res.
65:11287–11291. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mollerup S, Jørgensen K, Berge G and
Haugen A: Expression of estrogen receptors alpha and beta in human
lung tissue and cell lines. Lung Cancer. 37:1531592002. View Article : Google Scholar
|
|
43
|
Stabile LP, Davis AL, Gubish CT, Hopkins
TM, Luketich JD, Christie N, Finkelstein S and Siegfried JM: Human
non-small cell lung tumors and cells derived from normal lung
express both estrogen receptor alpha and beta and show biological
responses to estrogen. Cancer Res. 62:2141–2150. 2002.PubMed/NCBI
|
|
44
|
Carey MA, Card JW, Voltz JW, Germolec DR,
Korach KS and Zeldin DC: The impact of sex and sex hormones on lung
physiology and disease: Lessons from animal studies. Am J Physiol
Lung Cell Mol Physiol. 293:L272–L278. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Brandenberger AW, Tee MK, Lee JY, Chao V
and Jaffe RB: Tissue distribution of estrogen receptors alpha
(ER-alpha) and beta (ER-beta) mRNA in the midgestational human
fetus. J Clin Endocrinol Metab. 82:3509–3512. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Márquez-Garbán DC, Chen HW, Fishbein MC,
Goodglick L and Pietras RJ: Estrogen receptor signaling pathways in
human non-small cell lung cancer. Steroids. 72:135–143. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Baik CS and Eaton KD: Estrogensignaling in
lung cancer: An opportunity for novel therapy. Cancers (Basel).
4:969–988. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hsu LH, Liu KJ, Tsai MF, Wu CR, Feng AC,
Chu NM and Kao SH: Estrogen adversely affects the prognosis of
patients with lung adenocarcinoma. Cancer Sci. 106:51–59. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Heilbroner SP, Xanthopoulos EP, Buono D,
Huang Y, Carrier D, Shah A, Kim J, Corradetti M, Wright JD, Neugut
AI, et al: Impact of estrogen monotherapy on survival in women with
stage III–IV non-small cell lung cancer. Lung Cancer. 129:8–15.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Garon EB, Siegfried JM, Stabile LP, Young
PA, Marquez-Garban DC, Park DJ, Patel R, Hu EH, Sadeghi S, Parikh
RJ, et al: Randomized phase II study of fulvestrant and erlotinib
compared with erlotinib alone in patients with advanced or
metastatic non-small cell lung cancer. Lung Cancer. 123:91–98.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Giguere V, Yang N, Segui P and Evans RM:
Identification of a new class of steroid hormone receptors. Nature.
331:91–94. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Eudy JD, Yao S, Weston MD, Ma-Edmonds M,
Talmadge CB, Cheng JJ, Kimberling WJ and Sumegi J: Isolation of a
gene encoding a novel member of the nuclear receptor superfamily
from the critical region of Usher syndrome type IIa at 1q41.
Genomics. 50:382–384. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hong H, Yang L and Stallcup MR:
Hormone-independent transcriptional activation and coactivator
binding by novel orphan nuclear receptor ERR3. J Biol Chem.
274:22618–22626. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lata K and Mukherjee TK: Knockdown of
receptor for advanced glycation end products attenuate
17α-ethinyl-estradiol dependent proliferation and survival of MCF-7
breast cancer cells. Biochim Biophys Acta. 1840:1083–1091. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Riggins RB, Lan JP, Zhu Y, Klimach U,
Zwart A, Cavalli LR, Haddad BR, Chen L, Gong T, Xuan J, et al:
ERRgamma mediates tamoxifen resistance in novel models of invasive
lobular breast cancer. Cancer Res. 68:8908–8917. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Manna S, Bostner J, Sun Y, Miller LD,
Alayev A, Schwartz NS, Lager E, Fornander T, Nordenskjöld B, Yu JJ,
et al: ERRα is a marker of tamoxifen response and survival in
triple-negative breast cancer. Clin Cancer Res. 22:1421–1431. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lam SS, Mak AS, Yam JW, Cheung AN, Ngan HY
and Wong AS: Targeting estrogen-related receptor alpha inhibits
epithelial-to-mesenchymal transition and stem cell properties of
ovarian cancer cells. Mol Ther. 22:743–751. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yoriki K, Mori T, Kokabu T, Matsushima H,
Umemura S, Tarumi Y and Kitawaki J: Estrogen-related receptor alpha
induces epithelial-mesenchymal transition through cancer-stromal
interactions in endometrial cancer. Sci Rep. 9:66972019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fujimura T, Takahashi S, Urano T, Ijichi
N, Ikeda K, Kumagai J, Murata T, Takayama K, Horie-Inoue K, Ouchi
Y, et al: Differential expression of estrogen-related receptors
beta and gamma (ERRbeta and ERRgamma) and their clinical
significance in human prostate cancer. Cancer Sci. 101:646–651.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liang R, Lin Y, Yuan CL, Liu ZH, Li YQ,
Luo XL, Ye JZ and Ye HH: High expression of estrogen-related
receptor α is significantly associated with poor prognosis in
patients with colorectal cancer. Oncol Lett. 15:5933–5935.
2018.PubMed/NCBI
|
|
61
|
Li P, Wang J, Wu D, Ren X, Wu W, Zuo R,
Zeng Q, Wang B, He X, Yuan J and Xie N: ERRα is an aggressive
factor in lung adenocarcinoma indicating poor prognostic outcomes.
Cancer Manag Res. 11:8111–8123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ariazi EA and Jordan VC: Estrogen-related
receptors as emerging targets in cancer and metabolic disorders.
Curr Top Med Chem. 6:203–215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Busch BB, Stevens WC Jr, Martin R,
Ordentlich P, Zhou S, Sapp DW, Horlick RA and Mohan R:
Identification of a selective inverse agonist for the orphan
nuclear receptor estrogen-related receptor alpha. J Med Chem.
47:5593–5596. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Willey PJ, Murray IR, Qian J, Busch BB,
Stevens WC Jr, Martin R, Mohan R, Zhou S, Ordentlich P, Wei P, et
al: Regulation of PPARgamma coactivator 1alpha (PGC-1alpha)
signaling by an estrogen-related receptor alpha (ERRalpha) ligand.
Proc Natl Acad Sci USA. 101:8912–8917. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chisamore MJ, Cunningham ME, Flores O,
Wilkinson HA and Chen JD: Characterization of a novel small
molecule subtype specific estrogen-related receptor alpha
antagonist in MCF-7 breast cancer cells. PLoS One. 4:e56242009.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Teng CT, Beames B, Merrick BA, Martin N,
Romeo C and Jetten AM: Development of a stable cell line with an
intact PGC-1α/ERRα axis for screening environmental chemicals.
Biochem Biophys Res Commun. 444:177–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Teng CT, Hsieh JH, Zhao J, Huang R, Xia M,
Martin N, Gao X, Dixon D, Auerbach SS, Witt KL and Merick BA:
Development of novel cell lines for high-throughput screening to
detect estrogen-related receptor alpha modulators. SLAS Discov.
22:720–731. 2017.PubMed/NCBI
|
|
68
|
Wei W, Schwaid AG, Wang X, Wang X, Chen S,
Chu Q, Saghatelian A and Wan Y: Ligand activation of ERRα by
cholesterol mediates statin and bisphosphonate effects. Cell Metab.
23:479–491. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Nuclear Receptors Nomenclature Committee,
. A unified nomenclature system for the nuclear receptor
superfamily. Cell. 97:161–163. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tremblay AM and Giguere V: The NR3B
subgroup: An ovERRview. Nucl Recept Signal. 5:e0092007. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Heard DJ, Norby PL, Holloway J and Vissing
H: Human ERRgamma, a third member of the estrogen receptor-related
receptor (ERR) subfamily of orphan nuclear receptors:
Tissue-specific isoforms are expressed during development and in
the adult. Mol Endocrinol. 14:382–392. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Laudet V, Hënni C, Coll J, Catzeflis F and
Stéhelin D: Evolution of the nuclear receptor gene superfamily.
EMBO J. 11:1003–1013. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Huss JM, Garbacz W and Xie W: Constitutive
activities of estrogen related receptors: Transcriptional
regulation of metabolism by the ERR pathways in health and disease.
Biochim Biophys Acta. 1852:1912–1927. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Tremblay AM, Wilson BJ, Yang XJ and
Giguere V: Phosphorylation-dependent sumoylation regulates
estrogen-related receptor-alpha and-gamma transcriptional activity
through a synergy control motif. Mol Endocrinol. 22:570–584. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Vu EH, Kraus RJ and Mertz JE:
Phosphorylation-dependent sumoylation of estrogen-related
receptor-alpha1. Biochemistry. 46:9795–9804. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gearhart MD, Hombeck SM, Evans RM, Dyson
HJ and Wright PE: Monomeric complex of human orphan estrogen
related receptor-2 with DNA: A pseudo-dimer interface mediates
extended half-site recognition. J Mol Biol. 327:819–832. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Huppunen J and Aarnisalo P: Dimerisation
modulates the activity of the orphan nuclear receptor ERRgamma.
Biochem. Biophys Res Commun. 314:964–970. 2004. View Article : Google Scholar
|
|
78
|
Oka SI, Zhai P, Alcendor R, Park JY and
Sadoshima J: Suppression of ERR targets by a PPARα/Sirt1 complex in
the failing heart. Cell Cycle. 11:856–864. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Onofrio ND, Servillo L and Balestrieri ML:
SIRT1 and SIRt6 signaling pathways in cardiovascular diseases
protection. Antioxid Redox Signal. 28:711–732. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wang C, Fu M and Pestell RG: Estrogen
receptor acetylation and phosphorylation in hormone responses. Br
Cancer Online. 8:e462005. View Article : Google Scholar
|
|
81
|
Wilson BJ, Tremblay AM, Deblois G,
Sylvain-Drolet G and Giguere V: An acetylation switch modulates the
transcriptional activity of estrogen-related receptor alpha. Mol
Endocrinol. 24:1349–1358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chen S, Zhou D, Yang C and Sherman M:
Molecular basis for the constitutive activity of estrogen-related
receptor alpha-1. J Biol Chem. 276:28465–28470. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Xie W, Hong H, Yang NN, Lin RJ, Simon CM,
Stallcup MR and Evans RM: Constitutive activation of transcription
and binding of coactivator by estrogen-related receptors 1 and 2.
Mol Endocrinol. 13:2151–2162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Kallen J, Schlaeppi JM, Bitsch F,
Filipuzzi I, Schib A, Riou V, Graham A, Strauss A, Geiser M and
Foumier B: Evidence for ligand-independent transcriptional
activation of the human estrogen-related receptor alpha (ERRalpha):
Crystal structure of ERRalpha ligand binding domain in complex with
peroxisome proliferator-activated receptor coactivator-1alpha. J
Biol Chem. 279:49330–49337. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Greschik H, Flaig R, Renaud JP and Moras
D: Structural basis for the deactivation of the estrogen-related
receptor gamma by diethylstilbestrol or 4-hydroxytamoxifen and
determinants of selectivity. J Biol Chem. 279:33639–33646. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Greschik H, Wurtz JM, Sanglier S, Borguet
W, van Dorsselaer A, Moras D and Renaud JP: Structural and
functional evidence for ligand-independent transcriptional
activation by the estrogen-related receptor 3. Mol Cell. 9:303–313.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kallen J, Lattmann R, Beerli R,
Blechschmidt A, Blommers MJ, Geiser M, Ottl J, Schlaeppi JM,
Strauss A and Fournier B: Crystal structure of human
estrogen-related receptor alpha in complex with a synthetic inverse
agonist reveals its novel molecular mechanism. J Biol Chem.
282:23231–23239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang L, Zuercher WJ, Consler TG, Lambert
MH, Miller AB, Orband-Miller LA, McKee DD, Wilson TM and Nolte RT:
X-ray crystal structures of the estrogen-related receptor gamma
ligand binding domain in three functional states reveal the
molecular basis of small molecule regulation. J Biol Chem.
281:37773–37781. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Jin KS, Park JK, Yoon J, Rho Y, Kim JH,
Kim EE and Ree M: Small-angle X-ray scattering studies on
structures of an estrogen-related receptor α ligand binding domain
and its complexes with ligands and coactivators. J Phys Chem B.
112:9603–9612. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yaşar P, Ayaz G, User SD, Güpür G and
Muyan M: Molecular mechanism of estrogen-estrogen receptor
signaling. Reprod Med Biol. 16:4–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Couse JF and Korach KS: Estrogen receptor
null mice: What have we learned and where will they lead us? Endocr
Rev. 20:358–417. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Klinge CM, Brolly CL, Bambara RA and Hilf
R: Hsp70 is not required for high affinity binding of purified calf
uterine estrogen receptor to estrogen response element DNA in
vitro. J Steroid Biochem Mol Biol. 63:283–301. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Devin-Leclerc J, Meng X, Delahaye F,
Leclerc P, Baulieu EE and Catelli MG: Interaction and dissociation
by ligands of estrogen receptor and Hsp90: The antiestrogen RU
58668 induces a protein synthesis-dependent clustering of the
receptor in the cytoplasm. Mol Endocrinol. 12:842–854. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kumar S, Lata K, Mukhopadhyay S and
Mukherjee TK: Role of estrogen receptors in pro-oxidative and
anti-oxidative actions of estrogens: A perspective. Biochim Biophys
Acta. 1800:1127–1135. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Klinge CM: Estrogen receptor interaction
with co-activators and co-repressors. Steroids. 65:227–251. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Klein-Hitpass L, Ryffel GU, Heitlinger E
and Cato AC: A 13 bp palindrome is a functional estrogen responsive
element and interacts specifically with estrogen receptor. Nucleic
Acids Res. 16:647–663. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Cheung E and Kraus WL: Genomic analyses of
hormone signaling and gene regulation. Annu Rev Physiol.
72:191–218. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Carroll JS, Meyer CA, Song J, Li W,
Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC,
Hall GF, et al: Genome-wide analysis of estrogen receptor binding
sites. Nat Genet. 38:1289–1297. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hall JM, Couse JF and Korach KS: The
multifaceted mechanisms of estradiol and estrogen receptor
signaling. J Biol Chem. 276:36869–36872. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Li C, Briggs MR, Ahlborn TE, Kraemer FB
and Liu J: Requirement of Sp1 and estrogen receptor alpha
interaction in 17β-estradiol-mediated transcriptional activation of
the low density lipoprotein receptor gene expression. Endocrinol.
142:1546–1553. 2001. View Article : Google Scholar
|
|
101
|
Safe S: Transcriptional activation of
genes by 17 beta-estradiol through estrogen receptor Sp1
interactions. Vitam Horm. 62:231–252. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Paech K, Webb P, Kuiper GG, Nilsson S,
Gustafsson J, Kushner PJ and Scanlan TS: Differential ligand
activation of estrogen receptors ERalpha and ERbeta at AP1 sites.
Science. 277:1508–1510. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Webb P, Nguyen P, Valentine C, Lopez GN,
Kwok GR, McInerney E, Katzenellenbogen BS, Enmark E, Gustafsson JA,
Nilsson S and Kushner PJ: The estrogen receptor enhances AP-1
activity by two distinct mechanisms with different requirements for
receptor transactivation functions. Mol Endocrinol. 13:1672–1685.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Kushner PJ, Agard DA, Greene GL, Scanlan
TS, Shiau AK, Uht RM and Webb P: Estrogen receptor pathways toAP-1.
J Steroid Biochem Mol Biol. 74:311–317. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Marino M, Galluzzo P and Ascenzi P:
Estrogen signaling multiple pathways to impact gene transcription.
Curr Genomics. 7:497–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Klinge CM: Estrogen receptor interaction
with estrogen response elements. Nucleic Acids Res. 29:2905–2919.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Johnston SD, Liu XD, Zuo F, Eisenbraun TL,
Wiley SR, Kraus RJ and Mertz JE: Estrogen-related receptor alpha 1
functionally binds as a monomer to extended half-site sequences
including ones contained within estrogen-response elements. Mol
Endocrinol. 11:342–352. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhang Z and Teng CT: Estrogen
receptor-related receptor alpha 1 interacts with coactivator and
constitutively activates the estrogen response elements of the
human lactoferrin gene. J Biol Chem. 275:20837–20846. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Vanacker JM, Petterson K, Gustafsson JA
and Laudet V: Transcriptional targets shared by estrogen
receptor-related receptors (ERRs) and estrogen receptor (ER) alpha,
but not by ER beta. EMBO J. 18:4270–4279. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Mullen EM, Gu P and Cooney AJ: Nuclear
receptors in regulation of mouse ES cell pluripotency and
differentiation. PPAR Res. 2007:615632007. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Deblois G and Giguere V: Oestrogen-related
receptors in breast cancer: Control of cellular metabolism and
beyond. Nat Rev Cancer. 13:27–36. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Gravel SP: Deciphering the dichotomous
effects of PGC-1α on tumorigenesis and metastasis. Front Oncol.
8:752018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
LeBleu VS, O'Connell JT, Gonzalez Herrera
KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A,
Domingos Chinen LT, Rocha RM, et al: PGC-1α mediates mitochondrial
biogenesis and oxidative phosphorylation in cancer cells to promote
metastasis. Nat Cell Biol. 16:992–1003, 1-15. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Tan Z, Luo X, Xiao L, Tang M, Bode AM,
Dong Z and Cao Y: The role of PGC-1α in cancer metabolism and its
therapeutic implications. Mol Cancer Ther. 15:774–782. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Torrano V, Valcarcel-Jimenez L, Cortazar
AR, Liu X, Urosevic J, Castillo-Martin M, Fernandez-Ruiz S,
Morciano G, Caro-Maldonado A, Guiu M, et al: The metabolic
co-regulator PGC1α suppresses prostate cancer metastasis. Nat Cell
Biol. 18:645–656. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Deblois G, St-Pierre J and Giguere V: The
PGC-1/ERR signaling axis in cancer. Oncogene. 32:3483–3490. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Audet-Walsh É, Yee T, McGuirk S, Vernier
M, Ouellet C, St-Pierre J and Giguère V: Androgen-dependent
repression of ERRγ reprograms metabolism in prostate cancer. Cancer
Res. 77:378–389. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Sun P, Sehouli J, Denkert C, Mustea A,
Kongsen D, Koch I, Wei L and Lichtenegger W: Expression of estrogen
receptor-related receptors, a subfamily of orphan nuclear
receptors, as new tumor biomarkers in ovarian cancer cells. J Mol
Med. 83:457–467. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Kang MH, Choi H, Oshima M, Cheong JH, Kim
S, Lee JH, Park YS, Choi HS, Kweon MN, Pack CG, et al:
Estrogen-related receptor gamma functions as a tumor suppressor in
gastric cancer. Nat Commun. 9:19202018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Kim JH, Choi YK, Byun JK, Kim MK, Kang YN,
Kim SH, Lee S, Jang BK and Park KG: Estrogen-related receptor γ is
upregulated in liver cancer and its inhibition suppresses liver
cancer cell proliferation via induction of p21 and p27. Exp Mol
Med. 48:e2132016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Pons F, Varela M and Llovet JM: Sensing
systems in hepatocellular carcinoma. HPB (Oxford). 7:35–41. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhou Y, Jia Q, Meng X, Chen D and Zhu B:
ERRα regulates OTUB1 expression to promote colorectal cancer cell
migration. J Cancer. 10:5812–5819. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Luo C, Balsa E, Thomas A, Hatting M,
Jedrychowski M, Gygi SP, Widlung HR and Puigserver P: ERRα
maintains mitochondrial oxidative metabolism and constitutes an
actionable target in PGC1α-elevated melanomas. Mol Cancer Res.
15:1366–1375. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Bookout AL, Jeong Y, Downes M, Yu RT,
Evans RM and Mangelsdorf DJ: Anatomical profiling of nuclear
receptor expression reveals a hierarchical transcriptional network.
Cell. 126:789–799. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ranhotra HS: The orphan estrogen-related
receptor alpha and metabolic regulation: New frontiers. J Recept
Signal Transduct Res. 35:565–568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Giguère V: Transcriptional control of
energy homeostasis by the estrogen-related receptors. Endocr Rev.
29:677–696. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Ijichi N, Ikeda K, Horie-Inoue K, Yagi K,
Okazaki Y and Inoue S: Estrogen-related receptor alpha modulates
the expression of adipogenesis-related genes during adipocyte
differentiation. Biochem Biophys Res Commun. 358:813–818. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Kubo M, Ijichi N, Ikeda K, Horie-Inoue K,
Takeda S and Inoue S: Modulation of adipogenesis-related gene
expression by estrogen-related receptor gamma during adipocytic
differentiation. Biochim Biophys Acta. 1789:71–77. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lorke DE, Susens U, Borgmeyer U and
Hermans-Borgmeyer I: Differential expression of the estrogen
receptor related receptor gamma in the mouse brain. Brain Res Mol
Brain Res. 77:277–280. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Luo J, Sladek R, Bader JA, Matthyssen A,
Rossant J and Giguère V: Placental abnormalities in mouse embryos
lacking the orphan nuclear receptor ERR-beta. Nature. 388:778–782.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Borgmeyer-Hermans I, Süsens U and
Borgmeyer U: Developmental expression of the estrogen
receptor-related receptor gamma in the nervous system during mouse
embryogenesis. Mech Dev. 97:197–199. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Luo J, Sladek R, Carrier J, Bader JA,
Richard D and Giguère V: Reduced fat mass in mice lacking orphan
nuclear receptor estrogen-related receptor alpha. Mol Cell Biol.
23:7947–7956. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Huss JM, Imahashi K, Dufour CR, Weinheimer
CJ, Courtois M, Kovacs A, Giguère V, Murphy E and Kelly DP: The
nuclear receptor ERRalpha is required for the bioenergetic and
functional adaptation to cardiac pressure overload. Cell Metab.
6:25–37. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Villena JA and Kralli A: ERRalpha: A
metabolic function for the oldest orphan. Trends Endocrinol Metab.
19:269–276. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Alaynick WA, Kondo RP, Xie W, He W, Dufour
CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V and Evans
RM: ERRgamma directs and maintains the transition to oxidative
metabolism in the postnatal heart. Cell Metab. 6:13–24. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhou W, Liu Z, Wu J, Liu JH, Hyder SM,
Antoniou E and Lubahn DB: Identification and characterization of
two novel splicing isoforms of human estrogen-related receptor
beta. Clin Endocrinol Metab. 91:569–579. 2006. View Article : Google Scholar
|
|
137
|
Xu Z, Wang Y, Xiao ZG, Zou C, Zhang X,
Wang Z, Wu D, Yu S and Chan FL: Nuclear receptor ERRα and
transcription factor ERG form a reciprocal loop in the regulation
of TMPRSS2: ERGfusion gene in prostate cancer. Oncogene.
37:6259–6274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Bonnelye E, Vanacker JM, Dittmar T, Begue
A, Desbiens X, Denhardt DT, Aubin JE, Laudet V and Fournier B: The
ERR-1 orphan receptor is a transcriptional activator expressed
during bone development. Mol Endocrinol. 11:905–916. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Pettersson K, Svensson K, Mattsson R,
Carlsson B, Ohlsson R and Berkenstam A: Expression of a novel
member of estrogen response element-binding nuclear receptors is
restricted to the early stages of chorion formation during mouse
embryogenesis. Mech Dev. 54:211–223. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Wang J, Wang Y and Wong C:
Oestrogen-related receptor alpha inverse agonist XCT-790 arrests
A549 lung cancer cell population growth by inducing mitochondrial
reactive oxygen species production. Cell Prolif. 43:103–113. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Fouad YF and Aanei C: Revisiting the
hallmarks of cancer. Am. J Cancer Res. 7:1016–1036. 2017.
|
|
142
|
Pardee AB: G1 events and regulation of
cell proliferation. Science. 246:603–608. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Harper JV: Synchronization of cell
populations in G1/S and G2/M phases of the cell cycle. Methods Mol
Biol. 296:157–166. 2005.PubMed/NCBI
|
|
144
|
Makowiecki C, Nolte A, Sutaj B, Keller T,
Avci-Adali M, Stoll H, Schlensak C, Wendel HP and Walker T: New
basic approach to treat non-small cell lung cancer based on
RNA-interference. Thorac Cancer. 5:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Martin TA, Ye L, Sanders AJ, Lane J and
Jiang WG: Cancer invasion and metastasis: Molecular and cellular
perspective. Mad Curie Bioscience Database. Landes Bioscience;
2000-2013
|
|
146
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Craene BD and Berx G: Regulatory networks
defining EMT during cancer initiation and progression. Nat Rev
Cancer. 13:97–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Singh A and Settleman J: EMT, Cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Tiwari N, Gheldof A, Tatari M and
Christofori G: EMT as the ultimate survival mechanism of cancer
cells. Semin Cancer Biol. 22:194–207. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Krebs MG, Sloane R, Priest L, Lancashire
L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G,
et al: Evaluation and prognostic significance of circulating tumor
cells in patients with non-small cell lung cancer. J Clin Oncol.
29:1556–1563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Jakobsen KR, Demuth C, Sorensen BS and
Nielsen AL: The role of epithelial to mesenchymal transition in
resistance to epidermal growth factor receptor tyrosine kinase
inhibitors in non-small cell lung cancer. Transl Lung Cancer Res.
5:172–182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Li L, Gu X, Yue J, Zhao Q, Lv D, Chen H
and Xu L: Acquisition of EGFR TKI resistance and EMT phenotype is
linked with activation of IGF1R/NF-κB pathway in EGFR-mutant NSCLC.
Oncotarget. 8:92240–92253. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Byers LA, Diao L, Wang J, Saintigny P,
Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, et al: An
epithelial-mesenchymal transition gene signature predicts
resistance to EGFR and PI3K inhibitors and identifies Axl as a
therapeutic target for overcoming EGFR inhibitor resistance. Clin
Can Res. 19:279–290. 2013. View Article : Google Scholar
|
|
154
|
Aguilera TA and Giaccia AJ: Molecular
Pathways: Oncologic pathways and their role in T-cell exclusion and
immune evasion-A new role for the AXL receptor tyrosine kinase.
Clin Cancer Res. 23:2928–2933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Datar I and Schalper KA:
Epithelial-Mesenchymal transition and immune evasion during lung
cancer progression: The chicken or the egg? Clin Cancer Res.
22:3422–3424. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Lou Y, Diao L, Cuentas ER, Denning WL,
Chen L, Fan YH, Byers LA, Wang J, Papadimitrakopoulou VA, Behrens
C, et al: Epithelial-mesenchymal transition is associated with a
distinct tumor microenvironment including elevation of inflammatory
signals and multiple immune checkpoints in lung adenocarcinoma.
Clin Can Res. 22:3630–3642. 2016. View Article : Google Scholar
|
|
157
|
Chen L, Gibbons DL, Goswami S, Cortez MA,
Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al: Metastasis
is regulated via microRNA-200/ZEB1 axis control of tumour cell
PD-L1 expression and intratumoral immunosuppression. Nat Commun.
5:52412014. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Chae YK, Chang S, Ko T, Anker J, Agte S,
Iams W, Choi WM, Lee K and Cruz M: Epithelial-mesenchymal
transition (EMT) signature is inversely associated with T-cell
infiltration in non-small cell lung cancer (NSCLC). Sci Rep.
8:29182018. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Kwaśniak K, Czarnik-Kwaśniak J, Maziarz A,
Aebisher D, Zielińska K, Karczmarek-Borowska B and Tabarkiewicz J:
Scientific reports concerning the impact of interleukin 4,
interleukin 10 and transforming growth factor β on cancer cells.
Cent Eur J Immunol. 44:190–220. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Steen EH, Wang X, Balaji S, Butte MJ,
Bollyky PL and Keswani SG: The role of the anti-inflammatory
cytokine Interleukn-10 in tissue fibrosis. Adv Wound Care (New
Rochelle). 9:184–198. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Tsoukalas N, Aravantinou-Fatorou A, Tolia
M, Giaginis C, Galanapoulos M, Kiakou M, Kostakis ID, Dana E,
Vamvakaris I, Korogiannos A, et al: Epithelial-mesenchymal
transition in non small-cell lung cancer. Anticancer Res.
37:1773–1778. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Mahmood MQ, Ward C, Muller HK, Sohal SS
and Walters EH: Epithelial mesenchymal transition (EMT) and
non-small cell lung cancer (NSCLC): A mutual association with
airway disease. Med Oncol. 34:452017. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Thompson JC, Hwang WT, Davis C, Deshpande
C, Jeffries S, Rajpurohit Y, Krishna V, Smirnov D, Verona R,
Lorenzi MV, et al: Gene signatures of tumor inflammation and
epithelial-to-mesenchymal transition (EMT) predict responses to
immune checkpoint blockade in lung cancer with high accuracy. Lung
Cancer. 139:1–8. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Huang JW, Guan BZ, Yin LH, Liu FN, Hu B,
Zheng QY, Li FL, Zhong YX and Chen Y: Effects of estrogen-related
receptor alpha (ERRα) on proliferation and metastasis of human lung
cancer A549 cells. J Huazhong Univ Sci Technolog Med Sci.
34:875–881. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Zhang J, Guan X, Liang N and Li S:
Estrogen-related receptor alpha triggers the proliferation and
migration of human non-small cell lung cancer via interleukin-6.
Cell Biochem Funct. 36:255–262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Wang Y, Zhao M, Liu J, Ni J, Jiao Y and
Bai C: Up regulation of IL-6 is involved in di (2-ethylhexyl)
phthalate (DEHP) induced migration and invasion of non-small cell
lung cancer (NSCLC) cells. Biomed. Pharmacother. 89:1037–1044.
2017. View Article : Google Scholar
|
|
167
|
Kim JH: Di(2-ethylhexyl) phthalate
promotes lung cancer cell line A549 progression via Wnt/β-catenin
signaling. J Toxicol Sci. 44:237–244. 2019. View Article : Google Scholar : PubMed/NCBI
|