Introduction
Gliomas represent a serious clinical disease, with
high malignancy, common recurrence and poor prognosis. According to
statistics, in 2015, the prevalence of gliomas accounts for 4–5% in
the world (1,2). At present, the standard treatment is to
remove the tumor to the maximum extent using radiotherapy and
chemotherapy (3,4), but the 5-year survival rate is <5%
worldwide (5). Glioblastoma (GBM) is
one of the most malignant, recurrent and invasive tumor types, and
is classified as grade 4 according to the World Health Organization
nervous system tumor classification system (6). GBM is insidious and invasive, and its
clinical symptoms appear relatively late (7). Microsurgical resection, postoperative
chemotherapy and radiotherapy are the primary techniques to treat
GBM (7). Due to limitations of the
anatomical structure, highly invasive growth and high radiation
resistance, no marked positive effect on the overall prognosis of
patients has been achieved, despite numerous advances in treatment
methods in recent years (7). Novel
treatment strategies and drug research and development have become
increasingly urgent as this problem remains to be solved (8,9).
With the rapid development of high-throughput
sequencing and bioinformatics in recent years, some key biomarkers
and signaling pathways in the development of GBM have been
identified, providing a theoretical basis for the study of
GBM-targeting drugs (10). The
PI3K-Akt-mTOR signaling pathway is a key regulator of numerous
cellular processes, including cell proliferation, apoptosis,
migration and invasion (11). It is
one of the most commonly altered signal transduction networks in
human cancer (11). The PI3K family
of lipid kinases is a key component of this signaling pathway, and
therefore PI3K has become a research hotspot for targeted drugs
(12).
The PI3K/Akt/mTOR signaling pathway is composed of
three main acting molecules, namely PI3K, PKB/Akt and mTOR, and it
is a central regulatory mechanism that can promote the growth and
proliferation of tumor cells and inhibit autophagy (13). Neuroceroid lipofuscins (NCLs) are a
common cause of neurodegeneration in children (14). The sixth known NCL gene is CLN5,
which is predicted to encode a novel protein with two putative
transmembrane domains (15). The
CLN5 protein is a 407-amino acid, highly glycosylated protein with
unknown functions. It is localized to lysosomes through the
mannose-6-phosphate receptor pathway and other signaling pathways
(16). Pathogenic mutations result
in its retention in the ER/Golgi matrix (17). Lysosomes can effectively control
autophagy (18). Recently,
researchers examined the association between mTORC1 activation and
lysosomal localization through CRISPR/Cas9 knockout of lysosomal
localization-associated protein components, revealing that limbic
lysosomes could facilitate the activation of the mTORC1, mTORC2 and
Akt signaling pathways (19).
Therefore, the current study speculated that CLN5 may serve a role
in GBM. In the present study, the effect of CLN5 on GBM was
investigated at the molecular level.
Materials and methods
Agents
Antibodies against CDK4 (cat. no. 11026-1-AP;
1:1,000), CDK6 (cat. no. 14052-1-AP; 1:1,000), Akt (cat. no.
10176-2-AP; 1:1,000), mTOR (cat. no. 20657-1-AP; 1:1,000), GAPDH
(cat. no. 10494-1-AP; 1:1,000) and HRP-conjugated sheep
anti-rabbit/mouse (ES-0005, 1:5,000) were ordered from the
ProteinTech Group, Inc. Antibodies against CLN5 (cat. no. ab170899;
1:1,000), Bcl-2 (cat. no. ab32124; 1:1,000), Bax (cat. no. ab32503;
1:1,000), phosphorylated (p)-Akt (cat. no. ab38449; 1:1,000),
cyclin D1 (cat. no. ab134175; 1:1,000), pro-caspase-9 (cat. no.
ab138412; 1:1,000), activated-caspase-9 (cat. no. ab219590;
1:1,000), and p-mTOR (cat. no. ab109268; 1:1,000) were ordered from
Abcam.
Gene expression profiling interactive
analysis (GEPIA)
The expression of CLN5 and overall survival were
analyzed using GEPIA (http://gepia.cancer-pku.cn/), which include data from
The Cancer Genome Atlas (http://www.tcga.org/) and the Genotype-Tissue
Expression (http://commonfund.nih.gov/GTEx/) databases. For the
analysis of gene expression, Log2(Transcripts Per
Million + 1) was use for log-scale. The cut-off of |Log2
fold-change| was 1 and P<0.01 was considered to indicate a
statistically significant difference. The log-rank test, also known
as Mantel-Cox test, was used for the analysis of overall
survival.
Cell lines and cell culture
U251 and U87MG (glioblastoma of unknown origin) cell
lines were purchased from The Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences, Shanghai, China and
authenticated via STR profiling. The cells were cultured in DMEM
supplemented with 10% FBS (both from Gibco; Thermo Fisher
Scientific, Inc.), streptomycin (100 µg/ml) and penicillin (100
U/ml) in a 5% CO2 incubator at 37°C. The cells were
washed with PBS 3 times at the logarithmic growth stage, then
digested with 0.25% trypsin for 3–4 min and seeded in a 6-well
plate for subsequent experiments.
Transfection
Transfection was performed according to the
instructions for the Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). The complete culture medium was
replaced 2 h before transfection. The small interfering (si)-CLN5
(OriGene Technologies, Inc.) sequence was 5′-CCTTATTGTCAAGCTAAGT-3′
and the negative control (NC) siRNA (OriGene Technologies, Inc.)
sequence was 5′-GGCUGUAUGAGCACCGUUATT-3′. A total of 5 µl liposome
was dissolved into 125 µl serum-free and antibiotic-free medium,
then mixed gently and left at room temperature for 5 min.
Additionally, 5 µl si-CLN5 (50 nM) was dissolved into 125 µl
serum-free and antibiotic-free medium, mixed gently and left at
room temperature for 5 min. The liposome solution was mixed with
the si-CLN5 solution for 30 min. The cells were washed twice with
PBS. The mixture was added into the cells of each well (1×105
cells/well), gently mixed and cultured in the incubator. The cells
were incubated at 37°C for 24 h, then the complete culture medium
was replaced with fresh complete medium, and subsequent experiments
were performed after 24 h of continuous culture.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR detection
The Ultrapure RNA kit (CoWin Biosciences) was used
to extract the total RNA from cells. After reverse transcription at
85°C for 5 min using the Reverse Transcription Reaction kit (CoWin
Biosciences), real-time fluorescence qPCR was used to detect the
expression levels of CLN5. The thermocycling conditions were as
follows: 95°C for 5 min, followed by 40 cycles at 95°C for 30 sec,
60°C for 45 sec and 72°C for 30 min. The primers are listed as
follows: CLN5 forward, 5′-CAAGCGCTTTGACTTCCGTC-3′ and reverse,
5′-TCAAACCATGTCTCTGCCCC-3′; β-actin forward,
5′-CCCGAGCCGTGTTTCCT-3′ and reverse, 5′-GTCCCAGTTGGTGACGATGC-3′.
Time PCR system was performed using Super TaqMan Mixture (CoWin
Biosciences) according to the manufacturer's instructions. β-actin
was used as an internal control. The results were calculated using
the 2−ΔΔCq method (20).
Western blotting
For the immunoblot analysis, the NC and the
experimental groups underwent inference for 24 h, then the 6-well
plate was placed on ice. RIPA lysis buffer (supplemented with a
protease inhibitor; both from CoWin Biosciences) was used to
extract total protein, and the protein concentration was determined
using a BCA assay (CoWin Biosciences). A total of 20 µg
protein/lane was separated via 10% SDS-PAGE gel, and the proteins
were then transferred to a PVDF membrane and blocked with 5%
skimmed milk for 1 h at 37°C. The membrane was incubated with the
aforementioned primary antibodies overnight at 4°C, washed 3 times
(5 min each) with TBS-Tween-20 (0.05%; v/v) and then incubated with
the respective secondary antibodies for 1 h at room temperature.
ECL (EMD Millipore) was used to visualize the protein bands after
the membrane was washed. Quantity One 4.6 software (Bio-Rad
Laboratories, Inc.) was used to calculate the gray value.
Cell proliferation and viability
assays
Proliferation was assessed using the Cell Counting
Kit-8 (CCK-8; Beijing Solarbio Science & Technology, Co., Ltd.)
and colony formation assays. According to the manufacturer's
protocol, after 24 h of transfection, the cells were digested and
counted, and 2,000 cells/well were seeded in a 96-well plate. The
cells were then cultured in an incubator at 37°C for 1 h. A total
of 10 µl CCK-8 reagent was added and incubated at 37°C for 1 h
before detection. Every 24 h, the absorbance was measured at 450 nm
by a microplate reader.
For colony formation assays, cells were seeded into
a 6-cm plate after transfection and incubated for 2 weeks at 37°C.
Subsequently, cells were fixed with methanol for 15 min at room
temperature and stained with Giemsa for 20 min at room temperature.
Colonies with >10 cells were counted in 5 random fields.
Transwell assay for invasion
detection
Invasion was evaluated using Transwell chambers (EMD
Millipore). Matrigel® (cat. no. 356234, BD Biosciences)
was diluted in serum-free medium at the ratio of 1:6, and 100 µl
was added into the upper chamber, then incubated at 37°C for 4–6 h.
After transfection for 24 h, 5×104 cells in 500 µl
serum-free medium were added to the upper chamber, and 500 µl
complete medium was added to the lower chamber. After overnight
incubation at 37°C, a cotton swab was used to wipe off the
remaining cells in the upper chamber. After washing with PBS, cells
were fixed with 4% paraformaldehyde for 30 min at room temperature
and stained with 0.1% crystal violet for 20 min at 37°C, then
counted under the fluorescence microscope (magnification,
×100).
Cell migration assay
The migration of U251 and U87MG cells was detected
via scratch assay. After digestion, 5×104 cells/well
were seeded in a 6-well plate in 500 µl serum-free medium, and
cells were cultured until confluency overnight at 37°C. The cells
were scratched and cell migration was recorded every 24 h under the
fluorescence microscope (×40 magnification). The results were
processed using ImageJ version 1.8.0 software (National Institutes
of Health), and the cell migratory ability was assessed based on
the extent of the area of cell migration between the control group
and the experimental group.
Gelatinase spectrum
Gelatinase spectrum was used to detect MMP-2
expression. After transfection, the NC and experimental groups were
incubated at 37°C for 24 h, and were then washed with serum-free
DMEM 3 times and cultured for 24 h at 37°C. The supernatant was
collected via centrifugation in 5,000 × g for 5 min at 4°C, diluted
with serum-free DMEM without mercaptoethanol and added into each
well of the vertical electrophoresis tank. The samples were
separated using 10% SDS-PAGE (0.5 mg/ml gelatin). After
electrophoresis, the gel was dyed with 0.25% Coomassie Blue R-250
at room temperature for 4 h and decolorized with decolorizing
liquid (10% acetic acid) at room temperature for 1 h. Following
this process, the gel was imaged using a camera (Olympus
Corporation). The gray value was analyzed using Quantity One 4.6
software (Bio-Rad Laboratories, Inc.).
Cell apoptosis and cell cycle analysis
via flow cytometry
After 24 h of transfection, the cells were
serum-starved for 24 h and then digested with trypsin for 3–4 min
at 4°C without EDTA. Subsequently, they were collected into a
centrifuge tube, centrifuged at 300 × g for 5 min at 4°C and
resuspended in pre-cooled PBS at 4°C. After centrifugation, the
cell density was adjusted by adding 1X binding buffer to obtain
1–5×106 cells/ml. A total of 100 µl cell suspension and
5 µl Annexin-V/FITC (Beijing 4A Biotech Co., Ltd.) were added into
a 5-ml flow tube, incubated at room temperature in the dark for 5
min according to the manufacturer's protocol, and then 10 µl PI at
37°C for 5 min and 400 µl PBS were added to the mixture before flow
cytometry detection. FlowJo version 10.6.2 software (FlowJo LLC)
was used to analyze and process the flow cytometry (FACSCanto II;
BD Biosciences) results.
For cell cycle detection, ice-cold ethanol was used
to fix the cells for >24 h at 4°C. Subsequently, the procedure
for cell cycle detection was similar to the aforementioned
apoptosis detection. PI was used for single-staining at 37°C for 5
min and 400 µl PBS was added. FlowJo software was used to analyze
and process the flow cytometry results.
Statistical analysis
Results were analyzed using SPSS 18.0 software
(SPSS, Inc.). The comparison between two groups was performed using
unpaired Student's t-test. Results were expressed as the mean ± SD.
P<0.05 was considered to indicate a statistically significant
difference.
Results
CLN5 expression is upregulated in
GBM
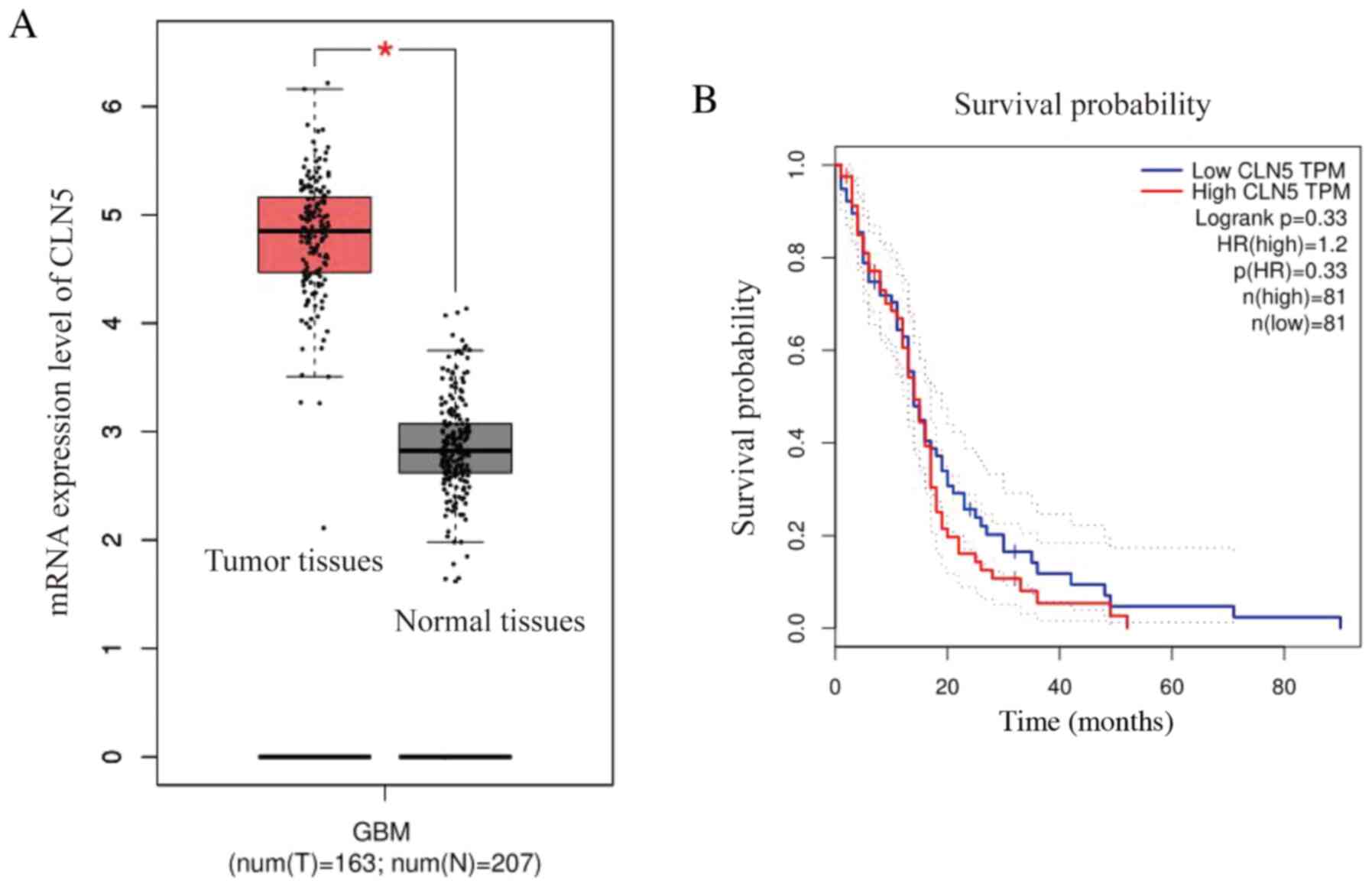
In order to study the association between CLN5
expression and GBM, GEPIA was used to extract RNA sequences from
The Cancer Genome Atlas and the Genotype-Tissue Expression
databases, in order to further understand the function of this gene
(21). The expression levels of CLN5
in GBM tissues were significantly higher compared with those in
normal tissues, indicating that CLN5 expression was upregulated in
GBM (Fig. 1A). Furthermore, the
prognostic survival curve revealed that the overall survival rate
of patients with GBM with high CLN5 expression was lower than that
of patients with low CLN5 expression; however, there was no
significant effect of CLN5 expression on survival (Fig. 1B).
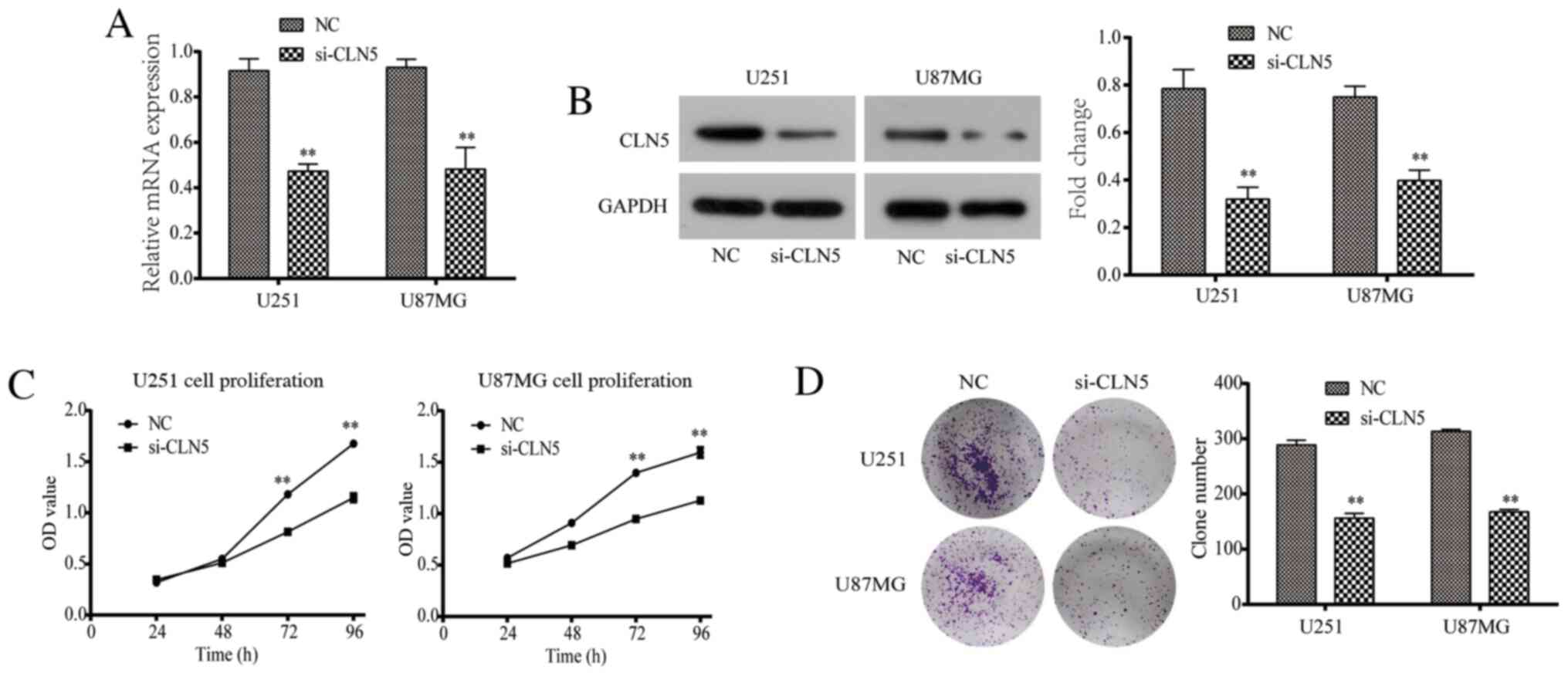
Knockdown of CLN5 decreases the
proliferation of GBM cells in vitro
RT-qPCR and western blotting were used to evaluate
the efficiency of CLN5-knockdown. The results revealed that CLN5
mRNA and protein expression levels in the si-CLN5 group were
significantly decreased in both U251 and U87MG cells compared with
those in the NC group (Fig. 2A and
B).
The CCK-8 assay was performed to detect the
proliferation of U251 and U87MG cells. The data revealed that after
72 and 96 h of culture, the proliferative ability of the si-CLN5
group was significantly decreased compared with that of the NC
group (P<0.05; Fig. 2C). In
addition, the colony assay demonstrated that the colony formation
ability of the si-CLN5 group was significantly lower than that of
the NC group (P<0.05; Fig. 2D).
Therefore, CLN5-knockdown significantly inhibited the proliferation
of GBM cells in vitro.
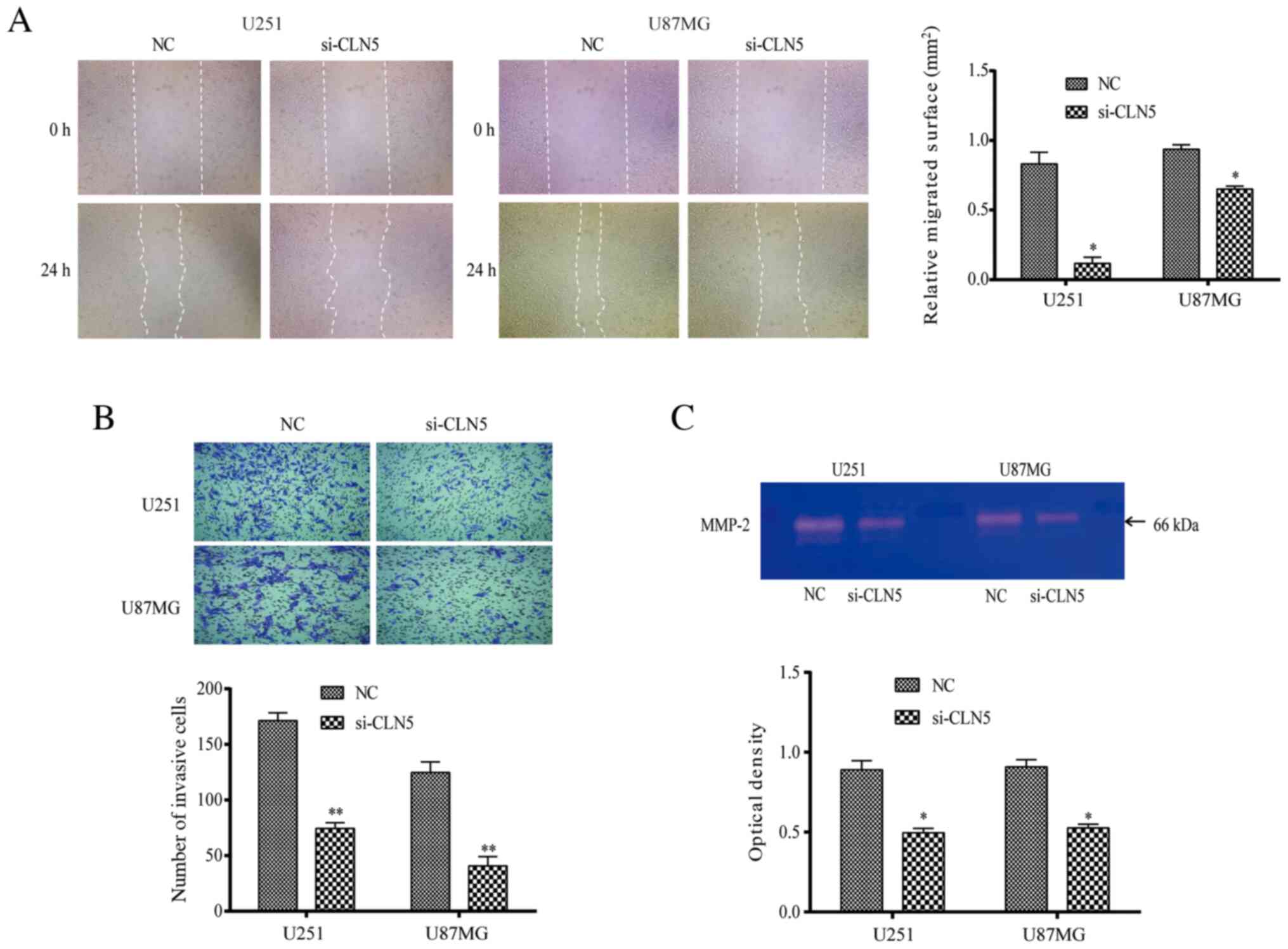
CLN5-knockdown inhibits the migration
and invasion of GBM cells
The migration of U251 and U87MG cells was detected
using the scratch test. The results revealed that the healing area
of the si-CLN5 group was decreased compared with that of the NC
group in both cell lines (P<0.05; Fig. 3A), indicating that migration of the
si-CLN5 group was inhibited. Transwell assays were performed to
evaluate the cell invasive ability. The invasive ability of the
si-CLN5 group was significantly decreased compared with that of the
NC group in both U251 and U87MG cells (P<0.05; Fig. 3B). Additionally, gelatinase spectrum
was performed to investigate MMP-2 expression, which is an
important factor affecting cell migration and invasion. MMP-2
expression in the si-CLN5 group was significantly lower than that
in the NC group (P<0.05; Fig.
3C). Therefore, the results revealed that CLN5-knockdown
inhibited the migration and invasion of GBM cells.
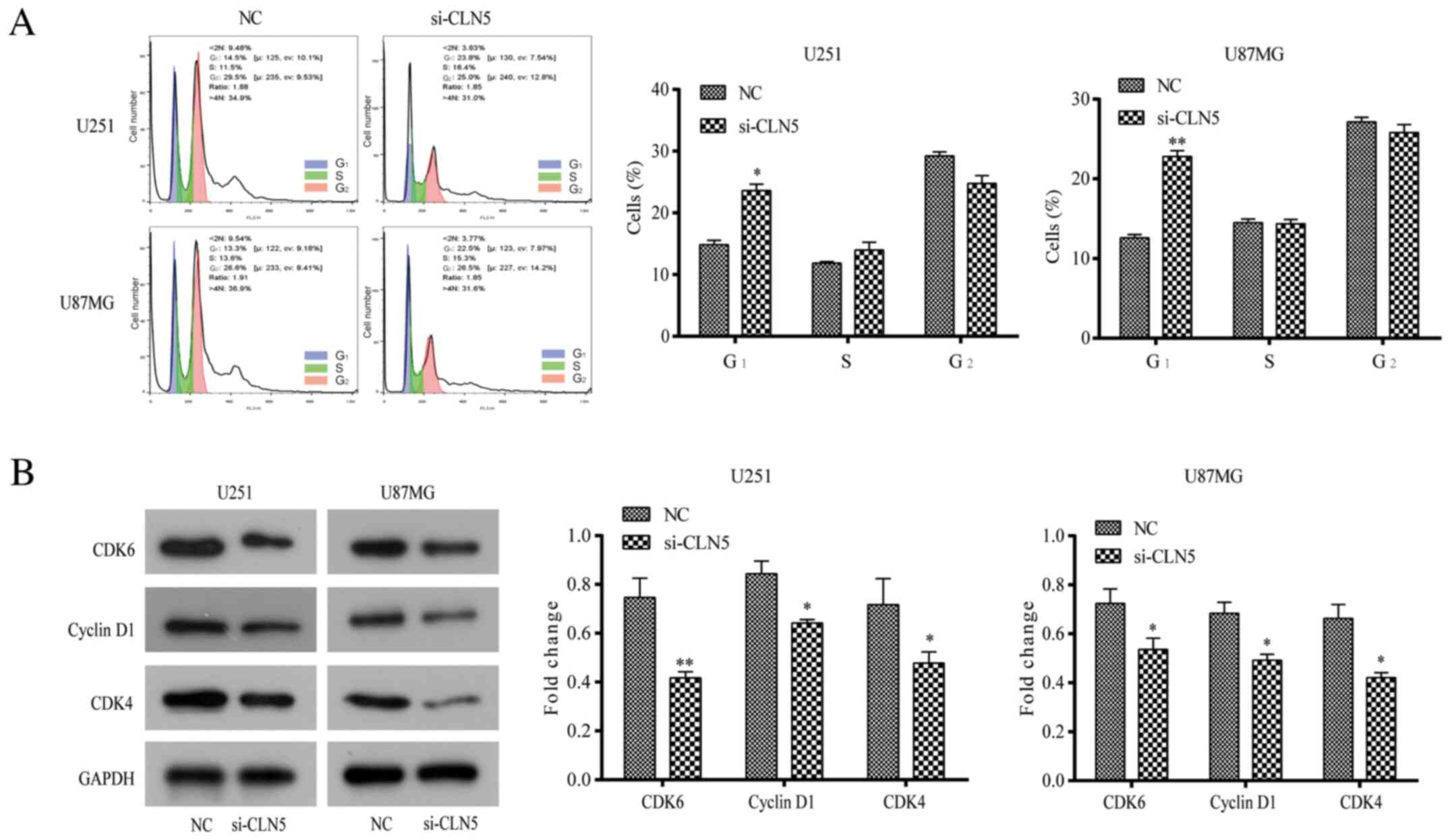
Depletion of CLN5 induces G1-phase
arrest in GBM cells
The cell cycle distribution of GBM CLN5-knockdown
cells was studied to determine the effect of CLN5 on the cell
cycle. Flow cytometry analysis revealed that the number of cells in
G1 phase transfected with si-CLN5 was significantly higher than
that of NC cells (P<0.05), while there was no significant
difference between S-phase and G2-phase cells (Fig. 4A). To further explore the regulatory
mechanism of G1-phase arrest, the expression levels of
cycle-associated molecules such as cyclin D1, CDK4 and CDK6, were
analyzed. Compared with NC cells, the expression levels of these
proteins in the si-CLN5 group were significantly decreased
(P<0.05; Fig. 4B), suggesting
that CLN5-knockdown may inhibit the proliferation of GBM cells by
blocking the G1 phase of the cell cycle.
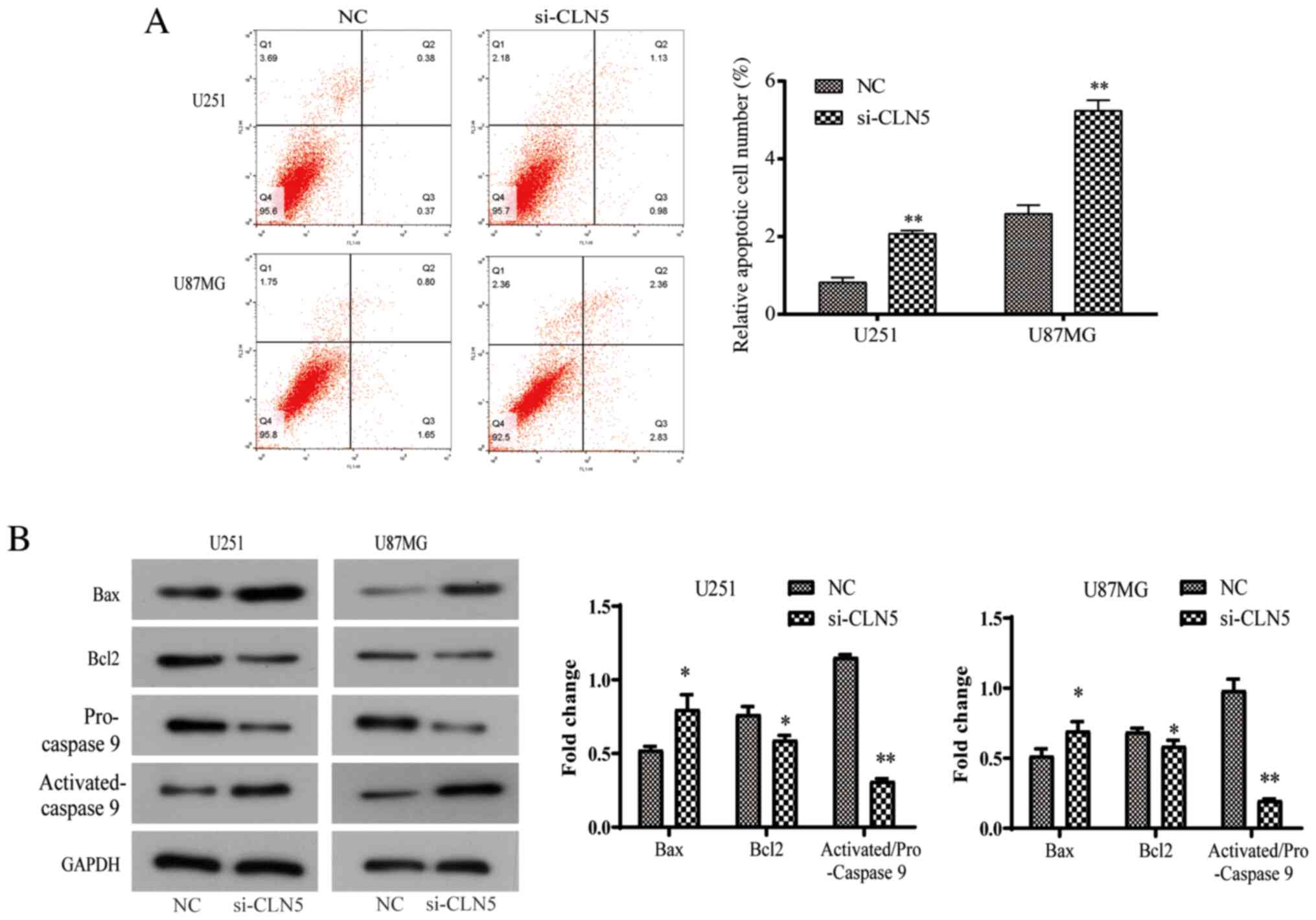
CLN5-knockdown promotes the apoptosis
of GBM cells
Due to the abnormal rate of cell proliferation and
irregular cell cycle, the present study further investigated the
potential mechanism of the effect of CLN5 on the life cycle of GBM
cells. Apoptosis is an important regulator of cell proliferation.
Flow cytometry was used to detect apoptosis after Annexin V-FITC
and PI double-staining. The results revealed that the apoptosis
rate increased significantly after si-CLN5 transfection (P<0.05;
Fig. 5A). In addition, changes in
the protein expression levels of Bcl-2, activated-caspase-9 and Bax
were investigated, which are closely involved in the process of
apoptosis. In the si-CLN5 group, the expression levels of Bax and
activated-caspase-9 were increased, while Bcl-2 expression was
decreased, indicating that CLN5-knockdown increased the Bax/Bcl-2
ratio (Fig. 5B). In conclusion, the
present results revealed that CLN5 may be an important factor in
the regulation of apoptosis in GBM cells.
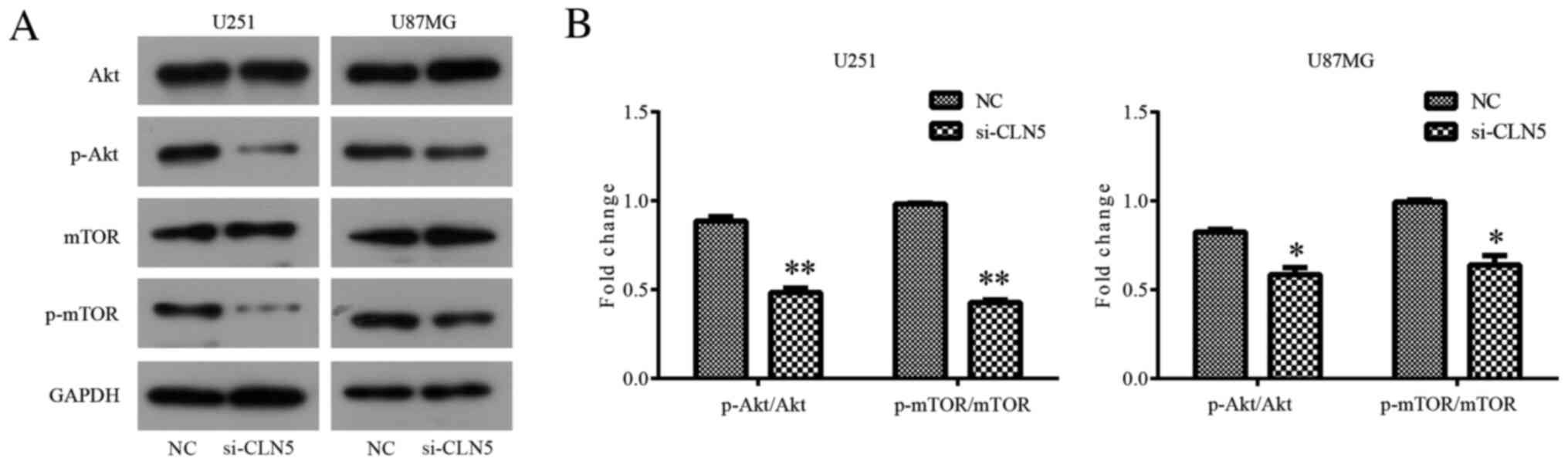
Knockdown of CLN5 inhibits the
Akt/mTOR signaling pathway in GBM cells
In order to further explore the mechanism of the
effect of CLN5 on GBM, western blotting was used to detect the
changes in the Akt/mTOR signaling pathway after CLN5-knockdown. The
results revealed that there was no significant change in total Akt
expression in si-CLN5 cells compared with in the NC group; however,
CLN5-knockdown significantly decreased the levels of p-Akt in both
U251 (P<0.01) and U87MG (P<0.05) cells (Fig. 6). Therefore, it was suggested that
CLN5-knockdown inhibited the activation of the Akt signaling
pathway in GBM. In addition, changes in the mTOR signaling pathway
were analyzed. The inhibitory effect of CLN5-knockdown on the mTOR
signaling pathway was similar to that in the Akt signaling pathway,
with significantly decreased levels of p-mTOR in the si-CLN5 group
compared with in the NC group (P<0.01 in U251 and P<0.05 in
U87MG; Fig. 6). The present results
suggested that CLN5 may affect the development of GBM by regulating
the Akt/mTOR signaling pathway.
Discussion
GBM is one of the most lethal types of cancer in
humans due to its high malignancy, strong invasion and poor
prognosis (22). The CLN5 protein is
a 407 amino acid-long glycosylated protein with unknown functions.
The present study detected CLN5 upregulation in GBM tissues using
GEPIA; therefore, the association between the CLN5 gene and GBM
cells was further investigated. The results revealed that
CLN5-knockdown effectively inhibited the proliferation and invasion
of GBM cells, and promoted apoptosis.
At first, CCK-8 and clone formation assays were used
to investigate cell proliferation. It was revealed that
CLN5-knockdown effectively inhibited the proliferation of GBM cells
in vitro. Subsequently, the cell migratory and invasive
abilities were tested, and the results demonstrated that migration
and invasion decreased significantly after CLN5-knockdown. In the
process of tumor migration and invasion, one of the most important
factors is the degradation of the basement membrane and
extracellular matrix (23). MMP-2 is
a type of multi-matrix metalloproteinase that is involved in the
migration and invasion of tumors via the degradation of the
extracellular matrix; it is widely involved in numerous
pathological processes of tumor development (24) and serves an important role in cancer
cells (25). The present results
revealed that MMP-2 expression decreased significantly after si-RNA
transfection, indicating that CLN5-knockdown may regulate MMP-2
expression and inhibit cell invasion and migration.
In order to further study the role of CLN5 in cell
proliferation and apoptosis, the association between CLN5 and the
Akt/mTOR signaling pathway was investigated. Cyclins are a family
of proteins related to chromosome condensation and mitotic spindle
involved in the regulation of the cell cycle (26,27). The
results of flow cytometry and western blotting revealed that
si-CLN5 downregulated the expression levels of cyclin D1, CDK4 and
CDK6, blocked the cell cycle at the G1-phase and induced early
apoptosis. In addition, si-CLN5 upregulated the expression levels
of Bax and downregulated Bcl-2 expression, which may trigger
mitochondrial apoptosis (28).
Furthermore, western blotting was used to detect molecules in the
Akt/mTOR signaling pathways, and CLN5-knockdown inhibited the
activation of the Akt and mTOR signaling pathways in GBM by
decreasing the levels of p-Akt and p-mTOR. However, the antitumor
effect of CLN5 has not been verified in mouse models. The mechanism
of CLN5 upregulation in glioma and the specific downstream target
of CLN5 remain unclear. In the future, further research is required
to clarify the regulatory mechanism of CLN5 in human GBM cells.
In conclusion, the present analysis revealed that
CLN5-knockdown inhibited proliferation and promoted apoptosis in
human GBM cells by potentially inhibiting the Akt and mTOR
signaling pathways.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JX and YL mainly performed the experiments. JX
analyzed the data and wrote the manuscript. HZ contributed to
analyzing the data and modifying the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brandsma D, Stalpers L, Taal W, Sminia P
and van den Bent MJ: Clinical features, mechanisms, and management
of pseudoprogression in malignant gliomas. Lancet Oncol. 9:453–461.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Davis FG, Freels S, Grutsch J, Barlas S
and Brem S: Survival rates in patients with primary malignant brain
tumors stratified by patient age and tumor histological type: An
analysis based on Surveillance, Epidemiology, and End Results
(SEER) data, 1973–1991. J Neurosurg. 88:1–10. 2008. View Article : Google Scholar
|
|
4
|
Penas-Prado M and Gilbert MR: Molecularly
targeted therapies for malignant gliomas: Advances and challenges.
Expert Rev Anticancer Ther. 7:641–661. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tachon G, Masliantsev K, Rivet P,
Petropoulos C, Godet J, Milin S, Wager M, Guichet PO and
Karayan-Tapon L: Prognostic significance of MEOX2 in gliomas. Mod
Pathol. 32:774–786. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Komori T, Sasaki H and Yoshida K: Revised
WHO classification of tumours of the central nervous system:
Summary of the revision and perspective. No Shinkei Geka.
44:625–635. 2016.(In Japanese). PubMed/NCBI
|
|
7
|
Wolf A, Agnihotri S and Guha A: Erratum:
Targeting metabolic remodeling in glioblastoma multiforme.
Oncotarget. 9:348552018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khosla D: Concurrent therapy to enhance
radiotherapeutic outcomes in glioblastoma. Ann Transl Med.
4:542016.PubMed/NCBI
|
|
9
|
Ohgaki H and Kleihues P: Genetic pathways
to primary and secondary glioblastoma. Am J Pathol. 170:1445–1453.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu P, Yang J, Liu J, Yang X, Liao J, Yuan
F, Xu Y, Liu B and Chen Q: Identification of glioblastoma gene
prognosis modules based on weighted gene co-expression network
analysis. BMC Med Genomics. 11:962018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao XF, He HQ, Zhu XB, Xie SL and Cao Y:
lncRNA SNHG20 promotes tumorigenesis and cancer stemness in
glioblastoma via activating PI3K/Akt/mTOR signaling pathway.
Neoplasma. 66:532–542. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hudson K, Hancox UJ, Trigwell C, McEwen R,
Polanska UM, Nikolaou M, Morentin Gutierrez P, Avivar-Valderas A,
Delpuech O, Dudley P, et al: Intermittent high dose scheduling of
AZD8835, a novel selective inhibitor of PI3Kα and PI3Kδ,
demonstrates treatment strategies for PIK3CA-dependent breast
cancers. Mol Cancer Ther. 15:877–889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu JZ, Ying Y, Liu Y, Sun CB, Dai C, Zhao
S, Tian SZ, Peng J, Han NP, Yuan JL, et al: Antifibrotic action of
Yifei Sanjie formula enhanced autophagy via PI3K-AKT-mTOR signaling
pathway in mouse model of pulmonary fibrosis. Biomed Pharmacother.
118:1092932019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mitchison HM and Mole SE:
Neurodegenerative disease: The neuronal ceroid lipofuscinoses
(Batten disease). Curr Opin Neurol. 14:795–803. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Savukoski M, Klockars T, Holmberg V,
Santavuori P and Peltonen L: CLN5, a novel gene encoding a putative
transmembrane protein mutated in Finnish variant late infantile
neuronal ceroid lipofuscinosis. Nat Genet. 19:286–288. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vesa J, Chin MH, Oelgeschläger K,
Isosomppi J, DellAngelica EC, Jalanko A and Peltonen L: Neuronal
Ceroid Lipofuscinoses Are Connected at Molecular Level: Interaction
of CLN5 Protein with CLN2 and CLN3. Mol Biol Cell. 13:2410–2420.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lebrun AH, Storch S, Rüschendorf F,
Schmiedt ML, Kyttälä A, Mole SE, Kitzmüller C, Saar K, Mewasingh
LD, Boda V, et al: Retention of lysosomal protein CLN5 in the
endoplasmic reticulum causes neuronal ceroid lipofuscinosis in
Asian sibship. Hum Mutat. 30:E651–E61. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bouhamdani N, Comeau D, Cormier K and
Turcotte S: STF-62247 accumulates in lysosomes and blocks late
stages of autophagy to selectively target von
Hippel-Lindau-inactivated cells. Am J Physiol Cell Physio.
316:C605–C620. 2019. View Article : Google Scholar
|
|
19
|
Jia R and Bonifacino JS: Lysosome
positioning influences mTORC2 and AKT signaling. Mol Cell.
75:26–38.e3. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Z, Li C, Kang B, Ge G, Li C and Zhang
Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bahadur S, Sahu AK, Baghel P and Saha S:
Current promising treatment strategy for glioblastoma multiform: A
review. Oncol Rev. 13:417:2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yilmaz M, Christofori G and Lehembre F:
Distinct mechanisms of tumor invasion and metastasis. Trends Mol
Med. 13:535–541. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schrump DS, Chen A and Consoli U:
Inhibition of lung cancer proliferation by antisense cyclin D.
Cancer Gene The. 3:131–135. 1996.
|
|
25
|
Stewart CJ and Crook ML: CD147 (EMMPRIN)
and matrix metalloproteinase-2 expression in uterine endometrioid
adenocarcinoma. Pathol Res Pract. 207:30–36. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hoshyar R, Bathaie SZ and Sadeghizadeh M:
Crocin Triggers the apoptosis through increasing the Bax/Bcl-2
ratio and caspase activation in human gastric adenocarcinoma, AGS,
cells. DNA Cell Biol. 32:50–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Viallard JF, Lacombe F, Belloc F,
Pellegrin JL and Reiffers J: Molecular mechanisms controlling the
cell cycle: Fundamental aspects and implications for oncology.
Cancer Radiother. 5:109–129. 2001.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang HL, Kuo YH, Tsai CT, Huang YT, Chen
SC, Chang HW, Lin E, Lin WH and Hseu YC: Anti-metastatic activities
of Antrodia camphorata against human breast cancer cells
mediated through suppression of the MAPK signaling pathway. Food
Chem Toxicol. 49:290–298. 2011. View Article : Google Scholar : PubMed/NCBI
|