Introduction
Lacrimal gland adenoid cystic carcinoma (LACC) is
the most common form of malignant epithelial lacrimal gland tumor.
It is characterized by a slow progression, local invasion and
metastasis, and usually has a poor prognosis due to osseous and
perineural invasion (1,2). A study in the University of Texas MD
Anderson Cancer Center shows the local recurrence rate is 35%, the
distant metastasis rate is 80% and the mortality is 65% (3,4). Local
recurrence and distant metastatic disease occur in ~50% of patients
within 5 to 10 years, and currently there is no effective treatment
(5,6). Despite classic histological types, ACC
can also show areas of classic ACC with a gradual or sharp
transition to the areas of high-grade carcinoma. Initially, this
atypical ACC was described as ‘hybrid tumors’, subsequently as
‘dedifferentiated’ and most recently as high-grade transformation
(HGT) (7–9). The histological features of ACC with
HGT are composed of two histological components: Conventional
low-grade ACC and high-grade ‘dedifferentiated’ tumor tissues
(10). LACC rarely undergoes HGT,
but there is evidence that histological subtypes are associated
with prognosis. A study shows the lymph node metastasis rate of
LACC-HGT is higher compared with LACC (43–57% vs. 5–25%) (11). According to a previous report,
LACC-HGT accelerates the process of tumor local recurrence and
distant metastasis, resulting in increased mortality, shortened
postoperative survival and poor outcomes of conventional treatment
(12).
Non-coding RNAs (ncRNAs) are generally defined as a
wide class of RNAs without protein-coding function, but these have
regulatory functions in the various biological processes, such as
post-transcriptional regulators, regulation of mitochondrial
biology and metabolic control. As a group of endogenous,
single-stranded, small non-coding RNA molecules, microRNAs
(miRNAs/miRs) negatively regulate gene expression at the
post-transcriptional level (13).
Previous studies have shown significant differences in miRNA
expression of ACC. For example, miR-24-3p was low expression in
high metastasis type ACC cells, and further research reported that
miR-24-3p is downregulated in LACC tissue and inhibits ACC cell
proliferation, migration and invasion through targeting protein
kinase C eta type (14). Meanwhile,
downregulated miR-24-3p also inhibits the epithelial-mesenchymal
transition (EMT) process of ACC. Most recently, Hao et al
(15) demonstrated that miR-93-5p is
overexpressed in tissues and plasma of patients with LACC compared
with healthy controls. miR-93-5p promotes LACC cell migration,
invasion and proliferation by targeting the downregulation of
breast cancer metastasis-suppressor 1-like protein through
regulation of the Wnt signaling pathway. Circular RNAs (circRNAs)
are novel non-coding RNAs that have been reported to be involved in
the progression of various cancers, such as non-small cell lung
carcinoma, bladder cancer and osteosarcoma (16). However, the regulatory mechanisms of
circRNAs in LACC remain unclear.
The hypothesis of ceRNA was defined by Salmena et
al (17) in 2011. ceRNA
introduced a new crosstalk theory of gene regulation that promotes
the physiology and development of diseases, such as cancer. Recent
studies about ceRNA have expanded our insights into LACC-HGT
pathogenesis (18).
Materials and methods
Patients and tissue samples
A total of patients with 12 LACC were originally
included in the study, all patients benefited from a surgical
resection. HGT occurred in six cases following the histological
analysis. All patient profiles were evaluated; their symptoms and
signs were assessed as two separate components, and two patients
were lost during follow-up in the LACC group. A total of six LACC
paraffin-embedded tissue samples and six LACC-HGT paraffin-embedded
tissue samples were provided by Tianjin Medical University Eye
Hospital (Tianjin, China) from June 2009 to May 2017. Informed
written consent for research purposes was obtained from the
patients before tissue collection. The Institutional of Human
Research Ethics Committee of the Tianjin Medical University Eye
Hospital approved the study [Tianjin, China; approval no.
2017KY(L)-23].
Primary cell line culture
Primary cell line culture was performed as described
previously (19) via tissue block
culture techniques, and the cells were cultured in RPMI-1640 medium
with 10% fetal bovine serum and 1% penicillin/streptomycin solution
(both HyClone; Cyvita) at 37°C in a 5% CO2 incubator.
Only LACC and LACC-HGT primary cells in passages 4–10 were used for
further experiments.
miRNA microarray analysis
Differentially expressed miRNAs (DEMs) of 12 LACC
samples were detected using the Agilent Human miRNA Microarray,
Release 21.0 (8*60K, Design ID: 070156). miRNA expression profiles
(GSE59700) of salivary adenoid cystic carcinoma (SACC) were
obtained from the GEO (http://www.ncbi.nlm.nih.gov/geo) (20). The dataset contains 12 SACC samples
with matched normal tissues.
Candidate miRNAs identification
In order to get raw data of miRNA microarray, the
array images were imported to Feature Extraction software (version
10.7.1.1, Agilent Technologies). Genespring software (version 13.1;
Agilent Technologies) was employed to finish the basic analysis
with the raw data. First, the raw data was normalized with the
quantile algorithm. DEMs were then identified through the ratio of
the expression quantity (fold-change; FC) as well as P-values
calculated using unpaired t-tests, these data are presented in
Table IV. The threshold set for up
and downregulated DEMs was a |log2FC|>1 and a P≤0.05. GEO2R is
one of the powerful tools to screen the DEMs (21). Datasets of SACC downloaded from the
GEO database were analyzed by GEO2R. Candidate miRNAs were selected
from the overlap between the miRNA microarray and the DEMs from the
GEO datasets.
 | Table IV.Expressions of differentially
expressed miRs. |
Table IV.
Expressions of differentially
expressed miRs.
| ID | Regulation | FC | Log FC | P-value |
|---|
| hsa-miR-140-3p | Down | −8.08 | −3.01 | 0.02 |
| hsa-miR-140-5p | Down | −6.35 | −2.67 | 0.04 |
|
hsa-miR-3653-3p | Down | −4.04 | −2.01 | 0.07 |
| hsa-miR-574-3p | Down | −2.20 | −1.14 | 0.03 |
| hsa-miR-29a-3p | Down | −2.09 | −1.07 | 0.01 |
|
hsa-miR-6785-5p | Up | 2.03 | 1.02 | 0.05 |
| hsa-miR-4651 | Up | 1.64 | 0.71 | 0.04 |
| hsa-miR-4459 | Up | 2.23 | 1.15 | 0.08 |
|
hsa-miR-7114-3p | Up | 3.50 | 1.81 | 0.04 |
| hsa-miR-636 | Up | 3.67 | 1.87 | 0.04 |
|
hsa-miR-6779-5p | Up | 5.35 | 2.42 | 0.02 |
|
hsa-miR-135a-3p | Up | 4.51 | 2.17 | 0.03 |
| hsa-miR-1469 | Up | 4.18 | 2.06 | 0.02 |
| hsa-miR-4488 | Up | 9.26 | 3.21 | <0.01 |
| hsa-miR-4322 | Up | 8.00 | 3.00 | 0.02 |
| hsa-miR-6075 | Up | 4.51 | 2.17 | 0.02 |
LACC-HGT differentially expressed
circRNAs (DECs) and genes screening
The DECs and differentially expressed genes (DEGs)
of LACC-HGT primary cell lines were detected using the Agilent
Human Microarray 2018 (4*180k, Design ID: 085630). To begin with,
the raw data was normalized with the quantile algorithm. DECs and
DEGs were then identified through fold-change as well as P-values
calculated using unpaired t-tests, these data are presented in
Tables SII and SIII. Upregulated and downregulated
circRNAs and genes were those with an absolute value of logarithmic
transformed fold-change (FC) [log2 FC≥log2(1.5)] and P≤0.05.
Functional enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) analyses were applied to determine the roles of
these DECs and DEGs. Afterwards, hierarchical clustering was
performed to display the distinguishable genes' expression pattern
among samples. The hypergeometric distribution algorithm was used
to calculate the significance of different gene enrichment in each
GO term and each pathway (22). The
result of this calculation gave P-values of enriched significance,
and the lower the value the more statistically significant it
is.
Gene Set Enrichment Analysis
(GSEA)
The expression profile of genes provided the ratio
of the expression quantity (log2FC) and then the genes were
arranged in descending order. The GSEA algorithm was used to
identify the more variable pathways and gene set of LACC-HGT
compared with LACC.
Interaction prediction of
circRNA-miRNA and miRNA-mRNA
To improve the predictive accuracy, CircInteractome
(23) and ENCORI (http://starbase.sysu.edu.cn/) were used to predict the
circRNA-miRNA targeting. The miRWalk (http://mirwalk.umm.uni-heidelberg.de/) database was
used to predict miRNA-mRNA targeting. In this analysis, as the DEGs
identified totaled >5,000, exceeding the limit of this tool, the
top 500 genes were selected for prediction.
Construction of ceRNA regulatory
network
According to the default screening criteria |log2FC|
>1 and P<0.05, only five downregulated miRNAs were obtained,
which did not meet the subsequent analysis requirements. Therefore,
the screening standard was adjusted to |log2FC| >1 and P<0.2,
and a total of 31 miRNAs with significant downregulation were
screened for ceRNA regulatory network construction. The previously
obtained circRNA-miRNA, miRNA-mRNA and downregulated miRNAs in
LACC-HGT were imported into Cytoscape 3.7.2 software (24) to map the ceRNA regulatory network. A
Sankey map was drawn.
Protein-protein interaction (PPI)
network of proteins in ceRNA regulatory network
The proteins in the ceRNA regulatory network were
imported into STRING (25) for PPI
network construction, and the interaction network was plotted using
Cytoscape 3.7.2 software.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
As a candidate miRNA, miR-140-3p was also involved
in ceRNA regulatory network, therefore the expression of miR-140-3p
in LACC-HGT tissues and primary cells was analyzed. miRNAs of
tissues and primary cells were extracted using the miRcute miRNA
Isolation kit (Tiangen Biotech Co., Ltd.). The RNA sample was
reversely transcribed into the first-strand cDNA using the miRcute
Plus miRNA First-Strand cDNA kit (Tiangen Biotech Co., Ltd.). The
temperature protocol was follows: 42°C For 3 min, 42°C for 15 min
and 95°C for 3 min. Quantification analysis was performed using
miRNA SYBR Green PCR master mix (Tiangen Biotech Co., Ltd.), the
temperature protocol was follows: 95°C For 15 min, 95°C for 10
sec), 60°C for 30 sec for 40 cycles. Total RNA of LACC and LACC-HGT
primary cells were extracted using the EZ-press RNA Purification
kit (EZBioscience). The RNA sample was reversely transcribed into
the first-strand cDNA using the FastQuant RT kit (Tiangen Biotech
Co., Ltd.). Quantification analysis was performed using SYBR Green
PCR master mix (Tiangen Biotech Co., Ltd.). and specific primers on
Roche LightCycler® 96 Sequence Detection system (Roche
Diagnostics). The forward primers for miR-140-3p was
5′-GCCCGTGGTTCTACCCT-3′; for 5 sec was 5′-GGAGACCGCCTGGGAATA-3′.
The reverse primers for these were universal primers included in
the kit. Primers are shown in Table
I. Expression levels were quantified using the
2−ΔΔCq method with and GAPDH as internal control
(26), these data are presented in
the Fig. 6.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primers | Sequence,
5′→3′ |
|---|
| miR-140-3p |
GCCCGTGGTTCTACCCT |
| 5s |
GGAGACCGCCTGGGAATA |
| CD200 |
|
|
Forward |
AAGTGGTGACCCAGGATGAAA |
|
Reverse |
AGGTGATGGTTGAGTTTTGGAG |
| PTHLH |
|
|
Forward |
ATTTACGGCGACGATTCTTCC |
|
Reverse |
GCTTGGAGTTAGGGGACACC |
| GAPDH |
|
|
Forward |
GATGCTGGCGCTGAGTACG |
|
Reverse |
GCTAAGCAGTTGGTGGTGC |
Statistical analysis
Each experiment was performed at least three times,
the data are presented as mean ± SD, and statistical analyses were
performed using SPSS version 21 (IBM Corp) and GraphPad Prism
version 7.00 (GraphPad Software). The analysis of clinical and
pathological dates was determined using Fisher's exact tests.
Independent samples unpaired t-tests were used to screen the DECs,
DEMs and DEGs. The results of RT-qPCR were also analyzed using
unpaired Student's t-test (Fig. 6).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patient characteristics
In this study, 12 patients clinically diagnosed with
LACC were enrolled. These included six cases of LACC and six cases
of LACC-HGT. There were differences between LACC group and LACC-HGT
group. Briefly, the mean age of patients with LACC was 55.5 years,
with the range between 29 and 68 years, and the mean age of the
patient with LACC-HGT was 38.5 years, with a range between 23 and
50 years. There were three cases in the right eye and three cases
in the left eye in LACC group and the same for the LACC-HGT group.
Main symptoms of LACC included pain or dysesthesia, ocular
proptosis, palpation, eye movement disorder and blurred vision
(Table II). The pathological grade
was higher in the LACC-HGT (all were grade III) compared with the
LACC group and the difference was statistically significant
(P<0.05; Table III). IHC showed
decreased expression of p63 and smooth muscle actin in LACC-HGT,
and an increased number of Ki-67 positive cells in LACC-HGT group,
but this was not significant (P>0.05). The mortality of LACC-HGT
group was higher compared with that of LACC group, the mortality of
LACC was 50% and the mortality of LACC-HGT was 83%. A summary of
clinical and pathological features of the 12 patients is listed in
Tables II and III. The results indicated that the
patients with HGT had a more aggressive disease compared with
conventional LACC, accompanied by a poorer prognosis. The recurrent
cases in 5 years and the death rate of the LACC-HGT group (3, 83%)
were higher compared with the LACC group (2, 50%). However, the
difference was not statistically significant.
 | Table II.Clinical features of the 12
patients. |
Table II.
Clinical features of the 12
patients.
| Clinical
features | LACC-HGT | LACC | P-value |
|---|
| Total number of
patients | 6 | 6 |
|
| Sex |
|
| 0.83 |
|
Male | 2 | 3 |
|
|
Female | 4 | 3 |
|
| Median age,
years | 38.5 | 55.5 | 0.07 |
| Tumor side |
|
| 0.83 |
|
Left | 3 | 2 |
|
|
Right | 3 | 4 |
|
| Duration of
symptoms, mean months | 5.0 | 10.5 |
|
| Symptom |
|
| 0.73 |
| Pain or
dysesthesia | 4 | 4 |
|
| Signs |
|
|
|
| Ocular
proptosis, mean mm | 3.67 | 3.17 |
|
|
Palpation | 5 | 5 | 0.78 |
| Eye
movement disorder | 4 | 5 | 0.50 |
| Blurred
vision | 3 | 4 | 0.50 |
| CT |
|
|
|
|
Periosteum involvement | 5 | 4 | 0.50 |
| Bony
involvement | 3 | 4 | 0.50 |
| MRI |
|
|
|
|
Brain | 1 | 2 | 0.50 |
|
Temporal fossa | 2 | 0 | 0.23 |
|
Sinus | 1 | 2 | 0.50 |
| Ocular
muscle | 3 | 5 | 0.28 |
| Follow up time,
months | 40.2 | 70.8a |
|
| Cases of death | 5 | 2 | 0.33 |
| Death rate, % | 83 | 50a | 0.67 |
| Cases of recurrence
in 5 years | 3 | 2 | 0.74 |
| Recurrence rate of
5 years, % | 50 | 50a | 0.83 |
| Survival time,
years |
|
|
|
| 2 | 3 | 3a | 0.45 |
| 5 | 1 | 3a | 0.23 |
 | Table III.Pathological features of the 12
patients. |
Table III.
Pathological features of the 12
patients.
| Clinical
features | LACC-HGT | LACC | P-value |
|---|
| Histological
grading |
|
| <0.01 |
| I | 0 | 4 |
|
| II | 0 | 2 |
|
|
III | 6 | 0 |
|
| TNM |
|
| 0.72 |
|
T2N0M0 | 1 | 2 |
|
|
T4aN0M0 | 2 | 1 |
|
|
T4bN0M0 | 1 | 2 |
|
|
T4cN0M0 | 2 | 1 |
|
| IHC, % |
|
|
|
|
SMA | 14.3 | 45 | <0.01 |
|
p63 | 16.8 | 53.3 | <0.01 |
|
p53 | 17 | 21 | 0.74 |
|
Ki-67 | 26.7 | 11.8 | 0.05 |
Differentially expressed miRNAs
The results showed that there were differences in
miRNA expression between LACC and LACC-HGT groups. A total of 16
DEMs in LACC-HGT paraffin-embedded tissues were screened by
microarray, including five downregulated miRNAs and 11 upregulated
miRNAs, their expressions are shown in Table IV. Overall, 704 DEMs of SACC tissues
were screened from GEO database (Table
SI).
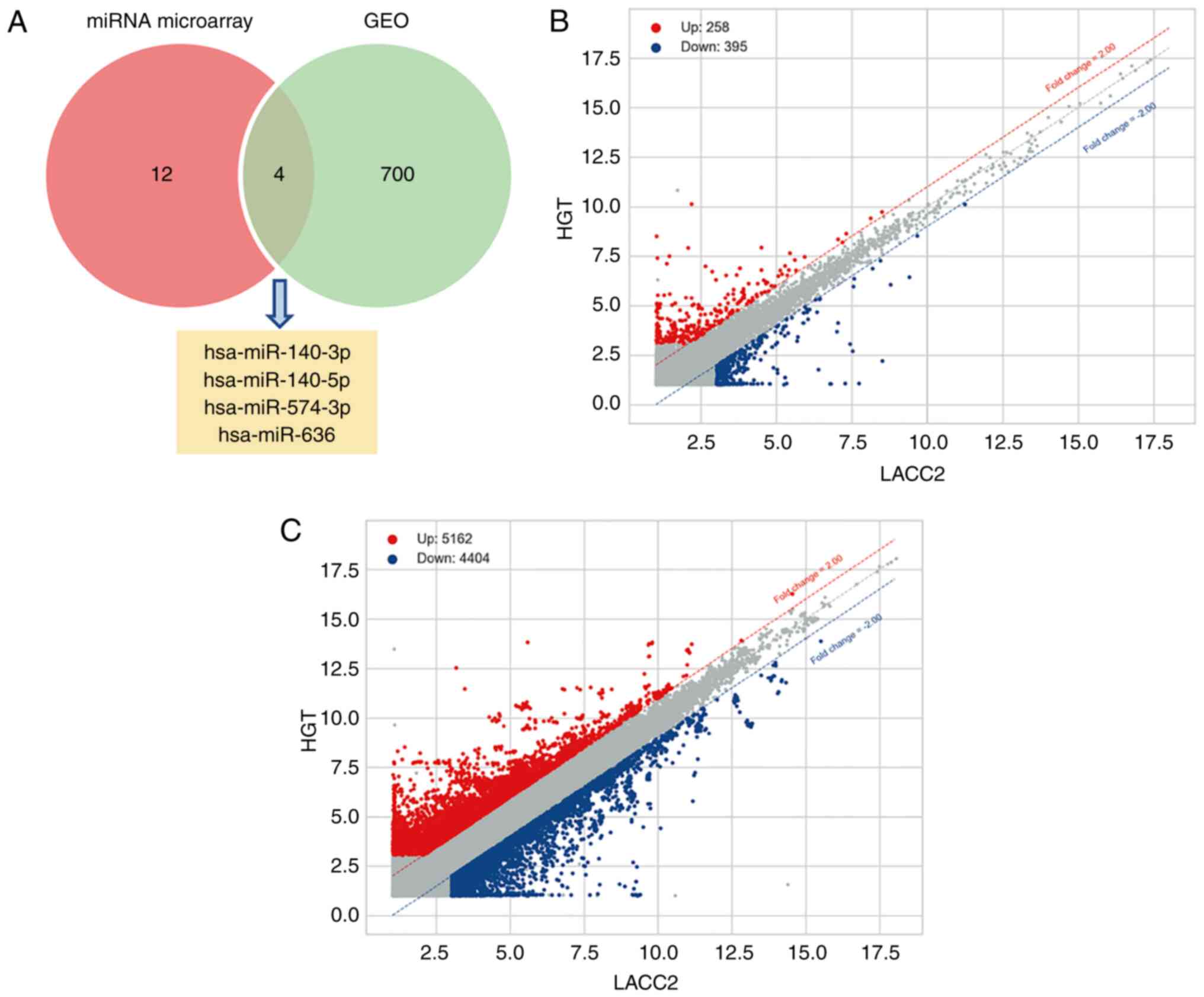
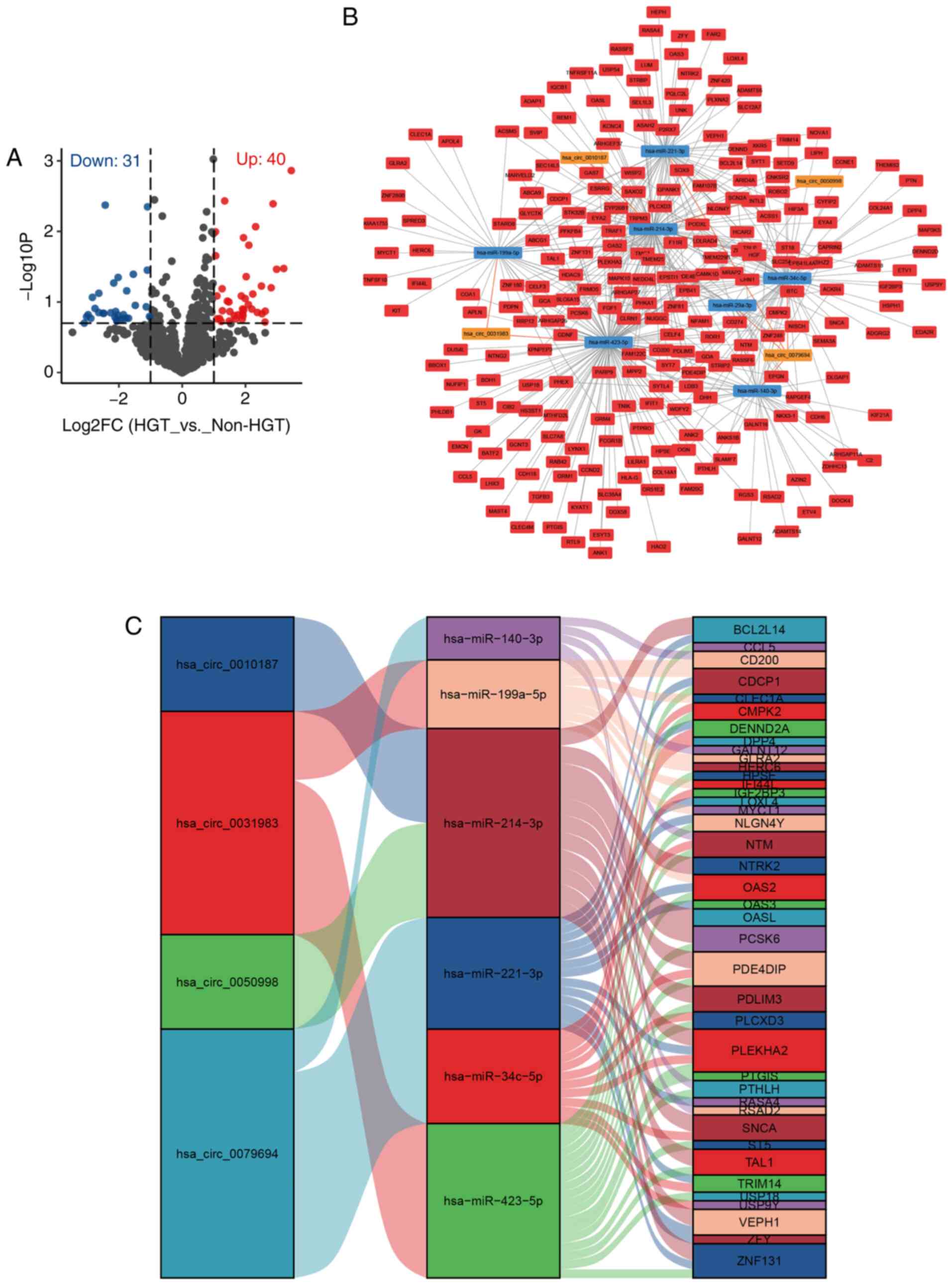
Candidate miRNAs of LACC-HGT
A total of four DEMs that could be candidate miRNAs
of LACC-HGT were selected from the overlap between the miRNA
microarray and GEO dataset (Fig.
1A). They were hsa-miR-140-3p, −140-5p, −574-3p and −636. Among
them, hsa-miR-140-3p, −140-5p and −574-3p were downregulated in the
LACC-HGT group and hsa-miR-636 was upregulated in LACC-HGT
group.
DECs and DEGs
The expression of circRNAs and genes in LACC and
LACC-HGT primary cell lines were significantly different. After
being compared with LACC primary cell lines, the results of circRNA
microarray showed that there were 258 upregulated circRNAs, 395
downregulated circRNAs (Fig. 1B;
Table SII, and 5,162 upregulated
and 4,404 downregulated genes (Fig.
1C) in LACC-HGT primary cell lines (Table SIII).
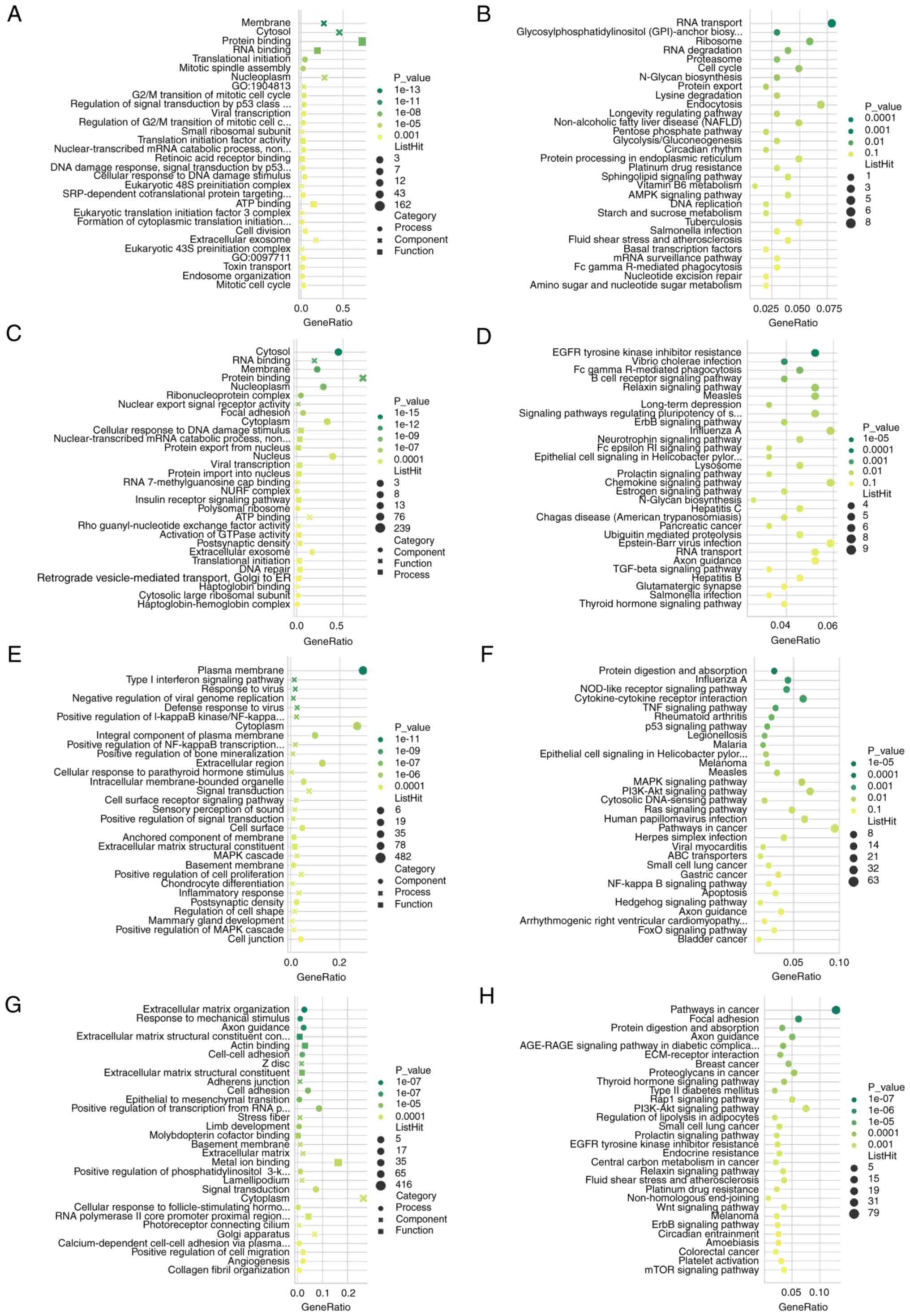
GO and pathway analysis of DECs and
DEGs
GO and KEGG pathway analyses were performed in DECs
and DEGs, respectively. The results indicated that the DECs are
mainly located in ‘cytosol’, ‘membrane’ and ‘nucleoplasm’, the
upregulated circRNAs are closely associated with ‘translational
initiation’, ‘RNA binding’, ‘regulation of G2/M
transition’ and ‘mitotic spindle assembly’ (Fig. 2A), and mainly involved in ‘RNA
transport’, ‘RNA degradation’ and ‘cell cycle’ processes (Fig. 2B). The downregulated circRNAs were
associated with ‘nuclear export signal receptor activity’, ‘focal
adhesion’ and ‘protein export from nucleus’ (Fig. 2C), and mainly involved in the
‘Relaxin signaling pathway’, ‘EGFR tyrosine kinase inhibitor
resistance’ and ‘B cell receptor signaling pathway’ (Fig. 2D). DEGs were mainly located in
‘plasma membrane’, ‘extracellular matrix’, ‘basement membrane and
cytoplasm’, the upregulated genes were associated with ‘type I
interferon signaling pathway’, ‘NF-κB transcription’,
‘extracellular matrix structural constituent’ and ‘cell
proliferation’ (Fig. 2E), and mainly
involved in ‘protein digestion and absorption’, ‘TNF signaling
pathway’, ‘p53 signaling pathway’ and ‘MAPK signaling pathway’
processes (Fig. 2F). The
downregulated genes were associated with ‘extracellular matrix
organization’, ‘cell-cell adhesion’ and ‘EMT’ (Fig. 2G), and mainly involved in ‘pathways
in cancer’, ‘focal adhesion’ and ‘Rap1 signaling pathway’ (Fig. 2H).
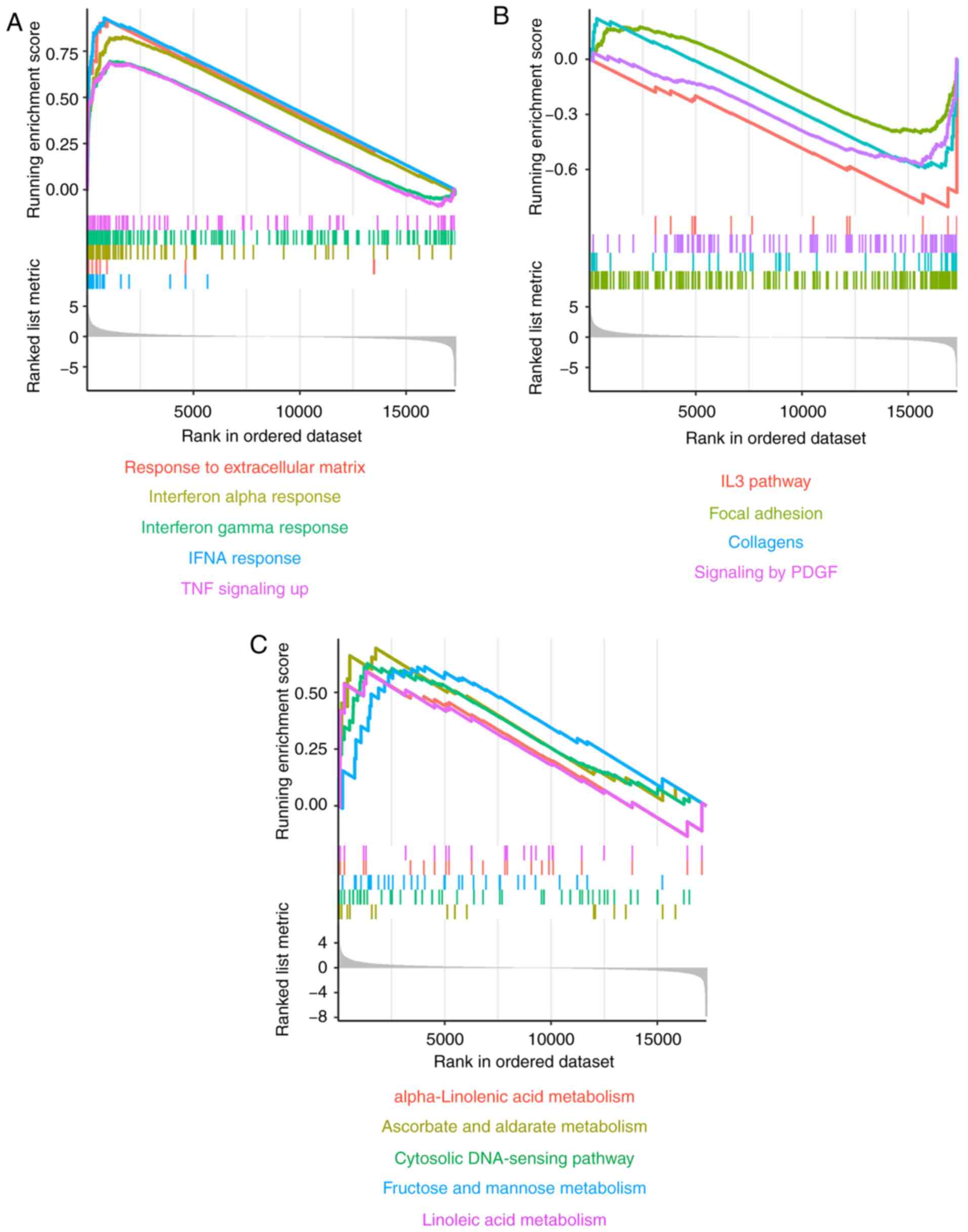
GSEA of DEGs
GSEA of DEGs revealed that the ‘interferon α
response pathway,’ ‘response to extracellular matrix’ and ‘TNF
signaling pathway’ were significantly upregulated in the LACC-HGT
group (Fig. 3A). However, the ‘IL-3
pathway’, ‘focal adhesion’, ‘collagens’ and ‘signaling by PDGF’
pathway were significantly downregulated (Fig. 3B). In addition, some metabolic
pathways, including ‘α-Linolenic acid metabolism’, ‘ascorbate and
alginate metabolism’, ‘fructose and mannose metabolism’ and
‘linoleic acid metabolism’ were upregulated, as was the ‘cytosolic
DNA-sensing pathway’ (Fig. 3C).
ceRNA regulatory network in
LACC-HGT
In order to construct ceRNA regulation networks, the
screening criteria of DEMs were adjusted to |log2FC|>1 and
P<0.2. Ultimately, 31 miRNAs were downregulated in LACC-HGT
group according to the microarray data (Fig. 4A). A total of eight circRNA-miRNA and
870 miRNA-mRNA interaction pairs were predicted from
CircInteractome, ENCORI and miRWalk databases. circRNA-miRNA-mRNA
ceRNA regulatory networks were constructed using the overlapping
results between the present 31 downregulated miRNAs and the
aforementioned circRNA-miRNA-mRNA interaction pairs, and 77
regulatory triplets (circRNA-miRNA-mRNA) were obtained (Fig. 4B). Finally, the ceRNA network that
involved four circRNAs (hsa_circ_0079694, _0031983, _0050998 and
_0010187), seven miRNAs (hsa-miR-140-3p, −199a-5p, −214-3p,
−221-3p, −29a-3p, −34c-5p and −423-5p) and 235 genes was visualized
using Cytoscape software. At the same time, in order to more
clearly demonstrate the circRNA-miRNA-mRNA interaction, ~100 genes
with the most significant upregulation were identified and a Sankey
map was drawn (Fig. 4C).
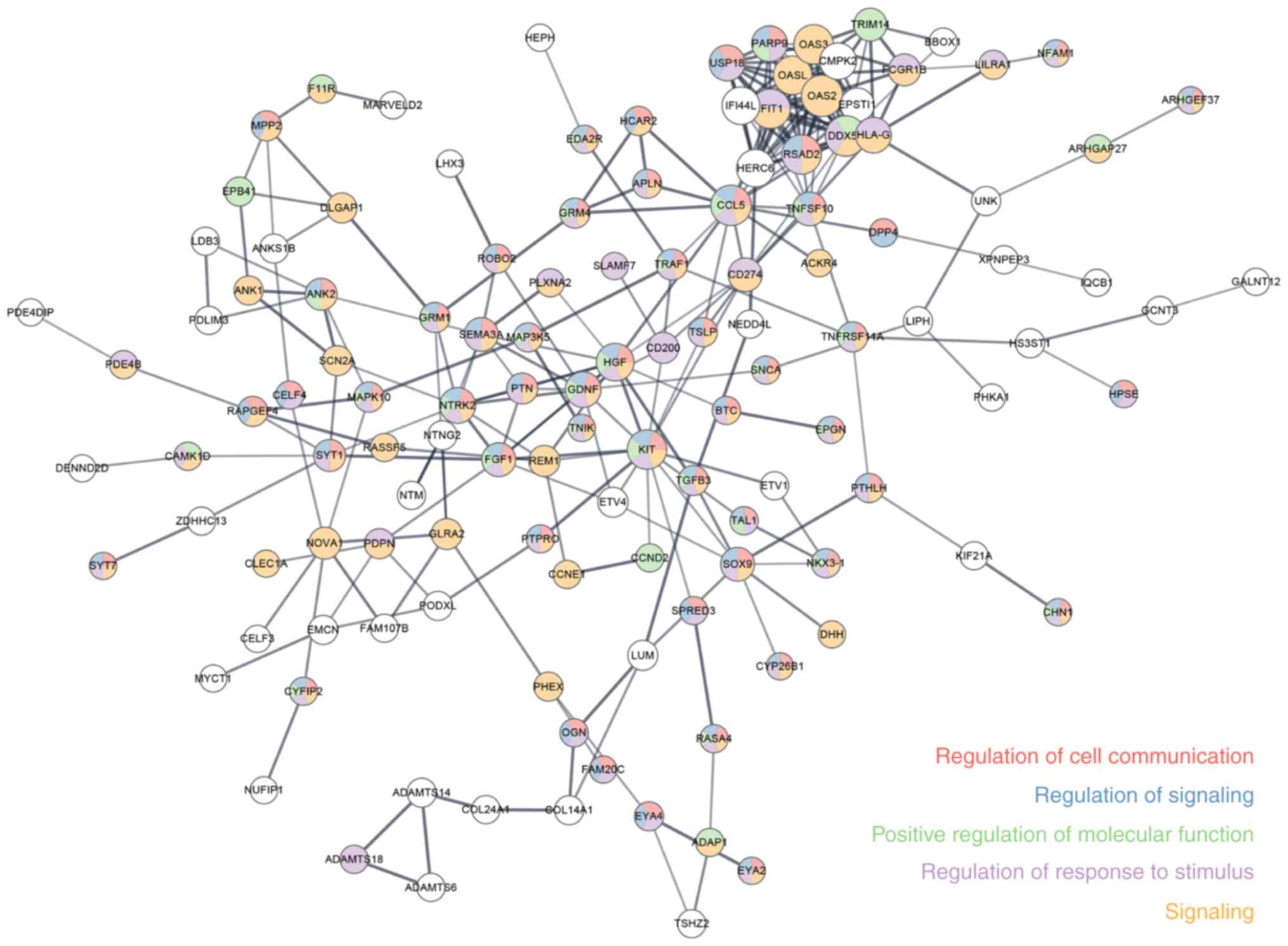
PPI network of proteins in ceRNA
regulatory network
PPI networks were established using the proteins
involved in the ceRNA regulatory network, 126 nodes and 261 edges
in total constituted the PPI network (Fig. 5). In a PPI network, each circle
represents a protein, and edges represent interactions. The GO
annotation of these proteins is marked in the figure to identify
proteins with similar effects. Different colors in the circle
represent different GO notes, and proteins of the same color have
similar functions. Proteins with a higher ‘connectivity degree’
(interaction with other proteins) may be more important in the
network. The interacting proteins of CD200 were identified such as
TNF receptor associated factor 1, receptor tyrosine kinase and
hepatocyte growth factor.
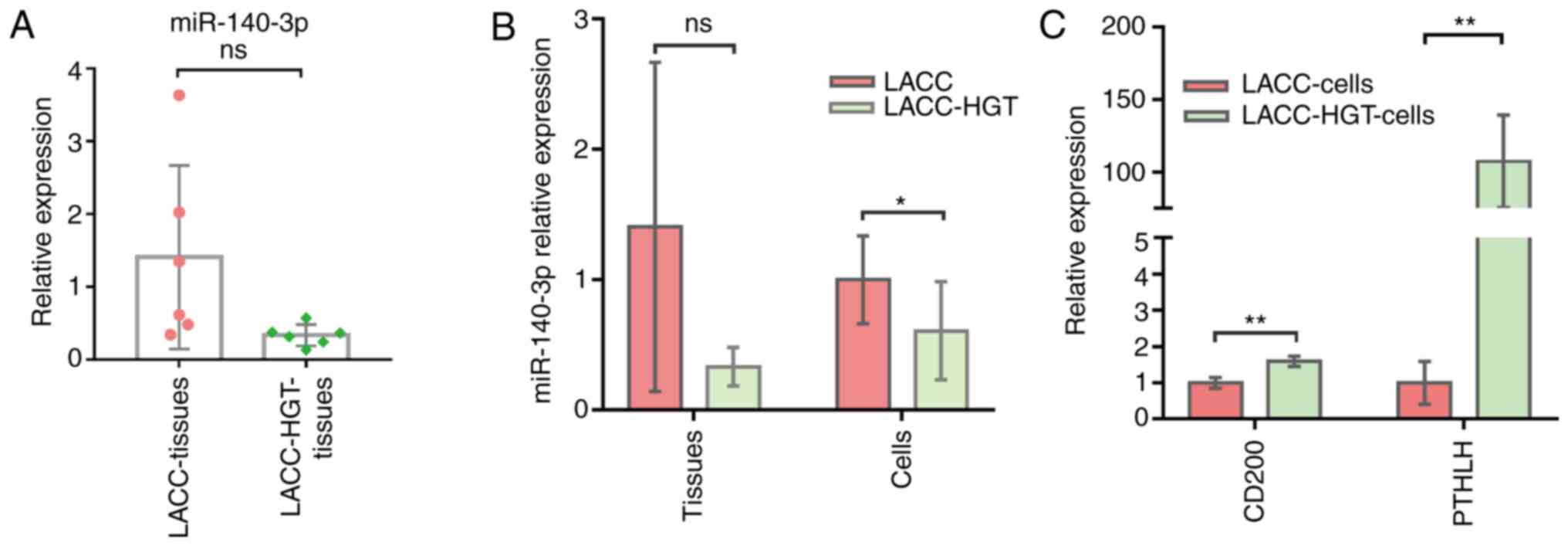
Expression of miR-140-3p and its
target genes
The data indicated that the expression of miR-140-3p
was not significantly downregulated in LACC-HGT tissues (P>0.05;
Fig. 6A), but was significantly
downregulated in LACC-HGT primary cells (P<0.05; Fig. 6B). miR-140-3p target genes CD200 and
parathyroid hormone-related protein (PTHLH) were significantly
upregulated (P<0.05; Fig. 6C).
GALNT12 has never been reported in lacrimal gland, and it was not
detected in LACC and LACC-HGT primary cells.
Discussion
As effective diagnostic and treatment strategies
have not been developed, the prognosis of patients with LACC
remains poor, mostly due to local or distant metastasis and
recurrence. Lacrimal gland carcinomas with HGT have been shown to
be more aggressive compared with conventional carcinomas with a
poorer prognosis, accompanied by higher local recurrence rate and
propensity for cervical lymph node metastasis (27). However, the molecular genetic
mechanisms responsible for the pathway of HGT in lacrimal gland
carcinomas largely remain to be elucidated.
ncRNAs have an essential role in the function of
oncogenes or tumor suppressor genes in cell signaling pathways.
Previous studies have shown that ncRNAs can affect gene expression
through RNA-RNA crosstalk-based ceRNA regulatory networks by
competing for the miRNA response elements of miRNAs through an
indirect post-transcriptional mechanism (28–30).
miRNAs and circRNAs not only promote tumor progression, but it can
also create the conditions that are not conducive to tumor
development, thereby inhibiting tumor progression. This competitive
mechanism theory is controversial, Han et al (31) reported that circMTO1 had no effect on
the expression level of miR-9; however, an increasing number of
studies have indicated that dysregulation of miRNAs is marker of
cancer. Previous studies have revealed that miRNAs are commonly
dysregulated in LACC, suggesting that these RNAs may play critical
roles in tumorigenesis and the progression of LACC-HGT (14,15).
circRNAs can act as miRNA sponges to bind miRNAs and indirectly
regulate mRNA expression (32).
However, little is known about the role of circRNAs in LACC, and
the present study is the first to report a ceRNA network of
LACC-HGT, to the best of our knowledge. Therefore, comprehensively
identifying the ncRNA profile and constructing ceRNA regulation
networks based on the interactions among circRNAs, miRNAs and genes
might provide novel insight into the pathogenesis of LACC-HGT.
The present study first analyzed the clinical and
pathological data of 12 patients with LACC with or without HGT, and
the results indicated that the patients with HGT had a more
aggressive disease compared with conventional LACC, accompanied by
a poorer prognosis, higher local recurrence rate and propensity for
cervical lymph node metastasis. In order to further study the
molecular mechanism of LACC-HGT, DEMs in LACC and LACC-HGT
paraffin-embedded tissues were identified. Meanwhile, the DECs and
DEGs in LACC primary cell lines with and without HGT were also
detected using a microarray. To the best of our knowledge, the
current study is the first to comprehensively investigate the
transcriptome circRNAs, miRNAs and genes profiles in LACC-HGT. ACC
is an epithelial malignant tumor, which occurs mostly in the
salivary glands and relatively rare in the lacrimal gland (33). The concept of ‘HGT’ was added to the
salivary gland tumors of the World Health Organisation
‘Classification of Head and Neck Tumors’ in the 4th edition of 2017
(34). HGT can also occur in LACC
(35). Therefore, it was
hypothesized that miRNAs involved in SACC may also participate in
LACC-HGT. Using combined bioinformatics analyses of GEO database,
four candidate miRNAs associated with LACC-HGT were identified. In
total, 16 DEMs were detected. Previous studies on ACC mainly
focused on the salivary gland, and miRNAs differentially expressed
of SACC may also be involved in the malignant progression of LACC
(36). Therefore, the overlapping
results from the microarray and GEO database were analyzed, and
four overlapping candidate miRNAs were identified; hsa-miR-140-3p,
−140-5p, −574-3p and −636 Among them, hsa-miR-140-5p mediates
inhibition of Smad2 and autophagy, inhibiting the survival and
invasion potential of colorectal cancer stem cells (37). The 5′ isomer miR with high expression
of hsa-miR-140-3p contributes to the tumor-suppressive effects of
miR-140 by reducing the proliferation and migration of breast
cancer cells (38). However, there
is no conclusive evidence that hsa-miR-574-3p and hsa-miR-636 are
associated with tumors. The present study detected DEMs through
miRNA microarrays, but further studies are needed to demonstrate
the accuracy of the results and their biological role. For example,
the expression of hsa-miR-140-3p and −140-5p in different tumors is
not the same and the current data indicated that they were
downregulated in LACC-HGT. Although candidate miRNAs were
identified through the GEO database, this does not mean that other
miRNAs do not play an important role in the occurrence and
development of LACC-HGT.
Ultimately, differential expression of 653 circRNAs
and 9,566 genes were detected. GO and KEGG pathway analyses of the
DECs showed that they are mainly located in ‘cytosol’, ‘membrane’
and ‘nucleoplasm’, closely associated with ‘translational
initiation’, ‘nuclear export signal receptor activity’, ‘regulation
of G2/M transition’ and ‘focal adhesion’ and mainly
involved in ‘RNA transport’, ‘RNA degradation’ and ‘cell cycle’
processes. DEGs were mainly located in ‘plasma membrane’,
‘extracellular matrix’, ‘basement membrane and cytoplasm’, closely
associated with ‘interferon signaling pathway’, ‘NF-κB signaling
pathway’, ‘extracellular matrix’ and ‘cell proliferation’ and
mainly involved in ‘protein digestion’ and ‘absorption’. DEG
processes included ‘cancer’, ‘p53 signaling pathway’ and ‘MAPK
signaling pathway’. GSEA based on the DEGs indicated that several
pathways may play important roles in LACC-HGT pathogenesis, mainly
including the ‘interferon α response pathway’, ‘IL-3 pathway’ and
some metabolic pathways. These signaling pathways are involved in
the development of tumors, such as NF-κB, p53 and MAPK signaling
pathways, which are canonical tumor-related signaling pathways
(39–42). Interferon is a protein involved in a
variety of functions, including antiviral and antimicrobial
responses, apoptosis, control of the cell cycle, and the interferon
signaling system plays an essential role in carcinogenic processes
such a triple-negative breast cancer (43) and Kaposi's sarcoma (44).
Next, a circRNA-miRNA-mRNA ceRNA network was
constructed using DECs, DEMs and DEGs. Ultimately, 77 ceRNA
triplets were matched with the DEMs of LACC-HGT. In total, four
circRNAs (hsa_circ_0079694, _0031983, _0050998 and _0010187), seven
miRNAs (hsa-miR-140-3p, −199a-5p, −214-3p, −221-3p, −29a-3p,
−34c-5p and −423-5p) and 235 genes were involved in the ceRNA
regulatory network. The function of these circRNAs is unclear.
However, these miRNAs play an important role in tumorigenesis,
among which miR-199a-5p is involved in the occurrence and
development of a variety of tumors, such as colon and rectal cancer
(45), uveal melanoma (46), oral cancer (47), glioma (48) and lung cancer (49). lncRNA XIST has anticancer effects on
epithelial ovarian cancer cells through inverse downregulation of
hsa-miR-214-3p (50). lncRNA
LINC00968 inhibits cell proliferation and migration and
angiogenesis in breast cancer through upregulation of prospero
homeobox protein 1 by reducing hsa-miR-423-5p expression (51). The proteins in the ceRNA regulatory
network were used to construct PPI regulation network, including
126 nodes and 261 edges. Candidate miRNAs (including miR-140-3p)
also participated in the ceRNA regulatory network, so RT-qPCR was
performed on miR-140-3p and its target genes in LACC-HGT tissues
and primary cells. The results showed that the expression of
miR-140-3p in LACC-HGT tissues was downregulated and its target
genes were upregulated. However, further exploration is needed to
verify the specific underlying mechanisms.
In summary, the present analyses revealed that ceRNA
regulation based on circRNA-miRNA-mRNA is a complex and
sophisticated mechanism, and may have an essential regulatory role
in the development of LACC-HGT. A ceRNA regulatory network in
LACC-HGT was reported, with the comprehensive elucidation of the
roles of ncRNAs in the pathogenesis of LACC-HGT. A limitation of
this study is that even though DECs, DEMs and DEGs were detected by
microarray, further investigation is still needed to validate the
results and elucidate their roles and mechanism in LACC-HGT.
Additional evidence should also be obtained to fully elucidate the
identified ceRNA regulatory networks in LACC. Future biological
studies may provide more detailed information to resolve the
complex regulation of ceRNAs in LACC-HGT. Transwell, wound-healing
assay and colony-formation assays may help identify the functions
of DECs, DEMs and DEGs. Biological markers of LACC-HGT could be
screened from a larger sample size. Fluorescence in situ
hybridization would aid with quantitative analysis.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from The
National Natural Science Foundation of China (grant no. 81570872)
and The Tianjin Key Clinical Discipline Construction Project (grant
nos. TJLCZDXKT006 and TJLCZDXKQ024), we acknowledge the support
received from Tianjin Medical University Eye Institute.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request. The datasets generated and/or analyzed during the present
study are available in the GEO repository, [http://www.ncbi.nlm.nih.gov/geo].
Authors' contributions
MJ, XL, CZ, LZ, HDW, LD, TW, TL, YH performed the
experiments. MJ, XL and TL confirmed the authenticity of all raw
data. MJ, XL and CZ conceived the project, drafted the initial
manuscript and revised it for important intellectual content. TL
and LZ collected the clinical data. HDW provided technical
guidance. YH made the final diagnosis and performed surgeries for
these patients. YH, LD and TL provided a critical review of the
content. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The Institutional of Human Research Ethics Committee
of the Tianjin Medical University Eye Hospital approved the study
(Tianjin, China; approval no. 2017KY(L)-23), and all procedures
were performed in accordance with the Declaration of Helsinki. All
patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ford JR, Rubin ML, Frank SJ, Ning J,
Debnam JM, Bell D, El-Naggar A, Ferrarotto R and Esmaeli B:
Prognostic factors for local recurrence and survival and impact of
local treatments on survival in lacrimal gland carcinoma. Br J
Ophthalmol. Jul 17–2020.(Epub ahead of print). doi:
10.1136/bjophthalmol-2020-316142. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sharma K, Rathi A, Khurana N, Mukherji A,
Kumar V, Singh K and Bahadur AK: A retrospective study of 18 cases
of adenoid cystic cancer at a tertiary care centre in Delhi. Indian
J Cancer. 47:424–429. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen TY, Keeney MG, Chintakuntlawar AV,
Knutson DL, Kloft-Nelson S, Greipp PT, Garrity JA, Salomao DR and
Garcia JJ: Adenoid cystic carcinoma of the lacrimal gland is
frequently characterized by MYB rearrangement. Eye (Lond).
31:720–725. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esmaeli B, Ahmadi M, Youssef A, Diba R,
Amato M, Myers JN, Kies M and El-Naggar A: Outcomes in patients
with adenoid cystic carcinoma of the lacrimal gland. Ophthalmic
Plast Reconstr Surg. 20:22–26. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wolkow N, Jakobiec F and Lee H: Long-term
outcomes of globe-preserving surgery with proton beam radiation for
adenoid cystic carcinoma of lacrimal gland. Am J Ophthalmol.
201:84–85. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han J, Kim Y, Woo K and Sobti D: Long-term
outcomes of eye-sparing surgery for adenoid cystic carcinoma of
lacrimal gland. Ophthalmic Plast Reconstr Surg. 34:74–78. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Snyder M and Paulino A: Hybrid carcinoma
of the salivary gland: Salivary duct adenocarcinoma adenoid cystic
carcinoma. Histopathology. 35:380–383. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagao T: ‘Dedifferentiation’ and
high-grade transformation in salivary gland carcinomas. Head Neck
Pathol. 7 (Suppl 1):S37–S47. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kamio N, Tanaka Y, Mukai M, Ikeda E,
Kuramochi S, Fujii M and Hosoda Y: A hybrid carcinoma: Adenoid
cystic carcinoma and salivary duct carcinoma of the salivary gland.
Virchows Archiv. 430:495–500. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagao T, Gaffey TA, Serizawa H, Sugano I,
Ishida Y, Yamazaki K, Tokashiki R, Yoshida T, Minato H, Kay PA and
Lewis JE: Dedifferentiated adenoid cystic carcinoma: A
clinicopathologic study of 6 cases. Mod Pathol. 16:1265–1272. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
White VA: Update on lacrimal gland
neoplasms: Molecular pathology of interest. Saudi J Ophthalmol.
26:133–135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pusztaszeri M and Brochu V: Metastatic
adenoid cystic carcinoma with high-grade transformation
(‘dedifferentiation’) in pleural effusion and neck lymph node: A
diagnostic challenge on cytology? Diagn Cytopathol. 48:679–683.
2020. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Panni S, Lovering RC, Porras P and Orchard
S: Non-coding RNA regulatory networks. Biochim Biophys Acta Gene
Regul Mech. 1863:1944172020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang M, Zhang J, Zhang H and Tang H:
miR-24-3p suppresses malignant behavior of lacrimal adenoid cystic
carcinoma by targeting PRKCH to regulate p53/p21 pathway. PLoS One.
11:e01584332016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hao J, Jin X, Shi Y and Zhang H: miR-93-5p
enhance lacrimal gland adenoid cystic carcinoma cell tumorigenesis
by targeting BRMS1L. Cancer Cell Int. 18:722018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi P: A ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smillie CL, Sirey T and Ponting CP:
Complexities of post-transcriptional regulation and the modeling of
ceRNA crosstalk. Crit Rev Biochem Mol Biol. 53:231–245. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin T, Zhu L, Zhou B, Xie L, Lv J, Dong L
and He Y: Establishment and characterization of a cell line from
human adenoid cystic carcinoma of the lacrimal glands and a nude
mouse transplantable model. Oncol Rep. 33:2797–2806. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao R, Cao C, Zhang M, Lopez MC, Yan Y,
Chen Z, Mitani Y, Zhang L, Zajac-Kaye M, Liu B, et al: A unifying
gene signature for adenoid cystic cancer identifies parallel
MYB-dependent and MYB-independent therapeutic targets. Oncotarget.
5:12528–12542. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar G, Kumar R, Pal MK, Pramanik N,
Lahiri T, Gupta A and Pandey S: APT: An Automated Probe Tracker
from gene expression data. IEEE/ACM Trans Comput Biol Bioinform.
Dec 9–2019.(Epub ahead of print). doi: 10.1109/TCBB.2019.2958345.
View Article : Google Scholar
|
|
22
|
Kim B: How to reveal magnitude of gene
signals: Hierarchical hypergeometric complementary cumulative
distribution function. Evol Bioinform Online.
14:11769343187973522018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Savithri V, Suresh R, Janardhanan M,
Aravind T and Mohan M: Primary intraosseous adenoid cystic
carcinoma with widespread skeletal metastases showing features of
high-grade transformation. Head Neck Pathol. Sep 21–2020.(Epub
ahead of print). doi: 10.1007/s12105-020-01228-x. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Falzone L, Grimaldi M, Celentano E,
Augustin L and Libra M: Identification of modulated MicroRNAs
associated with breast cancer, diet, and physical activity.
Cancers. 12:25552020. View Article : Google Scholar
|
|
29
|
Filetti V, Falzone L, Rapisarda V,
Caltabiano R, Eleonora Graziano AC, Ledda C and Loreto C:
Modulation of microRNA expression levels after naturally occurring
asbestiform fibers exposure as a diagnostic biomarker of
mesothelial neoplastic transformation. Ecotoxicol Environ Saf.
198:1106402020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Y, Di G, Hu S, Zhao T, Xu X, Wang X
and Chen P: Expression profiles of CircRNA and mRNA in lacrimal
glands of AQP5 mice with primary dry eye. Front Physiol.
11:10102020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nakaguro M, Faquin WC, Baloch ZW, Cantley
RL, Compton ML, Ely KA, Holmes BJ, Hu R, Kerr DA, Montone KT, et
al: Fine needle aspiration of salivary gland carcinomas with
high-grade transformation: A multi-institutional study of 22 cases
and review of the literature. Cancer Cytopathol. Nov 19–2020.(Epub
ahead of print). doi: 10.1002/cncy.22388. View Article : Google Scholar
|
|
34
|
Seethala R and Stenman G: Update from the
4th Edition of the World Health Organization Classification of Head
and Neck Tumours: Tumors of the salivary gland. Head Neck Pathol.
11:55–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seethala R, Hunt J, Baloch Z, Livolsi V
and Leon Barnes E: Adenoid cystic carcinoma with high-grade
transformation: A report of 11 cases and a review of the
literature. Am J Surg Pathol. 31:1683–1694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kiss O, Tőkés A, Spisák S, Szilágyi A,
Lippai N, Székely B, Szász AM and Kulka J: Breast- and salivary
gland-derived adenoid cystic carcinomas: potential
post-transcriptional divergencies. A pilot study based on miRNA
expression profiling of four cases and review of the potential
relevance of the findings. Pathol Oncol Res. 21:29–44. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhai H, Fesler A, Ba Y, Wu S and Ju J:
Inhibition of colorectal cancer stem cell survival and invasive
potential by hsa-miR-140-5p mediated suppression of Smad2 and
autophagy. Oncotarget. 6:19735–19746. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Salem O, Erdem N, Jung J, Münstermann E,
Wörner A, Wilhelm H, Wiemann S and Körner C: The highly expressed
5′isomiR of hsa-miR-140-3p contributes to the tumor-suppressive
effects of miR-140 by reducing breast cancer proliferation and
migration. BMC Genomics. 17:5662016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Joerger A and Fersht A: The p53 pathway:
Origins, inactivation in cancer, and emerging therapeutic
approaches. Ann Rev Biochem. 85:375–404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sanchez-Vega F, Mina M, Armenia J, Chatila
WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia
S, et al: Oncogenic signaling pathways in the cancer genome atlas.
Cell. 173:321–337.e10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang C and Zhu G: Classic and novel
signaling pathways involved in cancer: Targeting the NF-κB and Syk
signaling pathways. Curr Stem Cell Res Ther. 14:219–225. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo Y, Pan W, Liu S, Shen Z, Xu Y and Hu
L: ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med.
19:1997–2007. 2020.PubMed/NCBI
|
|
43
|
Park IA, Hwang SH, Song IH, Heo SH, Kim
YA, Bang WS, Park HS, Lee M, Gong G and Lee HJ: Expression of the
MHC class II in triple-negative breast cancer is associated with
tumor-infiltrating lymphocytes and interferon signaling. PLoS One.
12:e01827862017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Joo CH, Shin YC, Gack M, Wu L, Levy D and
Jung JU: Inhibition of interferon regulatory factor 7
(IRF7)-mediated interferon signal transduction by the Kaposi's
sarcoma-associated herpesvirus viral IRF homolog vIRF3. J Virol.
81:8282–8292. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Slattery M, Lundgreen A, Bondurant K and
Wolff R: Interferon-signaling pathway: Associations with colon and
rectal cancer risk and subsequent survival. Carcinogenesis.
32:1660–1667. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Falzone L, Romano GL, Salemi R, Bucolo C,
Tomasello B, Lupo G, Anfuso CD, Spandidos DA, Libra M and Candido
S: Prognostic significance of deregulated microRNAs in uveal
melanomas. Mol Med Rep. 19:2599–2610. 2019.PubMed/NCBI
|
|
47
|
Wei D, Wang W, Shen B, Zhou Y, Yang X, Lu
G, Yang J and Shao Y: MicroRNA-199a-5p suppresses migration and
invasion in oral squamous cell carcinoma through inhibiting the
EMT-related transcription factor SOX4. Int J Mol Med. 44:185–195.
2019.PubMed/NCBI
|
|
48
|
Zhang C, Chen Q, Zhu J and Liu Z:
MicroRNA-199a-5p regulates glioma progression via targeting MARCH8.
Eur Rev Med Pharmacol Sci. 23:7482–7487. 2019.PubMed/NCBI
|
|
49
|
Ahmadi A, Khansarinejad B, Hosseinkhani S,
Ghanei M and Mowla S: miR-199a-5p and miR-495 target GRP78 within
UPR pathway of lung cancer. Gene. 620:15–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang C, Qi S, Xie C, Li C, Wang P and Liu
D: Upregulation of long non-coding RNA XIST has anticancer effects
on epithelial ovarian cancer cells through inverse downregulation
of hsa-miR-214-3p. J Gynecol Oncol. 29:e992018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun X, Huang T, Zhang C, Zhang S, Wang Y,
Zhang Q and Liu Z: Long non-coding RNA LINC00968 reduces cell
proliferation and migration and angiogenesis in breast cancer
through up-regulation of PROX1 by reducing hsa-miR-423-5p. Cell
Cycle. 18:1908–1924. 2019. View Article : Google Scholar : PubMed/NCBI
|