Introduction
Pulmonary sarcomatoid carcinomas (PSCs) are a rare
subset of lung tumors, with an estimated incidence of <1% of all
primary lung neoplasms (1–3). PSCs are currently defined as poorly
differentiated non-small cell carcinomas containing a component
with sarcoma or sarcoma-like (spindle and/or giant cell) features
(4). Five major histological
variants have been described, namely pleomorphic carcinoma, spindle
cell carcinoma, giant cell carcinoma, carcinosarcoma, and pulmonary
blastoma. Carcinosarcomas contain ectopic components (bone,
chondrosarcoma, and rhabdomyosarcoma) as the non-epithelial
components. Patients with PSCs generally show an aggressive
clinical course. PSCs are considered to be in an ‘in transition’
status, at the crossroads of diverse pathways of clonal evolution.
The sarcomatous or sarcomatoid components in these tumors may be
derived from carcinoma cells through the activation of an
epithelial-mesenchymal transition (EMT) process that leads to
sarcomatous transformation or metaplasia of the carcinoma
cells.
The epithelial nature of sarcomatoid carcinomas is
highlighted by the expression of pan-epithelial markers, such as
pan-cytokeratin AE1/AE3, OSCAR and keratin, both in the
differentiated elements and the spindle or giant cells of these
tumors (5,6). In the rare cases in which the
sarcomatoid components of the tumors tested negative for
pan-epithelial markers, positive results are often obtained for
other markers associated with epithelial tumors (e.g., CK7, TTF-1
and p40). The majority of sarcomatoid carcinomas also express
markers of more specific differentiation. Expressions of markers
related to adenocarcinomatous differentiation, including CK7, TTF-1
and napsin A, have been demonstrated in up to 78, 61 and 39% of
PSCs, respectively (5,6). A subset of sarcomatoid carcinomas is
characterized by the expressions of p40, CK5/6, Sox2 and/or
desmocollin-3 (4,6,7) but
reactivity for these markers is generally lower than that of tumors
with an adenocarcinoma phenotype (5,6). A wider
panel of immune-markers is therefore required. The NanoString
digital spatial profiling technology allows identification using a
specially defined collection of oligonucleotide tags that are
cleaved from specific validated antibodies (8). The spots of interest may be
user-defined (drawn on an image) or molecularly defined using
fluorescence images of the same slide prior to collection. In the
present study, formalin-fixed paraffin-embedded (FFPE) tissue
sections of PSC were incubated with cocktails of 56 unique
oligonucleotide-conjugated antibodies and analyzed.
The precise molecular characteristics of sarcomatoid
carcinomas are largely unexplored. In this context, TP53 and
KRAS mutations are reported as being among the most common
genomic alterations in sarcomatoid carcinomas (up to 74 and 34% of
cases, respectively), the latter likely triggered by tobacco use
(2,5,9). In
addition, targetable oncogenic driver mutations, such as EGFR,
BRAF, HER2, RET, ALK, AKT1, JAK3, NRAS or PIK3CA, have
also been identified in a small but consistent subset of PSCs
(5,9). Recent progress in deep sequencing
technology has enabled concurrent analyses of gene mutation
profiling and transcriptome. Actionable mutations are somewhat less
frequent in sarcomatoid carcinomas as compared to non-small cell
lung cancer (NSCLC) (6). In this
study, we attempted to profile the molecular statuses of the same
PSC-FFPE samples at the genome, transcript and protein levels.
Materials and methods
Clinical specimens
Tumor specimens of PSCs and NSCLC were obtained from
patients seen at the Kindai University Faculty of Medicine, with
the approval of the Institutional Review Board (30-034). Written
informed consent was obtained from each patient.
Nucleic acid extraction
DNA and RNA were purified with the use of an Allprep
DNA/RNA FFPE kit (Qiagen, Inc.) according to the manufacturer's
instructions. The quality and quantity of the DNA/RNA were verified
using the NanoDrop 2000 device (Thermo Fisher Scientific, Inc.),
PicoGreen dsDNA assay kit (Life Technologies; Thermo Fisher
Scientific, Inc.) and the RiboGreen RNA assay kit (Life
Technologies; Thermo Fisher Scientific, Inc.). The extracted
DNA/RNA was stored at −80°C until the analysis.
Targeted DNA sequencing
For DNA sequencing, 40 ng of DNA were subjected to
multiplex PCR amplification with the use of an Ion AmpliSeq Library
Kit 2.0 and Ion AmpliSeq™ Comprehensive Cancer Panel (CCP, Thermo
Fisher Scientific, Inc.), covering all exon in 409 genes. After
multiplex PCR, Ion Xpress Barcode Adapters (Thermo Fisher
Scientific, Inc.) were ligated to the PCR products, which were then
purified with the use of Agencourt AMPure XP beads (Beckman
Coulter, Inc.). The purified libraries were pooled and then
sequenced with the use of an Ion Torrent S5 instrument and Ion 550
Chip Kit (all from Thermo Fisher Scientific, Inc.). DNA sequencing
data were accessed through the Torrent Suite v.5.10 program (Thermo
Fisher Scientific, Inc.). Reads were aligned against the hg19 human
reference genome, and variants were called with the use of Variant
Call Format ver. 5.10. Raw variant calls were filtered with quality
score of <100, depth of coverage of <19, and were manually
checked using the integrative genomics viewer (IGV; Broad
Institute, Cambridge, MA, USA). Germline mutations were excluded
with the use of the Genome Aggregation Database (gnomAD).
Whole transcriptome sequencing
For library preparation, a barcoded cDNA library is
first generated with SuperScript VILO cDNA Synthesis kit (Thermo
Fisher Scientific, Inc.) from 10 ng of total RNA. Then cDNA is
prepared using the AmpliSeq Transcriptome Human Gene Expression kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Pooled libraries were subjected to the Ion Chef
System (Thermo Fisher Scientific, Inc.) for template preparation.
Libraries were then loaded onto an Ion 550 chip and sequenced with
the Ion S5 sequencing system. The Ion Torrent Suite v5.10 software
(Thermo Fisher Scientific, Inc.) was used for base calling,
alignment to the human reference genome (hg19) and quality control.
Raw reads were then analyzed automatically using the AmpliSeqRNA
plugin to generate gene-level expression values for all 20802
RefSeq human genes. Genes with a fold change greater than 4 (the
absolute value of log2 fold change greater than 2) and an adjust
P-value <0.05 were considered as differentially expressed and
were investigated by Ingenuity® Pathway Analysis (IPA)
(Qiagen, Inc.). The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Digital spatial profiling
The NanoString digital spatial profiling technology
allows identification using a specially defined collection of
oligonucleotides tags that are cleaved from specific validated
antibodies (8,10). The regions of interest may be
user-defined (drawn on an image) or molecularly defined using a
fluorescence 152 image of the same slide prior to collection. The
FFPE tissue sections were incubated with cocktails of 56 unique
oligonucleotide-conjugated antibodies (Table SI). The selected compartments were
chosen for high-resolution multiplex profiling, and oligos from the
selected region were released upon exposure to UV light.
Photocleaved oligos were then collected via microcapillary tube
inspiration using an early version of the DSP platform (NanoString)
robotic system and transferred into a microwell plate with a
partial resolution of approximately 10 µm. Photocleaved oligos from
the spatially-resolved compartments in the microplate were then
hybridized to 4-color, 6-spot optical barcodes in the
nCounter® platform, enabling up to 800 distinctly labels
counts per compartment of the protein targets representing the
antibodies to which the tags were originally conjugated. Digital
counts from barcodes corresponding to the protein probes were first
normalized with internal spike-in controls (ERCCs) to account for
system variations, and then normalized to immunoglobulin G (IgG)
controls to correct for noise.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software (ver. 8, GraphPad Software Inc.). The data represent
the means ± standard deviation (SD). The difference between groups
was calculated using Mann-Whitney U test. Correlation between cell
component and 56 protein expression by DSP was calculated using
χ2 test. A P-value of <0.05 was considered
statistically significant.
Results
Somatic mutation analysis
DNA extracted from FFPE tissue samples obtained from
patients histopathologically diagnosed as having PSC at Kindai
University Hospital from 2016 to 2018 were analyzed. Samples from 6
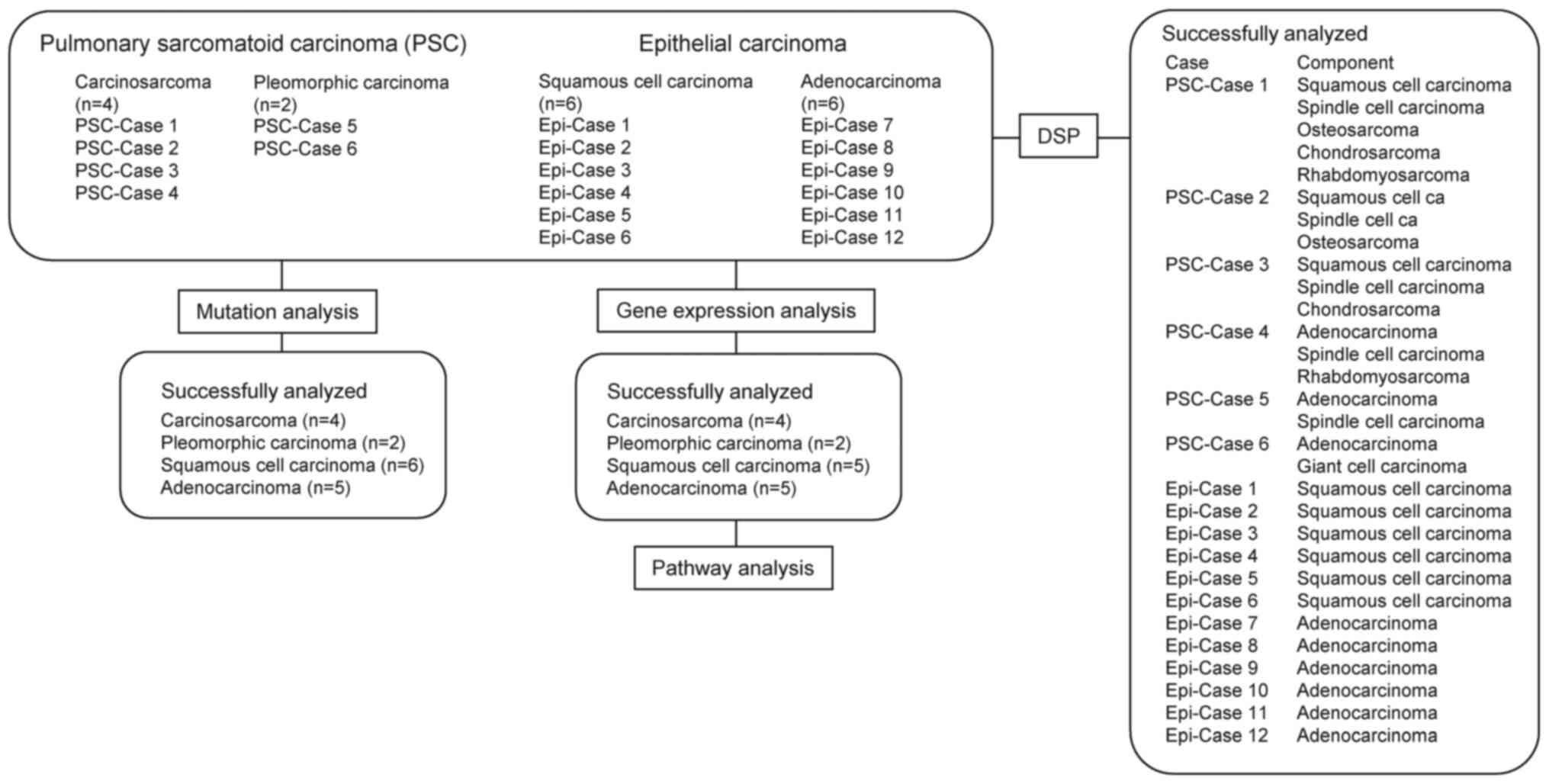
lung adenocarcinoma and 6 squamous cell lung cancer patients were
processed as the epithelial carcinoma samples (Fig. 1). No analysis was performed in one of
the lung adenocarcinoma case, because of the low quality of DNA
isolated. PSC-case 1 was a 63-year-old male patient, and the tumor
tissue contained squamous cell carcinoma, spindle cell carcinoma,
osteosarcoma, chondrosarcoma, and rhabdomyosarcoma components
(Fig. S1). PSC-case 2 was a
71-year-old female patient and her tumor tissue contained squamous
cell carcinoma, spindle cell carcinoma and osteosarcoma components.
PSC-case 3 was a 79-year-old male patient and his tumor tissue
contained squamous cell carcinoma, spindle cell carcinoma,
chondrosarcoma, and giant cell carcinoma components. PSC-case 4 was
an 86-year-old male patient, and his tumor tissue contained typical
adenocarcinoma, high-grade fetal carcinoma-like adenocarcinoma,
spindle cell carcinoma, and rhabdomyosarcoma components. All
patients had a history of smoking. Targeted deep sequencing with
CCP for 409 cancer-related genes revealed 7, 7, 6, and 5 pathogenic
mutations classified by Functional Analysis through Hidden Markov
Models (FATHMM) in PSC-cases 1–4, respectively (Fig. S2A). Mutations of TP53 (3/4),
SYNE1 (2/4), and APC (2/4) were detected in the
carcinosarcoma. These mutations have been previously recognized in
carcinosarcoma (5), but are not
found in pleomorphic carcinoma. On the other hand, TP53
mutation was also detected in 4/11 epithelial carcinoma
(adenocarcinoma and squamous cell carcinoma) cases. CCP panel
detected likely pathogenic mutations such as NTRK3 and
PIK3CA in PSC-case 1, AXL and KDR in PSC-case
2, PTEN in PSC-case 3, and BRAF in PSC-case 4. The
non-synonymous tumor mutation burden (TMB), which is a predictive
marker for response to immunocheckpoint inhibitor therapy (11–13), was
calculated from CCP panel (Fig.
S2B). The average TMB of the PSCs was 15.2 mutations/Mb, which
was nearly as high as that in epithelial carcinomas (adenocarcinoma
+ squamous cell carcinoma, 12.8 mutations/Mb), though the
difference was not statistically significant.
Gene expression profile
Whole-transcriptome analysis was performed for the 6
PSC (PSC-Cases 1–6), 6 adenocarcinoma and 6 squamous cell carcinoma
(Epi-Cases 1–6) specimens (Fig. 1).
No analysis was performed in two of the epithelial carcinomas (1
adenocarcinoma and 1 squamous cell carcinoma) cases, because of the
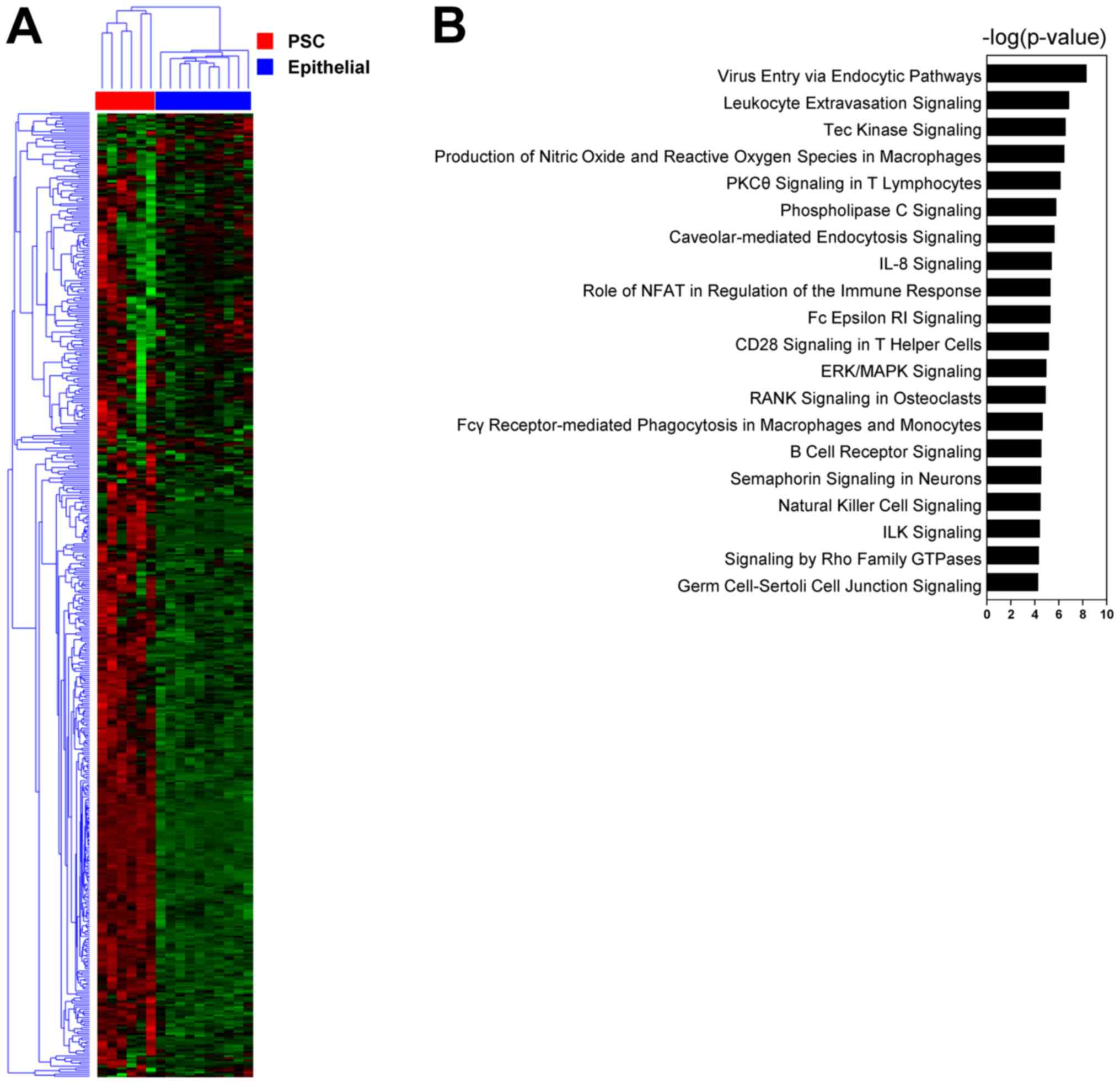
low quality of RNA isolated. Clustering analysis allowed the
specimens to be clearly classified into epithelial carcinoma and
PSC based on the gene expression profile (Fig. 2A). Pathway analysis (IPA) revealed
significant enrichment of pathways of PSCs (Fig. 2B). The top 20 pathways included the
integrin-linked kinase (ILK) signaling pathway, which is known to
be related to β-catenin-mediated EMT (14) through AKT and Gsk3β (15). ILK signaling activates the key
transcription factor SLUG (SNAI2), which upregulates the expression
of fibronectin as well as vimentin involved in the EMT process
(16). Fibronectin acts on cell
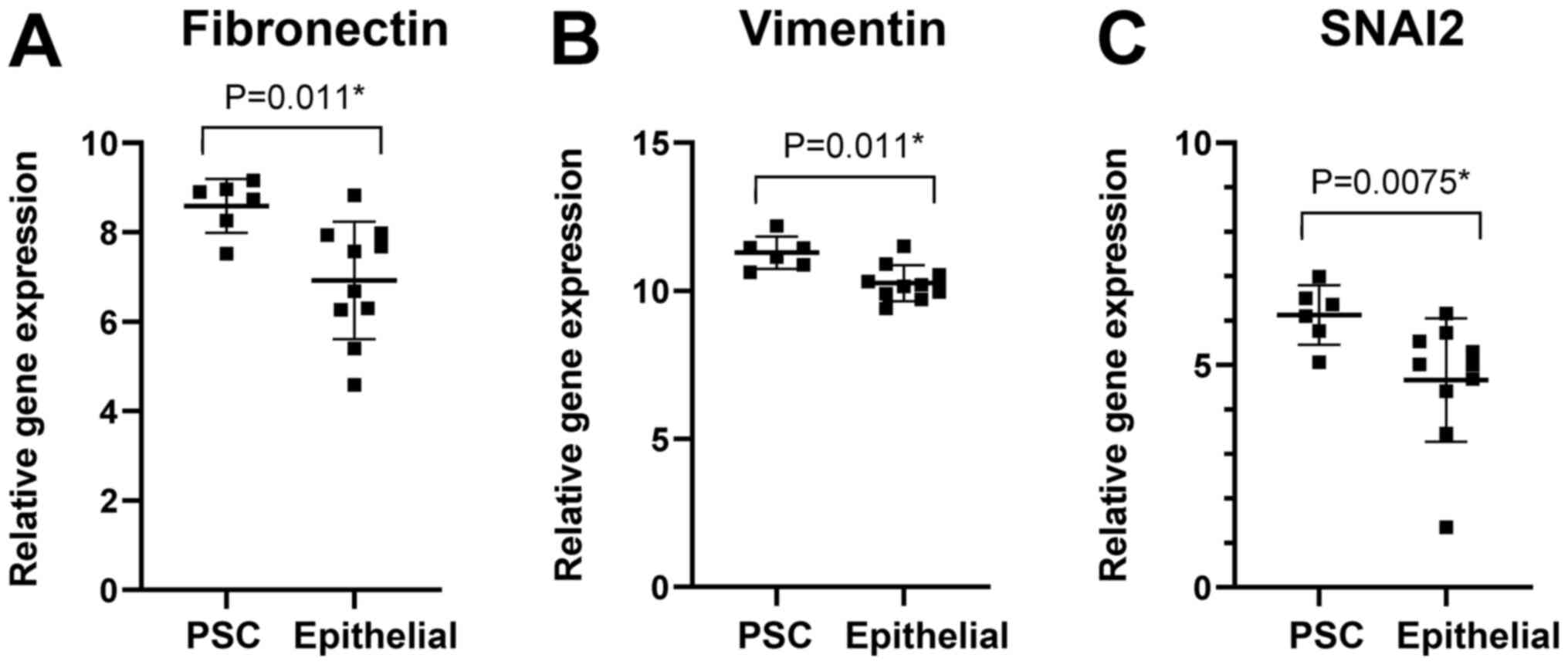
mobility and cell adhesion. The gene expression levels of
fibronectin as well as vimentin and SNAI2 were higher in the PSCs
than in the epithelial carcinomas (Fig.
3). The fibronectin gene expression levels were also higher in
the PSCs than in the squamous cell carcinomas.
Intratumor heterogeneity of
fibronectin expression in pulmonary sarcomatous carcinomas
Standard immunohistochemistry (IHC) was performed
for the PSC specimens (Table I).
With regards to E-cadherin expression, carcinoma components show
positive staining, whereas sarcomatous components, including
spindle cell carcinoma, osteosarcoma, chondrosarcoma,
rhabdomyosarcoma, in each sample are negative. E-cadherin
expression patterns were consistent across all cases. To further
investigate the biological features and intratumor heterogeneity of
PSCs, digital IHC analysis by DSP was performed for 60 selected
regions of interest (Fig. S3,
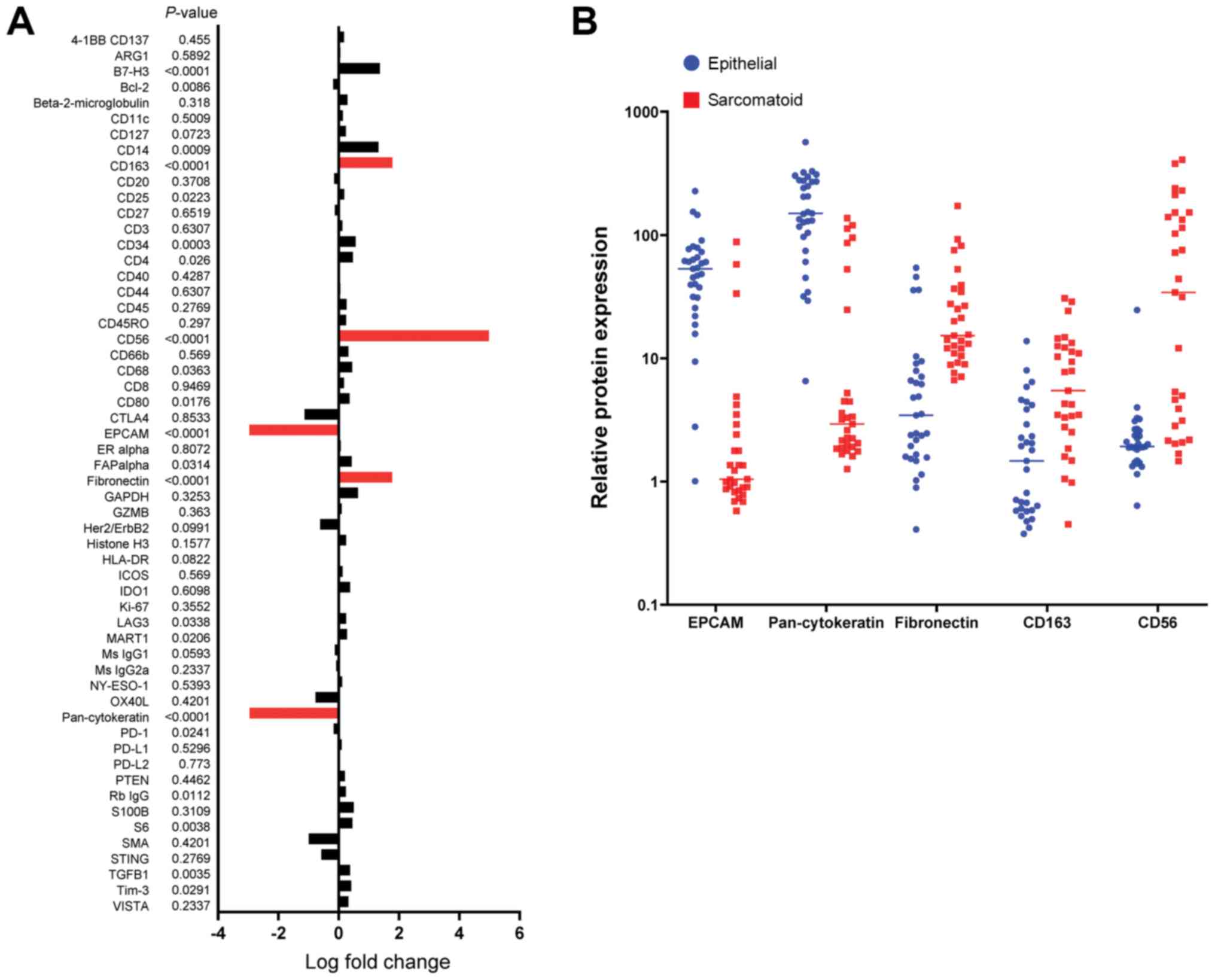
Table SII) using 56 antibodies.
Quantitative expression levels of 56 proteins in sarcomatoid (29
spots) and epithelial (31 spots) components were compared between
epithelial and sarcomatoid comportment (Fig. 4A). Higher expression levels of
fibronectin, CD163, and CD56 were observed in the sarcomatoid
components significantly (P<0.0001, Chi-squared test) (Fig. 4B). On the other hand, EPCAM, and
pan-cytokeratin expressed highly in the epithelial components.
Taken together, the fibronectin expression levels were
significantly higher in the sarcomatoid components than in the
epithelial components of the PSCs.
 | Table I.Detailed clinical features in 4
patients with carcinosarcoma. |
Table I.
Detailed clinical features in 4
patients with carcinosarcoma.
| A, PSC Case 1 |
|---|
|
|---|
| Component | IHC |
|---|
| Squamous cell
carcinoma | AE1/AE3(+),
E-cadherin(+), TTF-1(+), p40(+), SMA(−), Desmin(−),
Myogenin(−) |
| Spindle cell
carcinoma | AE1/AE3(focal +),
E-cadherin(−), TTF-1(+), p40(+), SMA(focal +), Desmin(−),
Myogenin(−) |
| Osteosarcoma | AE1/AE3(−),
E-cadherin(−), TTF-1(−), p40(−), SMA(−), Desmin(−),
Myogenin(−) |
| Chondrosarcoma | AE1/AE3(−),
E-cadherin(−), TTF-1(−), p40(−), SMA(−), Desmin(−),
Myogenin(−) |
|
Rhabdomyosarcoma | AE1/AE3(−),
E-cadherin(−), TTF-1(−), p40(−), SMA(−), Desmin(+),
Myogenin(+) |
|
| B, PSC Case
2 |
|
|
Component | IHC |
|
| Squamous cell
carcinoma | AE1/AE3(+),
E-cadherin(+), TTF-1(focal +), p40(+), CK5/6(+), SMA(−), CD56(−),
S100(−) |
| Spindle cell
carcinoma | AE1/AE3(−),
E-cadherin(−), TTF-1(−), p40(−), CK5/6(−), SMA(+), CD56(+),
S100(focal +) |
| Osteosarcoma | AE1/AE3(−),
E-cadherin(−), TTF-1(−), p40(−), CK5/6(−), SMA(−), CD56(+),
S100(−) |
|
| C, PSC Case
3 |
|
|
Component | IHC |
|
| Squamous cell
carcinoma | AE1/AE3(+),
E-cadherin(+), TTF-1(−), p40(+), SMA(−), CD56(−), Desmin(−),
S100(−) |
| Spindle cell
carcinoma | AE1/AE3(−),
E-cadherin(−), TTF-1(−), p40(+), SMA(−), CD56(−), Desmin(−),
S100(−) |
| Chondrosarcoma | AE1/AE3(−),
E-cadherin(−), TTF-1(−), p40(−), SMA(−), CD56(−), Desmin(−),
S100(+) |
|
| D, PSC Case
4 |
|
|
Component | IHC |
|
| Adenocarcinoma | AE1/AE3(+),
E-cadherin(+), TTF-1(+), p40(focal +), SMA(−), CD56(focal +),
AFP(−), SALL4(−), GPC3(−) |
| Spindle cell
carcinoma | AE1/AE3(−),
E-cadherin(−), TTF-1(focal +), p40(−), SMA(−), CD56(+), AFP(−),
SALL4(−), GPC3(−) |
|
Rhabdomyosarcoma | AE1/AE3(−),
E-cadherin(−), TTF-1(−), p40(−), SMA(−), CD56(−), Desmin(+),
Myogenin(+) |
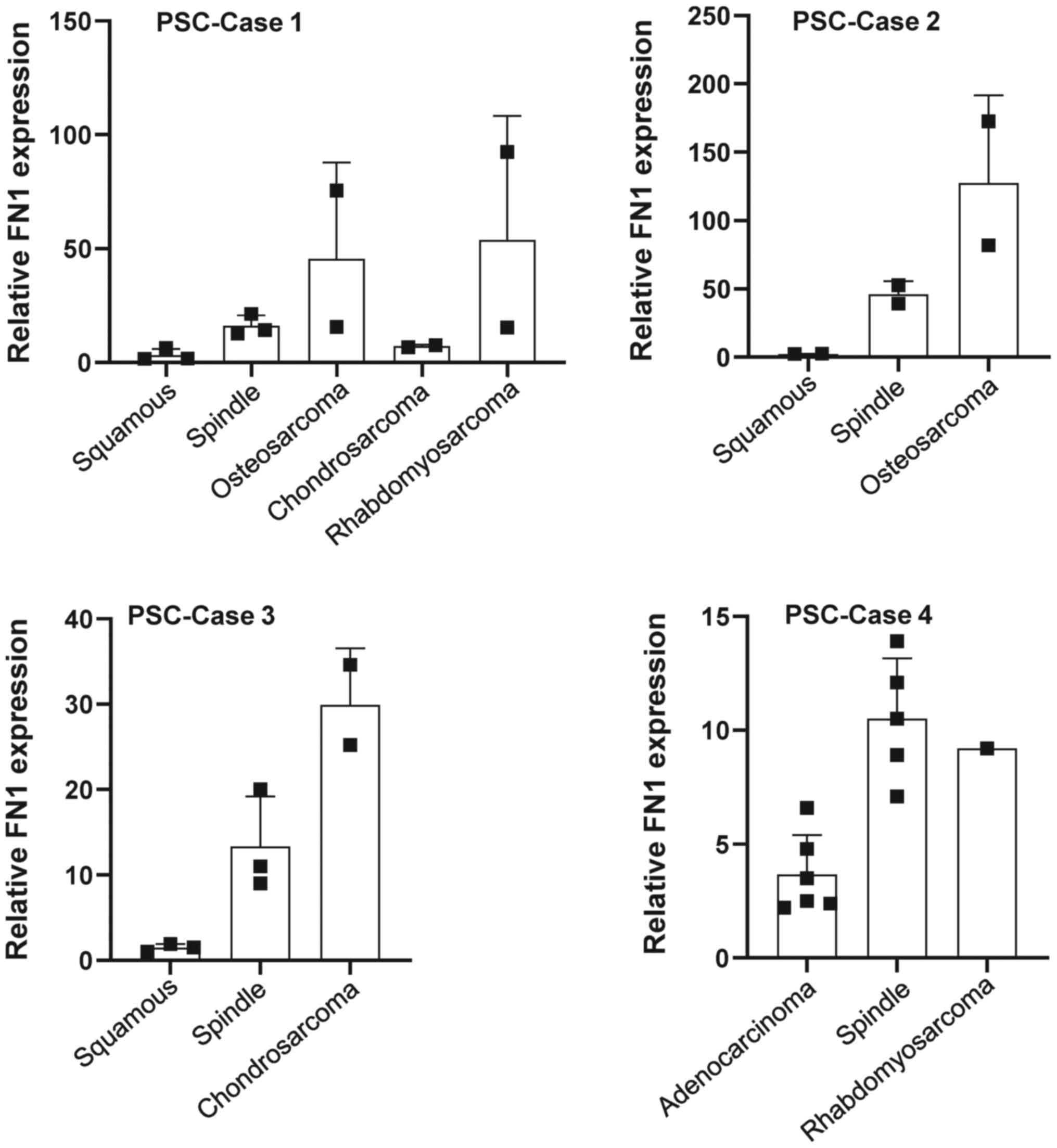
Differential intratumor expression of fibronectin
was observed in the different sarcoma components (spindle cell
sarcoma, osteosarcoma, chondrosarcoma, and rhabdomyosarcoma
components) in PSC cases 1–4 (Fig.
5); the highest expression of fibronectin was observed in the
osteosarcoma components, while modest expression of fibronectin was
observed in the spindle cell sarcoma, rhabdomyosarcoma, and
chondrosarcoma components. No fibronectin expression was observed
in the epithelial components in three of the four cases (Fig. 5). Differences in the intratumor
expression of fibronectin seemed to be related to the degree of EMT
in the tumors.
Discussion
PSCs are rare form of NSCLC cancers that are
characterized by their aggressive nature and difficulty to treat.
In this study, we have performed DNA mutation and whole
transcriptome analyses in a small cohort of PSC cases and compared
the patients' genomic and transcriptomic profiles to protein
expression profiles from digital multiplexed immunohistochemical
tissue sections. Molecular profiling of PSCs has been reported by a
few previous studies. Terra et al reported that PSCs,
consisting of pleomorphic carcinoma, spindle cell carcinoma,
carcinosarcoma, and giant cell carcinoma, exhibit mutations of
TP53 mutation (58%), JAK3 (3%), BRAF (3%),
NRAS (3%), and PIK3CA (3%) (5). According to the results of our
NGS-based mutation analysis, mutations of TP53 were detected
at a high frequency in the PSCs. TP53 mutations are also
detected in epithelial carcinomas. No difference in the frequency
of TP53 mutations were observed between the PSCs and
epithelial carcinoma specimens. Therefore, it is unlikely that they
contribute to PSC transformation. Liang et al reported that
a high frequency of RB1 mutations (25%) were detected as
well as that of TP53 (69%) (17). In our cases, no hot spot mutation of
RB1 was detected, although data for the full sequence of
RB1 was unavailable. NGS analysis also showed a high TMB of
PSCs. The TMB was as high in the PSCs as in the NSCLCs. NGS
analysis also showed a high TMB of PSCs. The TMB was as high in the
PSCs as in the NSCLCs. TMB as well as PD-L1 can be used to predict
efficacy of immune checkpoint inhibitors and has become a useful
biomarker to identify cancer patients that may likely benefit from
immunotherapy. Liang et al reported that 40.6% (13/32) of
Chinese patients with PSC had high TMB (17). This result thus suggests that PSCs
have a strong potential to present neoantigens, regardless of the
PD-L1 expression status. On the other hand, retrospective studies
reported a high incidence of PD-L1 expression in patients with PSC.
Velcheti et al reported that 69.2% (9/13) of patients were
positive for PD-L1 (18). Taking
these findings together, immunotherapy with immune-checkpoint
inhibitors could potentially be effective for PSCs, although
studies are required prove this assumption.
Pathway analysis (IPA) provided the signaling
pathway to identify molecular targets and biomarkers for PSCs. The
top 20 significant pathways included the ILK signaling pathway,
which is known to be related to beta-catenin-mediated EMT (14) through AKT and Gskβ (15). ILK signaling activates the key
transcription factor SLUG, which upregulates the expression of
fibronectin as well as vimentin involved in the EMT process
(16). In our PSC sample cohort,
higher expression levels of fibronectin were observed at both the
transcript and protein levels. Fibronectin has been shown to
promote cell motility, opsonization, and cell adhesion in
carcinomas (19,20) and to promote EMT (21).
EMT causes resistance to various types of
treatments, such as cytotoxic chemotherapy and tyrosine kinase
inhibitor therapy (22–24). The hypothesis that PSCs are caused by
fibronectin-mediated EMT of carcinomas is consistent with the
refractoriness of PSCs to treatment. Our findings allow us to
speculate on the involvement of EMT in the progression to sarcoma.
In a study of biphasic sarcomatoid carcinomas of the lung, Manzotti
et al found that PSC could originate from EMT based on
morphological characterization (25). In our study, a more detailed analysis
of the EMT status of PSC was yielded through transcriptomic and
digital spatial profiling, although focusing on pleomorphic
carcinoma of lung. The clinical relevance and possible applications
based on our findings can fall into several areas. From the
diagnosis and prediction of prognosis perspective, immunostaining
of EMT related molecules including fibronectin will be meaningful
for understanding the grade of malignancy and possibly predicting
of prognosis of PSC patients, although the association of
fibronectin expression and prognosis of PSC patients remains
unknown it will be meaningful to explore in future studies. From a
therapeutic perspective, EMT-targeted therapy is expected to
inhibit tumor growth, malignant and resistant transformation. For
other cancer types, Fresolimumab (26) targeting N-cadherin, ADH-1, TGFβ,
ZEB1, SNAIL2, TWIST, vimentin-targeted metformin (27), anti-EpCAM immunotoxin targeting EpCAM
(28), catumaxomab (29) are possible and other potential
candidates are being investigated in clinical trials.
We have also shown increased fibronectin expression
in the sarcomatoid components. Thus, targeting fibronectin is one
possible treatment approach. Most antibody approaches targeting
fibronectin utilized antibodies against the extra-domain B (EDB)
domain. A human EDB domain specific antibody (L19) was isolated by
Carnemolla et al (30). This
antibody has been used for targeted delivery of IL2 as a fused
L19-IL2 protein and has demonstrated efficacy in the mouse F9
teratocarcinoma as well as other preclinical tumor models (30). The strategy of targeting fibronectin
EDB at tumor sites has been extended with a fusion protein of the
L19 fragment with interleukin-12 (IL12) (31), a potent mediator of innate and cell
immunity with anti-tumor and anti-metastatic properties (32,33).
Other approaches with the L19 antibody have analyzed the use of
conjugated photosensitizers and liposomes. Fabbrini et al
conjugated L19 to a photosensitizer [bis(triethanolamine)Sn(IV)
chlorin e6] that generates toxic oxygen species after irradiation
with red light (34). Thus,
antibody-based approaches to target fibronectin in the tumor and
tumor vasculature and specifically deliver anti-tumor agents to
tumors are promising avenues in cancer therapy including the
management of PSC.
We also postulated that the ILK signaling pathway
may be implicated in the induction of EMT. Therefore, targeting ILK
may offer an additional novel therapeutic potential for managing
PSCs. Presently, ILK-targeted therapies are being investigated in
solid tumors. Preclinical studies have shown that knockdown of ILK
expression in cancer cells using siRNA or shRNA significantly
inactivates the PI3K/AKT pathway and suppresses EMT, tumor growth
and metastasis in tongue and prostate cancer cells in vitro
and in vivo (35,36). As an ILK inhibitor, Lee et al
found that
N-methyl-3-(1-(4-(piperazin-1-yl)phenyl)-5-(40-(trifluoromethyl)-[1,10-biphenyl]-4-yl)-1H-pyrazole-3-yl)
propanamide (compound 22) could potently inhibit the growth of
prostate and breast cancer cells via the inactivation of the AKT
pathway and inhibition of the transcription factor Y-box binding
protein-1 (YB-1). Compound 22 was used as a single agent to inhibit
the growth of prostate tumor xenografts in vivo (37). In addition, QLT0267, a novel ILK
inhibitor, has been reported to reduce tumor volume in thyroid
cancer and glioblastoma xenografts (38,39). It
was also shown that QLT0267 may inhibit EMT, which is involved in
5-Fluorouracil resistance in colorectal cancer (40). These findings suggest that ILK is an
effective therapeutic target for cancer treatment.
We previously showed that the tubulin binder
eribulin suppresses TGFβ- or 5-FU-induced EMT and have the
potential to induce mesenchymal epithelial transition (MET) in
triple-negative breast cancer cells (24,41,42).
Therefore, control of EMT or induction of mesenchymal-epithelial
transition are among the treatment strategies for conquering
PSCs.
One of the major limitations of this study is the
small number of PSC cases, which was unavoidable given the rare
nature of PSCs and the difficulty in obtaining surgically resected
tissue samples. Here, we focused on carcinosarcoma and pleomorphic
carcinoma as the main objective and used adenocarcinoma and
squamous carcinoma as a reference. Therefore, a confirmation study
using an additional sample cohort that includes pure pulmonary
blastoma, spindle cell carcinoma and giant cell carcinoma will be
needed to confirm the results of the present study. Unfortunately,
we were also unable to microscopically dissect intratumor
components for mutation and transcriptome analysis because the
samples were unsuitable.
In this study, we demonstrated that digital IHC and
NGS provided the molecular profiles of the FFPE specimens of PSCs
at the DNA, transcript and protein levels in this study. In
particular, DSP enabled comparison of different components in the
same tumor tissue and analysis of the intratumor heterogeneity. We
consider the results as being consistent with the hypothesis that
the ILK-fibronectin mediated EMT process is involved in the
pathogenesis of PSCs. Fibronectin may serve as a marker of
PSCs.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Keiko Obata
(Department of Thoracic Surgery, Kindai University Faculty of
Medicine), Mr. Yoshihiro Mine (Center for Instrumental Analyses
Central Research Facilities, Kindai University Faculty of
Medicine), and Ms. Ayaka Kitano (Department of Genome Biology,
Kindai University Faculty of Medicine) for technical assistance
provided during the study.
Funding
The current study was supported in part by a
Grant-in Aid for Scientific Research (C) from the Japan Society for
the Promotion of Science (grant no. JP19K07722) and in part by a
Grant-in Aid for Scientific Research on Innovative Areas ‘Frontier
Research on Chemical Communications’ (grant nos. JP17H06400 and
JP17H06404).
Availability of data and materials
Data analyzed and material used during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SS, KS and KN conceived and designed the study. SS,
KS, TC, and NS performed the experiments. TM and KN confirmed the
authenticity of raw data. SS, KS, TS, and TM performed the
statistical analysis. SS, KS, NS, TS, TM, and KN contributed to the
analysis and interpretation of data. SS, KS, and KN wrote the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Institutional
Review Board of Kindai University Faculty of Medicine, and written
informed consent was obtained from all patients for study
participation.
Patient consent for publication
Written informed consent was obtained from all
patients for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pelosi G, Sonzogni A, De Pas T, Galetta D,
Veronesi G, Spaggiari L, Manzotti M, Fumagalli C, Bresaola E, Nappi
O, et al: Review article: Pulmonary sarcomatoid carcinomas: A
practical overview. Int J Surg Pathol. 18:103–120. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schrock AB, Li SD, Frampton GM, Suh J,
Braun E, Mehra R, Buck SC, Bufill JA, Peled N, Karim NA, et al:
Pulmonary sarcomatoid carcinomas commonly harbor either potentially
targetable genomic alterations or high tumor mutational burden as
observed by comprehensive genomic profiling. J Thorac Oncol.
12:932–942. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yendamuri S, Caty L, Pine M, Adem S,
Bogner P, Miller A, Demmy TL, Groman A and Reid M: Outcomes of
sarcomatoid carcinoma of the lung: A surveillance, epidemiology,
and end results database analysis. Surgery. 152:397–402. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Travis WD; World Health Organization, :
International Agency for Research on Cancer, International
Association for the Study of Lung Cancer. And International Academy
of Pathology: Pathology and genetics of tumours of the lung,
pleura, thymus and heart. IARC Press, Oxford University Press
(distributor); Lyon, Oxford: 2004
|
|
5
|
Terra SB, Jang JS, Bi L, Kipp BR, Jen J,
Yi ES and Boland JM: Molecular characterization of pulmonary
sarcomatoid carcinoma: Analysis of 33 cases. Mod Pathol.
29:824–831. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weissferdt A: Pulmonary sarcomatoid
carcinomas: A review. Adv Anat Pathol. 25:304–313. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lucas DR, Pass HI, Madan SK, Adsay NV,
Wali A, Tabaczka P and Lonardo F: Sarcomatoid mesothelioma and its
histological mimics: A comparative immunohistochemical study.
Histopathology. 42:270–279. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Toki MI, Merritt CR, Wong PF, Smithy JW,
Kluger HM, Syrigos KN, Ong GT, Warren SE, Beechem JM and Rimm DL:
High-plex predictive marker discovery for melanoma
immunotherapy-treated patients using digital spatial profiling.
Clin Cancer Res. 25:5503–5512. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vieira T, Antoine M, Ruppert AM, Fallet V,
Duruisseaux M, Giroux Leprieur E, Poulot V, Rabbe N, Sclick L,
Beau-Faller M, et al: Blood vessel invasion is a major feature and
a factor of poor prognosis in sarcomatoid carcinoma of the lung.
Lung Cancer. 85:276–281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Decalf J, Albert ML and Ziai J: New tools
for pathology: A user's review of a highly multiplexed method for
in situ analysis of protein and RNA expression in tissue. J Pathol.
247:650–661. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alborelli I, Leonards K, Rothschild SI,
Leuenberger LP, Savic Prince S, Mertz KD, Poechtrager S, Buess M,
Zippelius A, Läubli H, et al: Tumor mutational burden assessed by
targeted NGS predicts clinical benefit from immune checkpoint
inhibitors in non-small cell lung cancer. J Pathol. 250:19–29.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao D, Xu H, Xu X, Guo T and Ge W: High
tumor mutation burden predicts better efficacy of immunotherapy: A
pooled analysis of 103078 cancer patients. Oncoimmunology.
8:e16292582019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heeke S, Benzaquen J, Long-Mira E, Audelan
B, Lespinet V, Bordone O, Lalvée S, Zahaf K, Poudenx M, Humbert O,
et al: In-house implementation of tumor mutational burden testing
to predict durable clinical benefit in non-small cell lung cancer
and melanoma patients. Cancers (Basel). 11:12712019. View Article : Google Scholar
|
|
14
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hannigan G, Troussard AA and Dedhar S:
Integrin-linked kinase: A cancer therapeutic target unique among
its ILK. Nat Rev Cancer. 5:51–63. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fenouille N, Tichet M, Dufies M, Pottier
A, Mogha A, Soo JK, Rocchi S, Mallavialle A, Galibert MD, Khammari
A, et al: The epithelial-mesenchymal transition (EMT) regulatory
factor SLUG (SNAI2) is a downstream target of SPARC and AKT in
promoting melanoma cell invasion. PLoS One. 7:e403782012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang X, Li Q, Xu B, Hu S, Wang Q, Li Y,
Zong Y, Zhang S and Li C: Mutation landscape and tumor mutation
burden analysis of Chinese patients with pulmonary sarcomatoid
carcinomas. Int J Clin Oncol. 24:1061–1068. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Velcheti V, Rimm DL and Schalper KA:
Sarcomatoid lung carcinomas show high levels of programmed death
ligand-1 (PD-L1). J Thorac Oncol. 8:803–805. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gopal S, Veracini L, Grall D, Butori C,
Schaub S, Audebert S, Camoin L, Baudelet E, Radwanska A,
Beghelli-de la Forest Divonne S, et al: Fibronectin-guided
migration of carcinoma collectives. Nat Commun. 8:141052017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang WY, Twu CW, Liu YC, Lin HH, Chen CJ
and Lin JC: Fibronectin promotes nasopharyngeal cancer cell
motility and proliferation. Biomed Pharmacother. 109:1772–1784.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rick JW, Chandra A, Dalle Ore C, Nguyen
AT, Yagnik G and Aghi MK: Fibronectin in malignancy:
Cancer-specific alterations, protumoral effects, and therapeutic
implications. Semin Oncol. 46:284–290. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park SY, Kim MJ, Park SA, Kim JS, Min KN,
Kim DK, Lim W, Nam JS and Sheen YY: Combinatorial TGF-β attenuation
with paclitaxel inhibits the epithelial-to-mesenchymal transition
and breast cancer stem-like cells. Oncotarget. 6:37526–37543. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sarkar FH, Li Y, Wang Z and Kong D:
Pancreatic cancer stem cells and EMT in drug resistance and
metastasis. Minerva Chir. 64:489–500. 2009.PubMed/NCBI
|
|
24
|
Terashima M, Sakai K, Togashi Y, Hayashi
H, De Velasco MA, Tsurutani J and Nishio K: Synergistic antitumor
effects of S-1 with eribulin in vitro and in vivo for
triple-negative breast cancer cell lines. Springerplus. 3:4172014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Manzotti G, Torricelli F, Benedetta D,
Lococo F, Sancisi V, Rossi G, Piana S and Ciarrocchi A: An
Epithelial-to-mesenchymal transcriptional switch triggers evolution
of pulmonary sarcomatoid carcinoma (PSC) and identifies dasatinib
as new therapeutic option. Clin Cancer Res. 25:2348–2360. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Singh M, Yelle N, Venugopal C and Singh
SK: EMT: Mechanisms and therapeutic implications. Pharmacol Ther.
182:80–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kordes S, Pollak MN, Zwinderman AH, Mathôt
RA, Weterman MJ, Beeker A, Punt CJ, Richel DJ and Wilmink JW:
Metformin in patients with advanced pancreatic cancer: A
double-blind, randomised, placebo-controlled phase 2 trial. Lancet
Oncol. 16:839–847. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Andersson Y, Inderberg EM, Kvalheim G,
Herud TM, Engebraaten O, Flatmark K, Dueland S and Fodstad Ø:
Immune stimulatory effect of anti-EpCAM immunotoxin-improved
overall survival of metastatic colorectal cancer patients. Acta
Oncol. 59:404–409. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mau-Sørensen M, Dittrich C, Dienstmann R,
Lassen U, Büchler W, Martinius H and Tabernero J: A phase I trial
of intravenous catumaxomab: A bispecific monoclonal antibody
targeting EpCAM and the T cell coreceptor CD3. Cancer Chemother
Pharmacol. 75:1065–1073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carnemolla B, Neri D, Castellani P,
Leprini A, Neri G, Pini A, Winter G and Zardi L: Phage antibodies
with pan-species recognition of the oncofoetal angiogenesis marker
fibronectin ED-B domain. Int J Cancer. 68:397–405. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Halin C, Rondini S, Nilsson F, Berndt A,
Kosmehl H, Zardi L and Neri D: Enhancement of the antitumor
activity of interleukin-12 by targeted delivery to neovasculature.
Nat Biotechnol. 20:264–269. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsung K, Meko JB, Peplinski GR, Tsung YL
and Norton JA: IL-12 induces T helper 1-directed antitumor
response. J Immunol. 158:3359–3365. 1997.PubMed/NCBI
|
|
33
|
Brunda MJ, Luistro L, Warrier RR, Wright
RB, Hubbard BR, Murphy M, Wolf SF and Gately MK: Antitumor and
antimetastatic activity of interleukin 12 against murine tumors. J
Exp Med. 178:1223–1230. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fabbrini M, Trachsel E, Soldani P, Bindi
S, Alessi P, Bracci L, Kosmehl H, Zardi L, Neri D and Neri P:
Selective occlusion of tumor blood vessels by targeted delivery of
an antibody-photosensitizer conjugate. Int J Cancer. 118:1805–1813.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xing Y, Qi J, Deng S, Wang C, Zhang L and
Chen J: Small interfering RNA targeting ILK inhibits metastasis in
human tongue cancer cells through repression of
epithelial-to-mesenchymal transition. Exp Cell Res. 319:2058–2072.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yuan Y, Xiao Y, Li Q, Liu Z, Zhang X, Qin
C, Xie J, Wang X and Xu T: In vitro and in vivo
effects of short hairpin RNA targeting integrin-linked kinase in
prostate cancer cells. Mol Med Rep. 8:419–424. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee SL, Hsu EC, Chou CC, Chuang HC, Bai
LY, Kulp SK and Chen CS: Identification and characterization of a
novel integrin-linked kinase inhibitor. J Med Chem. 54:6364–6374.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Edwards LA, Woo J, Huxham LA, Verreault M,
Dragowska WH, Chiu G, Rajput A, Kyle AH, Kalra J, Yapp D, et al:
Suppression of VEGF secretion and changes in glioblastoma
multiforme microenvironment by inhibition of integrin-linked kinase
(ILK). Mol Cancer Ther. 7:59–70. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Younes MN, Kim S, Yigitbasi OG, Mandal M,
Jasser SA, Dakak Yazici Y, Schiff BA, El-Naggar A, Bekele BN, Mills
GB and Myers JN: Integrin-linked kinase is a potential therapeutic
target for anaplastic thyroid cancer. Mol Cancer Ther. 4:1146–1156.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tsoumas D, Nikou S, Giannopoulou E,
Champeris Tsaniras S, Sirinian C, Maroulis I, Taraviras S, Zolota
V, Kalofonos HP and Bravou V: ILK expression in colorectal cancer
is associated with EMT, cancer stem cell markers and
chemoresistance. Cancer Genomics Proteomics. 15:127–141.
2018.PubMed/NCBI
|
|
41
|
Iwasa T, Tsurutani J, Watanabe S, Kato R,
Mizuno Y, Kojima Y, Takashima T, Matsunami N, Morimoto T, Yamamura
J, et al: Multicentre, phase II study of eribulin in combination
with S-1 in patients with advanced breast cancer. BMC Cancer.
19:9622019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sakiyama T, Tsurutani J, Iwasa T, Kawakami
H, Nonagase Y, Yoshida T, Tanaka K, Fujisaka Y, Kurata T, Komoike
Y, et al: A phase I dose-escalation study of eribulin and S-1 for
metastatic breast cancer. Br J Cancer. 112:819–824. 2015.
View Article : Google Scholar : PubMed/NCBI
|