Lymphomas are a heterogeneous group of cancers; more
specifically, they consist of a group of blood disorders derived
from lymphocytes and are presented with multiple variations in
clinical presentation, long-term prognosis and pathogenesis. They
represent one of the most common types of cancer worldwide and
affect numerous patients. According to the World Health
Organization (WHO) classification report, there are approximately
100 different types of lymphoma. The two main categories of
lymphomas are non-Hodgkin lymphoma (NHL) (consisting 9 out of 10 of
all lymphoma cases) and Hodgkin lymphoma (HL) (consisting 1 out of
10 of all lymphoma cases) (1–3).
Furthermore, non-Hodgkin lymphomas can be grouped into B- and
T-cell NHLs (TNHLs), which account for approximately 90 and 10% of
NHLs, respectively (4,5). According to the WHO, two other
categories are also considered as types of lymphoid tissue tumors:
Multiple myeloma (MM) and immunoproliferative diseases (3).
According to the WHO, there are the following
lymphoma subtypes (WHO 2016): i) Mature B-cell neoplasms; ii)
mature T-cell and natural killer (NK) cell neoplasms; iii)
precursor lymphoid neoplasms; iv) HL; and v)
immunodeficiency-associated lymphoproliferative disorders (4,5).
B-cell NHLs (BCNHLs) are tumors of B-cells that
exhibit a heterogeneity that is attributed to the fact that these
tumors are derived from different stages of mature B-cell
differentiation. The main subtypes of BCNHLs are the following: i)
diffuse large B-cell lymphoma (DLBCL); ii) chronic lymphocytic
leukemia (CLL); iii) follicular lymphoma (FL); iv) mantle cell
lymphoma (MCL); v) Burkitt's lymphoma (BL); vi) marginal zone
lymphoma (MZL); and vii) mucosa-associated lymphoid tissue (MALT)
(6). The majority of BCNHLs, such as
DLBCL and FL, have passed the germinal center (GC) reaction,
indicating that their immunoglobulin (IG) genes have been
hypermutated. Other subtypes, such as MCL and CLL, are derived from
GC-inexperienced B-cells (7).
The most common subtypes of TNHLs and NK-cell NHLS
(NK-NHLs) are the following: i) Cutaneous T-cell lymphomas (mycosis
fungoides, Sezary syndrome and others); ii) adult T-cell
leukemia/lymphoma; iii) angioimmunoblastic T-cell lymphoma; iv)
extranodal NK/T-cell lymphoma; nasal type; vi)
enteropathy-associated T-cell lymphoma; and vii) anaplastic large
cell lymphoma (ALCL) (4).
The most common risk factors for HL are Epstein-Barr
virus (EBV) infection and a family history of the disease (8). The most common risk factors for several
types of NHLs include the following: i) Autoimmune diseases, such
as Sjögren syndrome, celiac disease, rheumatoid arthritis and
systemic lupus erythematosus; ii) HIV/AIDS infection; iii) human
T-lymphotropic virus infection; iv) Helicobacter pylori
infection; v) HHV-8 infection; vi) hepatitis C virus infection;
vii) medical treatments (patients who have been previously treated
for Hodgkin lymphoma, methotrexate and the tumor necrosis factor-a
inhibitors); viii) genetic diseases; and ix) certain chemical
agents (benzene and certain herbicides and insecticides; weed- and
insect-killing substances) (9–15). Some
environmental agents, such as red meat consumption and tobacco
smoking may also play a role in increasing the risk of developing
NHL (11,12,16).
The diagnosis of lymphomas can be achieved due to
the enlargement of lymph nodes, which can be determined by
performing a lymph node biopsy (17). A lymph node biopsy commonly is
followed by performing immunophenotyping, flow cytometry,
fluorescence in situ hybridization testing, bone marrow
aspiration and bone marrow biopsy (18). Imaging via computed tomography of the
chest and upper-lower abdomen may then be performed to determine
the possible expansion of the lymphoma throughout the human body
(17).
ncRNAs are RNAs that are not translated to proteins.
Over the past ten years, a number of ncRNAs have been identified.
Any of the three RNA polymerases (RNA Pol I, RNA Pol II or RNA Pol
III) can perform the transcription of a ncRNA. The ncRNAs are
divided into the following two main categories: Small ncRNAs,
<200 bp in length and long ncRNAs (lncRNAs), >200 bp in
length (19).
In these two categories, several individual
categories of ncRNAs also exist. These include housekeeping ncRNAs
[transfer RNAs (tRNAs) and some ribosomal RNAs (rRNAs)], which are
essential for fundamental principles of cellular biology, small
nuclear RNAs (snRNAs), and a number of recently observed RNAs which
are associated with the transcription of genes into proteins
(20).
To date, miRNAs are the less extensively studied
ncRNAs for their roles in cancer. Over the past years, a number of
targeted reviews have been published (21–23),
which have described a complex basic mechanism through which miRNAs
can lead to the silencing of target gene expression; through the
formation of a silencing complex induced by RISC-induced RNA, which
uses proteins from the Argonaute family (such as AGO2) for the
splicing of target mRNAs or for the suspension of the translation
of these mRNAs (21). The patterns
of expression of miRNAs in different cancer types have been
well-observed, and studies have highlighted numerous miRNAs, such
as miR-10b, let-7, miR-101 and miR-15a-16 complex-1, which have
oncogenic or tumor-suppressive functions (22,23).
Recent observations of new species of lncRNAs have
led to the development of various possible candidates as lncRNAs.
Although a number of RNAs have a length of >200 bp, such as
repeat sequence transcripts and pseudogenes (24), the term lncRNA (also referred to as
lincRNAs, for long transgenic ncRNAs) is not used in the same
manner in all cases.
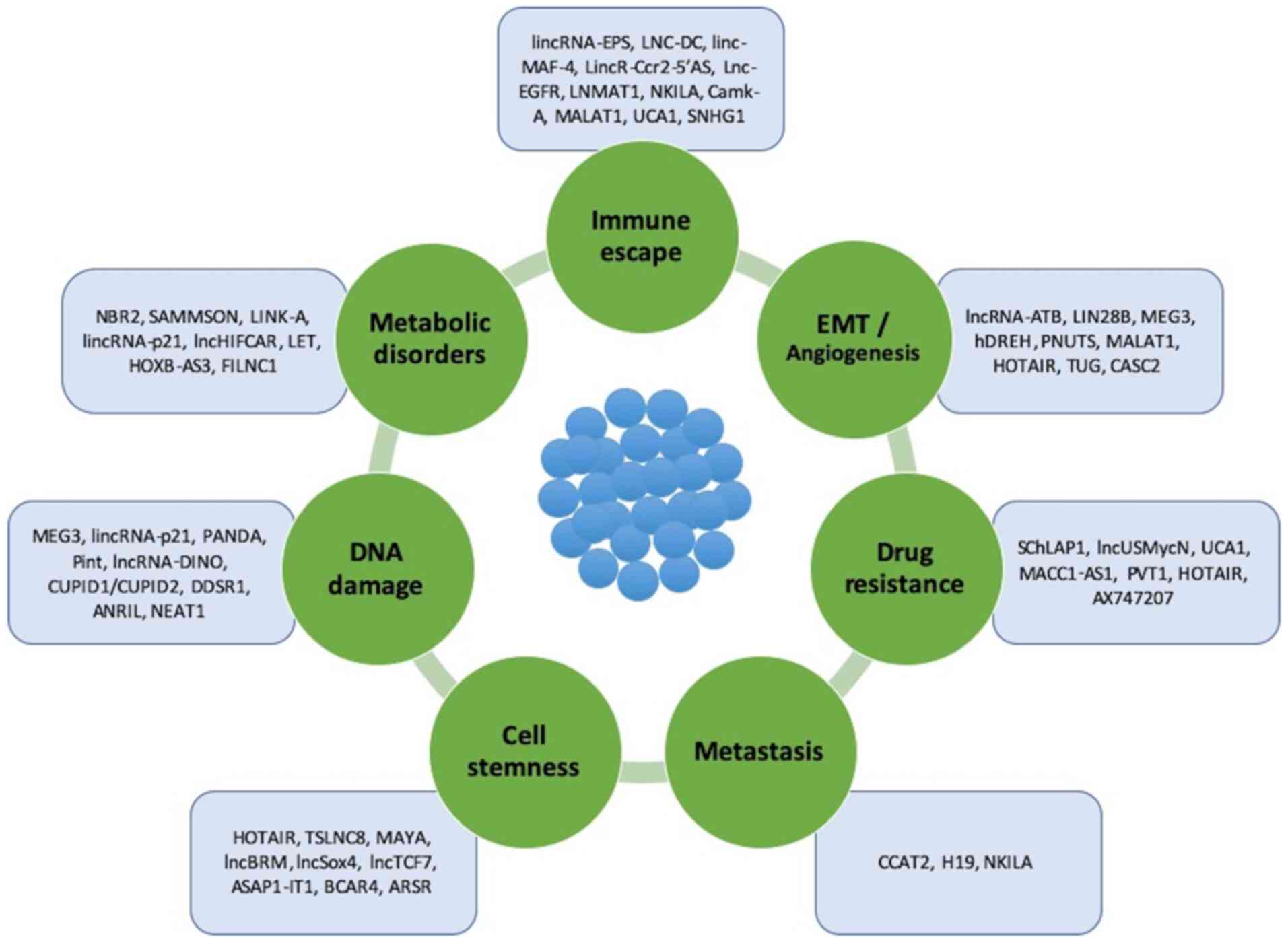
A number of common features of lncRNAs have been
indicated to confirm their biological identity, such as the
following: i) Epigenetic regulation as in a transcripted gene; ii)
transcription performed by RNA polymerase II; iii) poly-adenylation
to the 3′-untranslated region (3′-UTR); iv) frequent splicing of
multiple exons through specific molecular patterns; v) regulation
by classic transcription factors; and vi) frequent tissue-specific
expression (24) (Fig. 1).
B-cell differentiation in adult humans begins within
the bone marrow (BM) and is continued thereafter in the lymph
nodes, tonsils and spleen (25). On
the other hand, T-cells a derived from bone marrow hematopoietic
stem cells (HSCs), whose progenitors migrate to and colonize the
thymus (26).
The most common lncRNAs affecting normal B-cells are
the following: i) MYB-AS1, SMAD1-AS1 and LEF1-AS1, located on
6q23.3, 4q31.21 and 4q25, respectively, are involved in early
B-cell development; ii) CRNDE, located on 16q12.2-involved in
mitotic cell cycle related processes; and iii)
RP11-132N15.3/lnc-BCL6-3, located on 3q27.3, and involved in the
modulation of the GC reaction. However, data on the roles of
lncRNAs in normal T-cells are limited (27–31).
miRNAs are also involved in lymphocyte development,
as first described in 2004; it was demonstrated that miR-223,
miR-181 and miR-142 were highly expressed in B-cells (32). miR-181 can also contribute to the
regulation of the levels of CD69, BCL2 and TCR during T-cell
development. In addition, miR-155 and miR-181 play key roles in the
regulation of GC B-cell differentiation (33).
It is essential knowledge that all ncRNAs may play a
vital role as predictive and prognostic biomarkers in the
pathogenesis and progression of lymphomas and lymphoid malignancies
in general. The main aim of the present review was to provide an
up-to-date summary of available information on all the known miRNAs
and lncRNAs that participate in the development of all lymphoid
disorders, with a main focus on their connection to each lymphoma
subtype. Furthermore, these molecular biomarkers may be used, in
the near future, in the therapeutic management of the majority of
lymphomas. Thus, the present review summarizes all published data
to date on ncRNAs, in order to shed light on the future
perspectives of lymphoma management.
A literature search was performed, including studies
published up to August, 2020, using the following databases:
Medline (PubMed), Science Direct, Web of Science and Google
Scholar.
Systematic reviews, uncontrolled prospective,
retrospective and experimental studies were included for each
specific subject (total no. of studies, n=235). The following
inclusion criteria were applied: Studies concerning ncRNAs,
lncRNAs, miRNAs, cancer and lymphomas: HLs, and BCNHLs and TNHLs.
All studies concerning the association of lncRNAs and miRNAs with
NHLs and HLs were included.
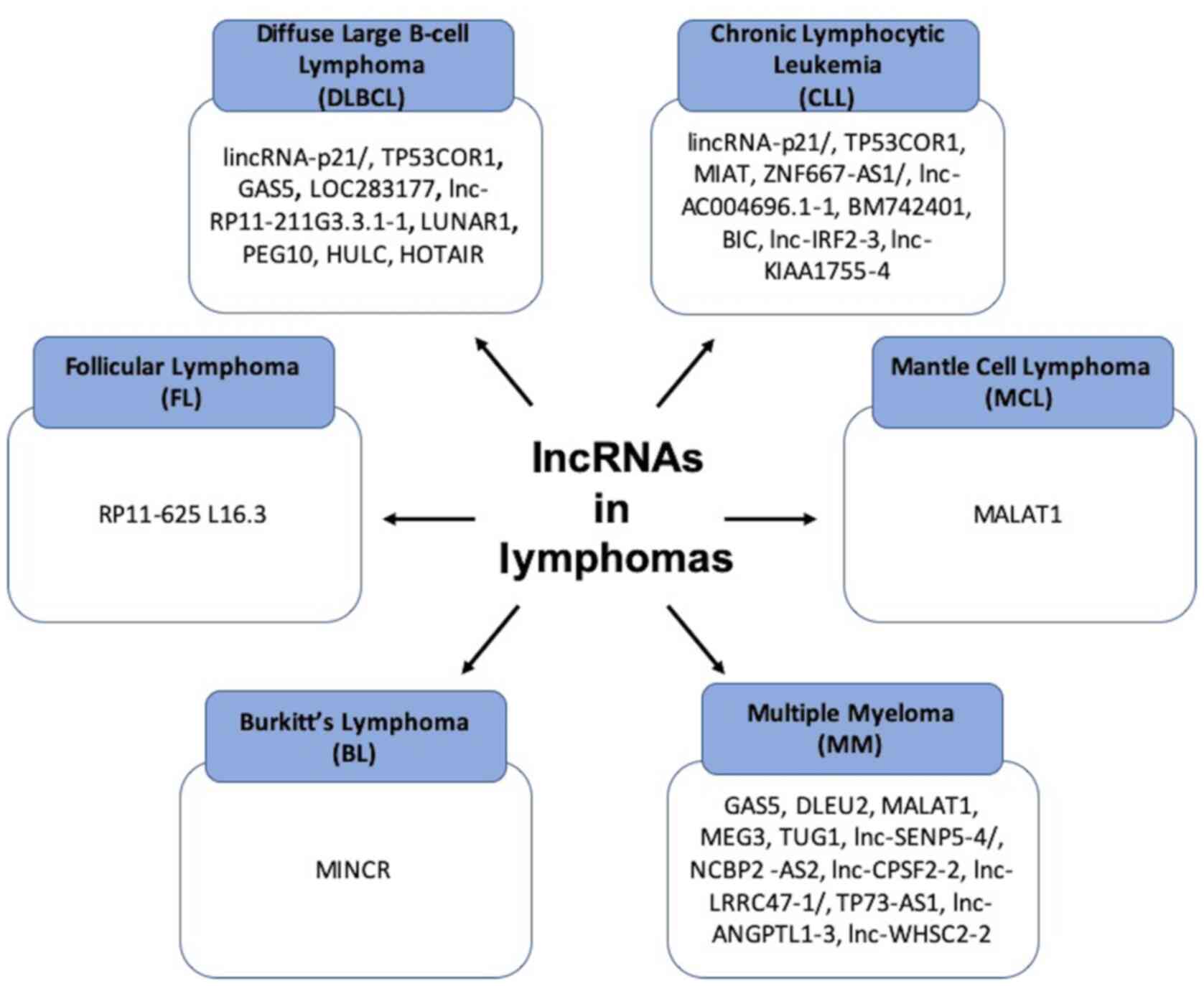
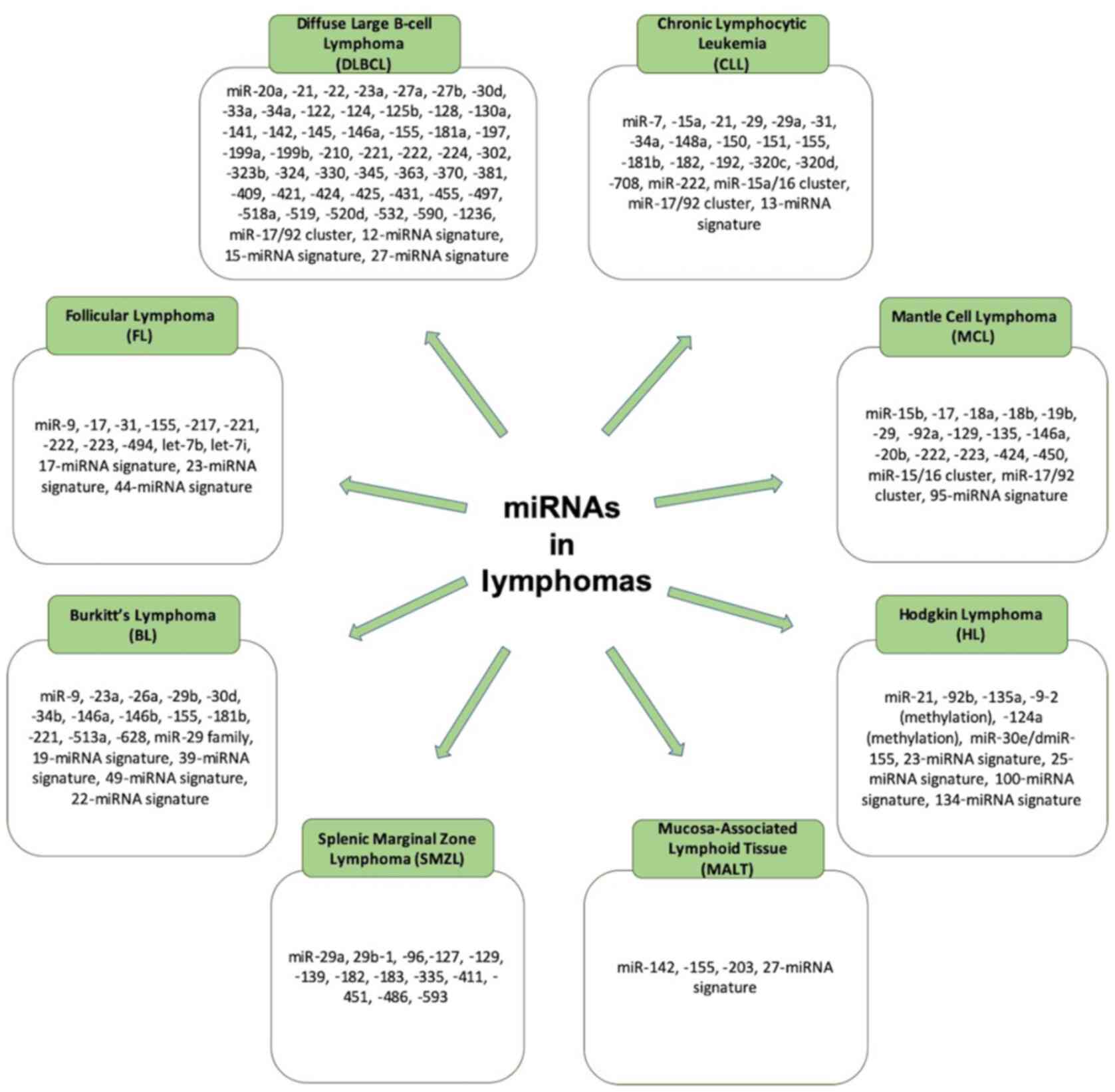
Over the past years, a number of studies have
referred to the significance of lncRNAs and miRNAs in the
pathophysiology of lymphomas, particularly B-cell NHLs. There are
different non-coding RNAs that play a role in each subtype of
lymphoma, and these are referred to the sections and tables below
(Figs. 2 and 3).
Subtypes of DLBCLs with a distinctive morphology or
immunophenotype are the following: i) T-cell/histiocyte-rich large
B-cell lymphoma; ii) ALK+ large B-cell lymphoma; iii)
plasmablastic lymphoma; iv) intravascular large B-cell lymphoma;
and v) large B-cell lymphoma with IRF4 rearrangement (38,39).
Subtypes of DLBCLs with distinctive clinical issues
are the following: i) Primary mediastinal large B-cell lymphoma;
ii) primary cutaneous DLBCL, leg type; iii) primary DLBCL of the
central nervous system; iv) DLBCL associated with chronic
inflammation; v) lymphomatoid granulomatosis; and vi) primary
effusion lymphoma (38,39).
Additionally, there are DLBCLs driven by viruses,
such as the following: i) EBV-positive DLBCL, not otherwise
specified; and ii) HHV8-positive DLBCL, NOS (Not otherwise
specified). There are also DLBCLs driven by disorders related to
DLBCL, such as: i) Helicobactor pylori-associated DLBCL; and
ii) EBV-positive mucocutaneous ulcer (40,41).
In order for a B-cell to be developed or to progress
into a DLBCL type, changes in the following genes need to occur:
BCL2 (42), BCL6 (42), MYC (36), EZH2 (43), MYD88 (42), CREBBP (44), CD79A and CD79B (44) and PAX5 (44). Therefore, the neoplastic cells in
DLBCL exhibit a pathologically overactivation of the nuclear factor
(NF)-κB, phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of
rapamycin (mTOR), Janus kinase (JAK)/signal transducer and
activator of transcription (STAT), mitogen-activated protein kinase
(MAPK)/extracellular signal-regulated kinase (ERK), B-cell receptor
and Toll-like receptor pathways (42).
Concerning miRNAs in DLBCLs, it has been shown than
in ABC-type DLBCL lymphoma, there is a high expression of miR-21,
miR-146a, miR155, miR-221 and miR-363, while in GCB-type DLBCL,
there is a high expression of miR-421 and the miR-17~92 cluster
(45–50) (Table
I). In serum samples of patients with DLBCL, increased levels
of miR-21, miR-155 and miR-210 have been identified, along with
increased levels of miR-124, miR-532-5p miR-15a, miR-16, miR-29c
and miR-155. Decreased levels of miR-122, miR-128, miR-141,
miR-145, miR-197, miR-345, miR-424 and miR-425 have been also found
in the serum of patients with DLBCL. miR-27a, miR-142, miR-199b,
miR-222, miR-302, miR-330, miR-425 and miR-519 seem to be
associated with the overall survival of patients with DLBCL
(50). In any case, miR-155,
miR-34a, miRNA-21, miRNA-23a, miRNA-27b, miRNA-34a, 12-miRNA
signature, 15-miRNA signature, 27-miRNA signature, miR-363,
miR-518a, miR-181a, miR-590, miR-421 and miR-324-either in serum
samples, or in tissue or cell line samples of patients with DLBCL,
can be used as diagnostic biomarkers in patients with DLBCL
(50–71) (Table I
and Fig. 3).
Concerning miRNAs in CLL, miR-15a/16-1 acts as a
tumor suppressor in patients with CLL and its expression is
therefore found to be decreased (105,106),
while miR-7-5p, miR-182-5p and miR-320c/d are regulated by p53, and
are increased in patients with CLL (107,108).
miR-181b expression is also low in patients with CLL (with a poor
outcome) (109). miR-155 is
overexpressed in patients with CLL and together with miR-21, lead
to higher mortality levels in patients with CLL (110) (Table
II and Fig. 3).
Additionally, patients with CLL are characterized by
higher levels of miR-34a, miR-31, miR-155, miR-150, miR-15a and
miR-29a; these can be used as diagnostic biomarkers (110–115).
In particular, miR-192 is expressed in low levels in patients with
CLL and can be thus used a diagnostic biomarker for CLL (116) (Table
II).
FL is the second most common type of NHL, and the
most common indolent NHL. It derives from the uncontrolled division
of centrocytes and centroblasts of the follicles in the GCs of
lymph nodes.
The genomic alterations that can be found in FL
include the following: i) the t(14:18)(q32:q21.3) translocation
(the majority of the cases); ii) 1p36 deletions (second most common
genomic alteration in FL) that lead to the loss of TNFAIP3; iii)
mutations in PRDM1; and iv) the same mutations observed in in
situ FL (ISFL), including KMT2D, CREEBP, BCL2 and EZH2, as well
as other mutations (45).
According to the WHO criteria, there are
differences, which can be observed under a microscope, which can be
used to diagnose and categorize FL into the following 3 grades,
with grade 3 comprising A and B subtypes (46): Grade 1, follicles with <5
centroblasts per high-power field (hpf); grade 2, follicles with 6
to 15 centroblasts per hpf; grade 3, follicles with >15
centroblasts per hpf; grade 3A, grade 3 in which the follicles
contain predominantly centrocytes; grade 3B, grade 3 in which the
follicles consist almost entirely of centroblasts.
Low-grade FLs are grades 1 and 2, as well as grade
3A. Grade 3B is regarded as a highly aggressive FL, which can be
easily transformed into a higher grade (46). The transformation of FL into a more
aggressive state or other type of aggressive lymphoma is associated
with specific genetic alterations, such as in the following genes:
CREEBP, KMT2D, STAT6, CARD11, CD79, TNFAIP3, CD58, CDKN2A or
CDKN2B, TNFRSF4 and c-MYC (45,46,136–138).
Concerning miRNAs in FLs, a number of studies have
demonstrated that there is an increase in the levels of 6
particular miRNAs: miR-223, miR-217, miR-222, miR221, let-7i and
let-7b in patients with FL, in which their lymphoma underwent a
transformation. In addition, the miR-17~92 cluster can be used as a
useful diagnostic biomarker found in patients with FL, while
miR-20a/b and miR-194 can also be found in patients with FL. Other
useful diagnostic biomarkers in patients with FL may be the
following: miR-9, miR-155, miR-31, miR-17, miR-217, miR-221,
miR-222, miR-223, let-7i, let-7b17-miRNA signature, 44-miRNA
signature, miR-494 23-miRNA signature (136–140)
(Table III and Fig. 3).
Concerning lncRNAs in patients with FL, studies have
demonstrated that there are 3-fold as many lncRNAs that are
upregulated than lncRNAs that are downregulated in patients with
FL3A stage disease, without their biological functions being
cleared yet. The only lncRNA that seems to be upregulated in
patients with FL3A grade disease is RP11-625 L16.3, located on
chromosome 12, with an uncharacterized mode of action (72) (Table
III and Fig. 2).
MCL is recognizable as an aggressive and incurable
small B-cell lymphoma. It predominantly affects older-aged males
(>60 years old), and sometimes it may be indolent in some
patients. MCLs arise from the mantle zone of early B-cells of the
lymph node follicle and they possess the t(11;14)(q13;q32)
translocation with an overexpression of cyclin D1.MCL cells also
exhibit CD5+ and CD23− and surface IgM/D
expression (141,142).
Concerning miRNAs in MCLs, a number of studies have
demonstrated the overexpression of miR-15/16 and miR-17~92 in MCL
and that this is associated with an aggressive form of the disease
(143,144). In addition, the inhibition of
miR-29 has been demonstrated to lead to the progression of MCL (a
potential prognostic marker for MCL) (143–145).
Additionally, the 95-miRNA signature can be a diagnostic biomarker
for MCL (145) (Table IV and Fig. 3).
Concerning lncRNAs in patients with MCL, it has been
demonstrated that MALAT1 is overexpressed in human MCL tissues and
cell lines compared to normal B-cells [a high international
prognostic index (IPI) is present], and is associated with the
lower overall survival of patients with MCL (146). Thus, MALAT1, located on 11q13
chromosome, can act as an oncogene in patients with MCL (regulation
of the bioavailability of TGF-β) (146–152)
(Table IV and Fig. 2).
BL is a type of aggressive B-NHL. It may be
presented with any of three main clinical variants: Endemic BL,
sporadic BL and the immunodeficiency-associated BL (158). In all types of BL, the
dysregulation of the c-myc gene is observed (the gene is found at
8q24), presented with any one of the three known chromosomal
translocations (159). The most
common variant is t(8;14)(q24;q32), which involves c-myc and IGH
(159).
Concerning miRNAs in patients with BL, it seems that
MYC regulates and is regulated by numerous miRNAs (Table V), the most common of which are the
following: miR-23a, miR-26a, miR-29b, miR-30d, miR-146a, miR-146b,
miR-155, and miR-221 (162–165) [widely used as diagnostic biomarkers
(166–173)] (Table
V and Fig. 3).
Concerning lncRNAs in patients with BL, 13 lncRNAs
have been identified thus far (173). The most well-identified lncRNA in
patients with BL is MINCR, located on chromosome 8q24.3, with an
uncharacterized role; but it seems that it causes the induction of
myc and modulates its transcriptional program (173).
There are also two other types of BCNHLs which
exhibit an indolent course. These are MALT lymphomas and MZL,
particularly the splenic type (SMZL).
None of the lncRNAs has been thus far identified as
playing a major role in the the activation or progression of a
B-cell to transform in any of these types of B-cell lymphomas.
Concerning miRNAs in SMZL, miR-96, miR-129, miR-29a,
miR-29b-1, miR-182, miR-183, miR-335 and miR-593 can be used as
diagnostic biomarkers, although without sufficient data to date
(174) (Table VI and Fig. 3).
As regards miRNAs in MALT lymphomas, miR-203
primarily, and secondly, miR-150, miR550, miR-124a, miR-518b and
miR-539, have been widely recognizable as being present in gastric
MALT lymphoma (175). Other miRNAs
identified in MALT lymphomas are the following: The 27-miRNA
signature, miR-142, miR-155, miR-203 miR-142 and miR-155 (176,177)
(Table VI and Fig. 3).
There are two main types of HL: Classical Hodgkin
lymphoma (9 out of 10 cases) and nodular lymphocyte predominant
Hodgkin lymphoma (1 out of 10 cases) (178,179).
There is a differentiation in morphology, phenotype and molecular
features between both these types. Furthermore, classical HL alone
can be subclassified into 4 more pathologic subtypes: i) Nodular
sclerosing HL; ii) mixed-cellularity subtype; iii) lymphocyte-rich;
and iv) lymphocyte-depleted HL (180–182).
Compared to B-NHLs, only limited data are available
on the expression of lncRNAs in HLs. As regards miRNAs in patients
with HL Hodgkin, there are studies which show that low miR-135a
levels lead to significantly poorer prognostic outcome in Hodgkin
patients (183,184). The inhibition of let-7 and miR-9
leads to the prevention of plasma cell differentiation (184). In particular, the inhibition of
miR-9 seems to lead to a decrease in cytokine production and a
reduced ability in attracting inflammatory cells (185). In addition, miR-155, the 23-miRNA
signature and 134- and 100-miRNA signature, 25-miRNA signature and
miR-9-2 (methylation) can be used as diagnostic biomarkers in
patients with HL (as they are presented in HL cell lines and
tissues) (186–193) (Table
VII and Fig. 3).
T-cell lymphomas affect T-cells and they are divided
into 4 major types: i) Extranodal T-cell lymphoma; ii) cutaneous
T-cell lymphomas: Sézary syndrome and Mycosis fungoides; iii)
anaplastic large cell lymphoma; and iv) angioimmunoblastic T-cell
lymphoma. There is also a clinical entity known as aggressive
NK-cell leukemia with an aggressive, systemic proliferation of NK
cells; it can also be termed aggressive NK-cell lymphoma (194,195).
As regards miRNAs in T-cell and NK-cell lymphomas,
very little is known so far. MiRNA-21, miRNA-155, miRNA-150,
miRNA-142 and miRNA-494 are present in various forms of cutaneous
T-cell lymphomas, compared to related benign disorders (196,197).
miRNA-146a and miRNA-155 are also present in patients with
cutaneous T-cell lymphomas (1).
miRNA-223, miRNA-BART-20, miRNA-BART-8, miRNA-BART-16 and
miRNA-BART-9 are EBV-encoded and are associated with the activation
of the EBV oncoprotein, LMP-1 (197–204)
(Table VIII).
Concerning lncRNAs in various types of T-cell and
NK-cell lymphomas, MALAT1, located on chromosome 11q13.1, has been
identified as being overexpressed and leads to the induction of
BMI1 activation (197); that is the
reason why MALAT1 can be used as prognostic marker and therapeutic
target in T- and NK-cell lymphomas (195) (Table
VIII).
MM, also known as plasma cell myeloma, is a fatal
malignant hematological disorder which lead to the proliferation of
monoclonal antibody-secreting plasma cells; the main criterion is
the presence of clonal plasma cells >10% in bone marrow biopsy
or in a biopsy from other tissues (plasmacytoma). MM accounts for
10% of all hematological malignancies (205).
Compared to all types of B-cell Lymphomas, very
little is known about miRNA expression in patients with MM. As
regards lncRNAs in patients with MM, the following have been
observed (Table IX): i) GAS5,
located on chromosome 1q25.1, acting as a tumor-suppressor by
regulating the mTOR pathway (77–86); ii)
DLEU2, located on chromosome 13q14.3, acting as a tumor-suppressor
by being a host of the miR-15a/16-1 cluster and targeting BCL2
(117–120); iii) 3) MALAT1, located on
chromosome 11q13, acting as an oncogene by regulating the
bioavailability of TGF-β (146–152);
iv) MEG3, located on chromosome 14q32.2, acting as a
tumor-suppressor by interacting with p53 and regulating p53 gene
expression (206–209); v) TUG1, located on chromosome
22q12.2, acting as an oncogene by being induced by p53 (150,152);
vi) lnc-SENP5-4/NCBP2-AS2, located on chromosome 3q29, with an
uncharacterized mode of action (85); vii) 7) lnc-CPSF2-2, located on
chromosome 14q32, with an uncharacterized mode of action (85); viii) lnc-LRRC47-1/TP73-AS1, located
on chromosome 1p36, with an uncharacterized mode of action
(85); ix) lnc-ANGPTL1-3, located on
chromosome 1q25, with an uncharacterized mode of action (85); x) lnc-WHSC2-2, located on chromosome
4p16.3, with an uncharacterized mode of action (85).
There are specific strategies that can be used in
order to target ncRNAs in tumor management. These are the
following: i) Antisense oligonucleotides (ASOs), which can trigger
RNaseH-mediated RNA degradation (210); ii) CRISPR/Cas9 genome editing
technique which can effectively silence the transcription of the
lncRNA-expressing loci (211,212);
iii) viral vectors (adenovirus, lentivirus and retrovirus) which
can be used as a RNA interference (RNAi) method and can lead to the
knockdown of gene expression by neutralizing the targeted RNA
through exogenous double-stranded RNA insertion (213–215);
and iv) nanomedicine, including lipid-based nanoparticles
(liposomes) (216), polymer-based
nanoparticles and micelles (217),
dendrimers (218), carbon-based
nanoparticles (219), and metallic
and magnetic nanoparticles, such as gold nanoparticles (220,221).
All these novel therapeutic strategies targeting
ncRNAs, have been tested to date in preclinical models with
lymphoid disorders. For example, a viral vector carrying miR-28 has
been delivered in DLBCL and BL xenografts and in murine models with
B-lymphoma, with acceptable prophylactic and therapeutic effects
(222).
Furthermore, the ASO strategy, such as
LNA-anti-miR-155, has been used in a B-cell lymphoma murine model,
exhibiting a significant effect in murine models (223). An anti-miR-155 oligonucleotide with
the trademark Cobomarsen is currently being clinically nowadays in
patients with cutaneous T-cell lymphoma (224).
Double-stranded RNAi and ASOs are the most commonly
used lncRNA-targeted therapies. When the target lncRNA is localized
in the nucleus, ASOs are the better therapeutic option (225).
The most important finding, by reviewing the
literature, is that either the lncRNA expression signature or miRNA
expression may help distinguish between the different lymphoma
entities. In addition, as certain ncRNAs may be associated with the
progression of lymphoma or drug resistance, these ncRNAs can be
used as predictive and prognostic markers (225). However, the ncRNA regulatory
network is complex and is not yet fully understood, as the majority
of ncRNAs have not yet been thoroughly investigated. Nevertheless,
ncRNA-based therapeutics can be combined, in the near future, with
other techniques, such as chimeric antigen receptor (CAR) T-cell
immunotherapy, the targeting of tumor cells, thus improving their
therapeutic efficacy (226,227).
Contemporary developments in biology have been
combined with insightful discoveries analyzing the role of ncRNAs,
either miRNAs or lncRNAs in human tumors, particularly lymphomas,
such as: BCNHLs, HLs, T-cell/NK cell NHLs (T-/NK-cell NHLs) and
other B-cell malignancies, such as MM.
The present review aimed to provide a thorough
summary of the current understanding of ncRNAs in lymphoid
malignancies by summarizing, for the first time, to the best of our
knowledge, the whole existing ncRNA (and not into different
categories), miRNAs and lncRNAs, which are associated with lymphoid
disorders.
The initial data suggest that mostly lncRNAs, play
key roles in lymphangiogenesis, as a great number of them are
deregulated in B-cell malignancies. However, this particular field
is still in its infancy, with insufficient data; thus, further
studies need to be performed.
Concerning the role of miRNAs as biomarkers in all
lymphoid malignancies, ample data are available, although without
immediate use in clinical practice. A number of miRNA biomarker
studies to date on B-NHLs, HLs, T-/NK-NHLs and MM are not based on
multi-center cooperations, and thus, in most cases, a number of
reviews are non-overlapping and even contradictory.
For all the above reasons, further multi-center
studies are warranted with the establishment of a standardized
approach and the use of the same techniques: RT-qPCR, microarrays
or next-generation sequencing (NGS). This is mandatory step in
order to explore more thoroughly the role and functions of lncRNAs
in normal B-cells and malignant B-cells; as well as to perform a
more in-depth miRNA biomarker analysis in order to ensure that
these molecules can be effectively used in daily practice. These
tasks are both compelling and challenging in the next future for
the prognosis and potential therapeutic targeting of all lymphoid
malignancies; leading to a better treatment plan.
Not applicable.
No funding was received.
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
GD developed, planned, supervised the review and
wrote the manuscript. MG created the figure and contributed to the
writing of the manuscript. VZ supervised the review, contributed to
the writing and revisions of the manuscript. DAS and SA collected
relevant literature. VZ and GD confirm the authenticity of all the
raw data All authors read and approved the final manuscript.
Not applicable.
Not applicable.
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
|
1
|
Bardia A and Seifter E: Johns Hopkins
Patients' Guide to Lymphoma. 1st edition. Jones and Bartlett
Publishers, Inc.; pp. 1382010
|
|
2
|
The Lymphoma Guide, . Information for
Patients and Caregivers. Leukemia and Lymphoma Society; New York,
USA: 2013
|
|
3
|
World Health Organization: World Cancer
Report 2014. IARC Publications; Lyon, France: pp. 348–528. 2014
|
|
4
|
Swerdlow SH, Campo E, Pileri SA, Harris
NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz
AD and Jaffe ES: The 2016 revision of the World Health Organization
classification of lymphoid neoplasms. Blood. 127:2375–2390. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coiffier B: Monoclonal antibody as therapy
for malignant lymphomas. C R Biol. 329:241–254. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Küppers R, Klein U, Hansmann ML and
Rajewsky K: Cellular origin of human B-cell lymphomas. N Engl J
Med. 341:1520–1529. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
National Cancer Institute, . General
information about adult Hodgkin Lymphoma. 2014.
|
|
9
|
National Cancer Institute, . General
Information about adult Non-Hodgkin Lymphoma. 2014.
|
|
10
|
Hu L, Luo D, Zhou T, Tao Y, Feng J and Mei
S: The association between non-Hodgkin lymphoma and organophosphate
pesticides exposure: A meta-analysis. Environ Pollut. 231:319–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang L, Dong J, Jiang S, Shi W, Xu X,
Huang H, You X and Liu H: Red and processed meat consumption
increases risk for Non-Hodgkin lymphoma: A PRISMA-compliant
meta-analysis of observational studies. Medicine. 94:e17292015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Solimini AG, Lombardi AM, Palazzo C and De
Giusti M: Meat intake and non-Hodgkin lymphoma: A meta-analysis of
observational studies. Cancer Causes Control. 27:595–606. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
National Cancer Institute, . Cancer Stat
Facts: Non-Hodgkin Lymphoma. 2014.
|
|
14
|
Marcus R, Sweetenham J and Williams L:
Lymphoma: Pathology, diagnosis and treatment (2nd edition).
Cambridge Medicine. 3262014.
|
|
15
|
Tepper JE, Niederhuber JO, Armitage JH,
Doroshow MB and Kastan JE: Childhood Lymphoma (5th edition):
Abeloff's Clinical Oncology. Chapter. 97. Elsevier Inc.; 2014
|
|
16
|
Kamper-Jørgensen M, Rostgaard KG, Zahm SH,
Cozen W, Smedby KE, Sanjosé S, Chang ET, Zheng T, La Vecchia C,
Serraino D, et al: Cigarette smoking and risk of Hodgkin lymphoma
and its subtypes: A pooled analysis from the International Lymphoma
Epidemiology Consortium (InterLymph). Ann Oncol. 24:2245–2255.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Manli JN, Bennani N and Feldman AL:
Lymphoma classification update: T-cell lymphomas, Hodgkin lymphoma,
and histiocytic/dendritic cell neoplasms. Expert Rev Hematol.
10:239–249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sheikhpour R, Pourhosseini F, Neamatzadeh
H and Karimi R: Immunophenotype evaluation of Non-Hodgkin's
lymphomas. Med J Islam Repub Iran. 31:1212017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao Q, Mani RS, Ateeq B, Dhanasekaran SM,
Asangani IA and Prensner JR: Coordinated regulation of Polycomb
Group Complexes through microRNAs in Cancer. Cancer Cell.
20:187–199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He Y, Vogelstein B, Velculescu VE,
Papadopoulos N and Kinzler KW: The antisense transcriptomes of
human cells. Science. 322:1855–1857. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Faulkner GJ, Kimura Y, Daub CO, Wani S,
Plessy C and Irvine KM: The regulated retrotransposon transcriptome
of mammalian cells. Nat Genet. 41:563–571. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pérez-Vera P, Reyes-León A and
Fuentes-Pananá EM: Signaling proteins and transcription factors in
normal and malignant early B cell development. Bone Marrow Res.
2011:5027512011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alberts B, Johnson A, Lewis J, Raff M,
Roberts K and Walter P: Molecular Biology of the Cell. T cells and
B cells derive their names from the organs in which they develop. T
cells develop in the thymus and B cells, in mammals, develop in the
bone marrow in adults or the liver in fetuses. Garland Science; New
York, NY: pp. pg13672002
|
|
27
|
Petri A, Dybkaer K, Bogsted M, Thrue CA,
Hagedorn PH, Schmitz A, Bodker JS, Johnsen HE and Kauppinen S: Long
noncoding RNA expression during Human B-cell development. PLoS One.
10:e01382362015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Herzog S, Reth M and Jumaa H: Regulation
of B-cell proliferation and differentiation by pre-B-cell receptor
signalling. Nat Rev Immunol. 9:195–205. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Graham LD, Pedersen SK, Brown GS, Ho T,
Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, et
al: Colorectal Neoplasia differentially expressed (CRNDE), a novel
gene with elevated expression in colorectal adenomas and
adenocarcinomas. Genes Cancer. 2:829–840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long NonCoding RNA involved in CanceR, neurobiology, and
development. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ellis BC, Graham LD and Molloy PL: CRNDE,
a long noncoding RNA responsive to insulin/IGF signaling, regulates
genes involved in central metabolism. Biochim Biophys Acta.
1843:372–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Yebenes VG, Belver L, Pisano DG,
Gonzalez S, Villasante A, Croce C, He L and Ramiro AR: miR-181b
negatively regulates activation-induced cytidine deaminase in B
cells. J Exp Med. 205:2199–206. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Teng G, Hakimpour P, Landgraf P, Rice A,
Tuschl T, Casellas R and Papavasiliou FN: MicroRNA-155 is a
negative regulator of activation-induced cytidine deaminase.
Immunity. 28:621–629. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Smith A, Howell D, Patmore R, Jack A and
Roman E: Incidence of haematological malignancy by sub-type: A
report from the Haematological malignancy research network. Br J
Cancer. 105:1684–1692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li S, Young KH and Medeiros LJ: Diffuse
large B-cell lymphoma. Pathology. 50:74–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lenz G, Wright GW, Emre NC, Kohlhammer H,
Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al:
Molecular subtypes of diffuse large B-cell lymphoma arise by
distinct genetic pathways. Proc Natl Acad Sci USA. 105:13520–13525.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sukswai N, Lyapichev K, Khoury JD and
Medeiros LJ: Diffuse large B-cell lymphoma variants: An update.
Pathology. 52:53–67. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shimada K, Hayakawa F and Kiyoi H: Biology
and management of primary effusion lymphoma. Blood. 132:1879–1888.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cheng Y, Xiao Y, Zhou R, Liao Y, Zhou J
and Ma X: Prognostic significance of Helicobacter
pylori-infection in gastric diffuse large B-cell lymphoma. BMC
Cancer. 19:8422019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kuo SH, Yeh KH, Chen LT, Lin CW, Hsu PN,
Hsu C, Wu MS, Tzeng YS, Tsai HJ, Wang HP and Cheng AL:
Helicobacter pylori-related diffuse large B-cell lymphoma of
the stomach: A distinct entity with lower aggressiveness and higher
chemosensitivity. Blood Cancer J. 4:e2202014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Abramson JS: Hitting back at lymphoma: How
do modern diagnostics identify high-risk diffuse large B-cell
lymphoma subsets and alter treatment? Cancer. 125:3111–3120. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chavez JC and Locke FL: CAR T cell therapy
for B-cell lymphomas. Best Pract Res Clin Haematol. 31:135–146.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu Y and Barta SK: Diffuse large B-cell
lymphoma: 2019 update on diagnosis, risk stratification, and
treatment. Am J Hematol. 94:604–616. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Roehle A, Hoefig KP, Repsilber D, Thorns
C, Ziepert M, Wesche KO, Thiere M, Loeffler M, Klapper W,
Pfreundschuh M, et al: MicroRNA signatures characterize diffuse
large B-cell lymphomas and follicular lymphomas. Br J Haematol.
142:732–744. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lawrie CH, Chi J, Taylor S, Tramonti D,
Ballabio E, Palazzo S, Saunders NJ, Pezzella F, Boultwood J,
Wainscoat JS and Hatton CS: Expression of microRNAs in diffuse
large B cell lymphoma is associated with immunophenotype, survival
and transformation from follicular lymphoma. J Cell Mol Med.
13:1248–1260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Caramuta S, Lee L, Ozata DM, Akçakaya P,
Georgii-Hemming P, Xie H, Amini RM, Lawrie CH, Enblad G, Larsson C,
et al: Role of microRNAs and microRNA machinery in the pathogenesis
of diffuse large B-cell lymphoma. Blood Cancer J. 3:e1522013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lawrie CH, Saunders NJ, Soneji S, Palazzo
S, Dunlop HM, Cooper CD, Brown PJ, Troussard X, Mossafa H, Enver T,
et al: MicroRNA expression in lymphocyte development and
malignancy. Leukemia. 22:1440–1446. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhong H, Xu L, Zhong JH, Xiao F, Liu Q,
Huang HH and Chen FY: Clinical and prognostic significance of
miR-155 and miR-146a expression levels in
formalin-fixed/paraffin-embedded tissue of patients with diffuse
large B-cell lymphoma. Exp Ther Med. 3:763–770. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chapuy B, Stewart C, Dunford AJ, Kim J,
Kamburov A, Redd RA, Lawrence MS, Roemer MGM, Li AJ, Ziepert M, et
al: Molecular subtypes of diffuse large B cell lymphoma are
associated with distinct pathogenic mechanisms and outcomes. Nat
Med. 24:679–690. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhou K, Feng X, Wang Y, Liu Y, Tian L, Zuo
Z, Yi S, Wei X, Song Y and Qiu L: miR-223 is repressed and
correlates with inferior clinical features in mantle cell lymphoma
through targeting SOX11. Exp Hematol. 58:27–34. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bouteloup M, Verney A, Rachinel N,
Callet-Bauchu E, Ffrench M, Coiffier B, Magaud JP, Berger F, Salles
GA and Traverse-Glehen A: MicroRNA expression profile in splenic
marginal zone lymphoma. Br J Haematol. 156:279–281. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fabbri M, Bottoni A, Shimizu M, Spizzo R,
Nicoloso MS, Rossi S, Barbarotto E, Cimmino A, Adair B, Wojcik SE,
et al: Association of a microRNA/TP53 feedback circuitry with
pathogenesis and outcome of B-cell chronic lymphocytic leukemia.
JAMA. 305:59–67. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hanke M, Hoefig K, Merz H, Feller AC,
Kausch I, Jocham D, Warnecke JM and Sczakiel G: A robust
methodology to study urine microRNA as tumor marker: microRNA-126
and microRNA-182 are related to urinary bladder cancer. Urol Oncol.
28:655–661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
He M, Gao L, Zhang S, Tao L, Wang J, Yang
J and Zhu M: Prognostic significance of miR-34a and its target
proteins of FOXP1, p53, and BCL2 in gastric MALT lymphoma and
DLBCL. Gastric Cancer. 17:431–441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jia YJ, Liu ZB, Wang WG, Sun CB, Wei P,
Yang YL, You MJ, Yu BH, Li XQ and Zhou XY: HDAC6 regulates
microRNA-27b that suppresses proliferation, promotes apoptosis and
target MET in diffuse large B-cell lymphoma. Leukemia. 32:703–711.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gu L, Song G, Chen L, Nie Z, He B, Pan Y,
Xu Y, Li R, Gao T, Cho WC and Wang S: Inhibition of miR-21 induces
biological and behavioral alterations in diffuse large B-cell
lymphoma. Acta Haematol. 130:87–94. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zheng Z, Li X, Zhu Y, Gu W, Xie X and
Jiang J: Prognostic significance of miRNA in patients with diffuse
large B-cell lymphoma: A meta-analysis. Cell Physiol Biochem.
39:1891–1904. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Pillar N, Bairey O, Goldschmidt N, Fellig
Y, Rosenblat Y, Shehtman I, Haguel D, Raanani P, Shomron N and
Siegal T: MicroRNAs as predictors for CNS relapse of systemic
diffuse large B-cell lymphoma. Oncotarget. 8:86020–86030. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lawrie CH, Gal S, Dunlop HM, Pushkaran B,
Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J,
Wainscoat JS, et al: Detection of elevated levels of
tumourassociated microRNAs in serum of patients with diffuse large
B-cell lymphoma. Br J Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bomben R, Gobessi S, Dal Bo M, Volinia M,
Marconi D, Tissino E, Benedetti D, Zucchetto A, Rossi D, Gaidano G,
et al: The miR-17~92 family regulates the response to Toll-like
receptor 9 triggering of CLL cells with unmutated IGHV genes.
Leukemia. 26:1584–1593. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Calin GA, Ferracin M, Cimmino A, Di Leva
GD, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M,
et al: A microRNA signature associated with prognosis and
progression in chronic lymphocytic leukemia. N Engl J Med.
353:1793–1801. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ferrajoli A, Shanafelt TD, Ivan C, Ivan C,
Shimizu M, Rabe KG, Nouraee N, Ikuo M, Ghosh AK, Lerner S, et al:
Prognostic value of miR-155 in individuals with monoclonal B-cell
lymphocytosis and patients with B chronic lymphocytic leukemia.
Blood. 122:1891–1899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Iqbal J, Shen Y, Huang X, Liu Y, Wake L,
Liu C, Deffenbacher K, Lachel CM, Wang C, Rohr J, et al: Global
microRNA expression profiling uncovers molecular markers for
classification and prognosis in aggressive B-cell lymphoma. Blood.
125:1137–1145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ferracin M, Zagatti B, Rizzotto L,
Cavazzini F, Veronese A, Ciccone M, Saccenti E, Lupini L, Grilli A,
De Angeli C, et al: MicroRNAs involvement in fludarabine refractory
chronic lymphocytic leukemia. Mol Cancer. 9:1232010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Khare D, Goldschmidt N, Bardugo A,
Gur-Wahnon D, Ben-Dov IZ and Avni B: Plasma microRNA profiling:
Exploring better biomarkers for lymphoma surveillance. PLoS One.
12:e01877222017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Meng Y, Quan L and Liu A: Identification
of key microRNAs associated with diffuse large B-cell lymphoma by
analyzing serum microRNA expressions. Gene. 642:205–211. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Song G, Gu L, Li J, Tang Z, Liu H, Chen B,
Sun X, He B, Pan Y, Wang S and Cho WC: Serum microRNA expression
profiling predict response to R-CHOP treatment in diffuse large B
cell lymphoma patients. Ann Hematol. 93:1735–1743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Leivonen SK, Icay K, Jäntti K, Siren I,
Liu C, Alkodsi A, Cervera A, Ludvigsen M, Hamilton-Dutoit SJ,
d'Amore F, et al: MicroRNAs regulate key cell survival pathways and
mediate chemosensitivity during progression of diffuse large B-cell
lymphoma. Blood Cancer J. 7:6542017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Thompson MA, Edmonds MD, Liang S,
McClintock-Treep S, Wang X, Li S and Eischen CM: miR-31 and
miR-17-5p levels change during transformation of follicular
lymphoma. Human Pathol. 50:118–126. 2016. View Article : Google Scholar
|
|
71
|
Leich E, Zamo A, Horn H, Haralambieva E,
Puppe B, Gascoyne RD, Chan WC, Braziel RM, Rimsza LM, Weisenburger
DD, et al: MicroRNA profiles of t(14;18)-negative follicular
lymphoma support a late germinal center B-cell phenotype. Blood.
118:5550–5558. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Peng W, Wu J and Feng J: LincRNA-p21
predicts favorable clinical outcome and impairs tumorigenesis in
diffuse large B cell lymphoma patients treated with R-CHOP
chemotherapy. Clin Exp Med. 17:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Blume CJ, Hotz-Wagenblatt A, Hullein J,
Sellner L, Jethwa A, Stolz T, Slabicki M, Lee K, Sharathchandra A,
Benner A, et al: p53-dependent non-coding RNA networks in chronic
lymphocytic leukemia. Leukemia. 29:2015–2023. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yoon JH, Abdelmohsen K, Srikantan S, Yang
X, Martindale JL, De S, Huarte M, Zhan M, Becker KG and Gorospe M:
LincRNA-p21 suppresses target mRNA translation. Mol Cell.
47:648–655. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Peponi E, Drakos E, Reyes G, Leventaki V,
Rassidakis GZ and Medeiros LJ: Activation of mammalian target of
rapamycin signaling promotes cell cycle progression and protects
cells from apoptosis in mantle cell lymphoma. The Am J Pathol.
169:2171–2180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Mourtada-Maarabouni M and Williams GT:
Role of GAS5 noncoding RNA in mediating the effects of rapamycin
and its analogues on mantle cell lymphoma cells. Clin Lymphoma
Myeloma Leuk. 14:468–473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Coccia EM, Cicala C, Charlesworth A,
Ciccarelli C, Rossi GB, Philipson L and Sorrentino V: Regulation
and expression of a growth arrest-specific gene (gas5) during
growth, differentiation, and development. Mol Cell Biol.
12:3514–3521. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nakamura Y, Takahashi N, Kakegawa E,
Yoshida K, Ito Y, Kayano H, Niitsu N, Jinnai I and Bessho M: The
GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a
result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer
Genet Cytogenet. 182:144–149. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F
and Song Y: A critical role for the long non-coding RNA GAS5 in
proliferation and apoptosis in non-small-cell lung cancer. Mol
Carcinog. 54 (Suppl 1):E1–E12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Williams GT, Mourtada-Maarabouni M and
Farzaneh F: A critical role for non-coding RNA GAS5 in growth
arrest and rapamycin inhibition in human T-lymphocytes. Biochem Soc
Trans. 39:482–486. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mourtada-Maarabouni M, Hasan AM, Farzaneh
F and Williams GT: Inhibition of human T-cell proliferation by
mammalian target of rapamycin (mTOR) antagonists requires noncoding
RNA growth-arrest-specific transcript 5 (GAS5). Mol Pharmacol.
78:19–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ronchetti D, Agnelli L, Taiana E, Galletti
S, Manzoni M, Todoerti K, Musto P, Strozzi F and Neri A: Distinct
lncRNA transcriptional fingerprints characterize progressive stages
of multiple myeloma. Oncotarget. 7:14814–14830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvationassociated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Conde L, Riby J, Zhang J, Bracci PM and
Skibola CF: Copy number variation analysis on a non-Hodgkin
lymphoma case-control study identifies an 11q25 duplication
associated with diffuse large B-cell lymphoma. PLoS One.
9:e1053822014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lu Z, Pannunzio NR, Greisman HA, Casero D,
Parekh C and Lieber MR: Convergent BCL6 and lncRNA promoters
demarcate the major breakpoint region for BCL6 translocations.
Blood. 126:1730–1731. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Peng W and Feng J: Long noncoding RNA
LUNAR1 associates with cell proliferation and predicts a poor
prognosis in diffuse large B-cell lymphoma. Biomed Pharmacother.
77:65–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Trimarchi T, Bilal E, Ntziachristos P,
Fabbri G, DallaFavera R, Tsirigos A and Aifantis I: Genome-wide
mapping and characterization of Notch-regulated long noncoding RNAs
in acute leukemia. Cell. 158:593–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Peng W, Fan H, Wu G, Wu J and Feng J:
Upregulation of long noncoding RNA PEG10 associates with poor
prognosis in diffuse large B cell lymphoma with facilitating
tumorigenicity. Clin Exp Med. 16:177–182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ono R, Kobayashi S, Wagatsuma H, Aisaka K,
Kohda T, Kaneko-Ishino T and Ishino F: A retrotransposon-derived
gene, PEG10, is a novel imprinted gene located on human chromosome
7q21. Genomics. 73:232–237. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Li CM, Margolin AA, Salas M, Memeo L,
Mansukhani M, Hibshoosh H, Szabolcs M, Klinakis A and Tycko B:
PEG10 is a c-MYC target gene in cancer cells. Cancer Res.
66:665–672. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Peng W, Wu J and Feng J: Long noncoding
RNA HULC predicts poor clinical outcome and represents
pro-oncogenic activity in diffuse large B-cell lymphoma. Biomed
Pharmacother. 79:188–193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Hammerle M, Gutschner T, Uckelmann H,
Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T,
Breuhahn K, et al: Posttranscriptional destabilization of the
liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding
protein 1 (IGF2BP1). Hepatology. 58:1703–1712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. Biomed Res Int. 2013:1361062013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Peng W, Gao W and Feng J: Long noncoding
RNA HULC is a novel biomarker of poor prognosis in patients with
pancreatic cancer. Med Oncol. 31:3462014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yan Y, Han J, Li Z, Yang H, Sui Y and Wang
M: Elevated RNA expression of long noncoding HOTAIR promotes cell
proliferation and predicts a poor prognosis in patients with
diffuse large B cell lymphoma. Mol Med Rep. 13:5125–5131. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hallek M, Shanafelt TD and Eichhorst B:
Chronic lymphocytic leukaemia. Lancet. 391:1524–1537. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Tresckow JV, Eichhorst B, Bahlo J and
Hallek M: The treatment of chronic lymphatic leukemia. Dtsch
Arztebl Int. 116:41–46. 2019.PubMed/NCBI
|
|
101
|
Choi SM and O'Malley DP: Diagnostically
relevant updates to the WHO classification of lymphoid neoplasms.
Ann Diagn Pathol. 37:67–74. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Matutes E, Owusu-Ankomah K, Morilla R,
Garcia Marco J, Houlihan A, Que TH and Catovsky D: The
immunological profile of B-cell disorders and proposal of a scoring
system for the diagnosis of CLL. Leukemia. 8:1640–1645.
1994.PubMed/NCBI
|
|
103
|
Hallek M: Chronic lymphocytic leukemia:
Update on diagnosis, risk stratification, and treatment. Am J
Hematol. 92:946–965. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Deans JP and Polyak MJ: FMC7 is an epitope
of CD20. Blood. 111:24922008. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Palumbo GA, Parrinello N, Fargione G,
Cardillo K, Chiarenza A, Berretta S, Conticello C, Villari L and Di
Raimondo F: CD200 expression may help in differential diagnosis
between mantle cell lymphoma and B-cell chronic lymphocytic
leukemia. Leuk Res. 33:1212–1216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Sampath D, Liu C, Vasan K, Sulda M,
Puduvalli VK, Wierda WG and Keating MJ: Histone deacetylases
mediate the silencing of miR-15a, miR-16, and miR-29b in chronic
lymphocytic leukemia. Blood. 119:1162–1172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Rossi S, Shimizu M, Barbarotto E, Nicoloso
MS, Dimitri F, Sampath D, Fabbri M, Lerner S, Barron LL, Rassenti
LZ, et al: MicroRNA fingerprinting of CLL patients with chromosome
17p deletion identify a miR-21 score that stratifies early
survival. Blood. 116:945–952. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Visone R, Veronese A, Balatti V and Croce
CM: MiR-181b: New perspective to evaluate disease progression in
chronic lymphocytic leukemia. Oncotarget. 3:195–202. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Cui B, Chen L, Zhang S, Mraz M, Fecteau
JF, Yu J, Ghia EM, Zhang L, Bao L, Rassenti LZ, et al: MicroRNA-155
influences B-cell receptor signaling and associates with aggressive
disease in chronic lymphocytic leukemia. Blood. 124:546–554. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Caivano A, La Rocca F, Simeon V, Girasole
M, Dinarelli S, Laurenzana I, De Stradis A, De Luca L, Trino S,
Traficante A, et al: MicroRNA-155 in serum-derived extracellular
vesicles as a potential biomarker for hematologic malignancies-a
short report. Cell Oncol (Dordr). 40:97–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Filip AA, Grenda A, Popek S, Koczkodaj D,
Wojnowska MM, Budzyński M, Szczepanek EW, Zmorzyński S,
Karczmarczyk A and Giannopoulos K: Expression of circulating miRNAs
associated with lymphocyte differentiation and activation in
CLL-another piece in the puzzle. Ann Hematol. 96:33–50. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Gaidano G, Foà R and Dalla-Favera R:
Molecular pathogenesis of chronic lymphocytic leukemia. J Clin
Invest. 122:3432–3438. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Dohner H, Stilgenbauer S, Benner A,
Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M and Lichter P:
Genomic aberrations and survival in chronic lymphocytic leukemia. N
Engl J Med. 343:1910–1916. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR16 induce apoptosis by targetinag BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Fathullahzadeh S, Mirzaei H, Honardoost
MA, Sahebkar A and Salehi M: Circulating microRNA-192 as a
diagnostic biomarker in human chronic lymphocytic leukemia. Cancer
Gene Ther. 23:327–332. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Klein U, Lia M, Crespo M, Siegel R, Shen
Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G and
Dalla-Favera R: The DLEU2/miR-15a/16-1 cluster controls B cell
proliferation and its deletion leads to chronic lymphocytic
leukemia. Cancer Cell. 17:28–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Lerner M, Harada M, Loven J, Castro J,
Davis Z, Oscier D, Henriksson M, Sangfelt O, Grander D and Corcoran
MM: DLEU2, frequently deleted in malignancy, functions as a
critical host gene of the cell cycle inhibitory microRNAs miR-15a
and miR-16-1. Exp Cell Res. 315:2941–2952. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Garding A, Bhattacharya N, Claus R, Ruppel
M, Tschuch C, Filarsky K, Idler I, Zucknick M, Caudron-Herger M,
Oakes C, et al: Epigenetic upregulation of lncRNAs at 13q14.3 in
leukemia is linked to the In Cis downregulation of a gene cluster
that targets NF-kB. PLoS Genet. 9:e10033732013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Baer C, Oakes CC and Ruppert AS:
Epigenetic silencing of miR-708 enhances NF-κB signaling in chronic
lymphocytic leukemia. Int J Cancer. 137:1352–1361. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Naganuma T, Nakagawa S, Tanigawa A, Sasaki
YF, Goshima N and Hirose T: Alternative 3′-end processing of long
noncoding RNA initiates construction of nuclear paraspeckles. EMBO
J. 31:4020–4034. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Sattari A, Siddiqui H, Moshiri F, Ngankeu
A, Nakamura T, Kipps TJ and Croce CM: Upregulation of long
noncoding RNA MIAT in aggressive form of chronic lymphocytic
leukemias. Oncotarget. 7:54174–54182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Ishii N, Ozaki K, Sato H, Mizuno H, Saito
S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al:
Identification of a novel noncoding RNA, MIAT, that confers risk of
myocardial infarction. J Hum Genet. 51:1087–1099. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ip JY and Nakagawa S: Long non-coding RNAs
in nuclear bodies. Dev Growth Differ. 54:44–54. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sheik Mohamed J, Gaughwin PM, Lim B,
Robson P and Lipovich L: Conserved long noncoding RNAs
transcriptionally regulated by Oct4 and Nanog modulate pluripotency
in mouse embryonic stem cells. RNA. 16:324–337. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Ferreira PG, Jares P, Rico D, Gomez-López
G, Martínez-Trillos A, Villamor N, Ecker S, Gonzalez-Perez A,
Knowles DG, Monlong J, et al: Transcriptome characterization by RNA
sequencing identifies a major molecular and clinical subdivision in
chronic lymphocytic leukemia. Genome Res. 24:212–226. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Ronchetti D, Manzoni M, Agnelli L and
Vinci C: lncRNA profiling in early-stage chronic lymphocytic
leukemia identifies transcriptional fingerprints with relevance in
clinical outcome. Blood Cancer J. 6:e4682016. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Park SM, Park SJ, Kim HJ, Kwon OH, Kang
TW, Sohn HA, Kim SK, Moo Noh S, Song KS, Jang SJ, et al: A known
expressed sequence tag, BM742401, is a potent lincRNA inhibiting
cancer metastasis. Exp Mol Med. 45:e312013. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Wang LQ, Wong KY, Li ZH and Chim CS:
Epigenetic silencing of tumor suppressor long non-coding RNA
BM742401 in chronic lymphocytic leukemia. Oncotarget.
7:82400–82410. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Eis PS, Tam W, Sun L, Chadburn A, Li Z,
Gomez MF, Lund E and Dahlberg JE: Accumulation of miR-155 and BIC
RNA in human B cell lymphomas. Proc Natl Acad Sci USA.
102:3627–3632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Tam W: Identification and characterization
of human BIC, a gene on chromosome 21 that encodes a noncoding RNA.
Gene. 274:157–167. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Elton TS, Selemon H, Elton SM and
Parinandi NL: Regulation of the MIR155 host gene in physiological
and pathological processes. Gene. 532:1–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Stamatopoulos B, Van Damme M, Crompot E,
Dessars B, Housni HE, Mineur P, Meuleman N, Bron D and Lagneaux L:
Opposite prognostic significance of cellular and serum circulating
MicroRNA-150 in patients with chronic lymphocytic leukemia. Mol
Med. 21:123–133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Georgiadis P, Liampa I, Hebels DG,
Krauskopf J, Chatziioannou A, Valavanis I, de Kok TMCM, Kleinjans
JCS, Bergdahl IA, Melin B, et al: Evolving DNA methylation and gene
expression markers of B-cell chronic lymphocytic leukemia are
present in pre-diagnostic blood samples more than 10 years prior to
diagnosis. BMC Genomics. 18:7282017. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Gascoyne RD, Nadel B, Pasqualucci L,
Fitzgibbon J, Payton JE, Melnick A, Weigert O, Tarte K, Gribben JG,
Friedberg JW, et al: Follicular lymphoma: State-of-the-art ICML
workshop in Lugano 2015. Hematological Oncol. 35:397–407. 2017.
View Article : Google Scholar
|
|
137
|
Boughan K and Caimi PF: Follicular
lymphoma: Diagnostic and prognostic considerations in initial
treatment approach. Curr Oncol Rep. 21:632019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Pan Y, Li H, Guo Y, Luo Y, Li H, Xu Y,
Deng J and Sun B: A pilot study of long noncoding RNA expression
profiling by microarray in follicular lymphoma. Gene. 577:132–139.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Wang W, Corrigan-Cummins M, Hudson J,
Maric I, Simakova O, Neelapu SS, Kwak LW, Janik JE, Gause B, Jaffe
ES and Calvo KR: MicroRNA profiling of follicular lymphoma
identifies microRNAs related to cell proliferation and tumor
response. Haematologica. 97:586–594. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Arribas AJ, Campos-Martín Y, Gómez-Abad C,
Algara P, Sánchez-Beato M, Rodriguez-Pinilla MS, Montes-Moreno S,
Martinez N, Alves-Ferreira J, Piris MA and Mollejo M: Nodal
marginal zone lymphoma: Gene expression and miRNA profiling
identify diagnostic markers and potential therapeutic targets.
Blood. 119:e9–e21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Jares P, Colomer D and Campo E: Molecular
pathogenesis of mantle cell lymphoma. J Clin Invest. 122:3416–3423.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Carvajal-Cuenca A, Sua LF, Silva NM,
Pittaluga S, Royo C, Song JY, Sargent RL, Espinet B, Climent F,
Jacobs SA, et al: In situ mantle cell lymphoma: Clinical
implications of an incidental finding with indolent clinical
behavior. Haematologica. 97:270–278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Navarro A, Beà S, Fernández V, Prieto M,
Salaverria I, Jares P, Hartmann E, Mozos A, López-Guillermo A,
Villamor N, et al: MicroRNA expression, chromosomal alterations,
and immunoglobulin variable heavy chain hypermutations in Mantle
cell lymphomas. Cancer Res. 69:7071–7078. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Chen RW, Bemis LT, Amato CM, Myint H, Tran
H, Birks DK, Eckhardt SG and Robinson WA: Truncation in CCND1 mRNA
alters miR-16-1 regulation in mantle cell lymphoma. Blood.
112:822–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Iqbal J, Shen Y, Liu Y, Fu K, Jaffe ES,
Liu C, Liu Z, Lachel CM, Deffenbacher K, Greiner TC, et al:
Genome-wide miRNA profiling of mantle cell lymphoma reveals a
distinct subgroup with poor prognosis. Blood. 119:4939–4948. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Wang X, Sehgal L, Jain N, Khashab T,
Mathur R and Samaniego F: LncRNA MALAT1 promotes development of
mantle cell lymphoma by associating with EZH2. J Transl Med.
14:3462016. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Yang F, Yi F, Han X, Du Q and Liang Z:
MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett.
587:3175–3181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Cho SF, Chang YC, Chang CS, Lin SF, Liu
YC, Hsiao HH, Chang JG and Liu TC: MALAT1 long non-coding RNA is
overexpressed in multiple myeloma and may serve as a marker to
predict disease progression. BMC Cancer. 14:8092014. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Isin M, Ozgur E, Cetin G, Erten N, Aktan
M, Gezer U and Dalay N: Investigation of circulating lncRNAs in
B-cell neoplasms. Clin Chim Acta. 431:255–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Li B, Chen P, Qu J, Shi L and Zhuang W, Fu
J, Li J, Zhang X, Sun Y and Zhuang W: Activation of LTBP3 gene by a
long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem
cells from multiple myeloma. J Biol Chem. 289:29365–29375. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Matsumoto T and Abe M: TGF-β-related
mechanisms of bone destruction in multiple myeloma. Bone.
48:129–134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Arakawa F, Kimura Y, Yoshida N, Miyoshi H,
Doi A, Yasuda K, Nakajima K, Kiyasu J, Niino D, Sugita Y, et al:
Identification of miR-15b as a transformation-related factor in
mantle cell lymphoma. Int J Oncol. 48:485–492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Roisman A, Huamán Garaicoa F, Metrebian F,
Narbaitz M, Kohan D, García Rivello H, Fernandez I, Pavlovsky A,
Pavlovsky M, Hernández L and Slavutsky I: SOXC and miR17-92 gene
expression profiling defines two subgroups with different clinical
outcome in mantle cell lymphoma. Genes Chromosomes Cancer.
55:531–540. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Zhao JJ, Lin J, Lwin T, Yang H, Guo J,
Kong W, Dessureault S, Moscinski LC, Rezania D, Dalton WS, et al:
microRNA expression profile and identification of miR-29 as a
prognostic marker and pathogenetic factor by targeting CDK6 in
mantle cell lymphoma. Blood. 115:2630–2639. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Di Lisio L, Gomez-López G, Sánchez-Beato
M, Gómez-Abad C, Rodrıguez ME, Villuendas R, Ferreira BI, Carro A,
Rico D, Mollejo M, et al: Mantle cell lymphoma: Transcriptional
regulation by microRNAs. Leukemia. 24:1335–1342. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Husby S, Ralfkiaer U, Garde C, Zandi R, Ek
S, Kolstad A, Jerkeman M, Laurell A, Räty R, Pedersen LB, et al:
miR-18b overexpression identifies mantle cell lymphoma patients
with poor outcome and improves the MIPI-B prognosticator. Blood.
125:2669–2677. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Molyneux E, Rochford R, Griffin B, Newton
R, Jackson G, Menon G, Harrison C, Israels T and Bailey S:
Burkitt's lymphoma. Lancet. 379:1234–1244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Hoffman R: Hematology: Basic Principles
and Practice (5th edition). Churchill Livingstone/Elsevier;
Philadelphia, PA: pp. 1304–1305. 2009
|
|
160
|
Smardova J, Grochova D, Fabian P, Moulis
M, Smarda J, Falkova I, Ravcukova B, Vankova J and Vasova I: An
unusual p53 mutation detected in Burkitt's lymphoma: 30 bp
duplication. Oncol Rep. 20:773–778. 2008.PubMed/NCBI
|
|
161
|
Liu D, Shimonov J, Primanneni S, Lai Y,
Ahmed T and Seiter K: t(8;14;18): A 3-way chromosome translocation
in two patients with Burkitt's lymphoma/leukemia. Mol Cancer.
6:352007. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Dorsett Y, McBride KM, Jankovic M,
Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, Reina San-Martin
B, Heidkamp G, Schwickert TA, et al: MicroRNA-155 suppresses
activation-induced cytidine deaminase-mediated Myc-Igh
translocation. Immunity. 28:630–638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Gao P, Tchernyshyov I, Chang TC, Lee YS,
Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT and
Dang CV: c-Myc suppression of miR-23a/b enhances mitochondrial
glutaminase expression and glutamine metabolism. Nature.
458:762–765. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Leucci E, Cocco M, Onnis A, De Falco G,
van Cleef P, Bellan C, van Rijk A, Nyagol J, Byakika B, Lazzi S, et
al: MYC translocation-negative classical Burkitt lymphoma cases: An
alternative pathogenetic mechanism involving miRNA deregulation. J
Pathol. 216:440–450. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Lenze D, Leoncini L, Hummel M, Volinia S,
Liu CG, Amato T, De Falco G, Githanga J, Horn H, Nyagol J, et al:
The different epidemiologic subtypes of Burkitt lymphoma share a
homogenous micro RNA profile distinct from diffuse large B-cell
lymphoma. Leukemia. 25:1869–1876. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Hezaveh K, Kloetgen A, Bernhart SH,
Mahapatra KD, Lenze D, Richter J, Haake A, Bergmann AK, Brors B,
Burkhardt B, et al: Alterations of microRNA and microRNA-regulated
messenger RNA expression in germinal center B-cell lymphomas
determined by integrative sequencing analysis. Haematologica.
101:1380–1389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Chang TC, Yu D, Lee YS, Wentzel EA, Arking
DE, West KM, Dang CV, Tikhonenko TA and Mendell JT: Widespread
microRNA repression by Myc contributes to tumorigenesis. Nat Genet.
40:43–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Bueno MJ, Gómez de Cedron M, Gomez-López
G, Pérez de Castro I, Di Lisio L, Montes-Moreno S, Martinez N,
Guerrero M, Sanchez-Martinez R, Santos J, et al: Combinatorial
effects of microRNAs to suppress the Myc oncogenic pathway. Blood.
117:6255–6266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Oduor CI, Kaymaz Y, Chelimo K, Otieno JA,
Ongecha JM, Moormann AM and Bailey JA: Integrative microRNA and
mRNA deep-sequencing expression profiling in endemic Burkitt
lymphoma. BMC Cancer. 17:7612017. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Ott G, Rosenwald A and Campo E:
Understanding MYC-driven aggressive B-cell lymphomas: Pathogenesis
and classification. Blood. 122:3884–3891. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Robaina MC, Mazzoccoli L, Arruda VO, de
Souza Reis FR, Apa AG, de Rezende LMM and Klumb CE: Deregulation of
DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel
contribution for disease pathogenesis. Exp Molec Pathol.
98:200–207. 2015. View Article : Google Scholar
|
|
172
|
Li JG, Ding Y, Huang YM, Chen WL, Pan LL,
Li Y, Chen XL, Chen Y, Wang SY and Wu XN: FAMLF is a target of
miR-181b in Burkitt lymphoma. Braz J Med Biol Res. 50:e56612017.
View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Doose G, Haake A, Bernhart SH, Lopez C,
Duggimpudi S, Wojciech F, Bergmann AK, Borkhardt A, Burkhardt B,
Claviez A, et al: MINCR is a MYC-induced lncRNA able to modulate
MYC's transcriptional network in Burkitt lymphoma cells. Proc Natl
Acad Sci USA. 112:E5261–5270. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Watkins AJ, Hamoudi RA, Zeng N, Yan O,
Huang Y, Liu H, Zhang J, Braggio E, Fonseca R, de Level L, et al:
An integrated genomic and expression analysis of 7q deletion in
splenic marginal zone lymphoma. PLoS One. 7:e449972012. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Thorns C, Kuba J, Bernard V, Senft A,
Szymczak S, Feller AC and Bernd HW: Deregulation of a distinct set
of microRNAs is associated with transformation of gastritis into
MALT lymphoma. Virchows Arch. 460:371–377. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Fernández C, Bellosillo B, Ferraro M,
Seoane A, Sánchez-González B, Pairet S, Pons A, Barranco L, Vela
MC, Gimeno E, et al: MicroRNAs 142-3p, miR-155 and miR-203 are
deregulated in gastric MALT lymphomas compared to chronic
gastritis. Cancer Genomics Proteomics. 14:75–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Liu TY, Chen SU, Kuo SH, Cheng AL and Lin
CW: E2A-positive gastric MALT lymphoma has weaker plasmacytoid
infiltrates and stronger expression of the memory B-cell-associated
miR-223: Possible correlation with stage and treatment response.
Modern Pathol. 23:1507–1517. 2010. View Article : Google Scholar
|
|
178
|
McKay P, Fielding P, Gallop-Evans E, Hall
GW, Lambert J, Leach M, Marafioti T and McNamara C: Guidelines for
the investigation and management of nodular lymphocyte predominant
Hodgkin lymphoma. Br J Haematol. 172:32–43. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Swerdlow SH, Campo E, Harris NL, Jaffe ES,
Pileri SA, Stein H and Thiele J: WHO classification of tumours of
haematopoietic and lymphoid tissues. World Health Organization,
International Agency for Research on Cancer (Revised 4th edition).
2018.
|
|
180
|
Re D, Roman TK, Behringer K and Diehl V:
From Hodgkin disease to Hodgkin lymphoma: Biologic insights and
therapeutic potential. Blood. 105:4553–4560. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Tan LP, Seinen E, Duns G, de Jong D, Sibon
OC, Poppema S, Kroesen BJ, Kok K and van den Berg A: A high
throughput experimental approach to identify miRNA targets in human
cells. Nucleic Acids Res. 37:e1372009. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Navarro A, Diaz T, Martinez A, Gaya A,
Pons A, Gel B, Codony C, Ferrer G, Martinez C, Montserrat E and
Monzo M: Regulation of JAK2 by miR-135a: Prognostic impact in
classic Hodgkin lymphoma. Blood. 114:2945–2951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Nie K, Gomez M, Landgraf P, Garcia JF, Liu
Y, Tan LH, Chadburn A, Tuschl T, Knowles DM and Tam W: MicroRNA
mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg
cells: A potential pathogenetic lesion in Hodgkin lymphomas. Am J
Pathol. 173:242–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Leucci E, Zriwil A, Gregersen LH, Jensen
KT, Obad S, Bellan C, Leoncini L, Kauppinen S and Lund AH:
Inhibition of miR-9 de-represses HuR and DICER1 and impairs Hodgkin
lymphoma tumour outgrowth in vivo. Oncogene. 31:5081–5089. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
185
|
van den Berg A, Kroesen BJ, Kooistra K, de
Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A,
Maggio E and Poppema S: High expression of B-cell receptor
inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes
Chromosomes Cancer. 37:20–28. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Metzler M, Wilda M, Busch K, Viehmann S
and Borkhardt A: High expression of precursor microRNA-155/BIC RNA
in children with Burkitt lymphoma. Genes Chromosomes Cancer.
39:167–169. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Gibcus JH, Tan LP, Harms G, Schakel RN, de
Jong D, Blokzijl T, Möller P, Poppema S, Kroesen BJ and van den
Berg A: Hodgkin lymphoma cell lines are characterized by a specific
miRNA expression profile. Neoplasia. 11:167–176. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Navarro A, Gaya A, Martinez A,
Urbano-Ispizua A, Pons A, Balagué O, Gel B, Abrisqueta P,
Lopez-Guillermo A, Artells R, et al: MicroRNA expression profiling
in classic Hodgkin lymphoma. Blood. 111:2825–2832. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Sanchez-Espiridion B, Martin-Moreno AM,
Montalban C, Figueroa V, Vega F, Younes A, Medeiros LJ, Alvés FJ,
Canales M, Estévez M, et al: MicroRNA signatures and treatment
response in patients with advanced classical Hodgkin lymphoma. Br J
Haematol. 162:336–347. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Ben Dhiab M, Ziadi S, Louhichi T, Ben
Gacem R, Ksiaa F and Trimeche M: Investigation of miR9-1, miR9-2
and miR9-3 methylation in Hodgkin lymphoma. Pathobiology.
82:195–202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Cao Z, Qiu J, Yang G, Liu Y, Luo W, You L,
Zheng L and Zhang T: MiR-135a biogenesis and regulation in
malignancy: A new hope for cancer research and therapy. Cancer Biol
Med. 17:569–582. 2020.PubMed/NCBI
|
|
192
|
Solé C, Arnaiz E and Lawrie CH: MicroRNAs
as biomarkers of B-cell lymphoma. Biomark Insights. Oct
16–2018.(Epub ahead of print). doi: 10.1177/1177271918806840.
View Article : Google Scholar
|
|
193
|
The Leukemia Lymphoma Society, . Non
Hodgkin Lymphoma. 2006.
|
|
194
|
Vose JM: Peripheral T-cell non-Hodgkin's
lymphoma. Hematol Oncol Clin North Am. 22:997–1005. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Mei M and Zhang M: Non-coding RNAs in
Natural Killer/T-cell lymphoma. Front Oncol. 9:5152019. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Ralfkiaer U, Hagedorn PH, Bangsgaard N,
Løvendorf MB, Ahler CB, Svensson L, Kopp KL, Vennegaard MT,
Lauenborg B, Zibert JR, et al: Diagnostic microRNA profiling in
cutaneous T-cell lymphoma (CTCL). Blood. 118:5891–5900. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Yamanaka Y, Tagawa H, Takahashi N,
Watanabe A, Guo YM, Iwamoto K, Yamashita Y, Saitoh H, Kameoka Y and
Shimizu N: Aberrant overexpression of microRNAs activate AKT
signaling via downregulation of tumor suppressors in natural
killer-cell lymphoma/leukemia. Blood. 114:3265–3275. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Zhang X, Ji W, Huang R, Li L, Wang X, Li
L, Fu X, Sun Z, Li Z, Chen Q and Zhang M: MicroRNA-155 is a
potential molecular marker of natural killer/T-cell lymphoma.
Oncotarget. 7:53808–53819. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Motsch N, Alles J, Imig J, Zhu J, Barth S,
Reineke T, Tinguely M, Cogliatti S, Dueck A, Meister G, et al:
MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell
lymphomas by deep sequencing. PLoS One. 7:e421932012. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Chen HH, Huang WT, Yang LW and Lin CW: The
PTEN-AKT-mTOR/RICTOR pathway in nasal natural killer cell lymphoma
is activated by miR-494-3p via PTEN but inhibited by miR-142-3p via
RICTOR. Am J Pathol. 185:1487–1499. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Liang L, Nong L, Zhang S, Zhao J, Ti H,
Dong Y, Zhang B and Li T: The downregulation of PRDM1/Blimp-1 is
associated with aberrant expression of miR-223 in extranodal
NK/T-cell lymphoma, nasal type. J Exp Clin Cancer Res. 33:72014.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Huang WT and Lin CW: EBV-encoded
miR-BART20-5p and miR-BART8 inhibit the IFN-γ-STAT1 pathway
associated with disease progression in nasal NK-cell lymphoma. Am J
Pathol. 184:1185–1197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Alles J, Menegatti J, Motsch N, Hart M,
Eichner N, Reinhardt R, Meister G and Grasser FA: miRNA expression
profiling of Epstein-Barr virus-associated NKTL cell lines by
Illumina deep sequencing. FEBS Open Bio. 6:251–263. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Ramakrishnan R, Donahue H, Garcia D, Tan
J, Shimizu N, Rice AP and Ling PD: Epstein-Barr virus BART9 miRNA
modulates LMP1 levels and affects growth rate of nasal NK T cell
lymphomas. PLoS One. 6:e272712011. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Raab MS, Podar K, Breitkreutz I,
Richardson PG and Anderson KC: Multiple myeloma. Lancet.
374:324–339. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista
DL, Gejman R, Ansell PJ, Zhao J, Weng C and Klibanski A: Activation
of p53 by MEG3 non-coding RNA. J Biol Chem. 282:24731–24742. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Benetatos L, Dasoula A, Hatzimichael E,
Georgiou I, Syrrou M and Bourantas KL: Promoter hypermethylation of
the MEG3 (DLK1/MEG3) imprinted gene in multiple myeloma. Clin
Lymphoma Myeloma. 8:171–175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Zhuang W, Ge X, Yang S, Huang M, Zhuang W,
Chen P, Zhang X, Fu J, Qu J and Li B: Upregulation of lncRNA MEG3
promotes osteogenic differentiation of mesenchymal stem cells from
multiple myeloma patients by targeting BMP4 transcription. Stem
Cells. 33:1985–1997. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Bennett CF, Baker BF, Pham N, Swayze E and
Geary RS: Pharmacology of antisense drugs. Annu Rev Pharmacol
Toxicol. 57:81–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Koch L: Functional genomics: Screening for
lncRNA function. Nat Rev Genet. 18:702017. View Article : Google Scholar
|
|
212
|
Gilbert LA, Horlbeck MA, Adamson B,
Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh
HL, Bassik MC, et al: Genome-Scale CRISPR-mediated control of gene
repression and activation. Cell. 159:647–661. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Brummelkamp TR, Bernards R and Agami R: A
system for stable expression of short interfering RNAs in mammalian
cells. Science. 296:550–553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Mattheolabakis G, Rigas B and
Constantinides PP: Nanodelivery strategies in cancer chemotherapy:
Biological rationale and pharmaceutical perspectives. Nanomedicine
(Lond). 7:1577–1590. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Webster DM, Sundaram P and Byrne ME:
Injectable nanomaterials for drug delivery: Carriers, targeting
moieties, and therapeutics. Eur J Pharm Biopharm. 84:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Fabbro C, Ali-Boucetta H, Da Ros T,
Kostarelos K, Bianco A and Prato M: Targeting carbon nanotubes
against cancer. Chem Commun (Camb). 48:3911–3926. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Libutti SK, Paciotti GF, Byrnes AA,
Alexander HR Jr, Gannon WE, Walker M, Seidel GD, Yuldasheva N and
Tamarkin L: Phase I and pharmacokinetic studies of CYT-6091, a
novel PEGylated colloidal gold-rhTNF nanomedicine. Clin Cancer Res.
16:6139–6149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Yang T, Choi MK, Cui FD, Lee SJ, Chung SJ,
Shim CK and Kim DD: Antitumor effect of paclitaxel-loaded PEGylated
immunoliposomes against human breast cancer cells. Pharm Res.
24:2402–2411. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Markman JL, Rekechenetskiy A, Holler E and
Ljubimova JY: Nanomedicine therapeutic approaches to overcome
cancer drug resistance. Adv Drug Deliv Rev. 65:1866–1879. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Bartolomé-Izquierdo N, de Yébenes VG,
Álvarez-Prado AF, Mur SM, Lopez Del Olmo JA, Roa S, Vazquez J and
Ramiro AR: miR-28 regulates the germinal center reaction and blocks
tumor growth in preclinical models of non-Hodgkin lymphoma. Blood.
129:2408–2419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Zhang Y, Roccaro AM, Rombaoa C, Flores L,
Obad S, Fernandes SM, Sacco A, Liu Y, Ngo H, Quang P, et al:
LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas.
Blood. 120:1678–1686. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Seto AG, Beatty X, Lynch JM, Hermreck M,
Tetzlaff M, Duvic M and Jackson AL: Cobomarsen, an oligonucleotide
inhibitor of miR-155, co-ordinately regulates multiple survival
pathways to reduce cellular proliferation and survival in cutaneous
T-cell lymphoma. Br J Haematol. 183:428–444. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Li J, Zou J, Wan X, Sun C, Peng F, Chu Z
and Hu Y: The role of noncoding RNAs in B-cell lymphoma. Front
Oncol. 10:5778902020. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Zhang X, Xie K, Zhou H, Wu Y, Li C, Liu Y,
Liu Z, Xu Q, Liu S, Xiao D and Tao Y: Role of non-coding RNAs and
RNA modifiers in cancer therapy resistance. Mol Cancer. 19:472020.
View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Lin R, Sampson JH, Li QJ and Zhu B:
miR-23a blockade enhances adoptive T cell transfer therapy by
preserving immune-competence in the tumor microenvironment.
Oncoimmunology. 4:e9908032015. View Article : Google Scholar : PubMed/NCBI
|