Introduction
Bladder cancer is a prevalent urinary system
malignancy, that seriously threatens human health and life.
According to the study ~430,000 new cases of bladder cancer were
reported worldwide in 2012, of which 165,000 were fatal (1). Bladder cancer is divided into
non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive
bladder cancer (MIBC) (2). In total,
~75% of patients with bladder cancer are diagnosed with NMIBC
(3). Transurethral resection of
bladder tumors is the major form of treatment for NMIBC. Regular
intravesical chemotherapy is administered following surgery to
prevent recurrence (4). However,
~50–70% of patients with NMIBC suffer from disease recurrences, and
30–40% of these patients suffer from disease progression (3,5,6). Data obtained from studies published
between 2006 and 2011 showed the 4-year survival rate of patients
with MIBC after progression from high-risk NMIBC was 35% (7). Pelvic lymph node dissection is not only
a treatment method, but also provides important information on
prognosis. Studies have shown that the risk of lymphatic metastasis
in muscular invasive bladder cancer was over 24% and was associated
with the depth of tumor invasion (pT2a 9–18%, pT2b 22–41%, pT3
41–50%, pT4 41–63%). Therefore, pelvic lymph node dissection is an
important part of radical cystectomy. Due to the high incidence and
recurrence rates, it is necessary to investigate the relevant
genetic pathways underlying the genesis and development of bladder
cancer.
Notch1 is involved in the proliferation and
differentiation of various cells, such as human dental pulp stem
cells (8,9) and neural stem cells (8). Studies have demonstrated that Notch1
expression in bladder cancer cells (BCCs) was associated with their
malignancy (10,11). Notch1 silencing, using shRNA,
could inhibit the proliferation of 5637 BCCs (12). However, the specific mechanism of
action of Notch1 in BCCs remains unclear.
Therefore, in the present study, Notch1 was
silenced in human T24 and 5637 BCCs to observe its effects on the
invasion potential, apoptosis, and epithelial-mesenchymal
transition (EMT) of these cells.
Materials and methods
Experimental materials
Human T24 and 5637 BCCs were purchased from Procell
Life Science & Technology Co., Ltd. The BCA protein assay kit
was purchased from Beyotime Institute of Biotechnology, while the
PLVX-shRNA2 vector was purchased from Addgene, Inc.
TRIzol® reagent and restriction endonucleases
(ExoRI and BamHI) were purchased from Thermo Fisher
Scientific, Inc. Reverse transcription (RT) and SYBR-Green PCR kits
were purchased from Takara Biotechnology, Co., Ltd. Rabbit
monoclonal antibodies against E-cadherin, N-cadherin, vimentin, and
Bcl2 were purchased from Cell Signaling Technology, Inc. Rabbit
polyclonal antibodies against Bad and Bid were purchased from
ProteinTech Group, Inc. and Abcam, respectively. The Transwell
chambers were purchased from BD Biosciences, while the apoptosis
detection and cell cycle detection kits were purchased from Nanjing
KeyGen Biotech Co., Ltd.
Cell culture
The T24 and 5637 cell lines were removed from liquid
nitrogen and thawed in a water bath at 37°C for up to 1 min. The
cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with penicillin (100 U/ml),
streptomycin (0.1 mg/l) and 10% FBS, and maintained at 37°C in a
humidified incubator with 5% CO2. The cells were
passaged when they reached ~80% confluence.
Plasmid construction and
transfection
The Notch1 scrambled negative control short hairpin
(sh) RNA (shRNA NC) and Notch1 interference (Notch1 shRNA)
sequences used were biosynthesized by Shanghai GenePharma Co.,
Ltd., and designed according to GeneID, 4851 (Table I). After annealing to form double
strands, the shRNA NC or Notch1 shRNA sequences were cloned into
the PLVX-shRNA2 vector pre-digested with ExoRI and
BamHI. After being transfected into competent Escherichia
coli DH5α cells, the plasmids were extracted from one single
colony and Sanger sequencing was performed to verify the presence
of the cloned plasmids. For shRNA (2 µg) and PLVX vector
transfection, the T24 and 5637 cell lines during the logarithmic
growth stage were seeded into 24-well plates to adjust the cell
density to 4×105/well, which corresponded to 70–80%
confluency. In accordance with the manufacturer's instructions,
Lipofectamine® 2000 transfection (Invitrogen; Thermo
Fisher Scientific, Inc.) reagent was used to transfect the plasmids
into the T24 and 5637 cell lines. After 8 h of transfection, at
37°C with 5% CO2, the RPMI 1640-medium containing 1%
ampicillin, used upon transfection (30 µg/ml) was replaced with
normal medium. The cells were cultured in a constant-temperature
incubator at 37°C with 5% CO2. After 48 h, the cells
were collected for subsequent experiments. Notch1 mRNA expression
levels in the T24 and 5637 transfected cells were detected using
RT-quantitative PCR.
 | Table I.shRNA sequence list. |
Table I.
shRNA sequence list.
| Name | Orientation | Sequence (5–3) |
|---|
| Notch1-shRNA | Forward |
GATCCCCAACATCCAGGACAACATTTCAAGAGAAT |
|
|
|
GTTGTCCTGGATGTTGGTTTTTTCTCGAGG |
|
| Reverse |
AATTCCTCGAGAAAAAACCAACATCCAGGACAA |
|
|
|
CATTCTCTTGAAATGTTGTCCTGGATGTTGGG |
| shRNA-NC | Forward |
GATCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGT |
|
|
|
GACACGTCGGAGAATTTTTTCTCGAGG |
|
| Reverse |
AATTCCTCGAGAAAAAATTCTCCGAACGTGT |
|
|
|
CACGTTCTCTTGAAACGTGACACGTCGGAGAAG |
MTT assay
The cells in the logarithmic growth phase were
cultured overnight in an incubator at 37°C with 5% CO2.
Then, 100 µl/well cell suspension (5×104 cells/ml) were
seeded in a 96-well plate and cultured at 37°C with 5%
CO2 for 4 h. After incubation with MTT (Nanjing
SenBeiJia Biological Technology Co., Ltd.), 0.15 ml DMSO (Beijing
Solarbio Science and Technology Co., Ltd.) was added and the
suspension was shaken for 10 min. Optical density (OD) at 568 nm
was measured using a microplate reader. The experiments were
performed independently 3 times.
Transwell assay
After 48 h of successful transfection, the cells in
each group were digested with 0.25% trypsin, collected and
centrifuged at 300 × g, at 4°C. The cells were then washed twice
with pre-cooled PBS. Cells were suspended in a serum-free RPMI-1640
medium and counted using the plate count method, by visually
observing the number of cells. Next, 0.8 ml medium with 10% FBS was
transferred into a 24-well plate, which was then placed in a
Transwell chamber. Then, 1 mg/ml Matrigel (100 µl) was added
vertically to the bottom of the upper Transwell chamber. After the
Matrigel solidified, 200 µl cell suspension(1×107) was
added to the upper Transwell chamber and cultured at 37°C in a
humidified incubator with 5% CO2 for 24 h. The Transwell
was then removed, the chamber was washed with PBS and the cells
were fixed in 10% methanol for 30 min at 4°C. Then, the membrane
was removed and the cells were stained with crystal violet (0.5%)
at room temperature for 20 min, followed by a wash with PBS. Images
were acquired and the cell numbers calculated using a counting
slide, under a light microscope (magnification, ×20; Olympus
Corporation). The experiments were performed independently 3
times.
Flow cytometry for apoptosis
After 2 days in culture, the cells were subjected to
trypsin digestion (0.25%) and then collected in a flow cytometry
tube. A total of ~1×105 suspended cells were centrifuged
at 300 × g, at 4°C. Detection was conducted following the
instructions of the Annexin V-APC/7-AAD detection kit. The cells
were resuspended in binding buffer (0.05 ml; 5×105/ml),
followed by the addition of the 7-AAD solution (5 µl), and
incubated for 15 min at room temperature. Finally, 0.45 ml binding
buffer and 1 µl Annexin V-APC were added at room temperature in the
dark for 15 min. The cell cycle distribution was evaluated using a
FACSVerse™ flow cytometer (Beckman Coulter, Inc.) and the data were
analyzed using FlowJo v10 software (FlowJo LLC). The experiments
were performed independently 3 times.
Cell cycle analysis
The cells were cultured for 48 h following
transfection, then digested with 0.25% non-EDTA trypsin, followed
by centrifugation at 300 × g, at room temperature for 5 min to
harvest the cells. The cells were then resuspended in 100 ml PBS,
and fixed by slowly adding 700 ml pre-cooled 80% ethanol to reach a
final concentration of 70%, following incubation at 4°C for 4 h
before centrifugation at 300 × g for 5 min, at 4°C. Then, RNase
(100 ml; 50 µg/ml) was added, and the cells were placed in a water
bath at 37°C for 30 min to permeabilize the cells. Finally, PI
solution (400 µl; 50 µg/ml) was added, and the cells were stained
at 4°C for 30 min in the dark. The cell cycle distribution was
evaluated using FACSVerse™ flow cytometer (Beckman Coulter, Inc.)
and the data were analyzed using FlowJo v10 software (FlowJo LLC).
The experiments were performed independently 3 times.
RT-qPCR
After the T24 and 5637 cells were transfected and
cultured for 48 h, total RNA was isolated using TRIzol®
following the manufacturer's instructions. RT was performed to
synthetize cDNA using a Prime Script RT kit (Takara, cat. no.
RR037A; Takara Bio, Inc.) at 65°C for 10 min [RNA, oligo(dT),
random primer and water], then on ice for 5 min. The samples were
then incubated at 25°C for 10 min, with the addition of dNTPs,
RNaseA, buffer, MTV reverse transcriptase. Next, the samples were
incubated at 37°C for 60 min, then heated to 70°C for 10 min.
Subsequently, qPCR was subsequently performed using the SYBR-Green
PCR kit, according to the manufacturer's instructions (cat. no.
RR430S; Takara Bio, Inc.), to analyze the mRNA expression levels of
vimentin, Bcl2, Bad, Bid, and E- and N-cadherin (primer sequences
are listed in Table II). The
following thermocycling conditions were used for the qPCR: Initial
denaturation at 95°C for 30 sec, then 40 cycles of 95°C for 30 sec
and 60°C for 30 sec. The 2−∆∆Cq method was used to
measure the relative mRNA expression levels (13); GAPDH was used as the internal
reference gene. The experiments were performed independently 3
times.
 | Table II.Primer sequences for E-cadherin,
N-cadherin, vimentin, Bcl-2, Bad, Bid and GAPDH. |
Table II.
Primer sequences for E-cadherin,
N-cadherin, vimentin, Bcl-2, Bad, Bid and GAPDH.
| Name | Primer | Sequence | Size, bp |
|---|
| E-cadherin | Forward |
5′-CGTAGCAGTGACGAATGTGG-3′ | 175 |
|
| Reverse |
5′-CTGGGCAGTGTAGGATGTGA-3′ |
|
| N-cadherin | Forward |
5′-CTTGCCAGAAAACTCCAGGG-3′ | 213 |
|
| Reverse |
5′-TGTGCCCTCAAATGAAACCG-3′ |
|
| Vimentin | Forward |
5′-CGCCAACTACATCGACAAGG-3′ | 166 |
|
| Reverse |
5′-GGCTTTGTCGTTGGTTAGCT-3′ |
|
| Bcl-2 | Forward |
5′-GCCTTCTTTGAGTTCGGTGG-3′ | 192 |
|
| Reverse |
5′-GAAATCAAACAGAGGCCGCA-3′ |
|
| Bad | Forward |
5′-GGGACGGAGGACGACG-3′ | 272 |
|
| Reverse |
5′-CACTGGGAGGGGGCGG-3′ |
|
| Bid | Forward |
5′-GGGAAGAATAGAGGCAGA-3′ | 304 |
|
| Reverse |
5′-GACATCACGGAGCAAGGA-3′ |
|
| GAPDH | Forward |
5′-TCAAGAAGGTGGTGAAGCAGG-3′ | 115 |
|
| Reverse |
5′-TCAAAGGTGGAGGAGTGGGT-3′ |
|
Western blot analysis
The T24 or 5637 cells were lysed with lysis buffer
(50 mM Tris-HCl, 0.5 mM EDTA, 1% SDS and 1 mM DTT) on ice. The
concentration of the extracted proteins from the cells were
determined using a BCA assay kit (cat. no. DEM183-1000T; Beijing
BioDee Biotechnology Co., Ltd.). A total of 20 µg protein was mixed
with 4 µl 2X SDS sample buffer. After denaturation, the samples
were separated using 10% SDS-PAGE, transferred to PVDF membranes
and blocked with 5% skimmed milk. Then, the membranes were
incubated with the following primary antibodies: Bad (cat. no.
ab32445; 1:1,000), Bid (cat. no. ab32060; 1:1,000), vimentin (cat.
no. ab92547; 1:1,000), E-cadherin (cat. no. ab40772; 1:1,000),
N-cadherin (cat. no. ab98952; 1:1,000), Bcl2 (cat. no. ab182858;
1:1,000) and GAPDH (cat. no. ab8245; 1:1,000) (all from Abcam)
overnight at 4°C. The membranes were then washed with PBS and
horseradish peroxidase-labeled sheep anti-rabbit IgG secondary
antibody (1:50,000) was added and incubated at room temperature for
60 min. Enhanced chemiluminescence assay (cat. no. WBKLS0500; EMD
Millipore) was used to detect the densitometry of the target
proteins, then the OD was measured using ImageJ software
(v1.8.0.112; National Institutes of Health). The experiments were
performed independently 3 times.
Statistical analysis
Statistical analyses were performed using SPSS
(version 22.0; IBM Corp.). Data were presented as mean ± SD, and
independent t-test was used to investigate intergroup distinctions.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Target gene Notch1 was successfully
constructed
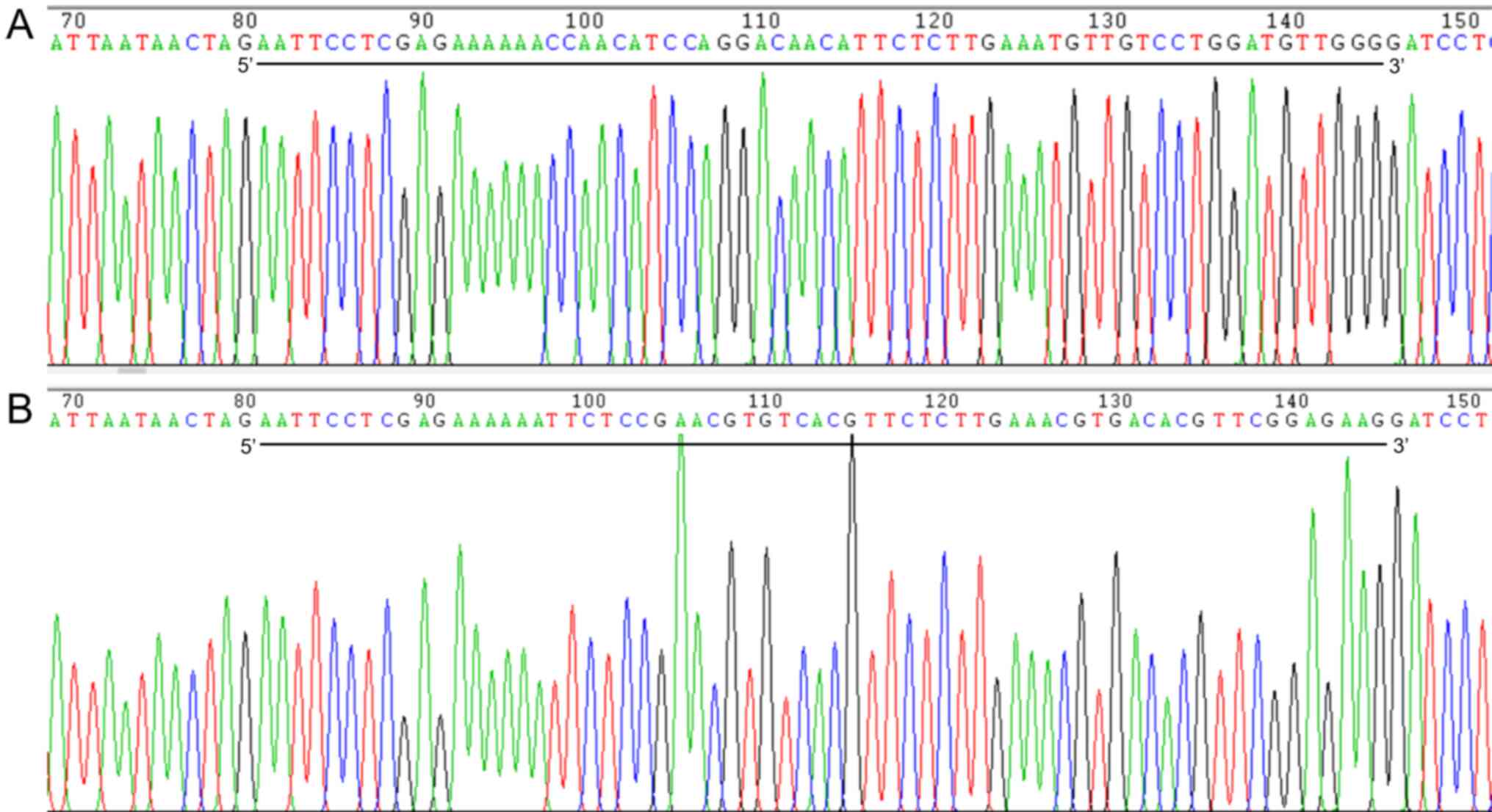
As shown in Fig. 1,
the constructed Notch1 shRNA vector was sequenced, and the inserted
sequence and locus were correct. The interfering vector plasmid of
the target gene Notch1 was successfully constructed.
Notch1 mRNA expression level in BCCs
following Notch1 shRNA
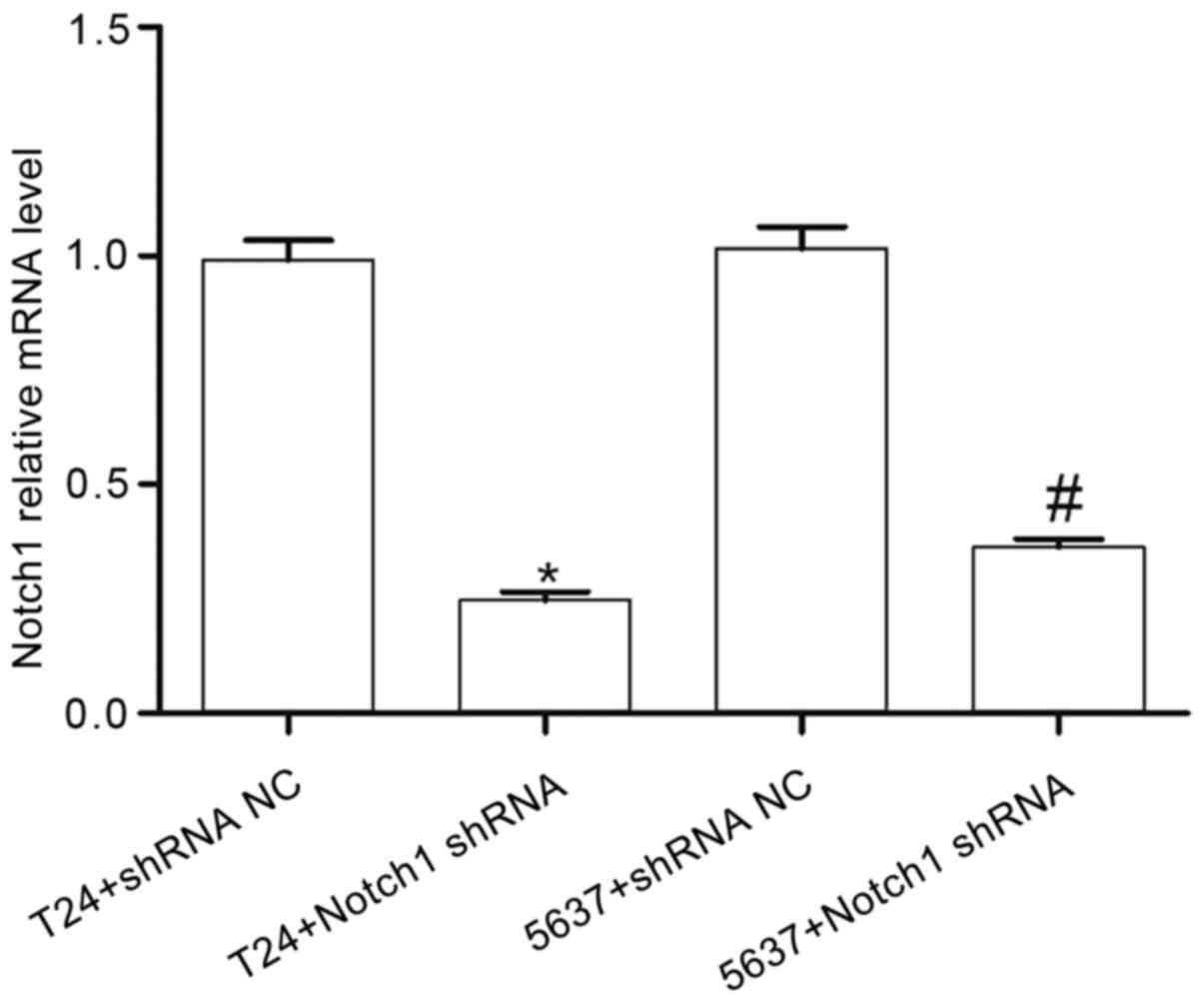
To investigate the role of Notch1 in the BCCs,
plasmid transfection was used to suppress Notch1 expression
in the T24 and 5637 cell lines. Notch1 expression was
detected using RT-qPCR. Compared with that in the T24+shRNA NC
group, the expression level of Notch1 in the T24+Notch1 shRNA group
was significantly decreased (t=16.310; P<0.001). Compared with
that in the 5637+shRNA NC group, the expression level of Notch1 in
the 5637+Notch1 shRNA group was also significantly decreased
(t=13.28; P<0.001) (Fig. 2).
These results indicated that the Notch1 gene was
successfully silenced and inhibited the expression of Notch1
mRNA.
Effect of Notch1 shRNA on the
proliferation of the T24 and 5637 cell lines
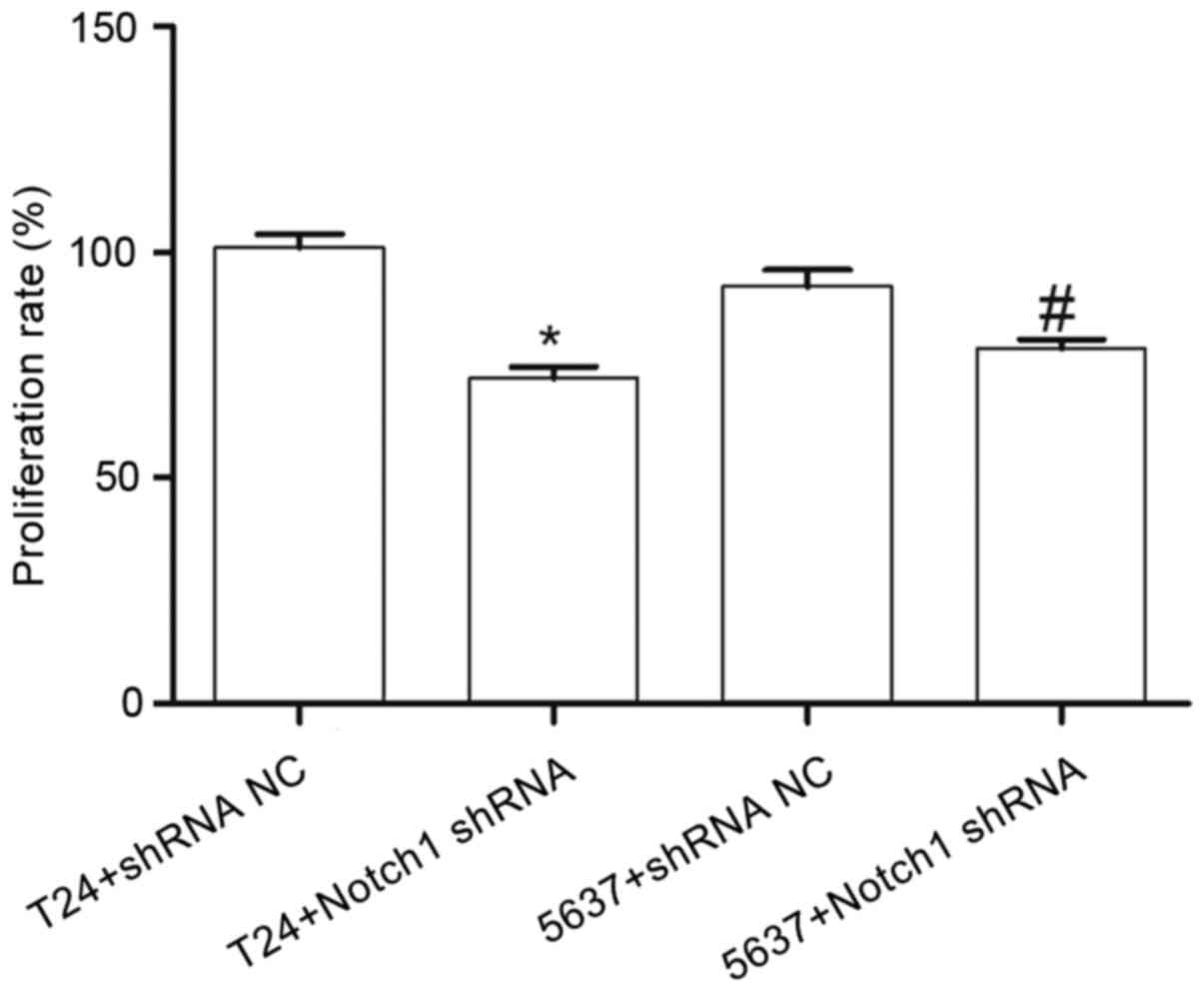
Compared with that in the T24+shRNA NC group, the
cell proliferation rate in the T24+Notch1 shRNA group was
significantly decreased (t=7.583; P<0.05). Similarly, compared
with that in the 5637+shRNA NC group, the cell proliferation rate
in the 5637+Notch1 shRNA group was also significantly decreased
(t=3.283; P<0.05) (Fig. 3). These
results indicated that by silencing the Notch1 gene, the
proliferation of the T24 and 5637 cells was decreased.
Effect of Notch1 shRNA on the invasive
ability of the T24 and 5637 cell lines
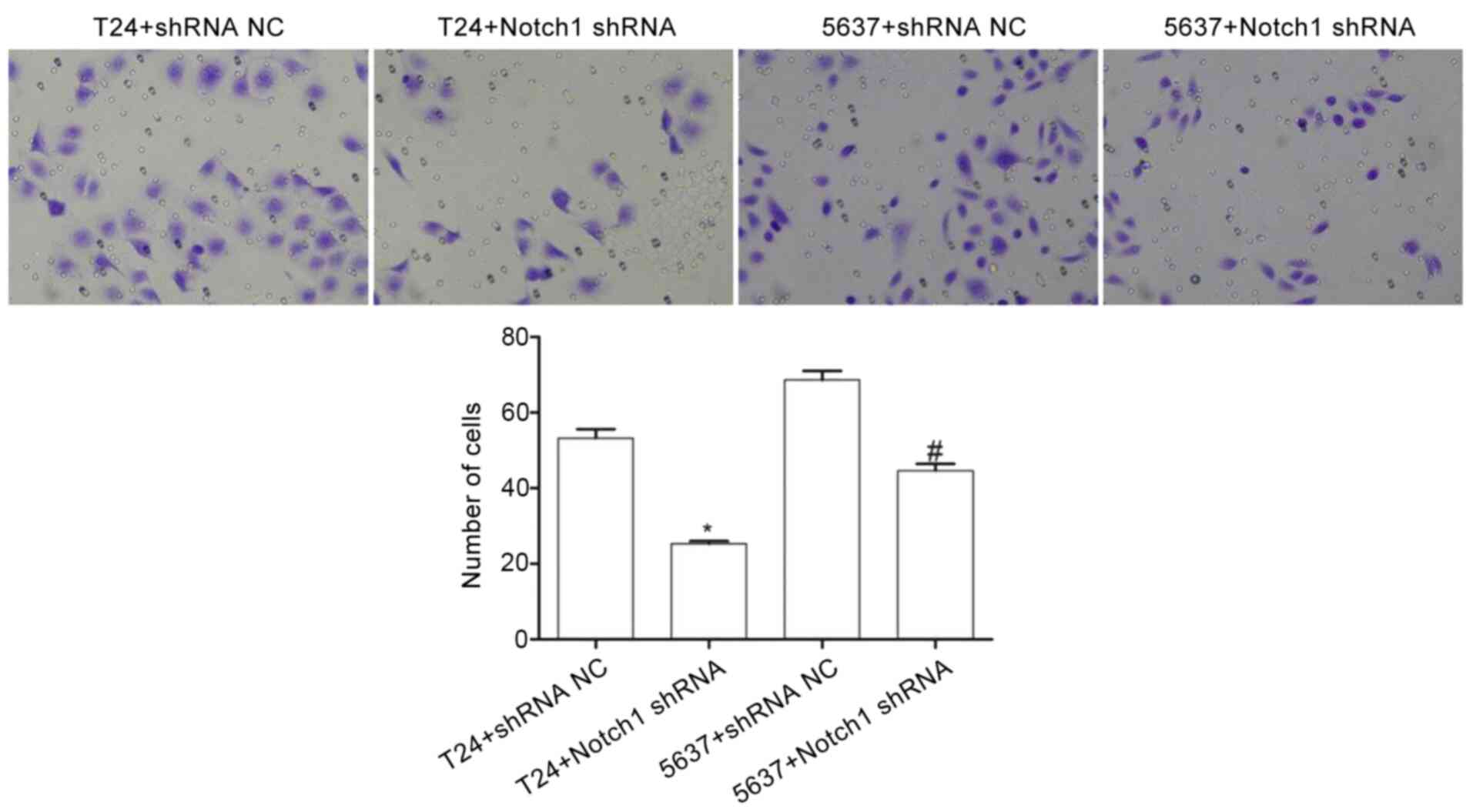
Compared with that in the T24+shRNA NC group, the
number of invasive cells in the T24+Notch1 shRNA group was
significantly decreased (t=11.538; P<0.001). Similarly, compared
with that in the 5637+shRNA NC group, the number of invasive cells
in the 5637+Notch1 shRNA group was significantly decreased
(t=8.205; P<0.05) (Fig. 4). These
results indicated that by silencing the Notch1 gene, the
invasive ability of the T24 and 5637 cells was decreased.
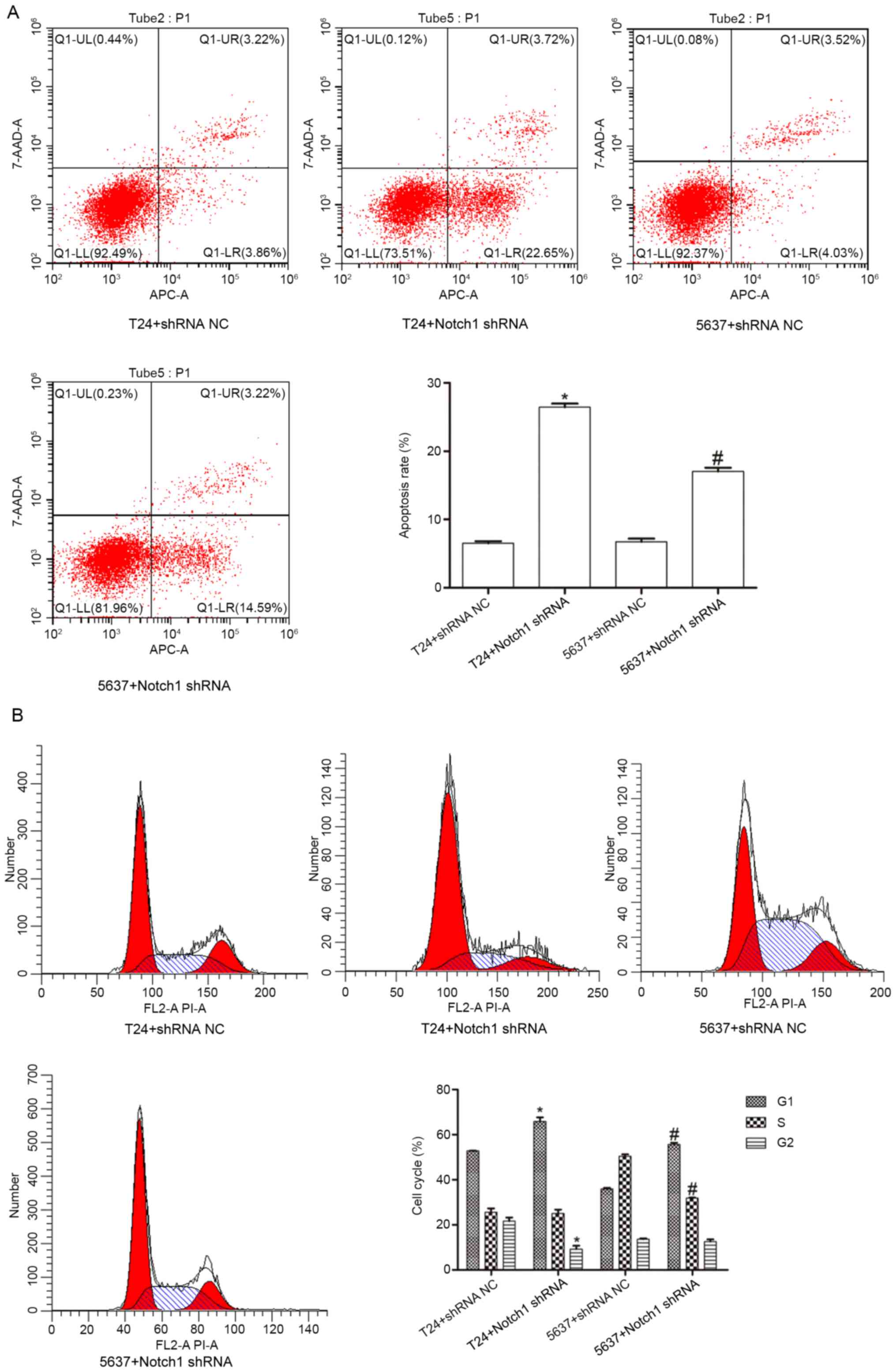
Effect of Notch1 shRNA on apoptosis
and the cell cycle in the T24 and 5637 cell lines
To investigate the effect of Notch1 on BCC
apoptosis, flow cytometry was used to detect T24 and 5637 cell
apoptosis upon Notch1 silencing. Compared with that in the
T24+shRNA NC group, the apoptosis rate in the T24+Notch1 shRNA
group was significantly increased (t=−34.083; P<0.001). Compared
with that in the 5637+shRNA NC group, the apoptosis rate in the
5637+Notch1 shRNA group was also significantly increased
(t=−14.316; P<0.001) (Fig. 5A).
These results indicated that by silencing the Notch1 gene,
the rate of apoptosis of the T24 and 5637 cells was increased.
Flow cytometry was also used to investigate the cell
cycle. The results revealed that compared with that in the
T24+shRNA NC group, the proportion of cells in the G1
phase was significantly increased (t=−6.929; P<0.05), but the
proportion of cells in the G2 phase was significantly
decreased (t=5.631; P<0.05) in the T24+Notch1 shRNA group; no
obvious differences were noted in the proportion of cells in S
phase (t=0.264; P=0.805). In addition, compared with that in the
5637+shRNA NC group, the proportion of cells in the G1
phase was significantly increased (t=−21.302; P<0.001), but the
proportion of cells in S phase was significantly decreased
(t=23.462; P<0.001) in the 5637+Notch1 shRNA group; no obvious
differences were noted in the proportion of cells in the
G2 phase (t=1.072; P=0.344) (Fig. 5B). These results indicated that by
silencing the Notch1 gene, the proportion of the T24 and
5637 cells in the G1 phase was increased.
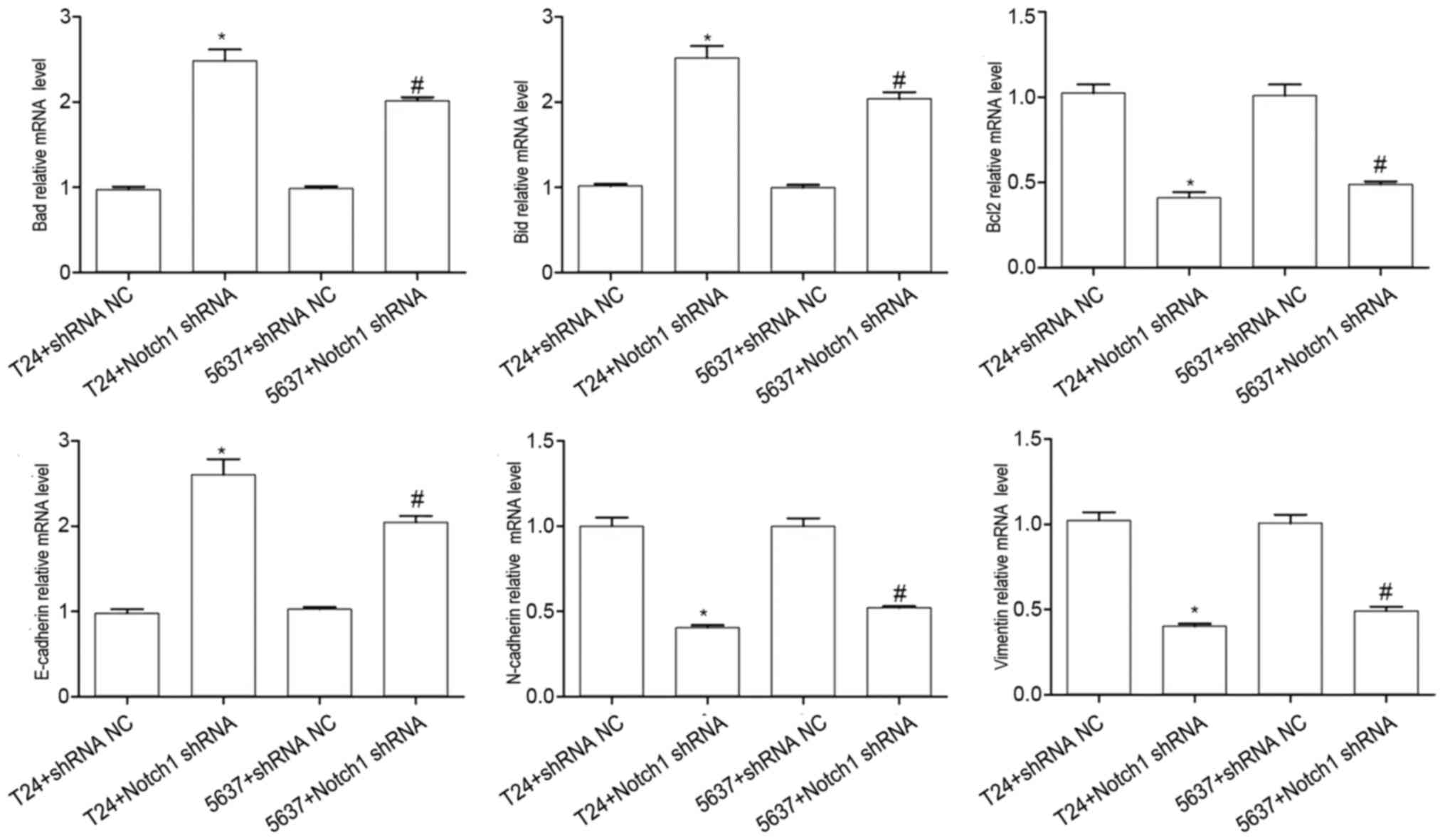
Effect of Notch1 on the mRNA
expression levels of N-cadherin, vimentin, Bcl2, E-cadherin, Bad
and Bid in the BCCs
Compared with that in the T24+shRNA NC, the mRNA
expression level of E-cadherin (t=−8.804; P<0.001), Bad
(t=−11.07; P<0.001), and Bid (t=−10.718; P<0.001) was
significantly increased, but the mRNA expression level of
N-cadherin (t=11.725; P<0.001), vimentin (t=12.597; P<0.001),
and Bcl2 (t=10.627; P<0.001) was significantly decreased in the
T24+Notch1 shRNA group (Fig. 6).
Compared with that in the 5637+shRNA NC, the mRNA
expression level of E-cadherin (t=−13.689; P<0.001), Bad
(t=−23.999; P<0.001), and Bid (t=−12.805; P<0.001) was
significantly increased, but the mRNA expression of N-cadherin
(t=0.105; P<0.001), vimentin (t=9.649; P<0.001), and Bcl2
(t=7.966; P<0.001) was significantly decreased in the
5637+Notch1 shRNA group (Fig. 6).
These results indicated that by silencing the Notch1 gene,
the mRNA expression levels of N-cadherin, vimentin and Bcl2 were
decreased, while E-cadherin, Bad and Bid were increased in the T24
and 5637 cells.
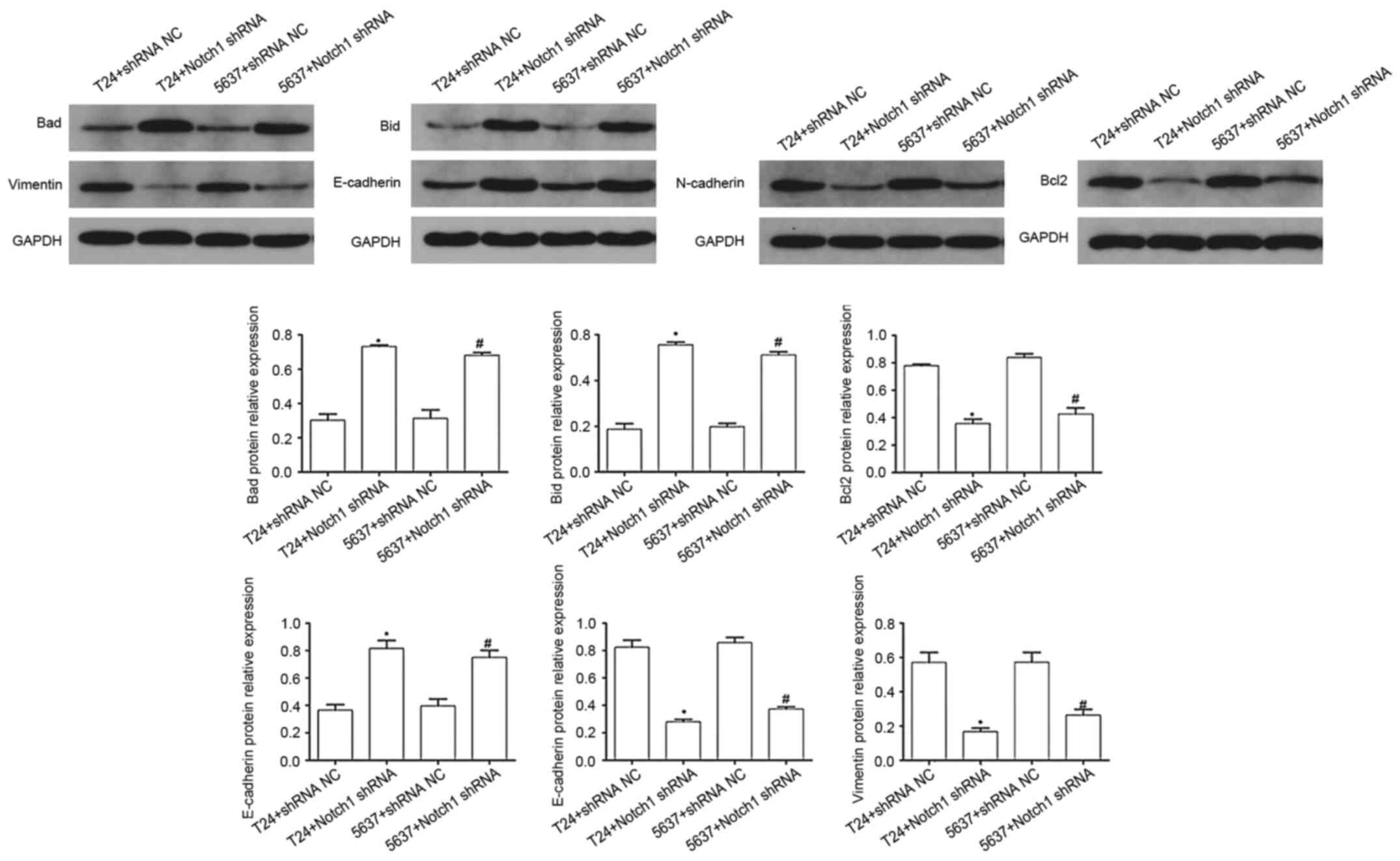
Effect of Notch1 on the protein
expression levels of vimentin, Bcl2, N-cadherin, Bad, Bid and
E-cadherin in the BCCs
The protein expression levels of apoptosis-related
proteins and EMT factors in the T24 and 5637 cells were examined.
Compared with that in the NC groups, the protein expression levels
of E-cadherin (T24 cells, t=−6.39; P<0.05; and 5637 cells,
t=−4.95; P<0.05), Bad (T24 cells, t=−11.372; P<0.001; and
5637 cells, t=−6.988; P<0.05), and Bid (T24 cells, t=−13.85;
P<0.001; and 5637 cells, t=−15.383; P<0.001) were
significantly increased, but the protein expression levels of
N-cadherin (T24 cells, t=9.957; P<0.05; and 5637 cells,
t=11.624; P<0.001), vimentin (T24 cells, t=6.502; P<0.05; and
5637 cells, t=4.681; P<0.05), and Bcl2 (T24 cells, t=12.576;
P<0.001; and 5637 cells, t=8.061; P<0.05) were significantly
decreased in the Notch1 shRNA group (Fig. 7). These results indicated that by
silencing the Notch1 gene, the protein expression levels of
N-cadherin, vimentin, and Bcl2 were decreased, while E-cadherin,
Bad and Bid were increased in the T24 and 5637 cells.
Discussion
Previous studies have confirmed that the Notch
signaling pathway and EMT play key roles in the occurrence,
development, and metastasis of human tumors, such as lung, breast,
prostate and colorectal cancers (14,15).
However, in bladder cancer, the association between the Notch
signaling pathway and EMT has not yet been fully elucidated. The
Notch signaling pathway is abundant in mammals and is involved in
the regulation of numerous life processes, including the
maintenance of the dynamic balance of cell differentiation,
proliferation, and apoptosis (16,17). The
Notch signaling pathway comprises Notch receptors and ligands, as
well as intracellular effector molecules (18). There are four Notch receptors,
including Notch1, Notch2, Notch3 and Notch4; the Notch ligands
include protein jagged (JAG)1, JAG2, δ-like protein (DLL)1, DLL3
and DLL4 (17). An increase in the
signaling levels in the Notch signaling pathway may induce the
development of a variety of tumors, such as lung and breast
cancers, and increase Notch1 mRNA expression level is the most
frequently observed abnormality in tumor tissues, such as lung and
breast (19–23). Notch1 has also been found to
participate in the occurrence and development of diverse tumors,
such as lung, breast, prostate and colorectal cancers by affecting
cell proliferation, differentiation and apoptosis (19–23).
Notch1 shRNA silencing could inhibit cell proliferation and
invasion of the SGC-7901 gastric cancer cell line (19). Notch1 suppression, by genetic
interference, could also inhibit the proliferation and invasion of
breast cancer cells (20). Goriki
et al (12) reported that
increased Notch1 protein expression level in bladder cancer was
associated with an increased pathological grade.
In the present study, the proliferation and invasion
of the BCCs upon Notch1 shRNA was significantly reduced, whereas
cell apoptosis was increased. The mRNA and protein expression level
of the apoptosis-related proteins, Bad and Bid in the Notch1 shRNA
group was significantly increased, whereas the expression level of
the anti-apoptotic protein, Bcl2 was significantly decreased,
suggesting that inhibition of Notch1 promoted apoptosis of
the BCCs. These findings suggested that suppression of
Notch1 inhibited T24 and 5637 cell proliferation and
invasion, and promoted apoptosis.
EMT refers to the ability of epithelial cells to
infiltrate and migrate (by losing polarity and loosening the tight
junctions between cells), which is stimulated by certain factors,
such as inflammation (24). Under
physiological conditions, EMT of epithelial cells can repair
injured normal tissues (25). In
addition, EMT assumes a critical role in tumor occurrence and
progression. EMT is a starting point and an important event in
metastasis cascade reactions, where epithelial cancer cells lose
polarity and cell-cell contact, and therefore the migratory ability
of cancer cells is enhanced (26).
The E-cadherin, N-cadherin, and vimentin proteins are related to
EMT. A decrease in E-cadherin expression has been considered to be
the most significant feature of EMT (27). E-cadherin maintains the tight
junctions between epithelial cells and plays an important role in
maintaining the morphological stability of cells (28). Increased vimentin and N-cadherin
expression ensures that cancer cells acquire mesenchymal
characteristics, that promote migration and invasion (27). N-cadherin mainly mediates the
adhesion between nerve tissue and fibroblasts, while vimentin is
widely distributed in interstitial cells (27). Notch1 can induce EMT by mediating the
expression of various EMT-related genes, such as Snail1 and Snail2
(29,30). Natsuizaka et al (31) reported that Notch1 protein expression
levels were increased in NCI-H2023 squamous cell carcinoma (SCC)
cell line and was associated with a poor prognosis for esophageal
SCC.
To date, there have been relatively few studies on
the role of the Notch1 signaling pathway in BCCs. Therefore, to
investigate whether Notch1 shRNA plays a role in bladder cancer by
inhibiting EMT of BCCs, the mRNA and protein expression level of
EMT-related factors were detected using RT-qPCR and western blot
analysis in BCCs after Notch1 silencing with shRNA.
E-cadherin expression level was found to be notably augmented at
both the mRNA and protein expression level, whereas vimentin and
N-cadherin was reduced upon Notch1 silencing. E-cadherin was
associated with BCC metastasis and invasion, whereas N-cadherin and
vimentin proteins can promote cell adhesion and signal transduction
(32). This suggests that the
inhibition of Notch1 may inhibit the expression level of
EMT-relevant proteins, thereby inhibiting BCC invasion and
metastasis.
In conclusion, inhibiting Notch1 expression
was found to inhibit the invasion of human BCCs, promote their
apoptosis, and inhibit EMT. However, there are still a number
experiments required to validate the results. Future studies should
focus on the Notch1 gene and its association with clinical
outcomes of patients with bladder cancer.
Acknowledgements
Not applicable.
Funding
This research was supported by the Zhejiang
Provincial Natural Science Foundation of China (grant no.
LQ17H050002), the Natural Science Foundation of Inner Mongolia
(grant no. 2017BS0805), the Doctoral Research Initiation Fund of
Baotou Medical College (grant no. BSJJ201707) and the Natural
Sciences-Sailing Plan of Baotou Medical College (grant no. BYJJ-YF
201616).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KWZ, XH, YS, QW, QM and KSZ conceived and designed
the project. KWZ and XH acquired the data. KWZ and XH confirm the
authenticity of all the raw data. KWZ, XH, YS, QW, QM and KSZ
analyzed and interpreted the data. QM and KSZ wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Antoni S, Ferlay J, Soerjomataram I, Znaor
A, Jemal A and Bray F: Bladder cancer incidence and mortality: A
global overview and recent trends. Eur Urol. 71:96–108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Breyer J, Wirtz RM, Otto W, Erben P,
Kriegmair MC, Stoehr R, Eckstein M, Eidt S, Denzinger S, Burger M,
et al BRIDGE Consortium, : In stage pT1 non-muscle-invasive bladder
cancer (NMIBC), high KRT20 and low KRT5 mRNA expression identify
the luminal subtype and predict recurrence and survival. Virchows
Arch. 470:267–274. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Babjuk M, Böhle A, Burger M, Capoun O,
Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M,
et al: EAU guidelines on non-muscle-invasive urothelial carcinoma
of the bladder: Update 2016. Eur Urol. 71:447–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou Z, Zhao S, Lu Y, Wu J, Li Y, Gao Z,
Yang D and Cui Y: Meta-analysis of efficacy and safety of
continuous saline bladder irrigation compared with intravesical
chemotherapy after transurethral resection of bladder tumors. World
J Urol. 37:1075–1084. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kinnaird A, Dromparis P and Evans H:
Recurrence and upstaging rates of T1 high-grade urothelial
carcinoma of the bladder on repeat resection in a Canadian,
resource-limited, healthcare system. Can Urol Assoc J. 12:267–269.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gordon PC, Thomas F, Noon AP, Rosario DJ
and Catto JW: Long-term outcomes from re-resection for high-risk
non-muscle-invasive bladder cancer: A potential to rationalize use.
Eur Urol Focus. 5:650–657. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van den Bosch S and Alfred Witjes J:
Long-term cancer-specific survival in patients with high-risk,
non-muscle-invasive bladder cancer and tumour progression: A
systematic review. Eur Urol. 60:493–500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu Y, Gao Q, Zhu S, Wu Q, Zhu R, Zhong H,
Xing C, Qu H, Wang D, Li B, et al: Low-intensity pulsed ultrasound
regulates proliferation and differentiation of neural stem cells
through notch signaling pathway. Biochem Biophys Res Commun.
526:793–798. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qiu Z, Lin S, Hu X, Zeng J, Xiao T, Ke Z
and Lv H: Involvement of miR-146a-5p/neurogenic locus notch homolog
protein 1 in the proliferation and differentiation of
STRO-1+ human dental pulp stem cells. Eur J Oral Sci.
127:294–303. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ai X, Jia Z, Liu S, Wang J and Zhang X:
Notch-1 regulates proliferation and differentiation of human
bladder cancer cell lines by inhibiting expression of Krüppel-like
factor 4. Oncol Rep. 32:1459–1464. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maraver A, Fernandez-Marcos PJ, Cash TP,
Mendez-Pertuz M, Dueñas M, Maietta P, Martinelli P, Muñoz-Martin M,
Martínez-Fernández M, Cañamero M, et al: NOTCH pathway inactivation
promotes bladder cancer progression. J Clin Invest. 125:824–830.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goriki A, Seiler R, Wyatt AW,
Contreras-Sanz A, Bhat A, Matsubara A, Hayashi T and Black PC:
Unravelling disparate roles of NOTCH in bladder cancer. Nat Rev
Urol. 15:345–357. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Ma J, Qian X, Wu Q, Xia J, Miele L,
Sarkar FH and Wang Z: Regulation of EMT by Notch signaling pathway
in tumor progression. Curr Cancer Drug Targets. 13:957–962. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Espinoza I and Miele L: Deadly crosstalk:
Notch signaling at the intersection of EMT and cancer stem cells.
Cancer Lett. 341:41–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
D'Angelo RC, Ouzounova M, Davis A, Choi D,
Tchuenkam SM, Kim G, Luther T, Quraishi AA, Senbabaoglu Y, Conley
SJ, et al: Notch reporter activity in breast cancer cell lines
identifies a subset of cells with stem cell activity. Mol Cancer
Ther. 14:779–787. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Henrique D and Schweisguth F: Mechanisms
of Notch signaling: A simple logic deployed in time and space.
Development. Feb 1–2019.(Epub ahead of print). doi:
10.1242/dev.172148. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kovall RA, Gebelein B, Sprinzak D and
Kopan R: The Canonical Notch Signaling pathway: Structural and
biochemical insights into shape, sugar, and force. Dev Cell.
41:228–241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei G, Chang Y, Zheng J, He S, Chen N,
Wang X and Sun X: Notch1 silencing inhibits proliferation and
invasion in SGC 7901 gastric cancer cells. Mol Med Rep.
9:1153–1158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hristova NR, Tagscherer KE, Fassl A,
Kopitz J and Roth W: Notch1-dependent regulation of p27 determines
cell fate in colorectal cancer. Int J Oncol. 43:1967–1975. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Q, Yuan Y, Cui J, Xiao T and Jiang
D: Paeoniflorin inhibits proliferation and invasion of breast
cancer cells through suppressing Notch-1 signaling pathway. Biomed
Pharmacother. 78:197–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lai XX, Li G, Lin B and Yang H:
Interference of Notch 1 inhibits the proliferation and invasion of
breast cancer cells: Involvement of the β catenin signaling
pathway. Mol Med Rep. 17:2472–2478. 2018.PubMed/NCBI
|
|
23
|
Lloyd-Lewis B, Mourikis P and Fre S: Notch
signalling: Sensor and instructor of the microenvironment to
coordinate cell fate and organ morphogenesis. Curr Opin Cell Biol.
61:16–23. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saitoh M: Involvement of partial EMT in
cancer progression. J Biochem. 164:257–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stone RC, Pastar I, Ojeh N, Chen V, Liu S,
Garzon KI and Tomic-Canic M: Epithelial-mesenchymal transition in
tissue repair and fibrosis. Cell Tissue Res. 365:495–506. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rodriguez-Aznar E, Wiesmüller L, Sainz B
Jr and Hermann PC: EMT and stemness-key players in pancreatic
cancer stem cells. Cancers (Basel). 11:11362019. View Article : Google Scholar
|
|
27
|
Pastushenko I and Blanpain C: EMT
Transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ko JH, Nam D, Um JY, Jung SH, Sethi G and
Ahn KS: Bergamottin suppresses metastasis of lung cancer cells
through abrogation of diverse oncogenic signaling cascades and
epithelial-to-mesenchymal transition. Molecules. 23:16012018.
View Article : Google Scholar
|
|
29
|
van Denderen BJ and Thompson EW: Cancer:
The to and fro of tumour spread. Nature. 493:487–488. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matsuno Y, Coelho AL, Jarai G, Westwick J
and Hogaboam CM: Notch signaling mediates TGF-β1-induced
epithelial-mesenchymal transition through the induction of Snai1.
Int J Biochem Cell Biol. 44:776–789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Natsuizaka M, Whelan KA, Kagawa S, Tanaka
K, Giroux V, Chandramouleeswaran PM, Long A, Sahu V, Darling DS,
Que J, et al: Interplay between Notch1 and Notch3 promotes EMT and
tumor initiation in squamous cell carcinoma. Nat Commun.
8:17582017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kang J, Kim E, Kim W, Seong KM, Youn H,
Kim JW, Kim J and Youn B: Rhamnetin and cirsiliol induce
radiosensitization and inhibition of epithelial-mesenchymal
transition (EMT) by miR-34a-mediated suppression of Notch-1
expression in non-small cell lung cancer cell lines. J Biol Chem.
288:27343–27357. 2013. View Article : Google Scholar : PubMed/NCBI
|