Introduction
Long non-coding RNAs (lncRNAs) are functional small
RNA molecules that cannot encode proteins (1). Increasing evidence suggest that lncRNAs
exert important effects on the physiological and pathological
processes of the human body, primarily through transcriptional or
post-transcriptional mechanisms (2,3). Long
intergenic non-protein coding RNA 52 (LINC00052, National Center
for Biotechnology Information reference sequence: NR_026869.1) is
1,786 base pairs in length and is located on human chromosome 15
(4). Abnormal expression of
LINC00052 has been observed in several tumors, such as liver cancer
(5–7), breast cancer (8) and colorectal cancer (9). The current research on LINC00052 mainly
focuses on the effect of LINC00052 on tumorigenesis, cancer
progression and metastasis (Table
I).
 | Table I.Targets of LINC00052 in tumors. |
Table I.
Targets of LINC00052 in tumors.
| Tumor type | miRNA | Target gene | Function | Refs. |
|---|
| HCC | miR-128,
miR-485-3p | NTRK3 | Cell invasion and
migration | (4) |
|
| miR-452-5p | EPB41L3 | Cell invasion and
migration | (5) |
|
| miR-101-3p | SOX9 | Cell proliferation
and metastasis | (6) |
| Breast cancer | – | HER3/ErbB3 | Cell
proliferation | (7) |
| CRC | miR-574-5p | CALCOCO1 | Cell
metastasis | (8) |
| Cervical
carcinoma | – | STAT3 | Cell metastasis and
invasion | (10) |
| GC | – | Wnt/β-catenin
pathway | Cell proliferation
and metastasis | (11) |
| HNSCC | miR-608 | EGFR | Cell proliferation,
migration and invasion | (52) |
| Glioma | – | KLF6 | Cell proliferation,
migration and invasion | (53) |
The effect of LINC00052 on tumors was first reported
by Xiong et al (5) in 2016,
with the help of the gene trapping technique (10). The capture plasmid pU21 was
transfected into hepatocellular carcinoma (HCC) cells through a
plasmid vector. Subsequently, several single cell clones (for
example A1 and A2) were obtained. The wound healing and Transwell
assays demonstrated that the migratory and invasive abilities of
the pU21 plasmid-obtained monoclonal cells were enhanced in the A26
and A55 cell lines, and slightly weakened in the A18 and A28 cells
lines compared with the controls (5). 5′RACE-PCR results demonstrated that the
gene trapped by pU21 in the A55 cell line was LINC00052 (5); functional tests demonstrated that
LINC00052 has the ability to regulate the invasion and migration of
HCC cells (5). Subsequently, several
studies have reported that LINC00052 is strongly associated with
the occurrence and development of several tumors, including breast
cancer (8), colorectal cancer (CRC)
(9), cervical carcinoma (11) and gastric carcinoma (GC) (12). However, there is no conclusive
article on the association between LINC00052 and these tumors. The
present review summarizes the biological functions of LINC00052
during the pathogenic process of certain tumors and discusses its
potential therapeutic targets.
LINC00052 and HCC
HCC is one of the most common types of cancer
worldwide. Bray et al (13)
reported that HCC ranked sixth among the most diagnosed cancers
(841,080 new cases) and was the fourth leading cause of
cancer-associated mortality (781,631 deaths) in 2018, among the 38
types of tumors evaluated in 185 countries.
It has been proven that LINC00052 inhibits the
invasion and migration of HCC cells (5). Through in-depth investigation of the
molecular mechanisms underlying the effects of LINC00052 on the
development of HCC, three target genes of LINC00052 were
identified: Neurotrophic receptor tyrosine kinase 3 (NTRK3)
(5), erythrocyte membrane protein
band 4.1-like 3 (EPB41L3) (6) and
SRY-related HMG-box gene 9 (SOX9) (7).
NTRK3 is a type of neurotrophin receptor that plays
an important role in the nervous system, and it may be involved in
the initiation, progression and metastasis of several human tumors,
such as breast, medullary thyroid, liver, colon, lung and prostate
cancers (14–16). Xiong et al (5) demonstrated that LINC00052 inhibits cell
proliferation, invasion and migration by upregulating NTRK3
expression through complementary base pairing with microRNA
(miRNA/miR)-128 and miR-485-3p. This is a key regulatory mechanism
through which lncRNAs regulate target gene expression in tumors by
sponging miRNAs, thus reducing their regulatory effect on mRNAs
(17) (Fig. 1).
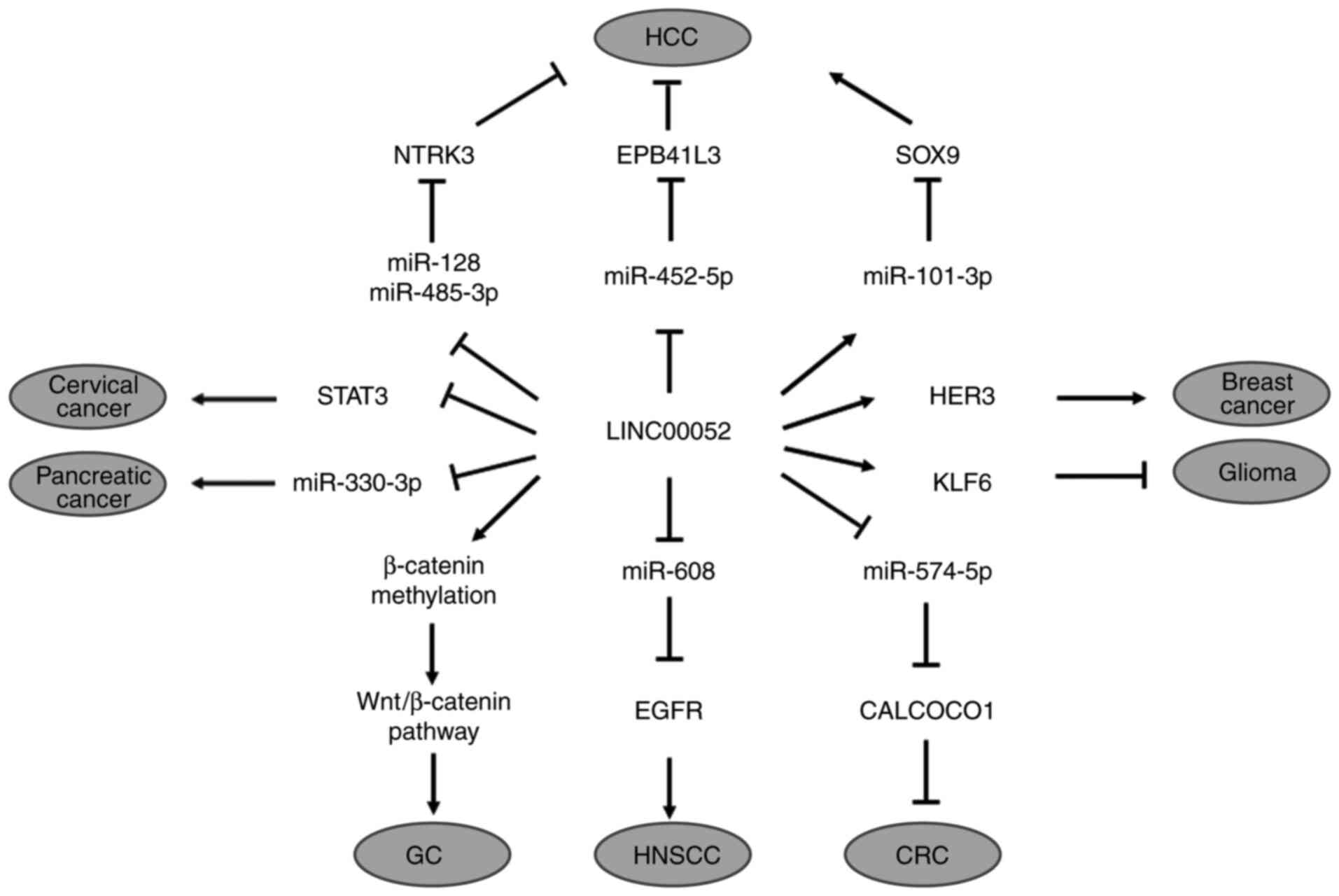 | Figure 1.Schematic overview of role of
LINC00052 in different tumors. LINC00052, long intergenic
non-protein coding RNA 52; HCC, hepatocellular carcinoma; NTRK3,
neurotrophic receptor tyrosine kinase 3; EPB41L3, erythrocyte
membrane protein band 4.1-like 3; SOX9, SRY-related HMG-box gene 9;
miR, microRNA; STAT3, signal transducer and activator of
transcription 3; HER3, human epidermal growth factor receptor 3;
KLF6, kruppel-like factor 6; EGFR, epidermal growth factor
receptor; CALCOCO1, calcium binding and coiled-coil domain 1; GC,
gastric carcinoma; HNSCC, head and neck squamous cell carcinoma;
CRC, colorectal cancer. |
Zhu et al (6)
reported that LINC00052 directly acts on EPB41L3 to affect the
occurrence and development of HCC by targeting miR-452-5p. EPB41L3,
a membrane skeletal protein that belongs to the protein 4.1 family
(18), functions as a tumor
suppressor in different tumors, such as GC, non-small cell lung
cancer and renal clear cell carcinoma (19–21). In
addition, Zhu et al (6)
confirmed that EPB41L3 is a downstream target gene of LINC00052 via
microarray, reverse transcription-quantitative PCR and western blot
analyses. miR-452-5p was predicted to have two binding sites in the
sequence of LINC00052 through the bioinformatics databases:
DIANA-LncBase, https://bigd.big.ac.cn; miRcode,
http://www.mircode.org and BiBiServ, https://bibiserv.cebitec.uni-bielefeld.de.
Furthermore, it was demonstrated that miR-452-5p can recover
LINC00052 function in HCC cells and inhibit EPB41L3 expression by
targeting the 3′-untranslated region of miR-452-5p (Fig. 1). Thus, Zhu et al (6) uncovered a novel
LINC00052/miR-452-5p/EPB41L3 regulatory network in HCC.
The third target gene of LINC00052 is SOX9. SOX9 is
a member of the SOX transcription factor family, which is
associated with early embryonic development (22). SOX9 expression is frequently
upregulated, and it is characterized as an oncogene in different
types of human cancer, including lung cancer, GC and HCC (23–25). Yan
et al (7) reported that
LINC00052 promotes miR-101-3p expression by enhancing its promoter
activity, and that miR-101-3p can interact with SOX9 to affect the
occurrence and development of HCC (Fig.
1).
LINC00052 and breast cancer
Breast cancer is the leading cause of
cancer-associated mortality among women worldwide (26). Bray et al (13) reported 2,088,849 new cases and
626,679 breast cancer-associated mortalities in 2018, and breast
cancer ranked second among the 38 types of tumors investigated in
185 countries. Thus, several studies continue to persistently
investigate the pathogenesis and pathogenic genes of breast cancer,
in the hope to identify biomarkers associated with its early
diagnosis and treatment (27–29). By
using RNA isolation, RNA-seq and microarray, Muñoz-Galindo et
al (29) identified 221 and 146
dysregulated lncRNAs after 12 and 20 days of three-dimensional
culture in breast cancer cells, respectively. It was demonstrated
that LINC00052 was the most extensively downregulated lncRNA at two
time points: Decreased by 47-fold on the 12th day and 69-fold on
the 20th day. These results suggest that LINC00052 is a key target
for intervention in tumor diagnosis and treatment.
Triple-negative breast cancer (TNBC), which is
characterized by the lack of estrogen receptor (ER), progesterone
receptor (PR) and human epidermal growth factor receptor 2
expression, is an extremely aggressive type of breast cancer
(30). Lv et al (31) demonstrated that LINC00052 is markedly
upregulated in TNBC tissues compared with non-TNBC tissues and may
serve as a powerful indicator for diagnosis of TNBC by analyzing
the result of the receiver operating characteristic curve (area
under the curve, 0.823; 95% confidence interval, 0.637–1.000).
These results suggest that LINC00052 may serve as a potential
biomarker for differentiating TNBC samples from non-TNBC
samples.
Human epidermal growth factor receptor 3
(HER3/ErbB3) is a member of the epidermal growth factor receptor
(EGFR) family of tyrosine kinase receptors (8). Upregulated HER3 expression has been
implicated in the development and progression of different types of
cancer (32,33). For example, Salameh et al
(8) confirmed that LINC00052
promotes the proliferation of breast cancer cells by increasing
HER3 signaling both in vitro and in vivo (Fig. 1). This suggests that LINC00052 may
act as a potential biomarker for HER3-targeted cancer
therapies.
LINC00052 and CRC
CRC is a common malignant gastrointestinal tumor
(34). Bray et al (13) reported that CRC ranked third among
the most diagnosed cancers and second in mortality rate among 38
types of tumors in 185 countries, with 1,096,601 new cancer cases
and 551,369 mortalities recorded in 2018. Previous studies have
confirmed that lncRNAs are implicated in the occurrence and
development of CRC, such as H19 (35), glycolysis-associated lncRNA of
colorectal cancer (36) and fez
family zinc finger protein 1-antisense RNA 1 (37). Yu et al (9) reported that downregulation of LINC00052
expression promotes the proliferative, migratory and invasive
abilities of CRC cells. It was also confirmed that LINC00052
affects the occurrence and development of CRC by modulating the
expression of calcium binding and coiled-coil domain 1 (CALCOCO1)
through interaction with miR-574-5p (Fig. 1). CALCOCO1 is located on chromosome
12p13 and its different domains can bind with glutamate receptor
interacting protein 1 and β-catenin, thereby affecting cell
division and apoptosis (38).
LINC00052 and cervical carcinoma
Epidemiological statistics indicate that cervical
cancer was the fourth most common gynecological malignancy
worldwide in 2012 (39). Bray et
al (13) reported that cervical
carcinoma was the fourth most diagnosed cancer and the fourth
leading cause of cancer-associated mortality in women among the 38
types of tumors in 185 countries, with 569,847 new cases and
311,365 mortalities reported in 2018. The use of cervical vaccines,
early detection and timely treatment have decreased the incidence
and mortality rates of cervical cancer (40). Increasing evidence suggest that
lncRNAs serve as important regulators in the progression of
cervical carcinoma (41). For
example, lncRNA papillary thyroid carcinoma susceptibility
candidate 3 has been demonstrated to suppress the invasion and
proliferation of cervical cancer cells by regulating miR-574-5p
expression (41), whereas lncRNA
nuclear paraspeckle assembly transcript 1 regulates the
proliferation and invasion of cervical cancer cells by targeting
the PI3K/AKT pathway (42). Lin
et al (11) demonstrated that
LINC00052 expression is significantly downregulated in both
cervical cancer tissues and cells. Overexpression of LINC00052
inhibits the proliferation, migration and invasion of cervical
cancer cells. In addition, signal transducer and activator of
transcription 3 (STAT3) protein expression is downregulated
following overexpression of LINC00052 (11), suggesting that LINC00052 inhibits
tumorigenesis of cervical cancer by suppressing the STAT3 pathway
(Fig. 1).
LINC00052 and GC
GC is the fifth most common cancer and the third
most common cause of cancer-associated mortality worldwide, with
over one million estimated new cases and 782,685 deaths in 2018
(13,43). Recent studies have reported that
lncRNAs are associated with the occurrence and development of GC
(44–46).
Shan et al (12) demonstrated that LINC00052 is highly
expressed in GC tissues, which is associated with tumor progression
and poor prognosis. The underlying molecular mechanisms of
LINC00052 on the occurrence and development of GC were
investigated, and the results demonstrated that LINC00052 maintains
the stability of β-catenin by promoting β-catenin methylation to
activate the Wnt/β-catenin pathway (Fig.
1). This suggests that LINC00052 may promote the proliferation
and metastasis of GC cells by activating the Wnt/β-catenin pathway.
However, some human GC cell lines (MGC-803, BGC-823 and SGC-7901)
may be contaminated with HeLa cells (47), thus authenticated cell lines or
animal models should be performed accordingly.
LINC00052 and other cancers
Tongue squamous cell carcinoma (TSCC) is the most
common type of oral squamous cell carcinoma, which is characterized
by marked invasiveness, early lymph node metastasis and poor
prognosis (48,49). To further investigate the molecular
mechanism underlying the development and progression of TSCC, Zhang
et al (50) identified a
total of 1,867 mRNAs, 828 lncRNAs and 81 miRNAs that are aberrantly
expressed in TSCC tissues compared with normal tissues via RNA
sequence data processing, expression profile analysis and
statistical analysis. It was predicted that LINC00052 is
significantly associated with the overall survival rate of TSCC
(50).
Pancreatic cancer is an aggressive malignant tumor
with a poor prognosis (51).
LINC00052 has been demonstrated to act as a tumor suppressor by
negatively modulating miR-330-3p expression to affect the
occurrence and progression of prostate cancer (52). In addition, LINC00052 may regulate
the miR-608/EGFR axis to promote the progression of head and neck
squamous cell carcinoma (53), and
suppresses the proliferation, migration and invasion of glioma
cells by upregulating kruppel-like factor 6 expression (54) (Fig.
1). Taken together, these results suggest that LINC00052
regulates a broad range of tumors.
Conclusions and perspectives
In the present review, the potential molecular
mechanisms of action of LINC00052 in the pathological process of
several tumors, such as HCC, breast cancer, CRC, prostate cancer
and GC, were described (Table I).
LINC00052 may represent a functional research hotspot in multiple
tumors. LINC00052 can affect the tumor biological processes by
regulating tumor cell proliferation, invasion, metastasis and
migration. LINC00052 may serve as a miRNA sponge, competing with
other genes for miRNA binding, and thus decrease mRNA transcription
of targeted genes. However, further studies are required to
validate the molecular mechanism of LINC00052. The tumorigenic
mechanism of lncRNA is intricate and more information on the
regulation of LINC00052 in response to malignant transformation is
required to elucidate its complicated role in different tumors. In
addition, the network of lncRNA/miRNA/target genes requires further
investigation to determine the association between lncRNA
expression and the occurrence and development of tumors. In
conclusion, the present review summarizes the potential biomarkers
or targets for novel therapeutic strategies.
Acknowledgements
Not applicable.
Funding
The present review was supported by the Chongqing
Basic Research and Exploration Project (grant no.
cstc2018jcyjAX0751).
Availability of data and materials
Not applicable.
Authors' contributions
DX, DW and YC were involved in the conception of the
present review. DX drafted the initial manuscript. DW and YC
critically revised the manuscript for important intellectual
content. DX and YC confirmed the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
LINC00052
|
long intergenic non-protein coding RNA
52
|
|
HCC
|
hepatocellular carcinoma
|
|
CRC
|
colorectal cancer
|
|
GC
|
gastric carcinoma
|
|
NTRK3
|
neurotrophic receptor tyrosine kinase
3
|
|
EPB41L3
|
erythrocyte membrane protein band
4.1-like 3
|
|
SOX9
|
SRY-related HMG-box gene 9
|
|
TNBC
|
triple-negative breast cancer
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
EGFR
|
epidermal growth factor receptor
|
|
HER3
|
human epidermal growth factor receptor
3
|
|
CALCOCO1
|
calcium binding and coiled-coil domain
1
|
|
STAT3
|
signal transducer and activator of
transcription 3
|
|
TSCC
|
tongue squamous cell carcinoma
|
References
|
1
|
Li T, Mo X, Fu L, Xiao B and Guo J:
Molecular mechanisms of long noncoding RNAs on gastric cancer.
Oncotarget. 7:8601–8612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing miRNA-lncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
National Center for Biotechnology
Information, . Homo sapiens long intergenic non-protein coding RNA
52 (LINC00052), long non-coding RNA [DB/OL]. [2021-02-09].
https://www.ncbi.nlm.nih.gov/nuccore/223634020
|
|
5
|
Xiong D, Sheng Y, Ding S, Chen J, Tan X,
Zeng T, Qin D, Zhu L, Huang A and Tang H: LINC00052 regulates the
expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC
cells invasion and migration. Oncotarget. 7:47593–47608. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu L, Yang N, Chen J, Zeng T, Yan S, Liu
Y, Yu G, Chen Q, Du G, Pan W, et al: LINC00052 upregulates EPB41L3
to inhibit migration and invasion of hepatocellular carcinoma by
binding miR-452-5p. Oncotarget. 8:63724–63737. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yan S, Shan X, Chen K, Liu Y, Yu G, Chen
Q, Zeng T, Zhu L, Dang H, Chen F, et al: LINC00052/miR-101-3p axis
inhibits cell proliferation and metastasis by targeting SOX9 in
hepatocellular carcinoma. Gene. 679:138–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salameh A, Fan X, Choi BK, Zhang S, Zhang
N and An Z: HER3 and LINC00052 interplay promotes tumor growth in
breast Cancer. Oncotarget. 8:6526–6539. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu GF, Xiong DM, Liu ZS, Li YG, Chen K and
Tang H: Long noncoding RNA LINC00052 inhibits colorectal cancer
metastasis by sponging microRNA-574-5p to modulate CALCOCO1
expression. J Cell Biochem. 120:17258–17272. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang H, Araki K, Li ZH and Yamamura K:
Characterization of Ayu17-449 gene expression and resultant kidney
pathology in a knockout mouse model. Transgenic Res. 17:599–608.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin J, Nong LL, Li MQ, Yang FC, Wang SH
and Liu MJ: LINC00052 inhibits tumor growth, invasion and
metastasis by repressing STAT3 in cervical carcinoma. Eur Rev Med
Pharmacol Sci. 23:4673–4679. 2019.PubMed/NCBI
|
|
12
|
Shan Y, Ying R, Jia Z, Kong W, Wu Y, Zheng
S and Jin H: LINC00052 promotes gastric cancer cell proliferation
and metastasis via activating the Wnt/β-catenin signaling pathway.
Oncol Res. 25:1589–1599. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin W, Kim GM, Kim MS, Lim MH, Yun C,
Jeong J, Nam JS and Kim SJ: TrkC plays an essential role in breast
tumor growth and metastasis. Carcinogenesis. 31:1939–1947. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Genevois AL, Ichim G, Coissieux MM,
Lambert MP, Lavial F, Goldschneider D, Jarrosson-Wuilleme L,
Lepinasse F, Gouysse G, Herceg Z, et al: Dependence receptor TrkC
is a putative colon cancer tumor suppressor. Proc Natl Acad Sci
USA. 110:3017–3022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Satoh F, Mimata H, Nomura T, Fujita Y,
Shin T, Sakamoto S, Hamada Y and Nomura Y: Autocrine expression of
neurotrophins and their receptors in prostate cancer. Int J Urol.
8:S28–S34. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Zhang J, Ye M, Zhu M, Zhang B, Roy
M, Liu J and An X: Tumor suppressor role of protein 4.1B/DAL-1.
Cell Mol Life Sci. 71:4815–4830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang H, Xu M, Cui X, Liu Y, Zhang Y, Sui
Y, Wang D, Peng L, Wang D and Yu J: Aberrant expression of the
candidate tumor suppressor gene DAL-1 due to hypermethylation in
gastric cancer. Sci Rep. 6:217552016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Xu R, Li G, Xie X, Long J and
Wang H: Loss of expression of the differentially expressed in
adenocarcinoma of the lung (DAL-1) protein is associated with
metastasis of non-small cell lung carcinoma cells. Tumour Biol.
33:1915–1925. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamada D, Kikuchi S, Williams YN,
Sakurai-Yageta M, Masuda M, Maruyama T, Tomita K, Gutmann DH,
Kakizoe T, Kitamura T, et al: Promoter hypermethylation of the
potential tumor suppressor DAL-1/4.1B gene in renal clear cell
carcinoma. Int J Cancer. 118:916–923. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao J, Zhang JY, Li YH and Ren F:
Decreased expression of SOX9 indicates a better prognosis and
inhibits the growth of glioma cells by inducing cell cycle arrest.
Int J Clin Exp Pathol. 8:10130–10138. 2015.PubMed/NCBI
|
|
23
|
Wang X, Ju Y, Zhou M, Liu X and Zhou C:
Upregulation of SOX9 promotes cell proliferation, migration and
invasion in lung adenocarcinoma. Oncol Lett. 10:990–994. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Choi YJ, Song JH, Yoon JH, Choi WS, Nam
SW, Lee JY and Park WS: Aberrant expression of SOX9 is associated
with gastrokine 1 inactivation in gastric cancers. Gastric Cancer.
17:247–254. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu C, Liu L, Chen X, Cheng J, Zhang H,
Shen J, Shan J, Xu Y, Yang Z, Lai M and Qian C: Sox9 regulates
self-renewal and tumorigenicity by promoting symmetrical cell
division of cancer stem cells in hepatocellular carcinoma.
Hepatology. 64:117–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fan L, Strasser KW, Li JJ, St Louis J,
Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast cancer
in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao W, Geng D, Li S, Chen Z and Sun M:
LncRNA HOTAIR influences cell growth, migration, invasion, and
apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer
Med. 7:842–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C,
Chen X and Han M: Activation of LncRNA TINCR by H3K27 acetylation
promotes Trastuzumab resistance and epithelial-mesenchymal
transition by targeting MicroRNA-125b in breast cancer. Mol Cancer.
18:32019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Muñoz-Galindo L, Melendez-Zajgla J,
Pacheco-Fernández T, Rodriguez-Sosa M, Mandujano-Tinoco EA,
Vazquez-Santillan K, Castro-Oropeza R, Lizarraga F, Sanchez-Lopez
JM and Maldonado V: Changes in the transcriptome profile of breast
cancer cells grown as spheroids. Biochem Biophys Res Commun.
516:1258–1264. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Agrawal LS and Mayer IA: Platinum agents
in the treatment of early-stage triple-negative breast cancer: Is
it time to change practice? Clin Adv Hematol Oncol. 12:654–658.
2014.PubMed/NCBI
|
|
31
|
Lv M, Xu P, Wu Y, Huang L, Li W, Lv S, Wu
X, Zeng X, Shen R, Jia X, et al: LncRNAs as new biomarkers to
differentiate triple negative breast cancer from non-triple
negative breast cancer. Oncotarget. 7:13047–13058. 2019. View Article : Google Scholar
|
|
32
|
Mujoo K, Choi BK, Huang Z, Zhang N and An
Z: Regulation of ERBB3/HER3 signaling in cancer. Oncotarget.
5:10222–10236. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Baselga J and Swain SM: Novel anticancer
targets: Revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer.
9:463–475. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wrobel P and Ahmed S: Current status of
immunotherapy in metastatic colorectal cancer. Int J Colorectal
Dis. 34:13–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen
S, Wang Y, Wang T and Hou Y: Carcinoma-associated fibroblasts
promote the stemness and chemoresistance of colorectal cancer by
transferring exosomal lncRNA H19. Theranostics. 8:3932–3948. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X,
Tian X, Guo F, Liang Q, Liu Q, et al: LncRNA GLCC1 promotes
colorectal carcinogenesis and glucose metabolism by stabilizing
c-Myc. Nat Commun. 10:34992019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bian Z, Zhang J, Li M, Feng Y, Wang X,
Zhang J, Yao S, Jin G, Du J, Han W, et al: LncRNA-FEZF1-AS1
promotes tumor proliferation and metastasis in colorectal cancer by
regulating PKM2 signaling. Cancer Res. 24:4808–4819. 2018.
|
|
38
|
Yang CK, Kim JH, Li H and Stallcup MR:
Differential use of functional domains by coiled-coil coactivator
in its synergistic coactivator function with beta-catenin or GRIP1.
J Biol Chem. 281:3389–3397. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Smith A, Baines N, Memon S, Fitzgerald N,
Chadder J, Politis C, Nicholson E, Earle C and Bryant H: Moving
toward the elimination of cervical cancer: Modelling the health and
economic benefits of increasing uptake of human papillomavirus
vaccines. Curr Oncol. 26:80–84. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang M, Song Y and Yu L: LncRNA PTCSC3
suppressed cervical carcinoma cell invasion and proliferation via
regulating miR-574-5p. Am J Transl Res. 11:7186–7194.
2019.PubMed/NCBI
|
|
42
|
Guo HM, Yang SH, Zhao SZ, Li L, Yan MT and
Fan MC: LncRNA NEAT1 regulates cervical carcinoma proliferation and
invasion by targeting AKT/PI3K. Eur Rev Med Pharmacol Sci.
22:4090–4097. 2018.PubMed/NCBI
|
|
43
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu B, Chen X, Li J, Gu Q, Zhu Z, Li C, Su
L and Liu B: microRNA-29c inhibits cell proliferation by targeting
NASP in human gastric cancer. BMC Cancer. 17:1092017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Arora N, Alsaied O, Dauer P, Majumder K,
Modi S, Giri B, Dudeja V, Banerjee S, Von Hoff D and Saluja A:
Downregulation of Sp1 by minnelide leads to decrease in HSP70 and
decrease in tumor burden of gastric cancer. PLoS One.
12:e01718272017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tian J, Hu X, Gao W, Zhang J, Chen M,
Zhang X, Ma J and Yuan H: Identification of the long non-coding RNA
LET as a novel tumor suppressor in gastric cancer. Mol Med Rep.
15:2229–2234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bian X, Yang Z, Feng H, Sun H and Liu Y: A
combination of species identification and STR profiling identifies
cross-contaminated cells from 482 human tumor cell lines. Sci Rep.
7:97742017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Abou-Elhamd KE and Habib TN: The role of
chromosomal aberrations in premalignant and malignant lesions in
head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol.
265:203–207. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zaman SU, Adeel M and Suhail A: Squamous
cell carcinoma of oral tongue in young patients-A 10 years tertiary
care experience. J Pak Med Assoc. 66:155–158. 2016.PubMed/NCBI
|
|
50
|
Zhang S, Cao R, Li Q, Yao M, Chen Y and
Zhou H: Comprehensive analysis of lncRNA-associated competing
endogenous RNA network in tongue squamous cell carcinoma. PeerJ.
7:e63972019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 36:215–233. 2009. View Article : Google Scholar
|
|
52
|
Xiong X, Shi Q, Yang X, Wang W and Tao J:
LINC00052 functions as a tumor suppressor through negatively
modulating miR-330-3p in pancreatic cancer. J Cell Physiol.
234:15619–15626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ouyang T, Zhang Y, Tang S and Wang Y: Long
non-coding RNA LINC00052 regulates miR-608/EGFR axis to promote
progression of head and neck squamous cell carcinoma. Exp Mol
Pathol. 111:1043212019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu CF, Liu P, Tan J and Hu DF: Long
noncoding RNA LINC00052 suppressed the proliferation, migration and
invasion of glioma cells by upregulating KLF6. Eur Rev Med
Pharmacol Sci. 23:4822–4827. 2019.PubMed/NCBI
|















