Introduction
Hepatocellular carcinoma (HCC) is the third most
common cause of cancer-associated mortality worldwide (1). Despite advances in diagnosis and
treatment, the prognosis of patients with HCC remains
unsatisfactory (2). Thus, the
pathogenesis of HCC must be further elucidated in order to identify
efficient targets for HCC therapy.
Long non-coding RNAs (lncRNAs) are a class of RNA
transcripts >200 nucleotides (nt) in length that have no
protein-coding ability (3,4). Emerging evidence has shown that lncRNAs
are involved in the tumorigenesis of multiple different cancer
types, including HCC (5). For
example, lncRNA DLEU2 promoted HCC cell proliferation and
metastasis by interacting with enhancer of zeste 2 polycomb
repressive complex 2 subunit (6).
lncRNA MEG3 suppressed the proliferation of HCC cells by regulating
the miR-9-5/SOX11 axis (7), and
lncRNA SNHG7 facilitated HCC progression by sponging miR-122-5p and
upregulating ribosomal protein L4 (8). Additionally, abnormal expression of
lncRNAs is reportedly involved in the resistance of various tumors
to chemotherapy, such as breast and gastric cancer (9,10).
Numerous studies have indicated that lncRNA LINC00173 functions as
an oncogene in the progression of human cancers. For example,
LINC00173 was found to regulate cervical cancer cell viability and
invasiveness via the miR-182-5p/F-box and the WD repeat
domain-containing 7 axes (11).
Furthermore, lncRNA LINC00173 accelerated breast cancer progression
by downregulating miR-490-3p (12),
and accelerated the development of melanoma by upregulating insulin
receptor substrate 4 via miRNA-493 sponging (13). However, the function of LINC00173 in
HCC chemoresistance remains unknown.
MicroRNAs (miRNAs/miRs) are non-coding RNAs that are
~22 nt in length and play key regulatory roles in the development
and chemoresistance of human cancers (14). For example, Niu et al
(15) revealed that miRNA-654-3p
enhanced cisplatin (DDP) sensitivity by downregulating quinolinate
phosphoribosyl transferase and suppressing the PI3K/AKT pathway in
ovarian cancer. Yang et al (16) suggested that miRNA-181a-5p promoted
the sensitivity of esophageal adenocarcinoma cells to DDP by
targeting Cbl proto-oncogene B. Furthermore, silencing of circRNA
CDR1as inhibited the stemness of DDP-resistant non-small cell lung
cancer cells by repressing homeobox A9 as a result of miR-641
upregulation (17), suggesting that
miR-641 may play an important role in the chemoresistance of cancer
cells. However, the mechanisms of miR-641 in the DDP-associated
resistance of HCC remain unclear.
The aim of the present study was to determine
whether LINC00173 is upregulated in DDP-resistant HCC tissues and
cell lines, and to investigate whether LINC00173 induced the DDP
resistance of HCC cells by sponging miR-641 and upregulating RAB14,
in order to determine whether the LINC00173/miR-641/RAB14 axis may
serve as a novel target for the treatment of HCC.
Materials and methods
Tissue samples
A total of 30 pairs of HCC and adjacent-normal
tissues were acquired from patients at the Weifang People's
Hospital (Weifang, China) between February 2018 and November 2019,
of which the clinical characteristics are presented in Table I. The inclusion criteria for the
patients were: i) Diagnosed with HCC; and ii) not received
preoperative radiotherapy, chemotherapy or other adjuvant
treatments. The exclusion criteria for the patients were: i)
Diagnosed with any other diseases apart from HCC; and ii) failure
to cooperate with researchers. The tumor tissues were then divided
into DDP-sensitive (n=11) and DDP-resistant (n=19) tissues
according to the Response Evaluation Criteria in Solid Tumors
(18). All fresh tissues were
preserved in liquid nitrogen until use. Written informed consent
was obtained from all participants and the study protocol was
approved by the Ethics Committee of Weifang People's Hospital.
 | Table I.Clinicopathological features of
patients with hepatocellular carcinoma. |
Table I.
Clinicopathological features of
patients with hepatocellular carcinoma.
| Variable | Cases, n |
|---|
| Age, years |
|
|
<60 | 19 |
| ≥60 | 11 |
| Sex |
|
|
Male | 17 |
|
Female | 13 |
| Liver
cirrhosis |
|
| No | 22 |
|
Yes | 8 |
| Pathological
stage |
|
|
I–II | 16 |
|
III–IV | 14 |
| Tumor size, cm |
|
|
<5 | 17 |
| ≥5 | 13 |
| TNM stage |
|
| T1+
T2 | 18 |
| T3+
T4 | 12 |
Cell culture
Human HCC cells (Huh7 and Hep3B) were purchased from
the American Type Culture Collection and were incubated with
gradually increasing concentrations of DDP for 12 months to obtain
DDP-resistant Huh7 (Huh7/DDP) and DDP-resistant Hep3B (Hep3B/DDP)
cell lines. The cells were subsequently cultured in RPMI-1640
medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C (5%
CO2).
Transfection
The small short hairpin (sh) RNA targeting LINC00173
(10 nM; shLINC00173; 5′-UCCGGGUAGCUAUAUCAUGCG-3′) and its negative
control (10 nM; shNC; 5′-GAUUAGUUCACAUGCGAUCCU-3′), the miR-641
mimics (10 nM; 5′-GACGAUCCUCUGCAUCGAGUA-3′) and scrambled negative
control (10 nM; NC mimics; 5′-UAGUUGACGCUUGACGCUAGU-3′), and the
miR-641 inhibitor (10 nM; 5′-GAUCGGACAUGGUGCUAGCUU-3′) and
scrambled negative control (NC inhibitor;
5′-AGUCGACGAUAUGCCAGGUCG-3′) were all obtained from Shanghai
GenePharma Co., Ltd. For LINC00173 and RAB14 overexpression,
LINC00173 or RAB14 cDNA were cloned into the pcDNA3.1 vector
(Shanghai GenePharma Co., Ltd.). Transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
transfected cells were used for subsequent experimentation 48 h
post-transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from HCC and adjacent-normal
tissues and Huh7 and Hep3B cells using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. A total of 1 µg RNA was reverse
transcribed into cDNA using the PrimeScript RT reagent kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. qPCR was performed using the ABI 7500
real-time PCR System (Thermo Fisher Scientific, Inc.) with the
SYBR-Green PCR Master Mix kit (Takara Biotechnology Co., Ltd.). The
PCR primers used were as follows: LINC00173 forward,
5′-GCCAGCTCTCGGTACCTGGA-3′ and reverse,
5′-GGATCGCAACATTCCTGCCAAG-3′; miR-641 forward,
5′-TTATACTCTCACCATTTGGATC-3′ and reverse,
5′-TGACAAGATTTTACATCAAGAA-3′; RAB14 forward,
5′-CGCTCGAGATGGCAACTGCACCATACAAC-3′ and reverse,
5′-CGGAATTCCTAGCAGCCACAGCCTTCTC-3′; GAPDH forward,
5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-ACCACCCTGTTGCTGTA-3′;
and U6 forward 5′-CTTCGGCAGCACATATACT-3′ and U6 reverse,
5′-AAAATATGGAACGCTTCACG-3′. The qPCR thermocycling conditions were
as follows: An initial predenaturation step at 94°C for 5 min,
followed by 40 cycles of denaturation at 95°C for 30 sec, annealing
at 60°C for 30 sec and extension at 72°C for 20 sec. The relative
expression of genes was calculated using the 2−ΔΔCq
method (19). GAPDH and U6 were used
as the internal controls for LINC00173 and RAB14, or miR-641,
respectively.
Drug sensitivity assay
DDP sensitivity was determined using the Cell
Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies Inc.)
based on the manufacturer's instructions to obtain the half maximal
inhibitory concentration (IC50) value. Briefly, Huh7/DDP
and Hep3B/DDP cells were seeded into 96-well plates at a density of
1×104 cells per well, and then treated with DDP for 48 h
at 37°C. Subsequently, 10 µl CCK-8 solution was added to each well
and incubated for a further 1 h. The absorbance was recorded using
a microplate reader at 450 nm.
Apoptosis analysis
Apoptosis was assessed using the One-Step TUNEL
Apoptosis Assay kit (Beyotime Institute of Biotechnology). Briefly,
Huh7/DDP and Hep3B/DDP cells (1×104) were seeded onto
coverslips and then fixed with 4% paraformaldehyde at 4°C for 30
min. The cells were then incubated with TUNEL enzyme for 60 min at
room temperature. Finally, the cells were counterstained with DAPI
for 10 min at room temperature to visualize the cell nuclei. Images
were captured from four fields of view using a fluorescence
microscope (magnification, ×200).
Bioinformatics analysis
The StarBase database (http://starbase.sysu.edu.cn/) was used to predict
interactions between miR-641 and LINC00173 or RAB14 by screening
for potential binding sites.
Luciferase reporter assay
The wild-type (wt) or mutant (mut) sequences of
LINC00173 and RAB14 were cloned into pmirGLO vectors (Promega
Corporation). Then, DDP-resistant HCC cells were co-transfected
with miR-641 mimics or NC mimics and the aforementioned reporter
vectors using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, the luciferase activity was measured
using the dual-luciferase reporter system (Promega Corporation).
Firefly luciferase activity was normalized to Renilla
luciferase gene activity.
Statistical analysis
The results are presented as the mean ± standard
deviation and all experiments were repeated in triplicate.
Statistical analysis was performed using SPSS software (version
19.0; IBM Corp.). Comparisons of parameters between HCC tissues and
adjacent normal tissues, or experimental and control groups, were
performed using the paired or unpaired Student's t-test,
respectively. Comparisons among multiple groups were performed by
one-way analysis of variance followed by Tukey's test. Pearson's
correlation analysis was used to determine correlations between
gene expression levels, and Kaplan-Meier analysis was used to
estimate overall survival. P<0.05 was considered to indicate a
statistically significant difference.
Results
LINC00173 expression is increased in
DDP-resistant HCC tissues and cell lines
RT-qPCR analysis demonstrated that LINC00173
expression was elevated in HCC tissues compared with adjacent
normal tissues (Fig. 1A). In
addition, LINC00173 expression in DDP-resistant HCC tissues was
markedly higher compared with that in DDP-sensitive HCC tissues
(Fig. 1B). The CCK-8 assay indicated
that the IC50 of DDP was markedly increased in Huh7/DDP
and Hep3B/DDP cells, suggesting that DDP-resistant HCC cell lines
were successfully established (Fig. 1C
and D). Furthermore, LINC00173 expression was markedly
upregulated in the DDP-resistant Huh7 and Hep3B cell lines
(Huh7/DDP and Hep3B/DDP) compared with that in parental Huh7 and
Hep3B cells (Fig. 1E and F).
Additionally, patients with HCC and high LINC00173 expression
levels exhibited shorter overall survival times (Fig. 1G). These data indicated that
LINC00173 upregulation may be associated with DDP resistance in
HCC.
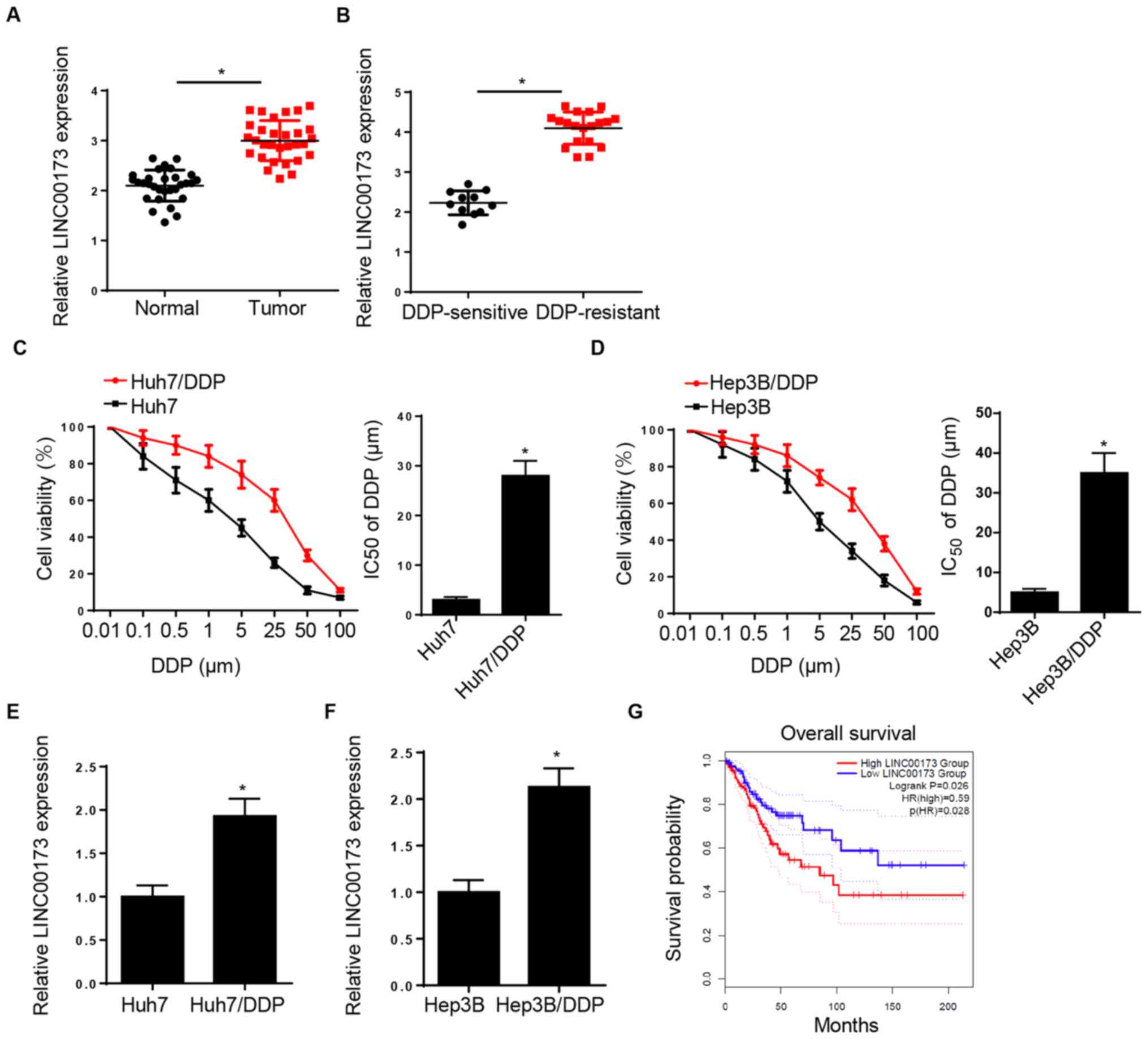 | Figure 1.LINC00173 is increased in
DDP-resistant HCC tissues and cells. (A) Relative LINC00173
expression levels in HCC tissues and adjacent-normal tissues were
determined by RT-qPCR; n=30. (B) RT-qPCR analysis of relative
LINC00173 expression of in DDP-resistant (Huh7/DDP and Hep3B/DDP)
and DDP-sensitive HCC tissues. Cell Counting Kit-8 analysis was
used to determine the IC50 value of DDP-resistant (C)
Huh7/DDP and (D) Hep3B/DDP cells and their parental cell lines
(Huh7 and Hep3B) treated with different concentrations of DDP
(0.01, 0.1, 0.5, 1, 5, 25, 50 and 100 µM). RT-qPCR analysis of
LINC0017 3expression in DDP-resistant (E) Huh7 and (F) Hep3B cells.
(G) Kaplan-Meier survival analysis of the association between
LINC00173 expression and the prognosis of patients with HCC.
*P<0.05. DDP, cisplatin; HCC, hepatocellular carcinoma; RT-qPCR,
reverse transcription-quantitative PCR; IC50, half
maximal inhibitory concentration; HR, hazard ratio. |
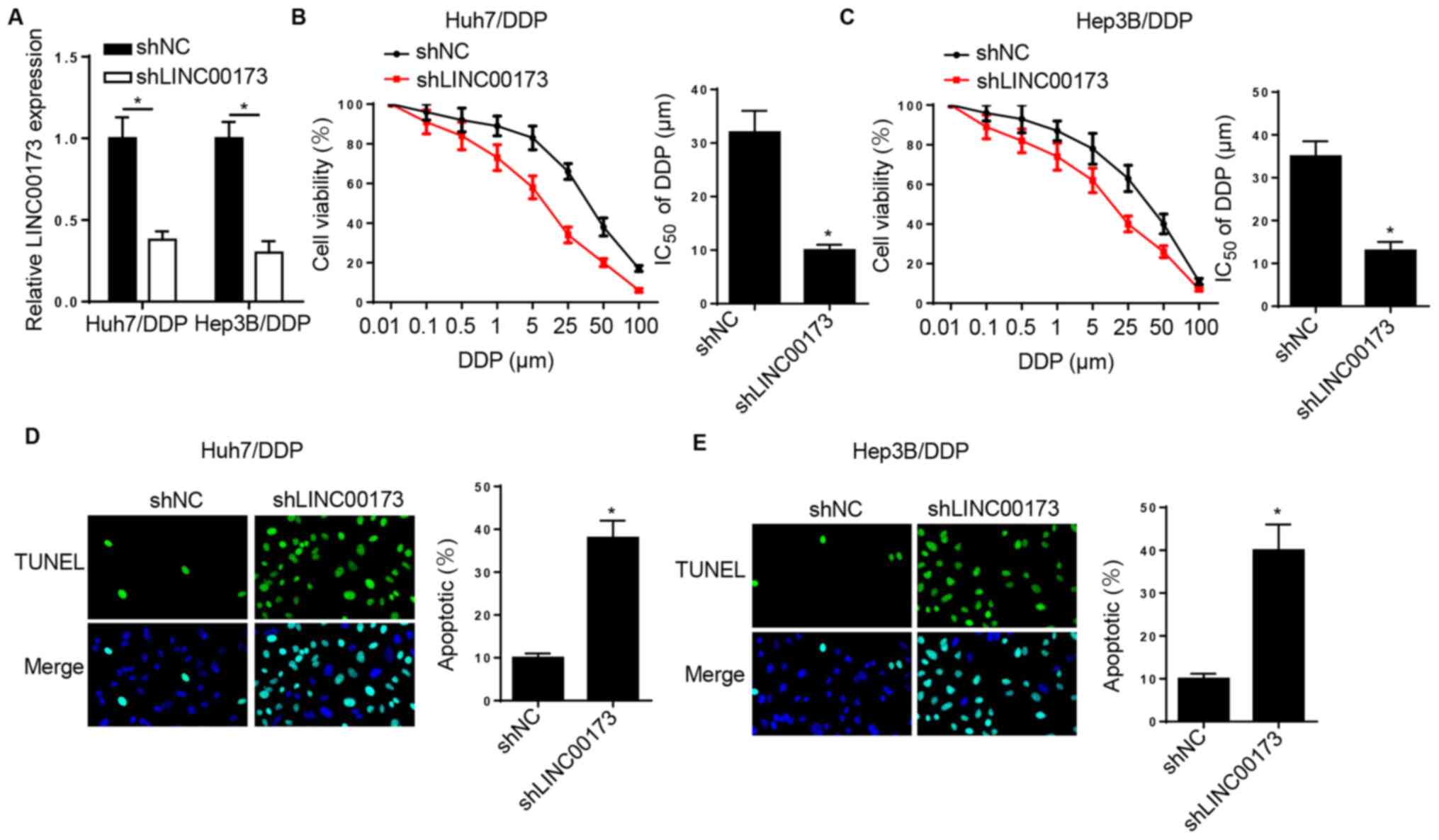
LINC00173 knockdown increases the DDP
sensitivity of DDP-resistant HCC cells
To determine the effects of LINC00173 on DDP
resistance in HCC, Huh7/DDP and Hep3B/DDP cells were transfected
with shNC and shLINC00173. RT-qPCR analysis indicated that
LINC00173 silencing downregulated LINC00173 expression in
DDP-resistant HCC cells (Fig. 2A).
Furthermore, the DDP sensitivity of DDP-resistant HCC cells was
enhanced by LINC00173 knockdown (Fig. 2B
and C). Additionally, TUNEL analysis revealed that silencing of
LINC00173 markedly increased the apoptotic rates of Huh7/DDP and
Hep3B/DDP cells (Fig. 2D and E).
Taken together, the aforementioned data indicated that LINC00173
knockdown increased the sensitivity of DDP-resistant HCC cells to
DDP.
miR-641 is a target of LINC00173 in
HCC cells
The StarBase website predicted that LINC00173
possessed a binding site for miR-641 (Fig. 3A). Then, RT-qPCR analysis revealed
that miR-641 expression was increased in Huh7/DDP and Hep3B/DDP
cells transfected with miR-641 mimics (Fig. 3B). A luciferase reporter assay was
performed to confirm the interaction between LINC00173 and miR-641,
and the results indicated that overexpression of miR-641 decreased
the luciferase activity of LINC00173-wt in Huh7/DDP and Hep3B/DDP
cells, but had no effect on the LINC00173-mut (Fig. 3C and D). Furthermore, the results of
the RT-qPCR analysis revealed that LINC00173 depletion increased
the expression of miR-641, while LINC00173 overexpression decreased
miR-641 expression in DDP-resistant HCC cells (Fig. 3E). In addition, the expression of
miR-641 was found to be low in HCC tissues compared with
adjacent-normal tissues, particularly in DDP-resistant HCC tissues
(Fig. 3F and G). Furthermore,
Pearson's correlation analysis revealed that LINC00173 expression
was inversely correlated with miR-641 expression in HCC tissues
(Fig. 3H). These results confirmed
that LINC00173 inhibited the expression of miR-641 by direct
interaction in DDP-resistant HCC cells.
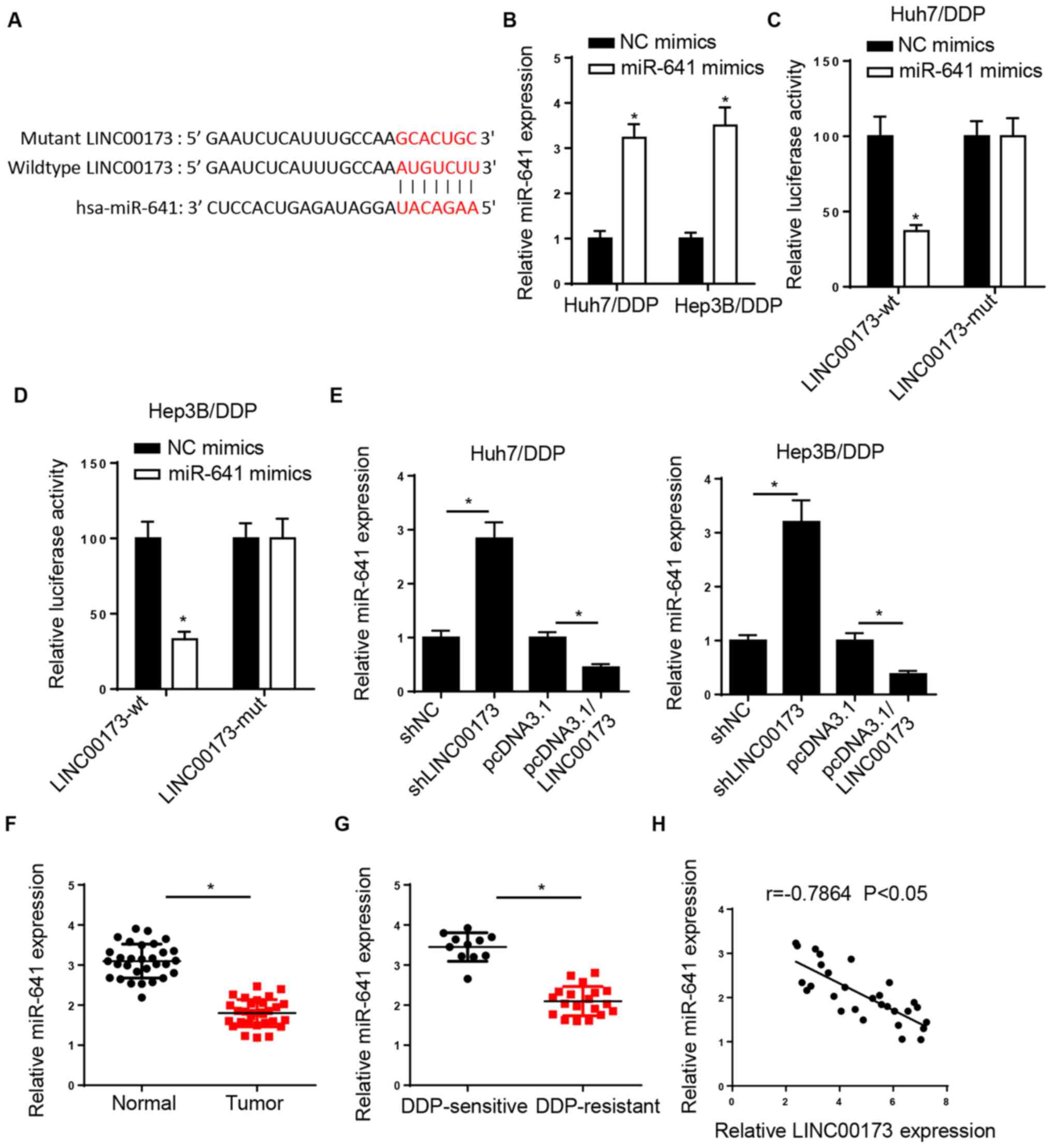 | Figure 3.miR-641 is a target of LINC00173 in
HCC cells. (A) StarBase was used to predict potential binding sites
between LINC00173 and miR-641. (B) RT-qPCR analysis of relative
miR-641 expression in Huh7/DDP and Hep3B/DDP cells transfected with
NC or miR-641 mimics. Luciferase activity of LINC00173-wt or
LINC00173-mut in (C) Huh7/DDP and (D) Hep3B/DDP cells transfected
with NC or miR-641 mimics. (E) RT-qPCR analysis of relative miR-641
expression in Huh7/DDP and Hep3B/DDP cells transfected with
shLINC00173, shNC, pcDNA3.1/LINC00173 and pcDNA3.1. Relative
miR-641 expression in (F) HCC tissues and (G) DDP-resistant HCC
tissues was determined by RT-qPCR. (H) Pearson's correlation
analysis revealed a negative correlation between miR-641 and
LINC00173 expression in HCC tissues. *P<0.05. miR, microRNA;
HCC, hepatocellular carcinoma; RT-qPCR, reverse
transcription-quantitative PCR; DDP, cisplatin; wt, wild-type; mut,
mutant; NC, negative control; sh, short hairpin. |
LINC00173-knockdown increases DDP
sensitivity in DDP-resistant HCC cells by targeting miR-641
Rescue experiments were performed to investigate the
role of miR-641 in LINC00173-mediated DDP resistance in HCC.
RT-qPCR analysis indicated that miR-641 expression was reduced in
Huh7/DDP and Hep3B/DDP cells transfected with the miR-641 inhibitor
(Fig. 4A). Furthermore, the level of
miR-641 was markedly elevated in Huh7/DDP and Hep3B/DDP cells
transfected with shLINC00173, which was reversed by the miR-641
inhibitor (Fig. 4B). Furthermore,
the CCK-8 assay revealed that LINC00173 interference decreased the
IC50 of DDP in Huh7/DDP and Hep3B/DDP cells, which was
reversed by miR-641 inhibition (Fig. 4C
and D). TUNEL analysis indicated that LINC00173-knockdown
promoted the apoptosis of Huh7/DDP and Hep3B/DDP cells, while
miR-641-knockdown counteracted this effect (Fig. 4E and F). In summary, these findings
indicated that LINC00173-knockdown increased the sensitivity of
DDP-resistant HCC cells to DDP by regulating miR-641
expression.
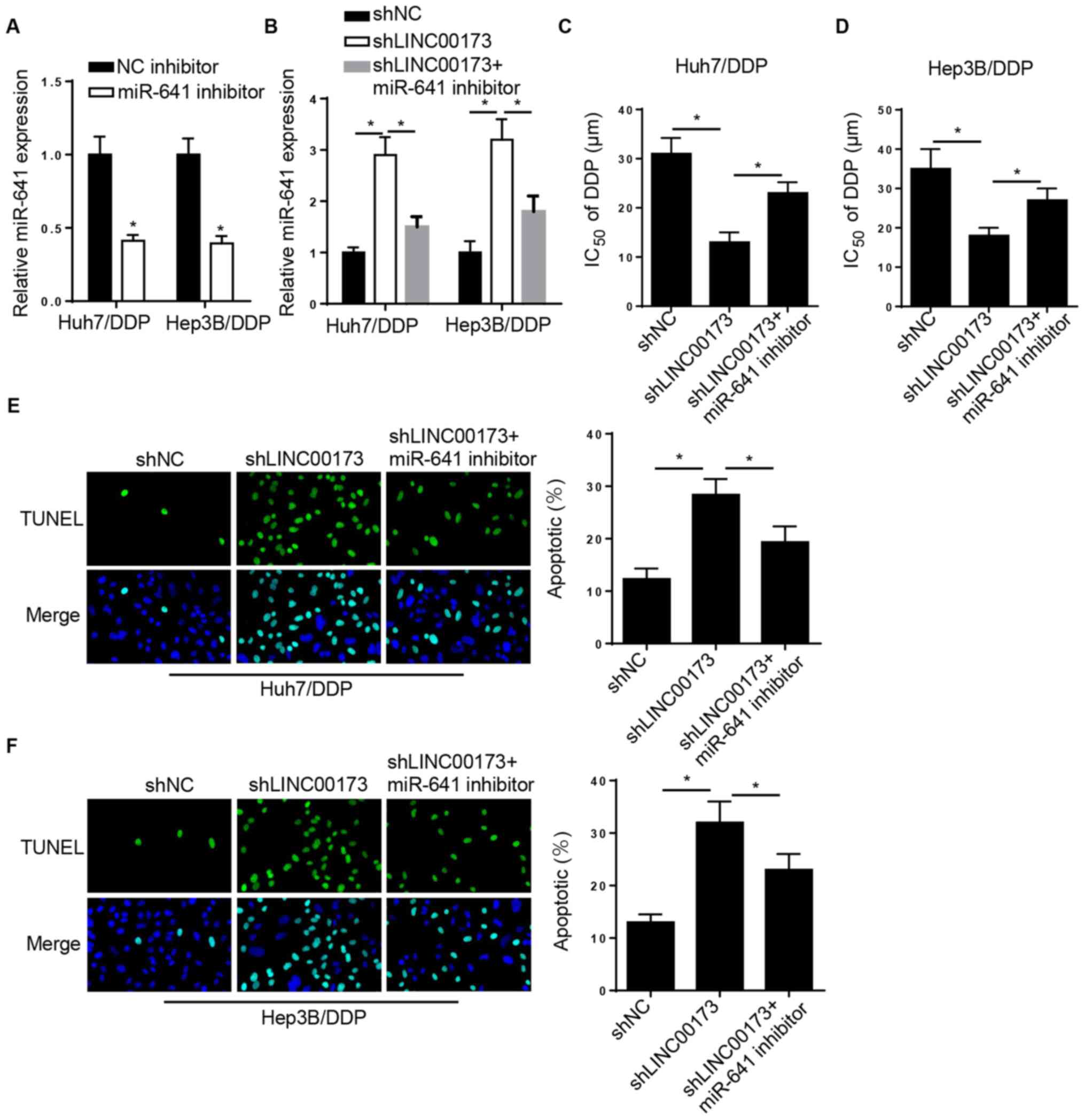 | Figure 4.LINC00173-knockdown decreases DDP
resistance in DDP-resistant HCC cells by targeting miR-641. (A)
RT-qPCR analysis of relative miR-641 expression in Huh7/DDP and
Hep3B/DDP cells transfected with the NC inhibitor or miR-641
inhibitor. (B) RT-qPCR analysis of relative miR-641 expression in
Huh7/DDP and Hep3B/DDP cells transfected with shNC, shLINC00173 or
shLINC00173+miR-641 inhibitor. Cell Counting Kit-8 analysis to
determine the IC50 value of (C) Huh7/DDP and (D)
Hep3B/DDP cells transfected with shNC, shLINC00173 or an
shLINC00173+miR-641 inhibitor. TUNEL analysis (magnification, ×100)
revealed the apoptosis of (E) Huh7/DDP and (F) Hep3B/DDP cells
transfected with shNC, shLINC00173 or shLINC00173+miR-641
inhibitor. *P<0.05. DDP, cisplatin; miR, microRNA; HCC,
hepatocellular carcinoma; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control; sh, short
hairpin; IC50, half maximal inhibitory
concentration. |
LINC00173 enhances DDP resistance in
HCC cells by regulating the miR-641/RAB14 axis
As shown in Fig. 5A,
the StarBase website predicted that RAB14 was a potential target of
miR-641. A luciferase reporter assay revealed that miR-641
overexpression decreased the luciferase activity of LINC00173-wt in
DDP-resistant HCC cells, while that of LINC00173-mut exhibited no
significant change (Fig. 5B and C).
Of note, RAB14 expression was reduced by miR-641 overexpression or
LINC00173-knockdown, and the changes caused by LINC00173-knockdown
were reversed by miR-641 inhibition (Fig. 5D and E). In addition, the level of
RAB14 was increased in HCC tissues and in DDP-resistant HCC tissues
(Fig. 5F and G). A positive
correlation between LINC00173 and RAB14 expression was also
observed in HCC tissues (Fig. 5H).
The CCK-8 assay demonstrated that transfection of pcDNA3.1/RAB14
recovered the inhibitory effect of LINC00173-knockdown on the
IC50 value of DDP in Huh7/DDP and Hep3B/DDP cells
(Fig. 5I and J). Addition of RAB14
neutralized the promoting effect of LINC00173-knockdown on the
apoptosis of Huh7/DDP and Hep3B/DDP cells (Fig. 5K and L). Collectively, these findings
indicated that LINC00173 contributed to DDP resistance in HCC cells
through sponging miR-641 and upregulating RAB14.
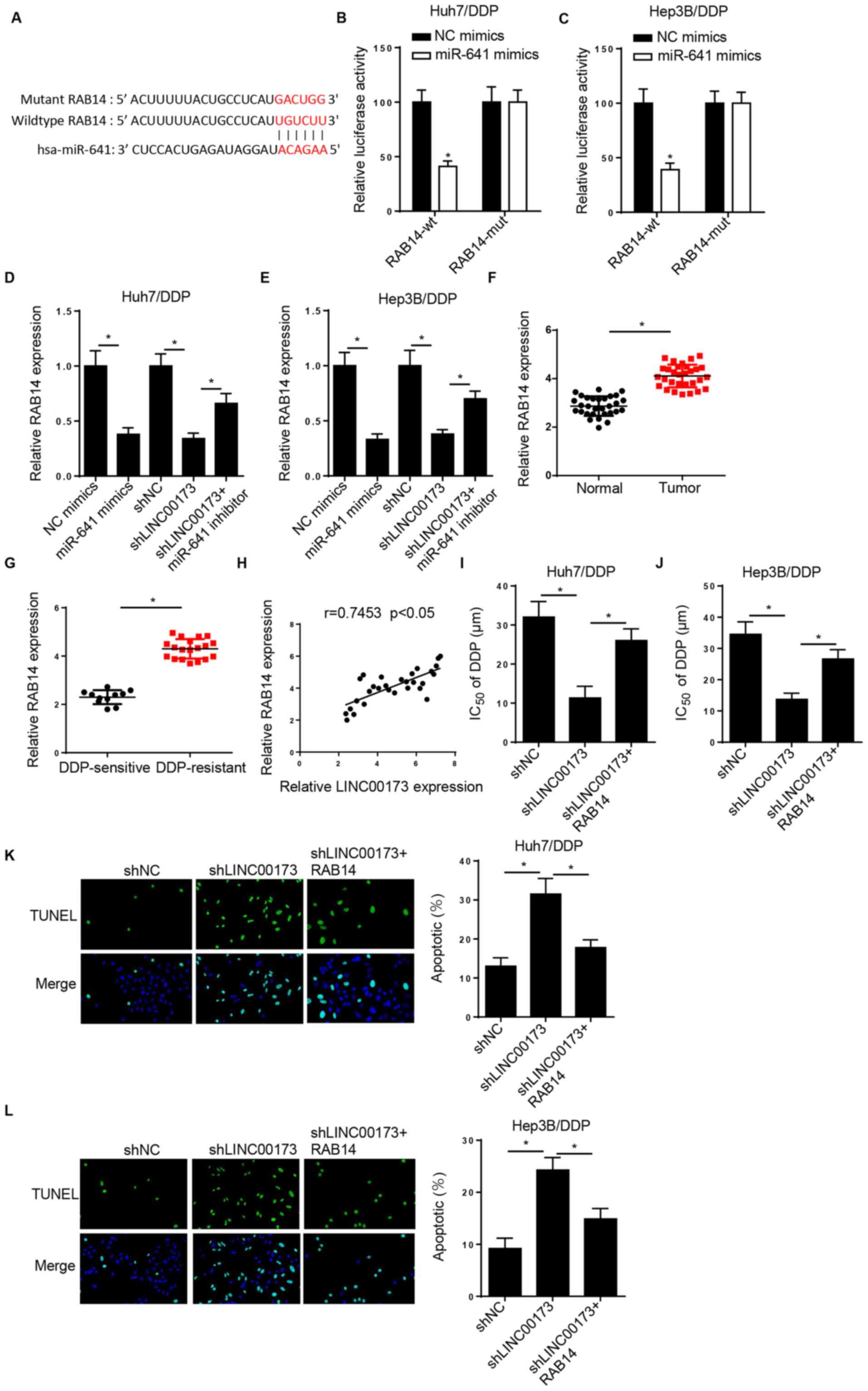 | Figure 5.LINC00173 induces DDP resistance in
HCC cells by modulating the miR-641/RAB14 axis. (A) StarBase was
used to predict potential binding sites between RAB14 and miR-641.
Luciferase activity of RAB14-wt or RAB14-mut in (B) Huh7/DDP and
(C) Hep3B/DDP cells transfected with NC mimics and miR-641 mimics.
RT-qPCR showed the relative RAB14 expression in (D) Huh7/DDP and
(E) Hep3B/DDP cells transfected with NC mimics, miR-641 mimics,
shNC, shLINC00173 or shLINC00173+miR-641 inhibitor. RT-qPCR showed
the relative miR-641 expression in (F) HCC tissues and (G)
DDP-resistant HCC tissues. (H) Pearson's correlation analysis
revealed a positive correlation between RAB14 and LINC00173
expression in HCC tissues. Cell Counting Kit-8 analysis was used to
determine the IC50 values of (I) Huh7/DDP and (J)
Hep3B/DDP cells transfected with shNC, shLINC00173 or
shLINC00173+pcDNA3.1/RAB14. TUNEL analysis (magnification, ×100) of
the apoptosis of (K) Huh7/DDP and (L) Hep3B/DDP cells transfected
with shNC, shLINC00173 or shLINC00173+pcDNA3.1/RAB14. *P<0.05.
DDP, cisplatin; HCC, hepatocellular carcinoma; miR, microRNA; wt,
wild-type; mut, mutant; NC, negative control; sh, short hairpin
(RNA); RT-qPCR, reverse transcription-quantitative PCR;
IC50, half maximal inhibitory concentration. |
Discussion
Chemoresistance severely compromises therapeutic
efficacy in patients with HCC. Thus, it is crucial to identify the
molecular mechanisms underlying HCC chemoresistance and to
investigate new therapeutic strategies for HCC. In the present
study, it was observed that silencing of LINC00173 improved the
sensitivity of DDP-resistant HCC cells by modulating the
miR-641/RAB14 axis, suggesting that LINC00173 may be an effective
treatment target to prevent or reverse drug resistance.
Numerous studies have reported that lncRNAs are key
regulators of chemoresistance in human cancers (20,21).
Dysregulated LINC00173 expression was found to be implicated in
drug resistance in cancer. For example, lncRNA LINC00173 promoted
colorectal cancer cell metastasis and chemoresistance via
modulation of the miR-765/PLP2 pathway (22). LINC00173 facilitated the
chemoresistance and progression of small-cell lung cancer by
targeting miR-218 and regulating Etk expression (23). The present study focused on the role
and mechanism of action of LINC00173 in DDP-resistant HCC. The
results indicated that LINC00173 expression was elevated in HCC
tissues and DDP-resistant HCC cells. Furthermore, LINC00173
knockdown enhanced the DDP sensitivity of DDP-resistant HCC cells.
These findings indicated that LINC00173 induced DDP resistance in
HCC cells.
A large number of studies have revealed that lncRNAs
function as competing endogenous RNAs, exerting their regulatory
effects on tumorigenesis by sponging miRNAs, which reverses the
inhibitory effects on their targets (24,25).
miR-641 has been reported to act as a tumor suppressor in various
cancer types. For example, miR-641 suppressed lung cancer cell
proliferation and induced apoptosis by regulating Programmed cell
death protein 4 (26). miR-641 also
inhibited cervical cancer progression and promoted apoptosis by
targeting zinc finger E-box-binding homeobox 1 (27). However, the biological function of
miR-641 in DDP-resistant HCC cells remained unclear. In the present
study, the level of miR-641 was reduced in DDP-resistant HCC
tissues and miR-641 was found to directly interact with LINC00173.
Furthermore, miR-641 inhibition abolished the promoting effect of
LINC00173 knockdown on the sensitivity of DDP-resistant HCC cells.
These results indicated that LINC00173 enhanced DDP resistance in
HCC by regulating miR-641.
RAB14, a member of the RAS oncogene family, has been
reported to be implicated in the development of several types of
cancer, such as cervical and gastric cancer (28,29).
Convincing evidence has suggested that RAB14 may promote
chemotherapy resistance in several tumors (30,31). For
example, miR-148a enhanced sensitivity to DDP by targeting RAB14 in
renal cancer cells (32). In the
present study, RAB14 expression was markedly enhanced in
DDP-resistant HCC tissues, and was confirmed to be a target of
miR-641. Furthermore, rescue assays revealed that RAB14
overexpression partially abolished the enhanced DDP sensitivity of
Huh7/DDP and Hep3B/DDP cells caused by LINC00173-knockdown.
Collectively, these data confirmed that LINC00173 contributed to
DDP resistance in HCC by modulating the miR-641/RAB14 axis.
However, the limitations of the present study must
be addressed in future research. For example, the downstream
effectors or signaling pathways associated with the
LINC00173/miR-641/RAB14 axis should be further investigated, and
in vivo experiments will help to fully elucidate the
associated underlying mechanisms.
In conclusion, the findings of the present study
demonstrated that LINC00173 promoted the DDP resistance of HCC
cells via sponging miR-641 to upregulate RAB14, providing a novel
therapeutic target for overcoming DDP resistance in HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
GZ and YD designed the study. AZ and SS performed
the experiments. GZ and AZ analysed the data and prepared the
figures. GZ and YD drafted, reviewed and revised the manuscript. GZ
and YD confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Weifang People's Hospital (Weifang, China), and
written informed consent was obtained from all subjects prior to
commencement.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Singal AG and El-Serag HB: Hepatocellular
carcinoma from epidemiology to prevention: Translating knowledge
into practice. Clin Gastroenterol Hepatol. 13:2140–2151. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan ST, Mau Lo C, Poon RT, Yeung C, Leung
Liu C, Yuen WK, Ming Lam C, Ng KK and Ching Chan S: Continuous
improvement of survival outcomes of resection of hepatocellular
carcinoma: A 20-year experience. Ann Surg. 253:745–758. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi Y, Yang X, Xue X, Sun D, Cai P, Song
Q, Zhang B and Qin L: HANR enhances autophagy-associated sorafenib
resistance through miR-29b/ATG9A axis in hepatocellular carcinoma.
Onco Targets Ther. 13:2127–2137. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo Y, Bai M, Lin L, Huang J, An Y, Liang
L, Liu Y and Huang W: LncRNA DLEU2 aggravates the progression of
hepatocellular carcinoma through binding to EZH2. Biomed
Pharmacother. 118:1092722019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Z, Chen JY, Zhong Y, Xie L and Li JS:
lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells
by sponging miR-9-5p to upregulate SOX11. Braz J Med Biol Res.
52:e86312019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang X, Sun L, Wang L, Yao B, Mo H and
Yang W: LncRNA SNHG7 accelerates the proliferation, migration and
invasion of hepatocellular carcinoma cells via regulating
miR-122-5p and RPL4. Biomed Pharmacother. 118:1093862019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu M, Zheng W, Zhang M, Dong X, Zhao Y,
Wang S, Jiang H and Zheng X: LncRNA NONHSAT141924 promotes
paclitaxel chemotherapy resistance through p-CREB/Bcl-2 apoptosis
signaling pathway in breast cancer. J Cancer. 11:3645–3654. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang J, Zhao B, Chen X, Wang Z, Xu H and
Huang B: Silence of long noncoding RNA NEAT1 inhibits malignant
biological behaviors and chemotherapy resistance in gastric cancer.
Pathol Oncol Res. 24:109–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Zhou M, Zhao X, Wang G and Li J:
Long noncoding RNA LINC00173 is downregulated in cervical cancer
and inhibits cell proliferation and invasion by modulating the
miR-182-5p/FBXW7 axis. Pathol Res Pract. 216:1529942020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan H, Yuan J and Li X, Ma Y, Wang X, Xu B
and Li X: LncRNA LINC00173 enhances triple-negative breast cancer
progression by suppressing miR-490-3p expression. Biomed
Pharmacother. 125:1099872020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang F, Lei P, Zeng W, Gao J and Wu N:
Long noncoding RNA LINC00173 promotes the malignancy of melanoma by
promoting the expression of IRS4 through competitive binding to
microRNA-493. Cancer Manag Res. 12:3131–3144. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao F, Li Y, Wan Y and Xue M:
MircroRNA-139 sensitizes ovarian cancer cell to cisplatin-based
chemotherapy through regulation of ATP7A/B. Cancer Chemother
Pharmacol. 81:935–947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Niu YC, Tong J, Shi XF and Zhang T:
MicroRNA-654-3p enhances cisplatin sensitivity by targeting QPRT
and inhibiting the PI3K/AKT signaling pathway in ovarian cancer
cells. Exp Ther Med. 20:1467–1479. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang S, Wang P, Wang S, Cong A, Zhang Q,
Shen W, Li X, Zhang W and Han G: miRNA-181a-5p enhances the
sensitivity of cells to cisplatin in esophageal adenocarcinoma by
targeting CBLB. Cancer Manag Res. 12:4981–4990. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Y, Zheng R, Chen J and Ning D:
CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes
to cisplatin resistance in non-small cell lung cancer (NSCLC).
Cancer Cell Int. 20:2892020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van Persijn van Meerten EL, Gelderblom H
and Bloem JL: RECIST revised: Implications for the radiologist. A
review article on the modified RECIST guideline. Eur Radiol.
20:1456–1467. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang SY, Wang X and Zhang CY: LncRNA SNHG7
enhances chemoresistance in neuroblastoma through cisplatin-induced
autophagy by regulating miR-329-3p/MYO10 axis. Eur Rev Med
Pharmacol Sci. 24:3805–3817. 2020.PubMed/NCBI
|
|
21
|
Xu Q, Lin YB, Li L and Liu J: LncRNA
TLR8-AS1 promotes metastasis and chemoresistance of ovarian cancer
through enhancing TLR8 mRNA stability. Biochem Biophys Res Commun.
526:857–864. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu Y, Lu X, Yang C and Yin F: Long
noncoding RNA LINC00173 contributes to the growth, invasiveness and
chemo-resistance of colorectal cancer through regulating
miR-765/PLP2 axis. Cancer Manag Res. 12:3363–3369. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zeng F, Wang Q, Wang S, Liang S, Huang W,
Guo Y, Peng J, Li M, Zhu W and Guo L: Linc00173 promotes
chemoresistance and progression of small cell lung cancer by
sponging miR-218 to regulate Etk expression. Oncogene. 39:293–307.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y
and Chen Y: The emerging function and mechanism of ceRNAs in
cancer. Trends Genet. 32:211–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou J, Li H, Li N, Li X, Zhang H, Song Q
and Peng M: MicroRNA-641 inhibits lung cancer cells proliferation,
metastasis but promotes apoptosis in cells by targeting PDCD4. Int
J Clin Exp Pathol. 10:8211–8221. 2017.PubMed/NCBI
|
|
27
|
Yao R, Zheng H, Wu L and Cai P: miRNA-641
inhibits the proliferation, migration, and invasion and induces
apoptosis of cervical cancer cells by directly targeting ZEB1. Onco
Targets Ther. 11:8965–8976. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang J, Liang B and Hou S: TMPO-AS1
promotes cervical cancer progression by upregulating RAB14 via
sponging miR-577. J Gene Med. 21:e31252019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo B, Wang W, Zhao Z, Li Q, Zhou K, Zhao
L, Wang L, Yang J and Huang C: Rab14 Act as oncogene and induce
proliferation of gastric cancer cells via AKT signaling pathway.
PLoS One. 12:e01706202017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhai Y, Liu M and Zheng Y: MicroRNA-451
dictates the anoikis resistance of osteosarcoma by targeting Rab14.
Int J Clin Exp Pathol. 10:10989–10997. 2017.PubMed/NCBI
|
|
31
|
Ge J and Ge C: Rab14 overexpression
regulates gemcitabine sensitivity through regulation of Bcl-2 and
mitochondrial function in pancreatic cancer. Virchows Arch.
474:59–69. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim EA, Kim TG, Sung EG, Song IH, Kim JY,
Doh KO and Lee TJ: miR-148a increases the sensitivity to cisplatin
by targeting Rab14 in renal cancer cells. Int J Oncol. 50:984–992.
2017. View Article : Google Scholar : PubMed/NCBI
|