Introduction
Renal cell carcinoma (RCC) is the most lethal
urological cancer, with an estimated 62,000 new cases diagnosed in
the United States in 2020 (1). The
most common histological subtype of RCC is clear cell (cc) RCC,
which accounts for ~80% of all cases (2). It is well-known that a germline
mutation in the von Hippel-Lindau (VHL) gene plays an
important role in ccRCC development, and the loss of function of
the VHL protein (pVHL) may also be associated with resistance to
cytotoxic chemotherapy (3,4). Currently, renal surgery is extensively
used for localized RCC, while the treatment options for patients
with advanced or metastatic RCC (mRCC) are limited. Targeted
therapy, such as sunitinib or sorafenib, is considered the standard
first-line treatment for mRCC, and is beneficial with regards to
long-term survival (5). However, the
development of drug resistance is considered inevitable following
targeted therapy for 6–12 months (6). Currently, several prognostic prediction
models have been used to assess the long-term outcomes of patients
with RCC, such as the Mayo Clinic Stage, Size, Grade and Necrosis
score (7) and the International
Metastatic Renal Cell Carcinoma Database Consortium (8). In addition, several studies have
reported that biomarker staining on postoperative pathology is an
effective method for predicting prognosis (9–11).
However, due to the heterogeneity of the molecular phenotype,
single biomarker staining may have limited prediction power
(12). Thus, a clinical outcome
prediction model of multiple combined biomarkers with high accuracy
is required for patients with RCC.
Previous studies have demonstrated that
hypoxia-inducible factor (HIF)1a and HIF2a are regulated by
pVHL-mediated ubiquitination. The loss of pVHL function may promote
HIFa accumulation and translocation, thus contributing to the
development of RCC (13,14). Targeted therapy with tyrosine kinase
inhibitors, which directly target the downstream factors of the
HIFa pathway, is beneficial in the treatment of patients with RCC,
while the HIF2a inhibitor exhibits limited efficiency (15). These findings suggest that there may
be other signaling pathways involved in VHL-deficient RCC. Zhang
et al (16) identified zinc
fingers and homeoboxes 2 (ZHX2) as a novel VHL substrate factor
that promotes the development of ccRCC. Subsequently, Zhu et
al (17) reported that ZHX2 can
directly target the MEK/ERK signaling pathway in ccRCC cell lines
and induce sunitinib resistance by overexpressing ZHX2. In
addition, scm-like with four malignant brain tumor domains (SFMBT1)
was also identified as a candidate VHL target by genome-wide
screening and SFMBT1 is considered an oncogenic driver in ccRCC
(18), however, its prognostic value
remains unclear.
The predictive ability of combining multiple novel
VHL substrate targets is yet to be investigated. Thus, the present
study aimed to investigate the clinical significance and prognostic
value of SFMBT1 combined with ZHX2, in the hope to better
understand the role of these novel VHL substrates in targeted
therapeutic intervention.
Materials and methods
Patients and tissues
A total of 97 patients, including 68 (70.1%) men and
29 (29.9%) women were enrolled in the present study. with confirmed
ccRCC pathology, who underwent radical nephrectomy at Putuo
hospital between January 2010 and December 2015, were enrolled in
the present study. Follow-up examinations (from January 2010 to
January 2020, including blood tests and CT/MRI, were performed
every 6 months in the first 3 years, and once a year after that.
All medical records and laboratory information were collected,
including age, surgery time, sex, tumor size and tumor stage. Those
who received chemoradiotherapy prior to selection, and had
incomplete clinical records or missed follow-up were excluded from
the present study.
A total of five paired fresh tumor tissues and
adjacent normal tissues were collected (>3 cm apart) for RNA
extraction and to detect mRNA expression. The present study was
approved by the Ethical Review Boards of Putuo Hospital (Shanghai,
China, approval no. 20200130) and written informed consent was
provided by all patients prior to the study start.
Database analysis and survival
data
The expression levels of SFMBT1 and ZHX2 in kidney
cancer were assessed using the Gene Expression Profiling
Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn). In addition, the GEPIA
database was used to retrieve information on the prognostic values
of SFMBT1 and ZHX2 in kidney cancer, including overall survival
(OS) and disease-free survival (DFS). The survival rates were
measured according to the data from 864 patients.
Tissue microarrays and
immunohistochemistry (IHC)
The tissue microarray was constructed based on the
tumor samples of the 97 patients with ccRCC. The protocol was as
follows: 1) Tissue chip was soaked in xylene for 10 min (three
times); 2) hydrated with 100% ethanol (three times), 95% ethanol
(once), 85% ethanol (once), 75% ethanol (once) and washed with
distilled water (three times); 3) washed with PBS (5 min each
time); 4) incubated with 3% H2O2 for 20 min
at room temperature to inhibit endogenous peroxidase activity and
washed with PBS (three times); 5) antigen repair: Incubated with 1
mmol/l EDTA antigen repair solution for 10 min; 6) Incubated with
10% goat serum at room temperature for 10 min; 7) incubated with
primary antibodies against SFMBT1 (1:200 dilution; cat. no.
A303-221A; Bethyl Laboratories, Inc.) and ZHX2 (1:50 dilution; cat.
no. GTX112232; GeneTex Inc.) overnight at 4°C, and washed with PBS
(three times); 8) incubated with secondary antibody at room
temperature for 1 h, washed with PBS (three times). DAB chromogen
was added to observe the chromogen state under the light microscope
(magnification, ×40), rinsed with distilled water. 10) Stained with
hematoxylin at room temperature: Soak the tissue chip in
hematoxylin solution (prepared at 1:20); 11) dehydrated with 75%
ethanol for 10 sec, 85% ethanol for 10 sec, 95% ethanol for 10 sec,
100% ethanol for 5 min (three times) and xylene for 5 min (three
times); 12) Sequestration. IHC analysis was performed as previously
described (19). IHC analysis was
performed to detect the expression levels of the novel VHL
substrate targets, SFMBT1 and ZHX2. The location and expression of
the targets were independently confirmed by two pathologists at
Putuo hospital.
The IHC staining score was the sum of the staining
percentage and the intensity degree. Staining percentage was
calculated as follows: 1, 0–25; 2, 26–50; 3, 51–75 and 4, >75%,
while intensity degree was calculated as follows: 0, negative; 1,
weak; 2, moderate and 3, strong. An IHC score <6 was classified
as the low expression group, while an IHC score ≥6 was classified
as the high expression group (12).
All patients were divided into two groups, according to the
combined expression levels of SFMBT1 and ZHX2. Patients with high
SFMBT1 and ZHX2 expression levels were classified into the SHZH
group.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from fresh tissue samples
using TRIzol® reagent (Thermo Fisher Scientific, Inc.,
cat. no. 15596018) and trichloromethane (Nanjing KeyGen Biotech
Co., Ltd., cat. no. C07615202). Total RNA was reverse transcribed
into cDNA using the HiScript II Q RT SuperMix RT kit (+gDNA wiper,
cat. no. R223-01; Vazyme Biotech Co., Ltd.) at 50°C for 15 min and
85°C for 5 sec. qPCR was subsequently performed using the SYBR
Green kit (Takara Biotechnology Co., Ltd.), according to the
manufacturer's protocol. The following thermocycling conditions
were used for qPCR: 95°C for 30 sec, cyclic reaction with 95°C for
10 sec, 60°C for 20 sec and 72°C for 20 sec in 40 rounds. The
following primer sequences were used for qPCR: SFMBT1 forward,
5′-TGCCACCATTTGCTGAT-3′ and reverse, 5′-TTTGTCCACCTCCATTCTG-3; ZHX2
forward, 5′-CTGCCTTAGCCCCACAC-3′ and reverse,
5′-TGCTACCCAGTTCTCCCA-3′; and GAPDH forward,
5′-ACAGTCAGCCGCATCTTCTT-3′ and reverse, 5′-GACAAGCTTCCCGTTCTCAG-3′.
Relative expression levels were calculated using the
2−ΔΔCq method (20)
normalized to the internal reference gene GAPDH.
Statistical analysis
Statistical analysis was performed using SPSS 19.0
software (IBM Corp.) and GraphPad Prism software (version 6;
GraphPad Software, Inc.). Paired Student's t-test or the
Mann-Whitney test were used to compare continuous variables, while
Pearson's χ2 or Fisher's exact tests were used to
compare categorical variables. OS and DFS analyses were performed
using the Kaplan-Meier method and log-rank test. Univariate Cox
regression analysis was performed to determine the prognostic
factors, and significant variables were further analyzed via
multivariate Cox regression analysis to determine the independent
prognostic factors. P<0.05 was considered to indicate a
statistically significant difference.
Results
SFMBT1 and ZHX2 expression in
ccRCC
The GEPIA database was searched, which indicated
that SFMBT1 expression was downregulated in ccRCC tissues, while
ZHX2 expression was relatively upregulated in ccRCC tissue
(Fig. S1). IHC analysis was
performed to detect the expression levels of SFMBT1 and ZHX2 in
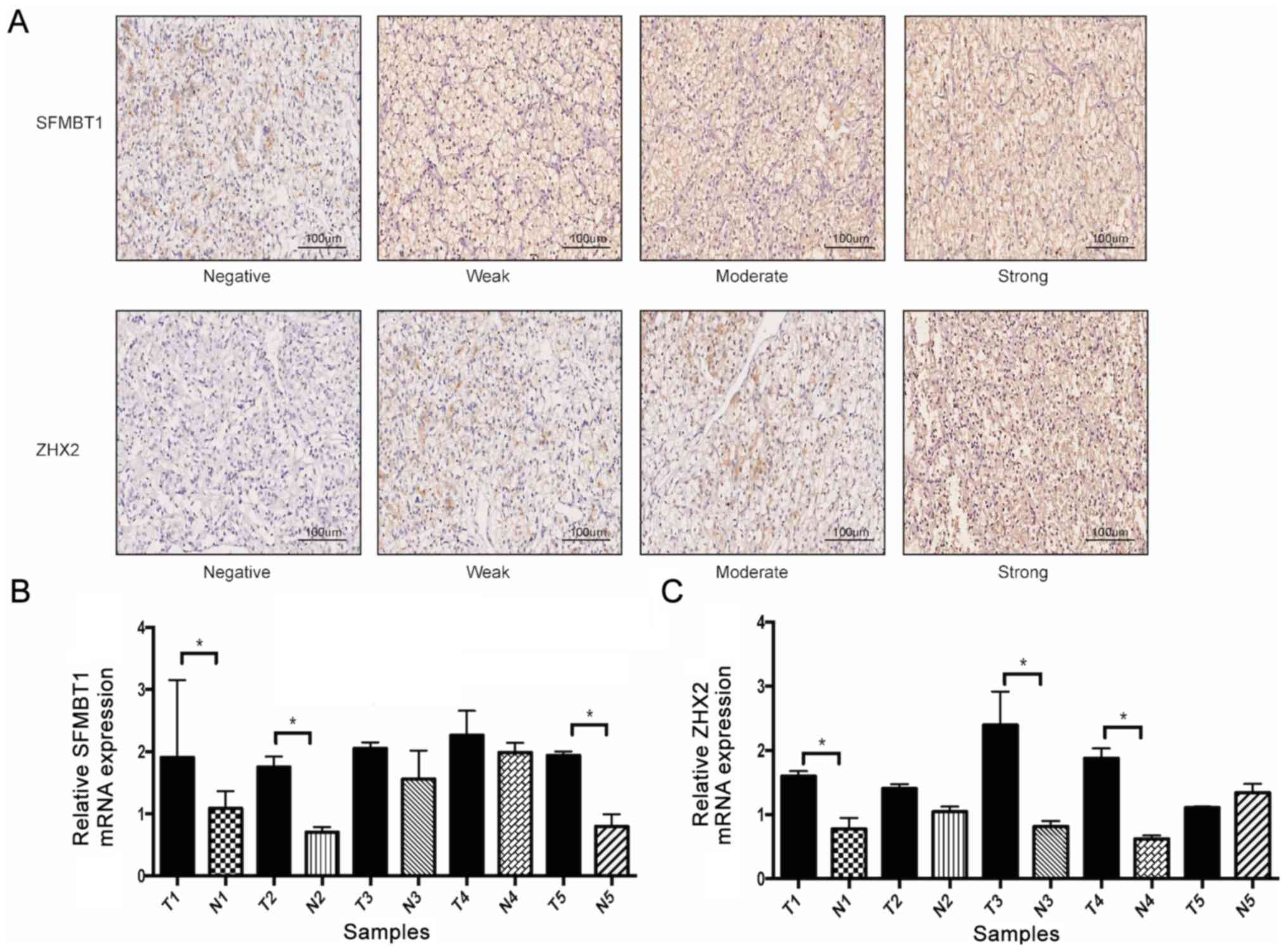
ccRCC samples. As presented in Fig.
1A, SFMBT1 was localized in the nucleus and cytoplasm, while
ZHX2 was predominantly localized in the nucleus, and both proteins
were upregulated in tumor tissues compared with adjacent normal
tissues. RT-qPCR analysis was subsequently performed to confirm
these results, which demonstrated that the mRNA expression levels
of SFMBT1 and ZHX2 were upregulated in tumor tissues compared with
adjacent normal tissues (Fig. 1B and
C).
The detailed clinicopathological characteristics of
all patients are presented in Table
I. A total of 68 (70.1%) men and 29 (29.9%) women were enrolled
in the present study. SFMBT1 expression was upregulated in 61.9%
(60/97) of patients with ccRCC, while ZHX2 expression was
upregulated in 52.6% (51/97) of patients. Notably, high SFMBT1
expression was significantly associated with advanced tumor status
(TNM stage (21) and Fuhrman grade
(22), while high ZHX2 expression
was significantly associated with advanced Fuhrman grade. Taken
together, these results suggest that SFMBT1 and ZHX2 act as
oncogenes in ccRCC.
 | Table I.Clinicopathological characteristics of
patients with clear cell renal cell carcinoma (n=97). |
Table I.
Clinicopathological characteristics of
patients with clear cell renal cell carcinoma (n=97).
|
|
| Tumoral SFMBT1
expression | Tumoral ZHX2
expression |
|---|
|
|
|
|
|
|---|
| Characteristic | Patients, n (%) | Low | High | P-value | Low | High | P-value |
|---|
| Patients | 97
(100.0) | 37 | 60 |
| 46 | 51 |
|
| Sex |
|
|
| 0.582 |
|
| 0.544 |
| Male | 68
(70.1) | 26 | 42 |
| 32 | 36 |
|
|
Female | 29
(29.9) | 11 | 18 |
| 14 | 15 |
|
| Age, years |
|
|
| 0.507 |
|
| 0.531 |
| ≤55 | 46
(47.4) | 18 | 28 |
| 24 | 22 |
|
|
>55 | 51
(52.6) | 19 | 32 |
| 22 | 29 |
|
| TNM stage |
|
|
| 0.035 |
|
| 0.237 |
| I/II | 87
(92.8) | 36 | 51 |
| 43 | 44 |
|
|
III/IV | 10 (7.2) | 1 | 9 |
| 3 | 7 |
|
| Fuhrman grade |
|
|
| 0.046 |
|
| 0.043 |
|
I/II | 76
(78.4) | 33 | 43 |
| 40 | 36 |
|
|
III/IV | 21
(21.6) | 4 | 17 |
| 6 | 15 |
|
| Tumor size, cm |
|
|
| 0.298 |
|
| 0.413 |
| ≤4 | 40
(41.2) | 17 | 23 |
| 20 | 20 |
|
|
>4 | 57
(58.8) | 20 | 37 |
| 26 | 31 |
|
Prognostic values of SFMBT1 and ZHX2
in patients with ccRCC
To determine the prognostic values of SFMBT1 and
ZHX2 in ccRCC, patients were divided into two groups, according to
the combined expression levels of SFMBT1 and ZHX2. Of the 97
patients, 32 patients were classified into the high expression
group (SHZH group), while 29 patients were classified into the low
expression group (SLZL group); the remaining 36 patients were
defined as other group (SLZH or SHZL groups).
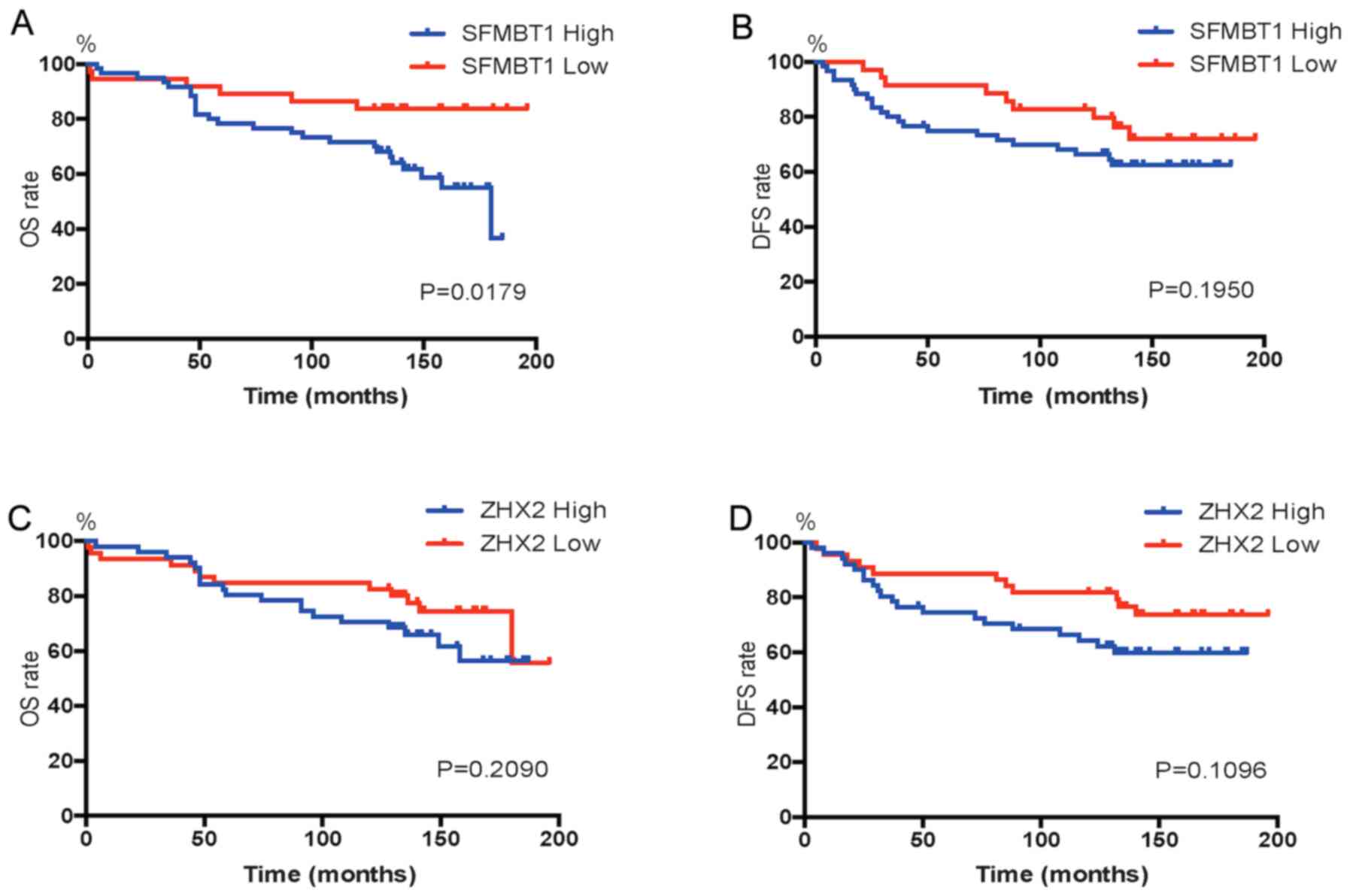
Kaplan-Meier survival analysis was performed to
determine the prognostic values of SFMBT1 and ZHX2. As presented in
Fig. 2, the 5-year survival rates
were 78.3 and 89.2% in patients with high and low SFMBT1
expression, respectively. In addition, patients with high SFMBT1
expression had a significantly lower OS rate (P=0.0179) than those
with low SFMBT1 expression. DFS was comparable between patients
with high and low SFMBT1 expression levels. Notably, no significant
differences were observed in the OS and DFS rates between patients
with high and low ZHX2 expression.
The GEPIA database was searched to retrieve
information on the prognostic values of SFMBT1 and ZHX2. As
presented in Fig. S2, no
significant differences in OS and DFS analyses were observed.
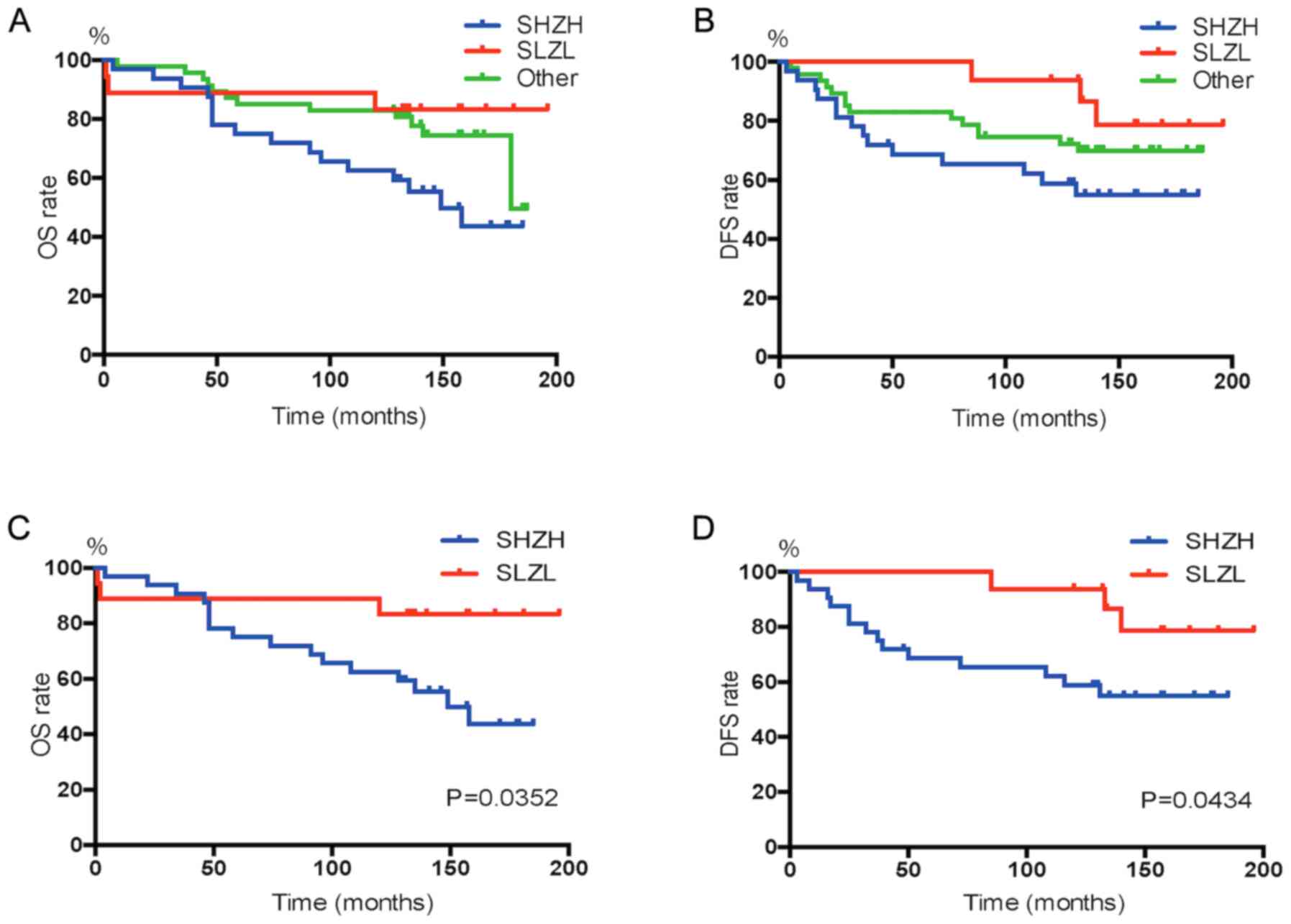
Subsequently, the prognostic value of SFMBT1 combined with ZHX2 in
ccRCC was assessed. The OS and DFS rates in different groups are
presented in Fig. 3A and B, there
was no significant difference between these groups, and patients in
the SHZH group had a worst outcome. As presented in Fig. 3C and D, patients in the SHZH group
had significantly worse OS (P=0.0352) and DFS (P=0.0434) rates
compared with patients in the SLZL group.
Combined expression of SFMBT1 and ZHX2
may be an independent factor in ccRCC prognosis
The prognostic value of different factors on OS and
DFS was determined via univariate and multivariate Cox regression
analyses (Tables II and III). Univariate analysis demonstrated
that SFMBT1 expression was significantly associated with OS rate
[hazard ratio (HR), 0.583; P=0.042; Table II], while SHZH (combined expression
group), age, TNM stage, Fuhrman grade and tumor size were
significantly associated with OS and DFS rates (Tables II and III). Multivariate analysis demonstrated
that SHZH was an independent predictive factor of OS (HR, 0.252;
P=0.021) and DFS (HR, 0.394; P=0.016) rates for patients with
ccRCC. Collectively, these results suggest that the combined
expression of SFMBT1 and ZHX2 may be used as a promising prognostic
factor in predicting the outcomes of patients with ccRCC.
 | Table II.Univariate and multivariate Cox
regression analyses based on overall survival. |
Table II.
Univariate and multivariate Cox
regression analyses based on overall survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| SFMBT1 expression
(Low vs. High) | 0.583 | 0.435–1.491 | 0.042 | 0.582 | 0.234–1.283 | 0.152 |
| ZHX2 expression
(Low vs. High) | 0.382 | 0.273–0.825 | 0.083 | − | − | − |
| SHZH expression
(SHZH vs. Other) | 0.343 | 0.213–0.628 | 0.003b | 0.252 | 0.203–0.755 | 0.021a |
| Age, years (≤55 vs.
>55) | 1.342 | 0.893–1.582 | 0.039a | 1.120 | 0.723–1.321 | 0.342 |
| TNM stage (I/II vs.
III/IV) | 5.823 | 2.783–10.809 |
<0.001c | 3.012 | 2.783–8.283 | 0.031a |
| Fuhrman grade (I/II
vs. III/IV) | 6.261 | 3.172–9.632 | 0.001b | 4.391 | 1.653–7.402 | 0.093a |
| Tumor size, cm (≤4
vs. >4) | 3.869 | 1.873–8.846 | 0.021a | 2.097 | 1.072–7.842 | 0.048a |
 | Table III.Univariate and multivariate Cox
regression analyses based on disease-free survival. |
Table III.
Univariate and multivariate Cox
regression analyses based on disease-free survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| SFMBT1 expression
(Low vs. High) | 0.472 | 0.235–1.145 | 0.148 | − | − | − |
| ZHX2 expression
(Low vs. High) | 0.455 | 0.334–0.893 | 0.273 | − | − | − |
| SHZH expression
(SHZH vs. Other) | 0.388 | 0.234–0.962 | 0.009b | 0.394 | 0.303–1.274 | 0.016a |
| Age, years (≤55 vs.
>55) | 1.538 | 0.712–1.863 | 0.041a | 1.684 | 0.592–1.702 | 0.281 |
| TNM stage (I/II vs.
III/IV) | 4.172 | 2.653–8.082 |
<0.001c | 3.712 | 2.301–9.729 | 0.030a |
| Fuhrman grade (I/II
vs. III/IV) | 6.261 | 3.172–9.632 | 0.001b | 4.391 | 1.601–8.784 | 0.086 |
| Tumor size, cm (≤4
vs. >4) | 3.142 | 1.421–6.429 | 0.039a | 2.537 | 1.694–8.893 | 0.072 |
Discussion
The present study investigated the expression levels
of SFMBT1 and ZHX2 in ccRCC tissues. The results demonstrated that
both genes were relatively upregulated in cancer tissues compared
with adjacent normal tissues. As both genes are considered VHL
substrate factors, their combined prediction value in ccRCC
prognosis was assessed in the present study. The association
between SFMBT1/ZHX2 expression and clinicopathological
characteristics demonstrated that both genes act as oncogenes in
ccRCC development. Furthermore, Kaplan-Meier survival analysis
indicated that the combined expression of SFMBT1 and ZHX2 was
significantly associated with OS and DFS, and patients in the SHZH
group had lower survival rates. Thus, SHZH was identified as an
independent predictor for ccRCC outcomes.
SFMBT1 is a member of the MBT domain-containing
protein family, which plays a critical role in chromatin regulation
(23). Tang et al (24) reported that SFMBT1 is an essential
part of LSD1 in genetic modification, and SFMBT1 is associated with
epithelial-to-mesenchymal transition and poor prognosis in human
breast cancer. Furthermore, Liu et al (18) demonstrated that SFMBT1 is regulated
by pVHL via a prolyl hydroxylation and proteasomal degradation
process, similar to HIFs and ZHX2. Their research found that
overexpression of SFMBT1 promotes cell proliferation and tumor
growth in ccRCC. Furthermore, the importance of the
pVHL-SFMBT1-SPHK1 signaling pathway in ccRCC development was
identified.
The role of ZHX2 in cancer remains controversial.
Previous studies have reported that ZHX2 plays a dual role as both
an oncogene and tumor suppressor (25–27).
Zhang et al (16) identified
ZHX2 as a VHL novel substrate factor, and ZHX2 is regulated by
pVHL-mediated degradation. The in vitro experiments
demonstrated that ZHX2 depletion can significantly downregulate
NF-κB activation, thereby inhibiting the proliferation and
tumor-forming ability of ccRCC cells. Zhu et al (17) used a ZHX2 overexpression lentivirus
to transfect 786-O and CAKI-1 cells for lineage reprogramming, and
the transcriptome analysis revealed that ZHX2 overexpression can
directly activate the MEK/ERK1/2 signaling pathway, which in turn
activates ccRCC angiogenesis and development.
The two novel VHL substrate factors, SFMBT1 and
ZHX2, appear to have the same mechanism of action; however, they
may activate different signaling pathways in ccRCC cell lines.
Further studies are required to confirm the prognostic value of
combining the transcription factors in RCC. To the best of our
knowledge, the present study was the first to investigate the
co-expression level of these two genes in ccRCC samples and assess
their association with prognostic outcomes. In the present study,
survival analysis demonstrated that combined high expression levels
of SFBMT1 and ZHX2 were associated with poor clinical outcomes in
patients with ccRCC. However, larger sample sizes and multicenter
data are required to verify the results presented here.
In conclusion, SHZH appears to be a promising
prognostic predictor in patients with ccRCC, and the result of the
present study provide novel insight into advanced ccRCC treatment
and follow-up.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YFC and LSZ drafted the initial manuscript. YFC and
LSZ conceived the present study and analyzed and interpreted the
data. YFC, LSZ, SX and JS collected the data and prepared the
figures and tables. CFH and QCZ performed the experiments and
designed and developed the database. YFC and QCZ confirmed the
authenticity of all the raw data. QCZ critically revised the
manuscript for important intellectual content. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical Review
Boards of Putuo Hospital (Shanghai, China; approval no. 20200130),
and written informed consent was provided by all patients prior to
the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Escudier B, Porta C, Schmidinger M,
Rioux-Leclercq N, Bex A, Khoo V, Grünwald V, Gillessen S and
Horwich A; ESMO Guidelines Committee. Electronic address, :
clinicalguidelines@esmo.org: Renal cell carcinoma: ESMO Clinical
Practice Guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 30:706–720. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of clear cell renal cell
carcinoma. Nature. 499:43–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaelin WG Jr: Molecular basis of the VHL
hereditary cancer syndrome. Nat Rev Cancer. 2:673–682. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Escudier B, Szczylik C, Porta C and Gore
M: Treatment selection in metastatic renal cell carcinoma: Expert
consensus. Nat Rev Clin Oncol. 9:327–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harshman LC, Xie W, Bjarnason GA, Knox JJ,
MacKenzie M, Wood L, Srinivas S, Vaishampayan UN, Tan MH, Rha SY,
et al: Conditional survival of patients with metastatic renal-cell
carcinoma treated with VEGF-targeted therapy: A population-based
study. Lancet Oncol. 13:927–935. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ficarra V, Novara G, Galfano A, Brunelli
M, Cavalleri S, Martignoni G and Artibani W: The ‘stage, size,
grade and necrosis’ score is more accurate than the University of
California los angeles integrated staging system for predicting
cancer-specific survival in patients with clear cell renal cell
carcinoma. BJU Int. 103:165–170. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ko JJ, Xie W, Kroeger N, Lee JL, Rini BI,
Knox JJ, Bjarnason GA, Srinivas S, Pal SK, Yuasa T, et al: The
international metastatic renal cell Carcinoma database consortium
model as a prognostic tool in patients with metastatic renal cell
carcinoma previously treated with first-line targeted therapy: A
population-based study. Lancet Oncol. 16:293–300. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu L, Wang J, Kong W, Huang J, Dong B,
Huang Y, Xue W and Zhang J: LSD1 inhibition suppresses the growth
of clear cell renal cell carcinoma via upregulating P21 signaling.
Acta Pharm Sin B. 9:324–334. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liao Y, Xiao H, Cheng M and Fan X:
Bioinformatics analysis reveals biomarkers with cancer stem cell
characteristics in lung squamous cell carcinoma. Front Genet.
11:4272020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liao Y, Wang Y, Cheng M, Huang C and Fan
X: Weighted gene coexpression network analysis of features that
control cancer stem cells reveals prognostic biomarkers in lung
adenocarcinoma. Front Genet. 11:3112020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu L, Ding R, Zhang J, Zhang J and Lin Z:
Cyclin-dependent kinase 5 acts as a promising biomarker in clear
cell renal cell carcinoma. BMC Cancer. 19:6982019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ivan M, Kondo K, Yang H, Kim W, Valiando
J, Ohh M, Salic A, Asara JM, Lane WS and Kaelin WG Jr: HIFalpha
targeted for VHL-mediated destruction by proline hydroxylation:
Implications for O2 sensing. Science. 292:464–468. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hon WC, Wilson MI, Harlos K, Claridge TD,
Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI and
Jones EY: Structural basis for the recognition of hydroxyproline in
HIF-1 alpha by pVHL. Nature. 417:975–978. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho H, Du X, Rizzi JP, Liberzon E,
Chakraborty AA, Gao W, Carvo I, Signoretti S, Bruick RK, Josey JA,
et al: On-target efficacy of a HIF-2α antagonist in preclinical
kidney cancer models. Nature. 539:107–111. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang J, Wu T, Simon J, Takada M, Saito R,
Fan C, Liu XD, Jonasch E, Xie L, Chen X, et al: VHL substrate
transcription factor ZHX2 as an oncogenic driver in clear cell
renal cell carcinoma. Science. 361:290–295. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu L, Ding R, Yan H, Zhang J and Lin Z:
ZHX2 drives cell growth and migration via activating MEK/ERK signal
and induces sunitinib resistance by regulating the autophagy in
clear cell renal cell carcinoma. Cell Death Dis. 11:3372020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu X, Simon JM, Xie H, Hu L, Wang J,
Zurlo G, Fan C, Ptacek TS, Herring L, Tan X, et al: Genome-wide
screening identifies SFMBT1 as an oncogenic driver in cancer with
VHL loss. Mol Cell. 77:1294–1306.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Matos LL, Trufelli DC, de Matos MG and da
Silva Pinhal MA: Immunohistochemistry as an important tool in
biomarkers detection and clinical practice. Biomark Insights.
5:9–20. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fuhrman SA, Lasky LC and Limas C:
Prognostic significance of morphologic parameters in renal cell
carcinoma. Am J Surg Pathol. 6:655–663. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bonasio R, Lecona E and Reinberg D: MBT
domain proteins in development and disease. Semin Cell Dev Biol.
21:221–230. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang M, Shen H, Jin Y, Lin T, Cai Q,
Pinard MA, Biswas S, Tran Q, Li G, Shenoy AK, et al: The malignant
brain tumor (MBT) domain protein SFMBT1 is an integral histone
reader subunit of the LSD1 demethylase complex for chromatin
association and epithelial-to-mesenchymal transition. J Biol Chem.
288:27680–27691. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kawata H, Yamada K, Shou Z, Mizutani T,
Yazawa T, Yoshino M, Sekiguchi T, Kajitani T and Miyamoto K:
Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX
family, functions as a transcriptional repressor. Biochem J.
373:747–757. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lv Z, Zhang M, Bi J, Xu F, Hu S and Wen J:
Promoter hypermethylation of a novel gene, ZHX2, in hepatocellular
carcinoma. Am J Clin Pathol. 125:740–746. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagel S, Schneider B, Meyer C, Kaufmann M,
Drexler HG and Macleod RA: Transcriptional deregulation of homeobox
gene ZHX2 in Hodgkin lymphoma. Leuk Res. 36:646–655. 2012.
View Article : Google Scholar : PubMed/NCBI
|