Introduction
Eukaryotic translation initiation factor 4A3
(EIF4A3), a member of the EIF4A Asp-Glu-Ala-Asp (DEAD)-box and
ATP-dependent RNA helicase families (1–4), serves
as a translation initiation factor by unraveling secondary
structures of the 5′-untranslated region (5). In 2004, EIF4A3 was reported as a novel
and vital element of the exon junction complex (EJC), which binds
spliced but not intronless mRNAs by anchoring the EJC to RNA
(6). The EJC is a complex that
contains multiple proteins and serves different functions,
including splicing and polyadenylation (7), mRNA export (8,9),
nonsense-mediated mRNA decay (NMD) (10), regulation of translation efficiency
(11) and the localization of mRNA
(12). EIF4A3 is situated ~24
nucleotides upstream of the exon junction, which is the canonical
EJC region, but can also be found in noncanonical regions within
the exons (13). NMD is an important
process that ensures accurate and efficient translation of
proteins, and prevents the synthesis of abnormal or defective
proteins by degrading incomplete or defective mRNA (14). The key event in inducing NMD is
phosphorylation of the trans-acting factor, UPF1 RNA helicase and
ATPase (UPF1), by SMG1 NMD associated PI3K related kinase (15). The translation termination release
factors, eukaryotic translation termination factor (ERF)1 and ERF3,
and the EJC are required for UPF1 phosphorylation and the
occurrence of NMD (15). RNA
interference experiments have demonstrated that nonsense
codon-induced partitioning shift depends on certain NMD factors,
including UPF1 and EIF4A3, but not the UPF3B regulator of NMD
(16). EIF4A3 is an essential
component in the physiological processes of humans. The present
review discusses current literature on the function of EIF4A,
particularly in tumors.
EIF4A3 and cell function
A previous study reported that the spliceosomal
factor, CWC22 spliceosome associated protein homolog (CWC22),
directly interacts with EIF4A3 to recruit the EJC to targeted mRNA
(17). Ryu et al (18) revealed the detailed mechanism,
demonstrating that threonine 163 in the RNA-binding motif of EIF4A3
can be phosphorylated by CDK1 and CDK2 in a cell cycle-dependent
manner, which prevents binding of EIF4A3 to spliced mRNAs and other
EJC members, and promotes the interaction between EIF4A3 and CWC22.
However, this phosphorylation inhibits NMD. The splicing of mRNA
relies on the RNA binding motif protein 8A (Y14)/mago homolog, EJC
subunit (Magoh) heterodimer, the core component of EJC, which is
located on the corresponding target site. The process also requires
the participation of EIF4A3. However, the location of other
components of the EJC is not affected by EIF4A3, which suggests
that the binding of different components of the EJC to mRNA
involves a different signaling pathway. Furthermore, EIF4A3 has
been reported to be associated with spliced mRNAs at the EJC,
suggesting that EIF4A3 may provide an association between splicing
and translation in the cytoplasm (6). Furthermore, computational screening has
demonstrated that EIF4A3 is a potential regulator for mRNA-protein
expression level discrepancy (19).
Thus, EIF4A3 is an indispensable molecule in protein
translation.
Previous studies have reported that EIF4A3 is a key
component of cell cycle and apoptosis regulation (20–23). In
addition, EJC-related components serve a vital role in the splicing
of apoptosis factor mRNA. For example, recombinant EIF4A3, Y14 and
Magoh proteins preferentially bind to the endogenous apoptotic
factor BCL2 like 1 (Bcl-x) precursor RNA, and specifically transfer
Bcl-x alternative splicing to the nuclear extract (20). Furthermore, EIF4A3 serves a role in
cell cycle monitoring. Inhibition of EIF4A3 using compounds or gene
interference technology decreases cell cycle arrest in the
G2/M phase, which in turn increases apoptosis (21).
EIF4A3 and tumors
Bioinformatics analyses have revealed that EIF4A3
expression is upregulated at the transcriptional level in common
malignant tumors (24). E59K/Q is
the most prevalent mutation in the DEAD domain, which influences
the TNF-α/NF-κB signaling pathway (24). Patients with tumors frequently
produce autoantibodies, and the identification of tumor
autoantigens may serve a role in the early diagnosis and
immunotherapy of cancer (25).
EIF4A3 reactivity has been observed in patients with pancreatic,
colorectal, gastric and hepatocellular cancers, but has not been
observed in patients with chronic pancreatitis or lung cancer, or
in healthy individuals, which suggests that EIF4A3 may serve as a
serum diagnostic marker for patients with cancer (26). In hepatocellular carcinoma (HCC),
phosphorylation of EIF4A3 is associated with metastasis by
regulating mRNA splicing, and other spliceosome proteins have also
been reported to be involved in the process (27). Lin et al (28) demonstrated that EIF4A3 expression is
upregulated in HCC tissues compared with healthy liver tissues, and
high EIF4A3 expression is associated with a poor prognosis. EIF4A3
is strongly associated with the expression of several types of cell
cycle regulatory genes (CDK1 and CDK2), tumor-associated
transcription factors, chemokine signaling pathways and spliceosome
signaling pathways (18,20–22). In
addition, EIF4A3 expression is upregulated in ovarian cancer
tissues compared with adjacent healthy ovarian tissues (23). However, in patients with
pregnancy-associated breast cancer, EIF4A3 expression is
downregulated, as determined via database analysis (29). Taken together, these findings suggest
that the pathogenic mechanisms of EIF4A3 vary in different types of
cancer.
EIF4A3, as an RNA binding protein (RBP), regulates
the expression of non-coding RNAs in tumors. In glioblastoma
multiforme (GBM), circ-matrix metallopeptidase (MMP)9 serves as an
oncogene and is associated with cell proliferation, migration and
invasion, among which EIF4A3 promotes circMMP9 expression by
interacting with the upstream region of the circMMP9 mRNA
transcript (30). In breast cancer,
EIF4A3 modulates the cell cycle by promoting the expression of
circ-septin 9 (SEPT9) by binding to SEPT9 pre-mRNA (22). In gastric cancer (GC),
hsa_circ_001988 attenuates GC progression, and EIF4A3 serves as an
RNA-binding protein to promote hsa_circ_001988 expression (31). Another study reported that VCAN
antisense RNA 1 interacts with EIF4A3 to prevent EIF4A3 from
recruiting tumor protein p53 mRNA, which downregulates p53
expression in GC cells (32). In
breast cancer and GC, bioinformatics analyses have demonstrated
that EIF4A3 is an RBP to circular RNA (33,34). In
addition, EIF4A3 has been identified as a long non-coding RNA
cancer susceptibility 2 (CASC2)-binding protein, which inhibits
SKOV3 ovarian cancer cell viability, migration and invasion, and
increases cell apoptosis by regulating CASC2 (23). Another study in GBM demonstrated that
EIF4A3 extends the half-life of long intergenic non-protein coding
RNA (LINC)00680 and TTN antisense RNA 1 (TTN-AS1) (35). Similar to LINC00680 and TTN-AS1
knockdown, EIF4A3 knockdown inhibits glioblastoma cell
proliferation, migration and invasion, and increases apoptosis
(35). In non-small cell lung
cancer, LINC00667 is recruited to EIF4A3 to stabilize vascular
endothelial growth factor A mRNA (36). Furthermore, in pancreatic
adenocarcinoma, LINC01232 is recruited to EIF4A3 to regulate the
mRNA stability of transmembrane 9 superfamily member 2 (TM9SF2),
which regulates TM9SF2 protein expression (37).
EIF4A3 inhibitors
In 2011, a pan EIF4A inhibitor, hippuristanol, was
reported to inhibit human T lymphotrophic virus type 1-infected
T-cell line and adult T-cell leukemia cell proliferation, but not
normal peripheral blood mononuclear cell proliferation, by inducing
cell cycle arrest at the G1 phase and decreasing the
expression of cell cycle protein and cyclin-dependent kinase, and
promoting apoptosis by decreasing the expression levels of Bcl-x,
baculoviral IAP repeat containing 3, X-linked inhibitor of
apoptosis and CASP8 and FADD like apoptosis regulator (38). Recent studies have demonstrated that
NMD inhibition induces tumor immunity and enhances cancer
chemotherapy (39,40). Based on the key role of EIF4A3 in NMD
and the therapeutic potential of targeting EIF4A3 in cancer,
several research groups are pursuing the development of EIF4A3
inhibitors.
There are three natural products that can be
combined with EIF4A, including hippuristanol, pateamine A and
rocaglates (41). Hippuristanol
displays decreased effectiveness toward EIF4A3, requiring 10-fold
higher concentrations compared with EIF4A1/2 (42). Nuclear magnetic resonance analysis
revealed that hippuristanol inhibits EIF4A1 by binding to its
allosteric site (42). Pateamine A
is a pan inhibitor for EIF4A, which blocks EIF4F complex (used for
translation initiation) formation by stabilizing the interaction
between EIF4A and targeted RNA (43). However, pateamine A can also induce
the ATPase activity of EIF4A3, and inhibit NMD by stabilizing UPF1
and the EJC complex (44).
Rocaglates displays a similar inhibitory mechanism against EIF4A1/2
as pateamine A (45). However, these
natural EIF4A inhibitors cannot specifically inhibit EIF4A3.
Therefore, selective EIF4A3 inhibitors are urgently required.
The current research on selective inhibitors of
EIF4A3 is primarily performed by the same research teams in Japan
and Canada. In April 2017, Ito et al (46) identified 1,4-diacylpiperazine
derivatives by chemical optimization via high-throughput screening
(HTS), and identified selective EIF4A3 inhibitors 53a and 52a for
the first time. The results demonstrated that 53a and 52a display
high selectivity for EIF4A3, but not for the EIF4A1/2 proteins or
other helicases. In addition, 53a and 52a display cellular NMD
inhibitory activity, and are associated with EIF4A3 ATPase
inhibitory activity with IC50 values of 0.20 µM
(0.16–0.25) and 0.26 µM (0.18–0.38), respectively (46). The binding sites of 53a and 52a to
EIF4A3 are in non-ATP binding sites (46). Another 1,4-diacylpiperazine
derivative, compound 2, was identified by performing HTS. Compound
2 is highly selective [IC50=0.11 µM (0.092–0.13)], but
non-competitively with ATP. Even at 100 µM, compound 2 displays
almost no inhibitory activity on EIF4A1 and EIF4A2 or DExH-box
helicase 29 (DHX29) and small nuclear ribonucleoprotein U5 subunit
200 (Brr2), which belong to the serine and arginine rich splicing
factor 1 (SF2) helicase family (47). Compound 2 binds to the allosteric
region of EIF4A3 and restrains ATPase, helicase and cellular NMD
activities in vitro by inducing a conformational alteration
without disrupting the association with the core components of EJC
(47). By performing chemical
optimization of compound 2, compound 18 was discovered, which is an
ATP-competitive EIF4A3 inhibitor with IC0 ATPase=0.97
µM, whereas compound 2 displays IC50 ATPase=27 µM
(48).
Using the novel
3-(4-chlorophenyl)-1,4-diacylpiperazine derivative 1a as a template
compound, two novel orally EIF4A3-selective inhibitors, 1o and 1q,
have been identified. Both 1o and 1q display highly selective
EIF4A3 inhibitory activity, but similar to compound 2, do not
display an inhibitory effect against other EIF4A family members or
other ATP-dependent RNA helicases, such as Brr2 and DHX29. The
IC50 values of inhibitors 1o and 1q are 0.1 µM
(0.06–0.15) and 0.14 µM (0.09–0.22), respectively. In addition,
they both display NMD inhibition activity, which was identified by
performing a luciferase based cellular NMD reporter assay (49). A HCT-116 ×enograft mouse model was
used to analyze the antitumor activity of 1o and 1q. The results
demonstrated that 1o and 1q significantly inhibit the growth of
transplanted tumors without severe weight loss of the xenograft
mouse models (49). Further studies
will identify additional EIF4A3 small molecule inhibitors for EJC
and NMD, and for cancer targeted EIF4A3 treatment.
Bioinformatics analysis of EIF4A3 in
gynecological tumors and breast cancer
Given the lack of understanding of the role of
EIF4A3 in gynecological cancer, as well as the controversial role
of EIF4A3 in breast cancer, the present review systematically
analyzed the expression difference and mutation status of EIF4A3 in
breast cancer and gynecological tumors via bioinformatics analysis.
Based on the differential expression of EIF4A3 between cancer and
matched healthy tissues, and the presence of mutations in some
diseases, three types of gynecological tumors, including uterine
corpus endometrial carcinoma, cervical squamous cell carcinoma and
ovarian cancer, and breast cancer were analyzed.
EIF4A3 expression in gynecological
tumors and breast cancer
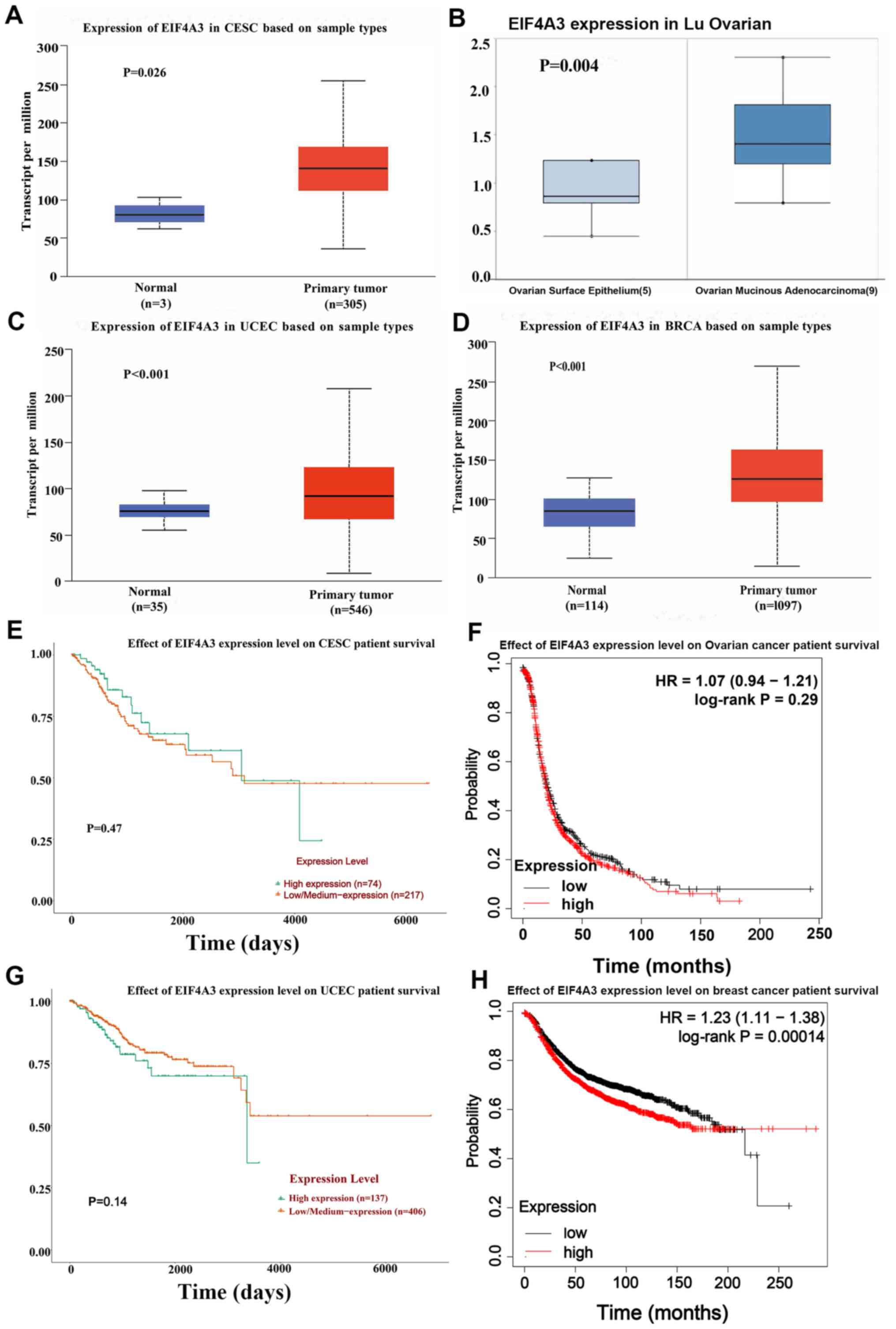
The gene expression levels of EIF4A3 were analyzed
in gynecological tumors using the University of Alabama Cancer
Database (UALCAN), an online website for The Cancer Genome Atlas
(ualcan.path.uab.edu), and Oncomine
(www.oncomine.org/resource/login.html) databases. The
results demonstrated that EIF4A3 expression was upregulated in
gynecological tumors and breast cancer (Fig. 1A-D). Furthermore, Kaplan-Meier
Plotter (https://kmplot.com/analysis) and
UALCAN were used to assess the association between EIF4A3
expression and the survival rate of patients with gynecological
tumors and breast cancer. In breast cancer, high EIF4A3 expression
was associated with poor survival rates (Fig. 1E-H).
EIF4A3 mutations in gynecological
tumors and breast cancer
Using cBioPortal (www.cbioportal.org) and Catalogue Of Somatic Mutations
In Cancer (cancer.sanger.ac.uk/cosmic), EIF4A3 mutations in
gynecological tumors and breast cancer were analyzed. The
mutations, such as substitution missense, nonsense, synonymous and
insertion frame shift, are presented in Fig. 2A-D. In all four types of cancers, the
most frequent mutation observed was substitution missense, and no
gene fusion was observed in all four types of cancers. The altered
frequencies of EIF4A3 in the four types of cancers are presented in
Fig. 2E-H. The results indicated
that 2.0–2.5% of cervical cancer, 3–7% of ovarian cancer, 1.5–3.5%
of endometrial carcinoma and 4–17% of breast cancer clinical
samples contained EIF4A3 mutations.
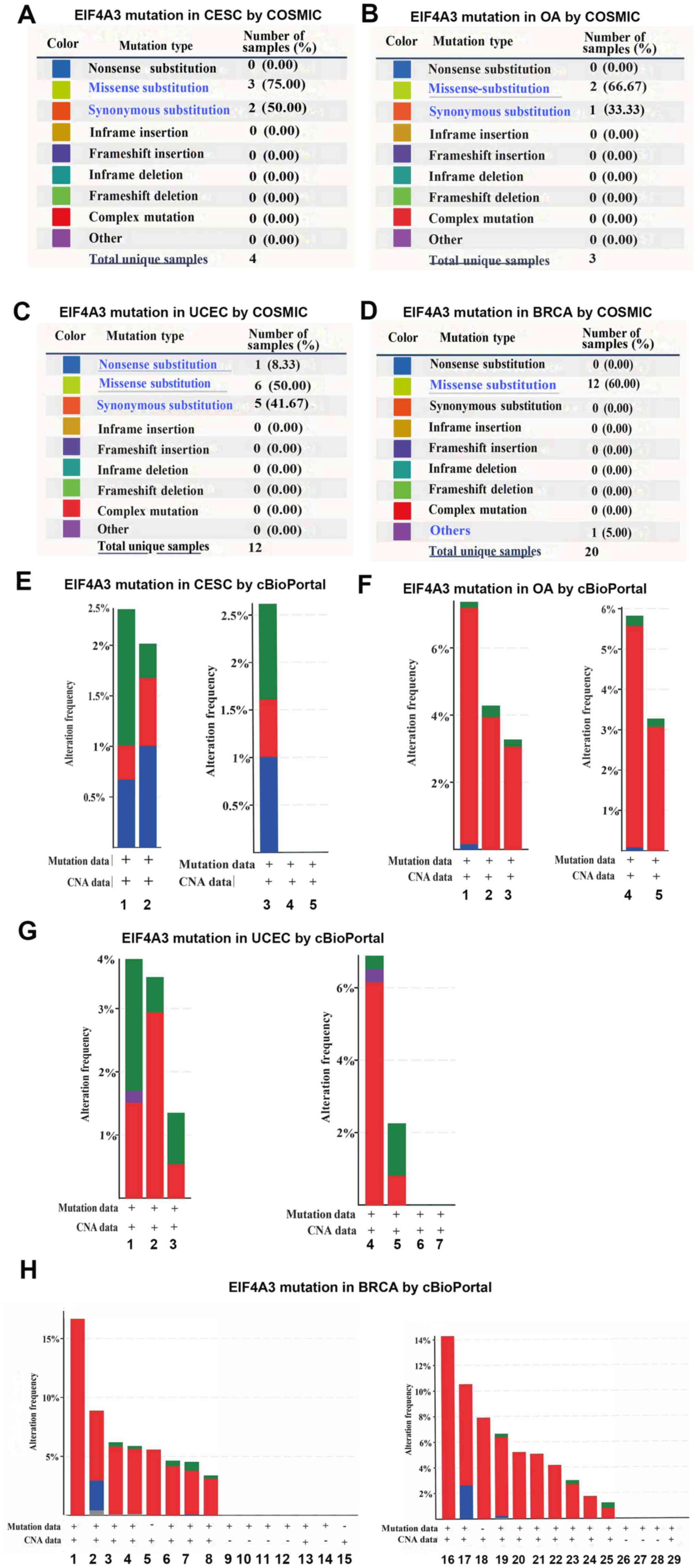 | Figure 2.COSMIC and cBioPortal analyses of
EIF4A3 mutations in gynecological tumors and breast cancer. EIF4A3
mutations in (A) CESC, (B) OA, (C) UCEC and (D) BRCA, using COSMIC
analyses. (E) Mutation analysis of EIF4A3 in CESC using cBioPortal
analysis; 1, cervical (TCGA PanCan); 2, cervical (TCGA); 3,
cervical squamous cell carcinoma; 4, endocervical adenocarcinoma
and 5, mucinous carcinoma. (F) Mutation analysis of EIF4A3 in OA
using cBioPortal analysis; 1, ovarian (TCGA); 2, ovarian (TCGA
PanCan 2018); 3, ovarian (TCGA pub); 4, serous ovarian cancer and
5, high-grade serous ovarian cancer. (G) Mutation analysis of
EIF4A3 in UCEC using cBioPortal analysis; 1, uterine (TCGA PanCan
2018); 2, uterine (TCGA); 3, uterine (TCGA pub); 4, uterine serous
carcinoma/uterine papillary serous carcinoma; 5, uterine
endometrioid carcinoma; 6, uterine mixed endometrial carcinoma and
7, endometrial carcinoma. (H) Mutation analysis of EIF4A3 in BRCA
using cBioPortal analysis; 1, ACBC (MSKCC/Breast 2015); 2, the MBC
project; 3, breast (TCGA); 4, breast (TCGA 2015); 5, breast
(METABRIC 2016); 6, BRCA (INSERM 2016); 7, breast invasive
carcinoma breast (TCGA PanCan 2018); 8, breast (TCGA pub); 9, BFN
(Duke-NUS); 10, breast (BCCRC 2012); 11, breast (BCCRC Xenograft);
12, breast (Broad 2012); 13, BRCA (MSKCC 2019); 14, breast
(Sanger); 15, BREAST (Alpelisib); 16, adenoid cystic breast cancer;
17, ‘Breast Invasive Cancer, NOS’; 18, breast cancer; 19, breast
invasive ductal carcinoma; 20, breast invasive carcinoma (NOS); 21,
breast mixed ductal and lobular carcinoma; 22, metaplastic breast
cancer; 23, invasive breast carcinoma; 24, breast invasive mixed
mucinous carcinoma; 25, breast invasive lobular carcinoma; 26,
benign phyllodes tumor of the breast; 27, invasive breast cancer;
28, infiltrating ductal carcinoma; 29, breast. EIF4A3, eukaryotic
translation initiation factor 4A3; CESC, cervical squamous cell
carcinoma; OA, ovarian cancer; UCEC, uterine corpus endometrial
carcinoma; BRCA, breast invasive carcinoma; TCGA, The Cancer Genome
Atlas. |
Conclusions
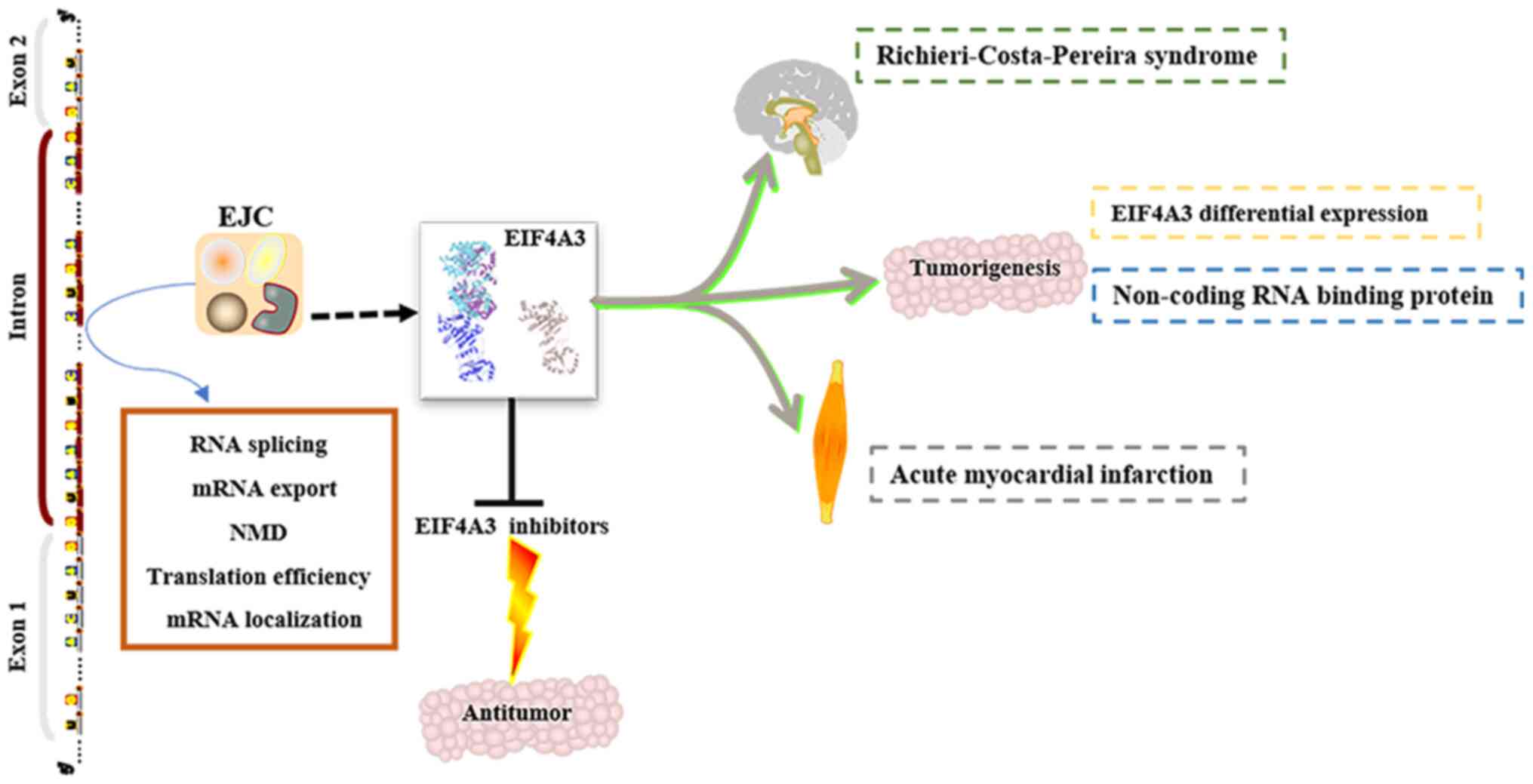
EIF4A3 has been identified as a key component in the
EJC, and is involved in benign and malignant disease progression
and development (Fig. 3). However,
the biological role of EIF4A3 remains unclear, thus, further
investigations on the EJC, NMD and tumors are required. EIF4A3
primarily functions via its role as a key component of the EJC, and
other key proteins of the EJC may serve similar roles, particularly
in neuromuscular development. For example, in the embryos of the
frog Xenopus laevis, EIF4A3 knockdown causes general
paralysis and defects in sensory neurons, pigment cells and heart
development (50). Following
knockdown of other core proteins of the EJC, similar phenotypes are
observed (50). Although there are
only a few studies on EIF4A3 in cancer (22,35,36), it
has been demonstrated that EIF4A3 is differentially expressed in
cancer tissues and healthy tissues. By performing bioinformatics
analysis, the present study demonstrated that EIF4A3 expression was
upregulated in gynecological tumors and breast cancer compared with
matched healthy tissues. In addition, EIF4A3 mutations were
observed in cancer, particularly in breast cancer. In a study on
the regulation of tumorigenesis, it was reported that EIF4A3, as an
RBP, regulates the expression of non-coding RNAs (30,37).
However, the roles and underlying molecular mechanisms of EIF4A3 in
EJC, NMD and tumorigenesis remain unclear. Thus, several medicinal
chemists are aiming to develop highly selective EIF4A3 inhibitors
to identify the role of EIF4A3. Although research on EIF4A3
inhibitors is still in the preclinical stage, several potent EIF4A3
inhibitors (EIF4A3 inhibitor 1a, 53a, 1o and 1q) have been
identified (46,48,49), and
with further research, the molecular mechanism underlying EIF4A3
will be revealed. Thus, EIF4A3 may serve as a novel therapeutic
target for cancer in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
YZ and CR contributed to the conception, literature
review, design and analysis of the study, and drafted the initial
manuscript. CR also revised the manuscript for important
intellectual content. LY contributed to the conception and
literature review of the study, and drafted the initial manuscript.
YZ and CR confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EIF4A3
|
eukaryotic translation initiation
factor 4A3
|
|
DEAD
|
Asp-Glu-Ala-Asp
|
|
EJC
|
exon junction complex
|
|
NMD
|
nonsense-mediated mRNA decay
|
|
HCC
|
hepatocellular carcinoma
|
|
GBM
|
glioblastoma multiforme
|
|
GC
|
gastric cancer
|
|
HTS
|
high-throughput screening
|
References
|
1
|
Linder P and Jankowsky E: From unwinding
to clamping- the DEAD box RNA helicase family. Nat Rev Mol Cell
Biol. 12:505–516. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tanner NK and Linder P: DExD/H box RNA
helicases: From generic motors to specific dissociation functions.
Mol Cell. 8:251–262. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jankowsky E: RNA helicases at work:
Binding and rearranging. Trends Biochem Sci. 36:19–29. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Andreou AZ and Klostermeier D: The
DEAD-box helicase eIF4A: Paradigm or the odd one out. RNA Biol.
10:19–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choe J, Ryu I, Park OH, Park J, Cho H, Yoo
JS, Chi SW, Kim MK, Song HK and Kim YK: eIF4AIII enhances
translation of nuclear cap-binding complex-bound mRNAs by promoting
disruption of secondary structures in 5′UTR. Proc Natl Acad Sci
USA. 111:E4577–E4586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chan CC, Dostie J, Diem MD, Feng W, Mann
M, Rappsilber J and Dreyfuss G: eIF4A3 is a novel component of the
exon junction complex. RNA. 10:200–209. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Le Hir H and Séraphin B: EJCs at the heart
of translational control. Cell. 133:213–216. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reed R: Coupling transcription, splicing
and mRNA export. Curr Opin Cell Biol. 15:326–331. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reed R and Hurt E: A conserved mRNA export
machinery coupled to pre-mRNA splicing. Cell. 108:523–531. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Le Hir H, Gatfield D, Izaurralde E and
Moore MJ: The exon-exon junction complex provides a binding
platform for factors involved in mRNA export and nonsense-mediated
mRNA decay. EMBO J. 20:4987–4997. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nott A, Le Hir H and Moore MJ: Splicing
enhances translation in mammalian cells: An additional function of
the exon junction complex. Genes Dev. 18:210–222. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giorgi C and Moore MJ: The nuclear nurture
and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol.
18:186–193. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Saulière J, Murigneux V, Wang Z, Marquenet
E, Barbosa I, Tonquèze OL, Audic Y, Paillard L, Crollius HR and Le
Hir H: CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of
the human exon junction complex. Nat Struct Mol Biol. 19:1124–1131.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fatscher T, Boehm V and Gehring NH:
Mechanism, factors, and physiological role of nonsense-mediated
mRNA decay. Cell Mol Life Sci. 72:4523–4544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kashima I, Yamashita A, Izumi N, Kataoka
N, Morishita R, Hoshino S, Ohno M, Dreyfuss G and Ohno S: Binding
of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction
complex triggers Upf1 phosphorylation and nonsense-mediated mRNA
decay. Genes Dev. 20:355–367. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bhalla AD, Gudikote JP, Wang J, Chan WK,
Chang YF, Olivas OR and Wilkinson MF: Nonsense codons trigger an
RNA partitioning shift. J Biol Chem. 284:4062–4072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barbosa I, Haque N, Fiorini F, Barrandon
C, Tomasetto C, Blanchette M and Le Hir H: Human CWC22 escorts the
helicase eIF4AIII to spliceosomes and promotes exon junction
complex assembly. Nat Struct Mol Biol. 19:983–990. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ryu I, Won YS, Ha H, Kim E, Park YK, Kim
MK, Kwon DH, Choe J, Song HK, Jung H and Kim YK: eIF4A3
phosphorylation by CDKs affects NMD during the cell cycle. Cell
Rep. 26:2126–2139.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu X, Zhao W, Cui Q and Zhou Y:
Computational screening of potential regulators for mRNA-protein
expression level discrepancy. Biochem Biophys Res Commun.
523:196–201. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Michelle L, Cloutier A, Toutant J, Shkreta
L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture
S, et al: Proteins associated with the exon junction complex also
control the alternative splicing of apoptotic regulators. Mol Cell
Biol. 32:954–967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mazloomian A, Araki S, Ohori M, El-Naggar
AM, Yap D, Bashashati A, Nakao S, Sorensen PH, Nakanishi A, Shah S
and Aparicio S: Pharmacological systems analysis defines EIF4A3
functions in cell-cycle and RNA stress granule formation. Commun
Biol. 2:1652019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng X, Huang M, Xing L, Yang R, Wang X,
Jiang R, Zhang L and Chen J: The circRNA circSEPT9 mediated by E2F1
and EIF4A3 facilitates the carcinogenesis and development of
triple-negative breast cancer. Mol Cancer. 19:732020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang S, Leng T, Zhang Q, Zhao Q, Nie X
and Yang L: Sanguinarine inhibits epithelial ovarian cancer
development via regulating long non-coding RNA CASC2-EIF4A3 axis
and/or inhibiting NF-κB signaling or PI3K/AKT/mTOR pathway. Biomed
Pharmacother. 102:302–308. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin Y, Zhang J, Cai J, Liang R, Chen G,
Qin G, Han X, Yuan C, Liu Z, Li Y, et al: Systematic analysis of
gene expression alteration and co-expression network of eukaryotic
initiation factor 4A-3 in cancer. J Cancer. 9:4568–4577. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gatenby PA, Basten A, Tattersall MH and
Fox RM: Autoantibodies in cancer patients given Corynebacterium
parvum/levamisole immunotherapy. Lancet. 1:10821980. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia Q, Kong XT, Zhang GA, Hou XJ, Qiang H
and Zhong RQ: Proteomics-based identification of DEAD-box protein
48 as a novel autoantigen, a prospective serum marker for
pancreatic cancer. Biochem Biophys Res Commun. 330:526–532. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tian M, Cheng H, Wang Z, Su N, Liu Z, Sun
C, Zhen B, Hong X, Xue Y and Xu P: Phosphoproteomic analysis of the
highly-metastatic hepatocellular carcinoma cell line, MHCC97-H. Int
J Mol Sci. 16:4209–4225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin Y, Liang R, Mao Y, Ye J, Mai R, Gao X,
Liu Z, Wainwright T, Li Q, Luo M, et al: Comprehensive analysis of
biological networks and the eukaryotic initiation factor 4A-3 gene
as pivotal in hepatocellular carcinoma. J Cell Biochem.
121:4094–4107. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang J, Zhou YJ, Yu ZH, Chen AX, Yu Y,
Wang X and Cao XC: Identification of core genes and clinical roles
in pregnancy-associated breast cancer based on integrated analysis
of different microarray profile datasets. Biosci Rep.
39:BSR201900192019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: EIF4A3-induced circular RNA MMP9 (circMMP9)
acts as a sponge of miR-124 and promotes glioblastoma multiforme
cell tumorigenesis. Mol Cancer. 17:1662018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun D, Wang G, Xiao C and Xin Y:
Hsa_circ_001988 attenuates GC progression in vitro and in vivo via
sponging miR-197-3p. J Cell Physiol. 236:612–624. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Feng L, Li J, Li F, Li H, Bei S, Zhang X
and Yang Z: Long noncoding RNA VCAN-AS1 contributes to the
progression of gastric cancer via regulating p53 expression. J Cell
Physiol. 235:4388–4398. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Afzali F and Salimi M: Unearthing
regulatory axes of breast cancer circRNAs networks to find novel
targets and fathom pivotal mechanisms. Interdiscip Sci. 11:711–722.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou
J, Jin H, Zhao A, Tang WW and Cao XF: Down-regulation of circPVRL3
promotes the proliferation and migration of gastric cancer cells.
Sci Rep. 8:101112018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang W, Wang D, Shao L, Liu X, Zheng J,
Xue Y, Ruan X, Yang C, Liu L, Ma J, et al: LINC00680 and TTN-AS1
stabilized by EIF4A3 promoted malignant biological behaviors of
glioblastoma cells. Mol Ther Nucleic Acids. 19:905–921. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang H, Yang W, Dai W, Ma Y and Zhang G:
LINC00667 promotes the proliferation, migration, and pathological
angiogenesis in non-small cell lung cancer through stabilizing
VEGFA by EIF4A3. Cell Biol Int. 44:1671–1680. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Q, Lei C, Lu C, Wang J, Gao M and Gao
W: LINC01232 exerts oncogenic activities in pancreatic
adenocarcinoma via regulation of TM9SF2. Cell Death Dis.
10:6982019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tsumuraya T, Ishikawa C, Machijima Y,
Nakachi S, Senba M, Tanaka J and Mori N: Effects of hippuristanol,
an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol.
81:713–722. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Popp MW and Maquat LE: Attenuation of
nonsense-mediated mRNA decay facilitates the response to
chemotherapeutics. Nat Commun. 6:66322015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pastor F, Kolonias D, Giangrande PH and
Gilboa E: Induction of tumour immunity by targeted inhibition of
nonsense-mediated mRNA decay. Nature. 465:227–230. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shen L and Pelletier J: Selective
targeting of the DEAD-box RNA helicase eukaryotic initiation factor
(eIF) 4A by natural products. Nat Prod Rep. 37:609–616. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lindqvist L, Oberer M, Reibarkh M, Cencic
R, Bordeleau ME, Vogt E, Marintchev A, Tanaka J, Fagotto F, Altmann
M, et al: Selective pharmacological targeting of a DEAD box RNA
helicase. PLoS One. 3:e15832008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Low WK, Dang Y, Schneider-Poetsch T, Shi
Z, Choi NS, Merrick WC, Romo D and Liu JO: Inhibition of eukaryotic
translation initiation by the marine natural product pateamine A.
Mol Cell. 20:709–722. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dang Y, Low WK, Xu J, Gehring NH, Dietz
HC, Romo D and Liu JO: Inhibition of nonsense-mediated mRNA decay
by the natural product pateamine A through eukaryotic initiation
factor 4AIII. J Biol Chem. 284:23613–23621. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bordeleau ME, Robert F, Gerard B,
Lindqvist L, Chen SMH, Wendel HG, Brem B, Greger H, Lowe SW, Porco
JA Jr and Pelletier J: Therapeutic suppression of translation
initiation modulates chemosensitivity in a mouse lymphoma model. J
Clin Invest. 118:2651–2660. 2008.PubMed/NCBI
|
|
46
|
Ito M, Tanaka T, Cary DR,
Iwatani-Yoshihara M, Kamada Y, Kawamoto T, Aparicio S, Nakanishi A
and Imaeda Y: Discovery of novel 1,4-diacylpiperazines as selective
and cell-active eIF4A3 inhibitors. J Med Chem. 60:3335–3351. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Iwatani-Yoshihara M, Ito M, Ishibashi Y,
Oki H, Tanaka T, Morishita D, Ito T, Kimura H, Imaeda Y, Aparicio
S, et al: Discovery and characterization of a eukaryotic initiation
factor 4A-3-selective inhibitor that suppresses nonsense-mediated
mRNA decay. ACS Chem Biol. 12:1760–1768. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ito M, Iwatani M, Kamada Y, Sogabe S,
Nakao S, Tanaka T, Kawamoto T, Aparicio S, Nakanishi A and Imaeda
Y: Discovery of selective ATP-competitive eIF4A3 inhibitors. Bioorg
Med Chem. 25:2200–2209. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mizojiri R, Nakata D, Satoh Y, Morishita
D, Shibata S, Iwatani-Yoshihara M, Kosugi Y, Kosaka M, Takeda J,
Sasaki S, et al: Discovery of novel
5-(Piperazine-1-carbonyl)pyridin-2(1H)-one derivatives as orally
eIF4A3-selective inhibitors. ACS Med Chem Lett. 8:1077–1082. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Haremaki T, Sridharan J, Dvora S and
Weinstein DC: Regulation of vertebrate embryogenesis by the exon
junction complex core component Eif4a3. Dev Dyn. 239:1977–1987.
2010. View Article : Google Scholar : PubMed/NCBI
|