Introduction
Vulvar squamous cell carcinoma (VSCC) constitutes
90% of all vulvar malignancies, and its incidence has risen over
the past decades (1,2). Approximately 25% of VSCCs arise in
association with a human papillomavirus (HPV)-infection, via the
precursor lesion, high grade squamous intraepithelial lesion (HSIL)
(3). The majority (75%) of VSCCs,
however, is postulated to develop on the background of chronic
dermatoses, via the precursor lesion, differentiated vulvar
intraepithelial neoplasia (dVIN) (3).
The dual pathogenesis of VSCC has been recognized
several years ago, however, molecular mechanisms of the
carcinogenesis have not been well characterized (4). This is largely because the genomic
profiles of VSCC or its precursor lesions have been investigated in
only a few studies so far (1,5–11). These studies identified somatic
mutations of TP53 to be the pivotal oncogenic driver of
HPV-independent VSCC, and also detected genomic alterations of
PIK3CA, HRAS, or FGFR3 in both subtypes of VSCC
(7–9,11).
Nevertheless, limited sample sizes and dissimilar methodologies of
these studies have prevented significant advancement of knowledge
of VSCC carcinogenesis (4).
A better understanding of the molecular pathways
involved in VSCC carcinogenesis can enable identification of
biomarkers that may be used to improve the diagnosis, for
prognostic stratification, or as targets for precision treatment.
Currently, the mainstay of VSCC treatment is surgical excision,
which is often associated with post-operative morbidities due to
the anatomical complexity of the vulvar region. Discovering novel
biomarkers for targeted treatment may help improve personalization
of treatment for patients with VSCC.
A key method for discovering candidate biomarkers is
through identifying genes that are differentially expressed in
cancer tissue and normal tissue (12). To this end, we analyzed datasets of
gene expression microarray on VSCC and normal vulvar tissue, from
two independent studies, using the latest bioinformatics tools. We
further investigated the expression of some of the differentially
expressed genes (DEGs) identified thereby, by performing
immunohistochemistry (IHC) on VSCC, HSIL, dVIN, and normal vulvar
tissue.
Materials and methods
Identification and analysis of
datasets
A publicly available dataset (GSE38228) was
identified and downloaded from gene expression omnibus (GEO)
(13). This dataset consists of
VSCCs (n=14) and normal vulvar tissues (n=5), for which gene
expression microarray had been performed using the gene-chip
platform Illumina HumanHT-12 V4.0. A 2nd dataset was obtained from
a study previously conducted by researchers at our center. This
dataset consists of VSCCs (n=5), for which gene expression
microarray was performed using the gene-chip platform Affymetrix HG
U133 Plus 2.0.
The datasets were imported into OmniViz (version
6.1.13.0, BioWisdomLtd.). Statistical analysis of microarrays (SAM)
was performed to identify DEGs using the following cutoff-values-a
false discovery rate (FDR) of ≤0.01 and a fold change of 1.5.
P-value <0.05 was considered as statistically significant.
Functional annotations of the SAM results were done using Ingenuity
Pathway Analysis (IPA, Qiagen, Inc.). Expression levels of p16
(CDKN2A), which is known to be overexpressed in HPV-related
VSCC, were used to distinguish the samples as HPV-related or
HPV-independent VSCC. For both subtypes of VSCC, DEGs that were
upregulated or downregulated in both datasets with statistical
significance were identified. The Database for Annotation,
Visualization and Integrated Discovery (DAVID; version 6.8) was
used to identify the most significantly enriched functional genes
(14,15). Gene ontology (GO) enrichment analyses
were performed using the DAVID online tool to annotate biological
process, cellular component, and molecular function of DEGs.
Additional information on the DEGs was obtained from IPA,
cBioPortal, and Gene Expression Profiling Interactive Analysis
(GEPIA). Protein-protein interaction (PPI) networks of the DEGs
were constructed using Search Tool for the Retrieval of Interacting
Genes (STRING) (16–19).
Immunohistochemistry (IHC)
Formalin fixed paraffin embedded (FFPE)-tissues of
VSCC, HSIL, dVIN, and normal vulva were retrieved from the archives
of Department of Pathology, Erasmus MC. Histology of all tissues
was reviewed by two pathologists (SDG and PCEG). Patient data were
anonymized and patient materials were handled following the
guidelines of World Medical Association Declaration of
Helsinki.
For performing IHC, DEGs were selected-i) that were
expressed in the cytoplasm or nucleus and ii) for which primary
antibodies were commercially available. In addition, for all
samples, IHC was performed with p16 to determine the HPV-status,
and with p53 to confirm the histological diagnoses. Sequential
sections of 4 µm-thickness were prepared from the FFPE-tissues and
automated IHC was performed using the Ventana Benchmark ULTRA
(Ventana Medical Systems Inc.), following the manufacturer's
instructions (Data S1 and Table SI).
The IHC markers were scored as follows: For the IHC
markers of DEGs, the percentage of cells showing staining,
irrespective of the intensity of staining, was assessed manually.
In addition, the intensity of staining (weak, moderate, and strong)
and the distribution of staining within the epithelium was
recorded. p16-expression patterns were scored as block-type or
non-block-type (patchy), following the guidelines of Lower
Anogenital Squamous Terminology Standardization Project (LAST)
(16). Block-type p16-expression,
i.e. diffuse, continuous, moderate-to-intense nuclear and/or
cytoplasmic staining in ≥1/3rd of the epithelial thickness is
considered to be a reliable surrogate marker of high-risk
HPV-infection (20). p53-expression
patterns were scored as p53-mutant or p53-wild-type following
descriptions in recent literature (10,21).
p53-mutant patterns have been reported to accurately reflect the
presence of TP53 mutations (10). p53-mutant patterns include basal to
para-basal/diffuse overexpression, basal overexpression, or
aberrant negative/null-pattern. p53-wild-type patterns include
scattered heterogeneous basal and/or para-basal expression, and
scattered mid-epithelial expression with basal sparing. The latter
p53-wild-type pattern is associated with HPV-related lesions
(10,22).
Ethics statement
This study was conducted in accordance with the
guidelines of the Dutch Federation of Biomedical Scientific
Societies (www.federa.org/codes-conduct), which state that no
separate ethical approval is required for the use of anonymized
residual tissue procured during regular treatment.
Results
Dataset analyses
From GSE38228, 3 samples were identified as
HPV-related VSCC and 3 samples were identified as HPV-independent
VSCC. A total of 342 genes (244 upregulated and 98 downregulated)
were found to be differentially expressed with statistical
significance only in HPV-related VSCC. A total of 382 genes (203
upregulated and 179 downregulated) were found to be differentially
expressed with statistical significance only in HPV-independent
VSCC. From the 2nd dataset, 3 samples were identified as
HPV-related VSCC and 2 samples as HPV-independent VSCC. A total of
7005 genes were differentially expressed with statistical
significance in HPV-related VSCC, and 4,283 genes were
differentially expressed with statistical significance in
HPV-independent VSCC.
Combining both datasets, for HPV-related VSCC, 88
DEGs were identified that were similarly regulated with statistical
significance. This comprised 69 upregulated and 19 downregulated
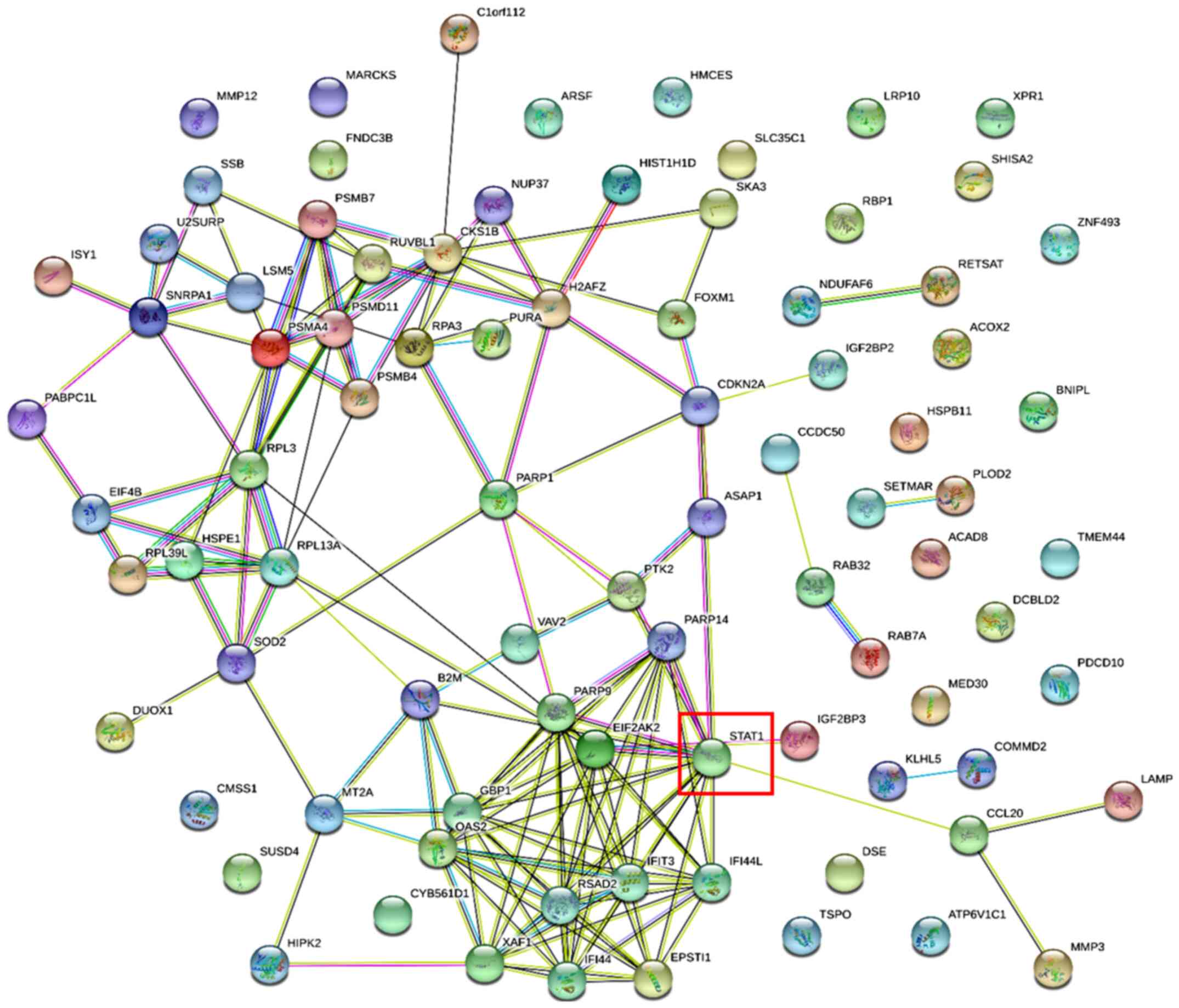
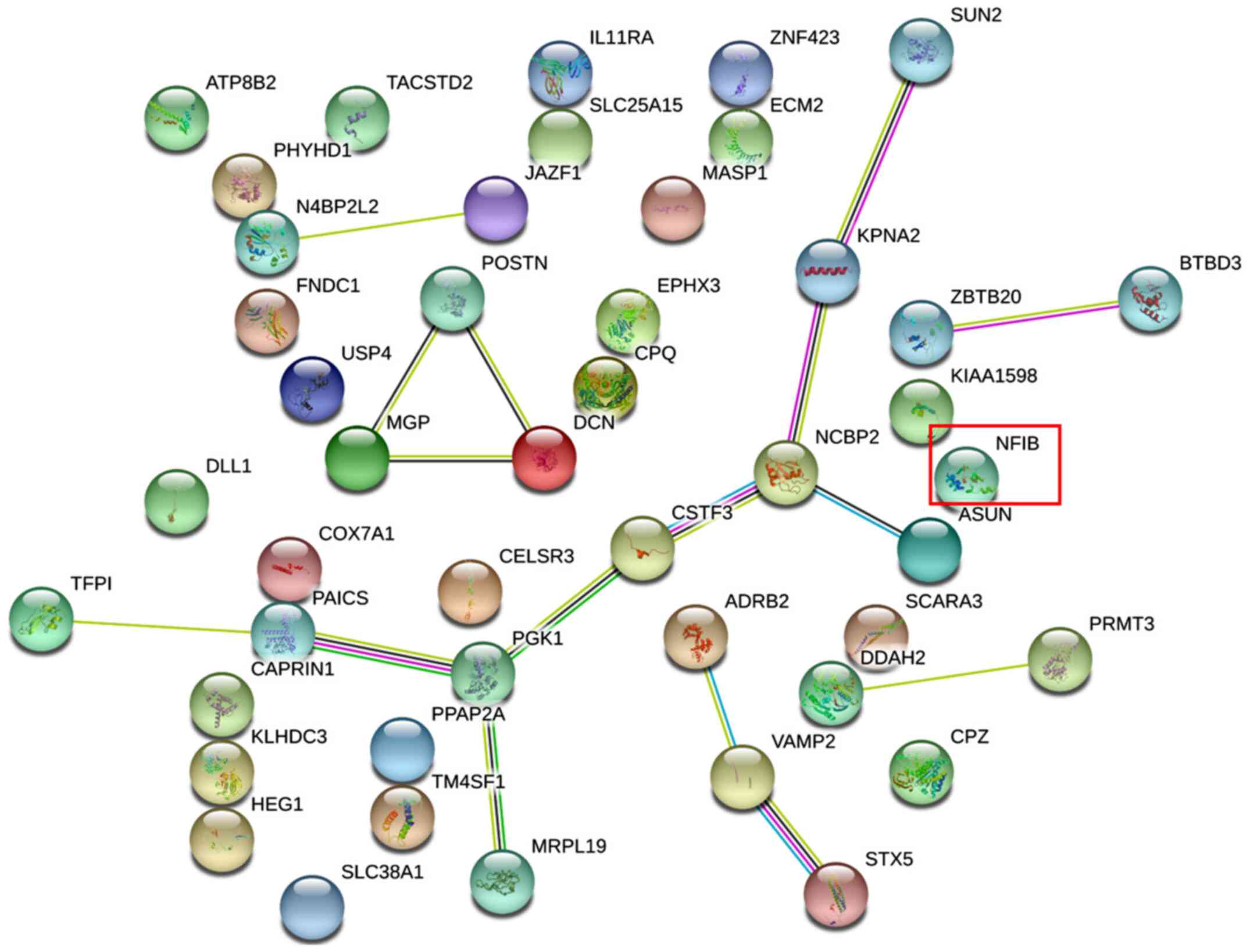
DEGs; signal transducer and activator of transcription 1 (STAT1)
was one of the upregulated DEGs. For HPV-independent VSCC, 46 DEGs
were identified that were similarly regulated with statistical
significance. This comprised 16 upregulated and 30 downregulated
DEGs; nuclear factor IB (NFIB) was one of the downregulated DEGs.
The PPI networks of these DEGs are visualized in Figs. 1 and 2, and the DEGs along with their subcellular
locations, functions, and related canonical pathways are listed in
Tables SII and SIII.
The DEGs identified for HPV-related VSCC mainly
participate in response to stimulus and regulation of cellular and
biological processes (Table I). As
for the molecular function, these DEGs are mainly involved in
binding with ions or signaling receptors (Table II). The cellular component of these
DEGs include cytoplasm and extracellular region.
 | Table I.GO enrichment analysis of the
differentially expressed genes for human papilloma virus-related
vulvar squamous cell carcinoma. |
Table I.
GO enrichment analysis of the
differentially expressed genes for human papilloma virus-related
vulvar squamous cell carcinoma.
| A, Biological
processes |
|---|
|
|---|
| Term | Description | Gene count | P-value |
|---|
| GO:0050896 | Response to
stimulus | 35 |
2.89×10−10 |
| GO:0050794 | Regulation of
cellular process | 34 |
2.92×10−05 |
| GO:0050789 | Regulation of
biological process | 34 |
1.19×10−04 |
| GO:0032501 | Multicellular
organismal process | 33 |
6.85×10−11 |
| GO:0007275 | Multicellular
organism development | 26 |
1.39×10−08 |
| GO:0048856 | Anatomical structure
development | 26 |
7.60×10−08 |
| GO:0042221 | Response to
chemical | 25 |
8.56×10−09 |
| GO:0007154 | Cell
communication | 24 |
1.09×10−05 |
| GO:0006950 | Response to
stress | 23 |
8.86×10−09 |
| GO:0007165 | Signal
transduction | 23 |
6.01×10−06 |
|
| B, Molecular
functions |
|
| Term |
Description | Gene
count | P-value |
|
| GO:0043167 | Ion binding | 29 |
2.59×10−08 |
| GO:0005102 | Signaling receptor
binding | 21 |
2.35×10−13 |
| GO:0097367 | Carbohydrate
derivative binding | 19 |
4.81×10−09 |
| GO:0043169 | Integrin
binding | 19 |
1.11×10−04 |
| GO:0098772 | Molecular function
regulator | 18 |
6.66×10−05 |
| GO:0008201 | Heparin
binding | 15 |
1.60×10−21 |
| GO:1901681 | Sulfur compound
binding | 15 |
9.93×10−19 |
| GO:0030545 | Receptor regulator
activity | 12 |
2.24×10−10 |
| GO:0005198 | Structural molecule
activity | 9 |
5.41×10−06 |
| GO:0005509 | Growth factor
binding | 8 |
5.60×10−05 |
|
| C, Cellular
component |
|
| Term |
Description | Gene
count | P-value |
|
| GO:0005737 | Cytoplasm | 33 |
4.88×10−04 |
| GO:0005576 | Extracellular
region | 30 |
1.54×10−13 |
| GO:0012505 | Endomembrane
system | 24 |
1.73×10−07 |
| GO:0031982 | Vesicle | 20 |
5.59×10−06 |
| GO:0043230 | Extracellular
organelle | 14 |
1.66×10−05 |
| GO:0031410 | Cytoplasmic
vesicle | 14 |
8.38×10−05 |
| GO:0097708 | Intracellular
vesicle | 14 |
8.57×10−05 |
| GO:0062023 | Collagen-containing
extracellular Matrix | 13 |
6.10×10−13 |
| GO:0070062 | Extracellular
exosome | 13 |
7.18×10−05 |
| GO:1903561 | Extracellular
vesicle | 13 |
7.96×10−05 |
 | Table II.Functional annotation analysis of the
differentially expressed genes for human papilloma virus-related
vulvar squamous cell carcinoma. |
Table II.
Functional annotation analysis of the
differentially expressed genes for human papilloma virus-related
vulvar squamous cell carcinoma.
| Functional
annotation cluster | Gene count | P-value |
|---|
| Immunity | 8 |
5×10−06 |
| Antiviral
defense | 7 |
9×10−09 |
| Defense response to
virus | 7 |
2×10−07 |
| Host-virus
interaction | 6 |
2×10−05 |
| Protease | 6 |
8×10−05 |
| Type I interferon
signaling | 5 |
3×10−07 |
| Innate
immunity | 5 |
2×10−05 |
| Perinuclear region
of cytoplasm | 5 |
3×10−05 |
| Antigen processing
and presentation of exogenous peptide antigen via MHC class I,
TAP-dependent | 5 |
2×10−07 |
| Tumor necrosis
factor-mediated signaling | 5 |
3×10−06 |
The DEGs identified for HPV-independent VSCC mainly
participate in regulation of cellular and metabolic processes
(Table III). As for the molecular
function, these DEGs are mainly involved in protein and ion binding
(Table IV). The cellular component
of these DEGs include membrane-bound organelles and the
cytoplasm.
 | Table III.GO enrichment analysis of
differentially expressed genes for human papilloma
virus-independent vulvar squamous cell carcinoma. |
Table III.
GO enrichment analysis of
differentially expressed genes for human papilloma
virus-independent vulvar squamous cell carcinoma.
| A, Biological
processes |
|---|
|
|---|
| Term | Description | Gene count | P-value |
|---|
| GO:0050794 | Regulation of
cellular process | 33 |
5.27×10−12 |
| GO:0008152 | Metabolic
process | 31 |
1.02×10−11 |
| GO:0016043 | Cellular component
organization | 25 |
7.37×10−09 |
| GO:0032502 | Developmental
process | 22 |
3.69×10−11 |
| GO:0032501 | Multicellular
organismal process | 19 |
4.55×10−11 |
| GO:0065008 | Regulation of
biological quality | 19 |
6.93×10−12 |
| GO:0007275 | Multicellular
organism development | 19 |
7.01×10−08 |
| GO:0007154 | Cell
communication | 18 |
9.79×10−06 |
| GO:0030154 | Cell
differentiation | 18 |
1.04×10−06 |
| GO:0042221 | Transport | 17 |
8.87×10−10 |
|
| B, Molecular
functions |
|
| Term |
Description | Gene
count | P-value |
|
| GO:0005515 | Protein
binding | 38 |
5.43×10−07 |
| GO:0043167 | Ion binding | 20 |
7.70×10−10 |
| GO:0003824 | Catalytic
activity | 13 |
1.00×10−11 |
| GO:0097159 | Organic cyclic
compound binding | 12 |
7.47×10−09 |
| GO:1901363 | Heterocyclic
compound binding | 11 |
6.24×10−09 |
| GO:0003676 | Nucleic acid
binding | 8 |
8.53×10−05 |
| GO:0016787 | Hydrolase
activity | 7 |
5.01×10−12 |
| GO:0043168 | Anion binding | 7 |
8.30×10−10 |
| GO:0098772 | Molecular function
regulator | 7 |
7.05×10−06 |
| GO:0097367 | Carbohydrate
derivative binding | 6 |
6.35×10−05 |
|
| C, Cellular
component |
|
| Term |
Description | Gene
count | P-value |
|
| GO:0043227 | Membrane-bounded
organelle | 36 |
2.20×10−09 |
| GO:0005737 | Cytoplasm | 31 |
1.82×10−09 |
| GO:0016020 | Cell membrane | 26 |
2.41×10−11 |
| GO:0005576 | Extracellular
region | 20 |
7.74×10−05 |
| GO:0031224 | Intrinsic component
of membrane | 20 |
3.26×10−06 |
| GO:0005634 | Nucleus | 18 |
7.60×10−11 |
| GO:0031982 | Vesicle | 14 |
5.76×10−05 |
| GO:0071944 | Cell periphery | 14 |
7.46×10−07 |
| GO:0043233 | Organelle
lumen | 14 |
8.72×10−12 |
| GO:0012505 | Endomembrane
system | 13 |
3.75×10−10 |
 | Table IV.Functional annotation analysis of
differentially expressed genes for human papilloma
virus-independent vulvar squamous cell carcinoma. |
Table IV.
Functional annotation analysis of
differentially expressed genes for human papilloma
virus-independent vulvar squamous cell carcinoma.
| Functional
annotation cluster | Gene count | P-value |
|---|
| Developmental
protein | 21 |
5×10−07 |
| Calcium ion
binding | 16 |
2×10−05 |
| EGF-like
domain | 12 |
1×10−04 |
| EGF-like
calcium-binding, conserved site | 11 |
7×10−06 |
| Cell-cell
adhesion | 7 |
2×10−04 |
| Integral component
of membrane | 6 |
5×10−05 |
| Cell
differentiation | 5 |
1×10−04 |
| Acetylation | 4 |
1×10−05 |
| Extracellular
matrix organization | 3 |
2×10−05 |
| Transmembrane
helix | 3 |
6×10−04 |
Immunohistochemistry
Primary antibodies for performing IHC were
commercially available for i) STAT1, one of the upregulated DEGs,
and ii) NFIB, one of the downregulated DEGs. IHC was performed on
11 VSCCs, 6 dVINs, 6 HSILs, and 7 normal vulva tissues; these were
from women with a median age of 72.5 years (range, 26–90 years).
Immunohistochemical expression of p53, p16, NFIB, and STAT1 are
presented in Tables V and VI. For NFIB and STAT1, the IHC patterns
observed in the tissues are described below, and the distribution
of expression is depicted in Fig.
S1.
 | Table V.Immonohistochemical expression
patterns of p53 and p16. |
Table V.
Immonohistochemical expression
patterns of p53 and p16.
|
| Diagnosis |
|---|
|
|
|
|---|
| Marker and
expression pattern | Normal vulvar
tissue (n=7) | HSIL (n=6) | HPV-related VSCC
(n=5) | dVIN (n=6) | HPV-independent
VSCC (n=6) |
|---|
| p53-mut |
|
|
|
|
|
|
Parabasal/diffuse | 0 (0) | 0 (0) | 1 (20) | 5 (83) | 3 (50) |
|
Basal | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
|
Absent/null | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (33) |
| p53-wt |
|
|
|
|
|
|
Wild-type scattered | 7 (100) | 0 (0) | 0 (0) | 1 (17) | 1 (17) |
|
Wild-type mid-epithelial | 0 (0) | 6 (100) | 4 (80) | 0 (0) | 0 (0) |
| p16 |
|
|
|
|
|
|
Block-type | 0 (0) | 6 (100) | 5 (100) | 0 (0) | 0 (0) |
|
Non-block-type/patchy | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 0 (0) |
| No
expression | 7 (100) | 0 (0) | 0 (0) | 5 (83) | 6 (100) |
 | Table VI.Immunohistochemical expression of
NFIB and STAT1. |
Table VI.
Immunohistochemical expression of
NFIB and STAT1.
|
| Diagnosis |
|---|
|
|
|
|---|
| Immunohistochemical
marker | Normal vulvar
tissue (n=7) | HSIL (n=6) | HPV-related VSCC
(n=5) | dVIN (n=6) | HPV-independent
VSCC (n=6) |
|---|
| NFIB | 18 (10.3–23.1) | 12.5
(9.1–17.6) | 5 (1.1–9.2) | 6 (4.3–10.1) | 2.5 (0.9–11.3) |
| STAT1 | 65 (50.8–87.5) | 67.5
(38.8–81.2) | 80 (68.5–91.5) | 85 (69.1–92.5) | 90 (53.1–95.6) |
STAT1
Normal vulvar tissue (n=7): Five showed diffuse,
cytoplasmic STAT1-expression of moderate-to-strong intensity,
across full epithelial thickness; two showed focal STAT-1
expression of moderate-to-strong intensity.
dVIN (n=6), HSIL (n=6), HPV-related VSCC (n=5),
HPV-independent VSCC (n=6): All showed diffuse, cytoplasmic
STAT1-expression of moderate-to-strong intensity, across full
epithelial thickness.
NFIB
Normal vulvar tissue (n=7): All showed strong,
diffuse, nuclear NFIB-expression, predominantly along the basal
layers, which occasionally extended to the para-basal layers.
HSIL (n=6): All showed strong nuclear
NFIB-expression along the basal layers and occasionally in the
para-basal layers. Staining in the basal layer was discontinuous,
and expression in the para-basal layers was primarily seen only at
the tips of rete ridges.
HPV-related VSCC (n=5): Two were completely
negative, and 2 were predominantly negative, showing only focal,
weak, nuclear NFIB-expression along the periphery of the tumor cell
nests. One VSCC showed NFIB-expression of moderate intensity along
the periphery of the tumor cell nests.
dVIN (n=6): One dVIN was completely negative and 5
showed only focal, weak, nuclear NFIB-expression.
HPV-independent VSCC (n=6): One was completely
negative, and 5 were predominantly negative, showing only focal,
weak, nuclear NFIB-expression along the periphery of the tumor cell
nests.
Immunohistochemical expressions of p53, p16, STAT1,
and NFIB in normal vulvar tissue, HSIL, dVIN, and VSCC (both
subtypes) are demonstrated in Figs.
3–7.
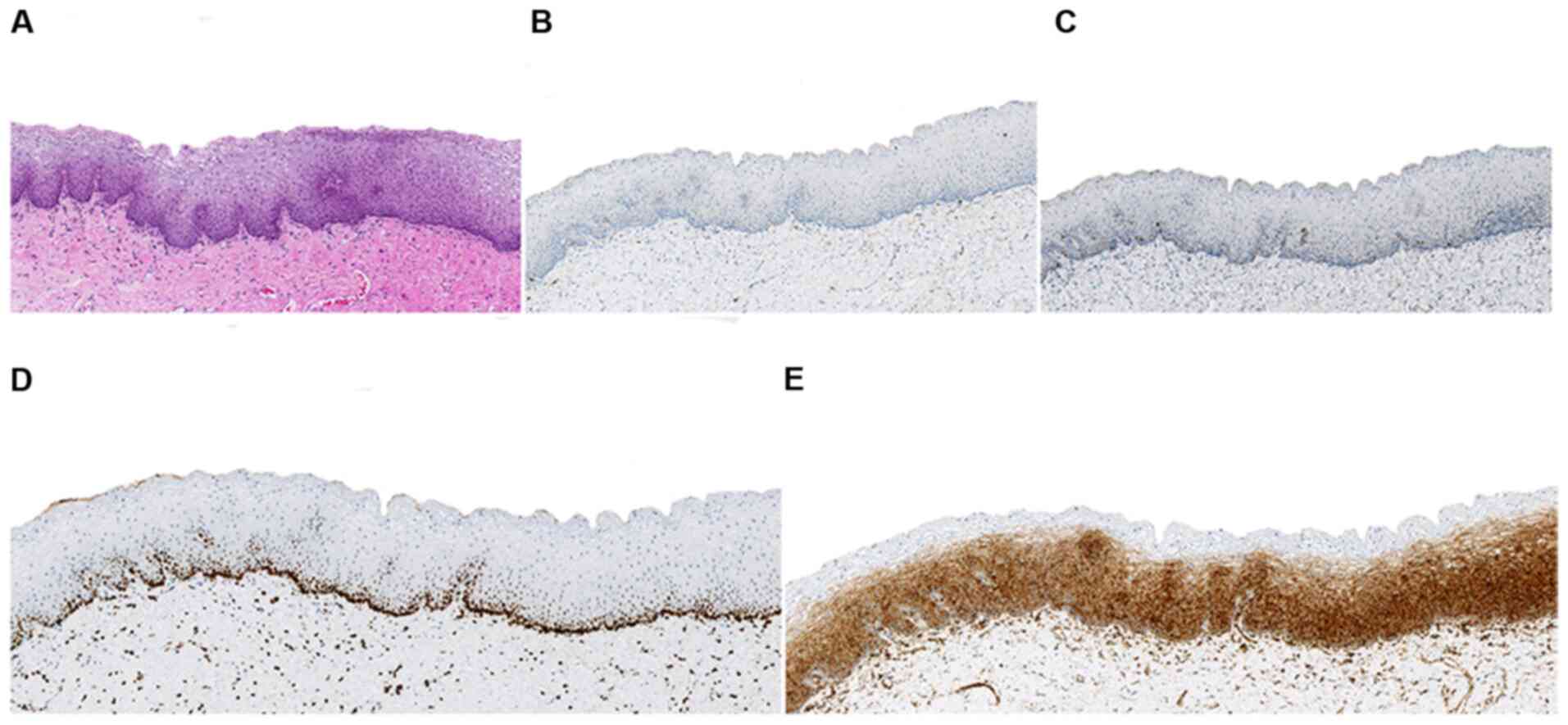 | Figure 3.Normal vulvar tissue histology and
IHC. (A) Histological appearance (hematoxylin and eosin stain). (B)
p16-IHC was negative. (C) p53-IHC revealed wild-type expression.
(D) NFIB-IHC exhibited strong, diffuse, nuclear expression,
predominantly along the basal layers and occasionally in the
para-basal layers. (E) signal transducer and activator of
transcription 1-IHC demonstrated diffuse, cytoplasmic expression of
moderate-to-strong intensity, across full epithelial thickness
(A-C, magnification, ×100; D and E, magnification, ×200). IHC,
immunohistochemistry. |
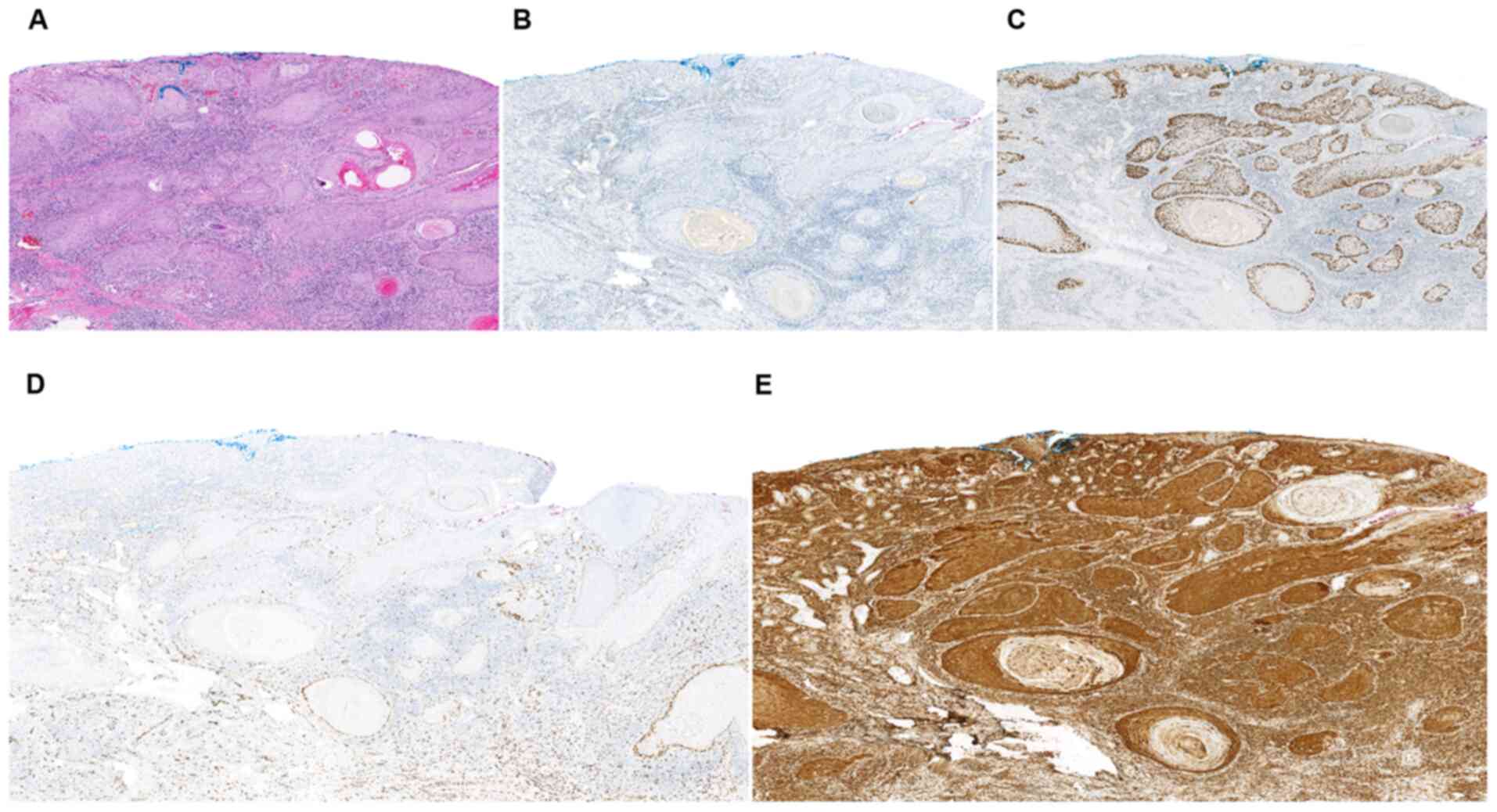 | Figure 7.Human papillomavirus independent
vulvar squamous cell carcinoma: Histology and IHC. (A) Histological
appearance (hematoxylin and eosin staining). (B) p16-IHC was
completely negative. (C) p53-IHC demonstrated a mutation-pattern.
(D) NFIB-IHC was negative in certain tumor nests, demonstrating
focal, weak, nuclear expression along the periphery of some of the
tumor nests. (E) STAT1-IHC revealed diffuse, cytoplasmic expression
of moderate-to-strong intensity, across full epithelial thickness
(A-C, magnification, ×100; D and E, magnification, ×200). IHC,
immunohistochemistry; NFIB, nuclear factor IB; STAT1, signal
transducer and activator of transcription 1. |
Discussion
In this study, we utilized bioinformatics tools to
gain insight into VSCC carcinogenesis, and to identify potential
biomarkers that may have diagnostic, prognostic, or therapeutic
applications. For both subtypes of VSCC (i.e., HPV-related and
HPV-independent) we identified a set of DEGs that appeared to be
similarly regulated (up or down) in two independent gene expression
microarray datasets. We found that the majority of DEGs that were
identified for HPV-related VSCC are involved in the immune
response, whereas those identified for HPV-independent VSCC were
involved in second messenger signaling-this provides support for
the dual pathogenesis of VSCC.
We studied the expression of two of the DEGs, i.e.
NFIB and STAT1, that were found to be similarly regulated in both
datasets, in whole tissue sections of VSCCs, dVINs, HSILs, and
normal vulvar tissues, by performing IHC. NFIB was identified to be
downregulated in HPV-independent VSCC, and STAT1 was identified to
be upregulated in HPV-related VSCC. Neither of these markers has
been previously studied for VSCC or its precursor lesions.
NFIB showed strong, nuclear expression in the basal
and para-basal epithelial layers in normal vulvar tissue, whereas,
in dVIN and both subtypes of VSCC, NFIB was either completely
negative or minimally expressed. NFIB expression was also reduced
in HSIL in comparison with normal vulvar tissue, but to a lesser
extent than that in dVIN and VSCC.
NFIB is a transcription factor which has tumor
suppressive, as well as, oncogenic potential (23). In cervical SCC and head-and-neck SCC
(HNSCC), NFIB-expression has been observed to be lower than in
normal tissues from the corresponding sites (23). Furthermore, lower levels of
NFIB-expression have been reported to correlate significantly with
worse prognosis for both of these malignancies (23). Interestingly, NFIB is a key regulator
of the aryl hydrocarbon pathway, which we previously identified to
be involved in HPV-independent VSCC (2). In addition, high-confidence proximity
interactions have been reported between NFIB and SOX2 (24); SOX2 is a cancer-stemness related
transcription factor that is overexpressed in dVIN and VSCC
(25). In view of these
observations, we believe that the role of NFIB in VSCC and its
potential as a therapeutic target deserve further investigation. In
addition to SCCs, genomic alterations of NFIB have been detected in
several other malignancies, as shown in Fig. S2.
Unlike NFIB, no discernable difference was observed
in immunohistochemical expression of STAT1 between normal vulvar
tissue, dVIN, HSIL, or VSCC (both subtypes). For all tissue types,
diffuse, cytoplasmic STAT1-expression of moderate-to-strong
intensity was noted across full epithelial thickness. STAT1 is a
component of the Janus kinase (JAK)-STAT signaling pathway, and can
act as an antimicrobial mediator, a tumor suppressor, or a promotor
of tumor progression (26). Aberrant
expression of STAT1 in HPV-related lesions is considered to reflect
activation of the JAK-STAT pathway as a consequence of the
inflammatory response induced by HPV (26).
Our results regarding IHC-expression of STAT1 were
in contrast to those of a recent study, which reported a higher
STAT1-expression in cervical intraepithelial neoplasia (CIN) than
in normal cervical epithelium, and deduced an association of
increased STAT1-expression with malignant progression of CIN
(26). Since STAT-1 expression is
regulated by a complex network of interferons, we speculate that
the difference in expression between vulvar and cervical tissue
could be ascribed to the dissimilar microenvironments of these
anatomical sites. Similarly to NFIB, genomic alterations of STAT1
have been detected in several malignancies, as shown in Fig. S3.
This study was an attempt to leverage bioinformatics
to identify DEGs in VSCC. We identified NFIB as a downregulated
gene in VSCC, and observed that its immunohistochemical expression
was reduced in both subtypes of VSCC. Hence, we believe that the
relevance of NFIB as a diagnostic/prognostic biomarker deserves
further exploration. However, an apparent limitation of this study
is that the DEGs were identified from datasets consisting of small
sample sizes, and IHC was also performed on a limited set of
tissues. Further experiments are needed to confirm the function of
these DEGs in VSCC and to validate their immunohistochemical
expression in vulvar tissues.
Nevertheless, we hope that our results will
instigate further research into VSCC carcinogenesis and pave the
path for unravelling novel biomarkers of VSCC and its precursor
lesions.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Leen J. Blok
(Erasmus MC) for the use of data on gene expression analysis.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SD, PCEG, TPPVDB, SMAS, LAMS, PJVDS, SK and FJVK
conceived and designed the methodology of the current study. SMAS,
PJVDS analyzed the data. SD, SMAS, PJVDS and FJVK confirm the
authenticity of all the raw data. TPPVDB and LAMS performed the
experiments. SD, PCEG, TPPVDB, SS, LAMS, HCVD and PJVDS curated the
pathological and clinical data. SD drafted the manuscript,
constructed the graphs and arranged the histology figures. PCEG, SK
and FJVK supervised the current study. All authors reviewed and
edited the manuscript and read and approved the final
manuscript.
Ethics approval and consent to
participate
The current study was conducted in accordance with
the guidelines of the Dutch Federation of Biomedical Scientific
Societies (www.federa.org/codes-conduct), which state that no
separate ethical approval is required for the use of anonymized
residual tissue procured during regular treatment. This is also
part of the standard treatment agreement at Erasmus MC.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Prieske K, Alawi M, Oliveira-Ferrer L,
Jaeger A, Eylmann K, Burandt E, Schmalfeldt B, Joosse SA and
Woelber L: Genomic characterization of vulvar squamous cell
carcinoma. Gynecol Oncol. 158:547–554. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dasgupta S, Ewing-Graham PC, Swagemakers
SMA, van der Spek PJ, van Doorn HC, Noordhoek Hegt V, Koljenović S
and van Kemenade FJ: Precursor lesions of vulvar squamous cell
carcinoma-histology and biomarkers: A systematic review. Crit Rev
Oncol Hematol. 147:1028662020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Martel C, Georges D, Bray F, Ferlay J
and Clifford GM: Global burden of cancer attributable to infections
in 2018: A worldwide incidence analysis. Lancet Glob Health.
8:e180–e190. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zeimet AG: Molecular characterization of
vulvar squamous cell cancer: High time to gain ground. Gynecol
Oncol. 158:519–520. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han MR, Shin S, Park HC, Kim MS, Lee SH,
Jung SH, Song SY, Lee SH and Chung YJ: Mutational signatures and
chromosome alteration profiles of squamous cell carcinomas of the
vulva. Exp Mol Med. 50:e4422018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weberpals JI, Lo B, Duciaume MM, Spaans
JN, Clancy AA, Dimitroulakos J, Goss GD and Sekhon HS: Vulvar
squamous cell carcinoma (VSCC) as two diseases: HPV status
identifies distinct mutational profiles including oncogenic
fibroblast growth factor receptor 3. Clin Cancer Res. 23:4501–4510.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nooij LS, Ter Haar NT, Ruano D, Rakislova
N, van Wezel T, Smit VTHBM, Trimbos BJBMZ, Ordi J, van Poelgeest
MIE and Bosse T: Genomic characterization of vulvar (Pre)cancers
identifies distinct molecular subtypes with prognostic
significance. Clin Cancer Res. 23:6781–6789. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zieba S, Pouwer AW, Kowalik A, Zalewski K,
Rusetska N, Bakuła-Zalewska E, Kopczyński J, Pijnenborg JMA, de
Hullu JA and Kowalewska M: Somatic mutation profiling in
premalignant lesions of vulvar squamous cell carcinoma. Int J Mol
Sci. 21:48802020. View Article : Google Scholar
|
|
9
|
Williams EA, Werth AJ, Sharaf R, Montesion
M, Sokol ES, Pavlick DC, McLaughlin-Drubin M, Erlich R, Toma H,
Williams KJ, et al: Vulvar squamous cell carcinoma: Comprehensive
genomic profling of HPV+ versus HPV-forms reveals
distinct sets of potentially actionable molecular targets. JCO
Precis Oncol. 4:647–661. 2020. View Article : Google Scholar
|
|
10
|
Tessier-Cloutier B, Kortekaas KE, Thompson
E, Pors J, Chen J, Ho J, Prentice LM, McConechy MK, Chow C, Proctor
L, et al: Major p53 immunohistochemical patterns in in situ and
invasive squamous cell carcinomas of the vulva and correlation with
TP53 mutation status. Mod Pathol. 33:1595–1605. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tessier-Cloutier B, Pors J, Thompson E, Ho
J, Prentice L, McConechy M, Aguirre-Hernandez R, Miller R, Leung S,
Proctor L, et al: Molecular characterization of invasive and in
situ squamous neoplasia of the vulva and implications for
morphologic diagnosis and outcome. Mod Pathol. 34:508–518. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ge Y, Zhang C, Xiao S, Liang L, Liao S,
Xiang Y, Cao K, Chen H and Zhou Y: Identification of differentially
expressed genes in cervical cancer by bioinformatics analysis.
Oncol Lett. 16:2549–2558. 2018.PubMed/NCBI
|
|
13
|
Micci F, Panagopoulos I, Haugom L,
Dahlback HS, Pretorius ME, Davidson B, Abeler VM, Tropé CG,
Danielsen HE and Heim S: Genomic aberration patterns and expression
profiles of squamous cell carcinomas of the vulva. Genes
Chromosomes Cancer. 52:551–563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35((Web Server Issue)): W169–W175. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47D:D607–D613. 2019.
View Article : Google Scholar
|
|
19
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res.
45W:W98–W102. 2017. View Article : Google Scholar
|
|
20
|
Darragh TM, Colgan TJ, Cox JT, Heller DS,
Henry MR, Luff RD, McCalmont T, Nayar R, Palefsky JM, Stoler MH, et
al: The lower anogenital squamous terminology standardization
project for HPV-associated lesions: Background and consensus
recommendations from the college of American pathologists and the
American society for colposcopy and cervical pathology. Arch Pathol
Lab Med. 136:1266–1297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kortekaas KE, Solleveld-Westerink N,
Tessier-Cloutier B, Rutten TA, Poelgeest MIE, Gilks CB, Hoang LN
and Bosse T: Performance of the pattern-based interpretation of p53
immunohistochemistry as a surrogate for TP53 mutations in vulvar
squamous cell carcinoma. Histopathology. 77:92–99. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Heller DS, Day T, Allbritton JI, Scurry J,
Radici G, Welch K and Preti M; ISSVD Difficult Pathologic Diagnoses
Committee, : Diagnostic criteria for differentiated vulvar
intraepithelial neoplasia and vulvar aberrant maturation. J Low
Genit Tract Dis. 25:57–70. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Sun C, Tan Y, Li L, Zhang H, Liang
Y, Zeng J and Zou H: Transcription levels and prognostic
significance of the NFI family members in human cancers. PeerJ.
8:e88162020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim BR, Coyaud E, Laurent EMN, St-Germain
J, Van de Laar E, Tsao MS, Raught B and Moghal N: Identification of
the SOX2 interactome by BioID reveals EP300 as a mediator of
SOX2-dependent squamous differentiation and lung squamous cell
carcinoma growth. Mol Cell Proteomics. 16:1864–1888. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brustmann H and Brunner A:
Immunohistochemical expression of SOX2 in vulvar intraepithelial
neoplasia and squamous cell carcinoma. Int J Gynecol Pathol.
32:323–328. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu S, Wu Y, Lu Y, Yue Y, Cui C, Yu M, Wang
S, Liu M, Zhao Y and Sun Z: STAT1 expression and HPV16 viral load
predict cervical lesion progression. Oncol Lett.
20:282020.PubMed/NCBI
|