Introduction
Triple-negative breast cancer (TNBC) accounts for
~15% of all breast cancer cases and is defined by a lack of
estrogen and progesterone receptors' expression and the absence of
human epidermal growth factor receptor-2 (1). Molecular studies have demonstrated that
TNBCs are a heterogeneous group of tumors with different clinical
features, prognoses, genetic-molecular alterations, and responses
to treatment (2). TNBCs are more
likely to recur earlier after surgery than other subtypes, and many
patients have poor prognosis (3). To
increase success rate during TNBC treatment, personalized medicine
designed for each patient's tumor profile individually taking
genetic alterations in each patient into account is necessary.
Currently, for recurrence detection, serum tumor
marker evaluation or computed tomography (CT) are used; however,
more sensitive strategies should be developed to improve detection
reliability. To identify relevant mutations, more accurate and
cost-effective tools, such as strategies that use cell-free DNA
(cfDNA), are required in clinical settings. However, the analysis
of cfDNA using next-generation sequencing (NGS) is expensive and
time-consuming. Among new technologies for cfDNA quantification,
droplet digital PCR (ddPCR) provides the highest sensitivity for
detecting and tracking actionable mutations at frequencies as low
as 0.01–0.05% (4). In hormone
receptor-positive breast cancer, the utility of liquid biopsy to
analyze hormone therapy-resistant mutations has been studied
(5,6). In case of TNBC, liquid biopsy for
detecting minimal residual disease during neoadjuvant chemotherapy
was also studied and reported by some groups. These studies have
demonstrated that circulating tumor DNA (ctDNA) level during
neoadjuvant chemotherapy was associated with patient survival
(7,8). However, there is still no valid
protocol that can be used for diagnosis of disease recurrence,
which is focusing on specific mutations.
Here, we explore the practical utility of liquid
biopsy using ddPCR-based detection of frequently mutated genes in
samples of TNBC patients. We also describe the clinical course of
metastatic TNBC patients whose genome harbors AKT1 E17K and
PIK3CA H1047R mutations, which have been reported to be
oncogenic driver mutations in breast cancer that lead to PI3K/AKT
pathway activation and tumor progression.
Materials and methods
Patients and breast cancer
samples
We examined 57 consecutive TNBC patients who
underwent mastectomy or breast-conserving surgery at the Asahikawa
Medical University Hospital between April 2000 and March 2017.
Thirty-six patients who relapsed within 2 years postoperatively
were enrolled in the study. Tissue resected metastatic specimens of
these patients were also evaluated. All cases were reviewed by two
experienced pathologists and categorized according to the WHO
classification (9). After surgical
resection, patients underwent follow-up examination every 3 months
for 2 years and every 6 months thereafter. The diagnosis of
recurrence was made based on abnormal findings in the conserved
mammary glands or by imaging diagnosis during the follow-up period.
When patients had tumor recurrence, they were treated with
chemotherapy (e.g., eribulin, tegafur-uracil, bevacizumab,
paclitaxel, or vinorelbine).
This study protocol, including tumor sample
collection and genetic analysis, was approved by the Asahikawa
Medical University Research Ethics Committee (approval nos. 17043
and 18118). Written informed consent was obtained from all the
patients before their enrollment for genetic analysis.
Sample collection and processing
Genomic DNA was extracted from formalin-fixed
paraffin-embedded (FFPE) primary and recurrent tumor samples. Two
cases were excluded because the specimens contained too few tumor
cells. The tumor cell content ranged widely (from 20 to 90%), with
44 (94%) samples containing >60% tumor cells. Viable tumor areas
were dissected with a razor blade, and DNA was extracted using the
GeneRead DNA FFPE Kit (Qiagen).
Plasma samples were collected from AKT1 E17K
(c.49G>A)- and PIK3CA H1047R (c.3140A>G)-positive
patients using PAXgene Blood ccfDNA Tube (Qiagen). Plasma was then
isolated by centrifuging the samples at 1,400 × g for 10 min at
room temperature. The cell-free plasma was transferred into 2-ml
collection tubes and stored at −80°C until purification. For cfDNA
purification, QIAamp Circulating Nucleic Acid kit (Qiagen) was
utilized. The genomic DNA concentration was measured using the
Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, Inc.) and the
Qubit 3.0 fluorometer (Thermo Fisher Scientific, Inc.).
Targeted amplicon sequencing
To identify frequently mutated in our TNBCs and to
determine which genetic alteration should be screening for cfDNA
targeted amplicon sequencing was employed.
Between 20 and 100 nanograms of cfDNA were used for
library construction with the Ion AmpliSeq Cancer Panel v2, which
targets thousands of mutational hotspot regions in 50
cancer-associated genes using the Ion AmpliSeq Library Kit (Thermo
Fisher Scientific, Inc.) (10). The
sequencing was performed on an Ion PGM System according to the
manufacturer's protocol, and sequencing leads were multiplexed,
quality-filtered, and aligned to the human reference genome
(GRCh37) using the Torrent Suite software (ver. 5.0.4; Thermo
Fisher Scientific, Inc.). Variants were identified with the Variant
Caller software (ver. 5.0.4.0; Thermo Fisher Scientific, Inc.) and
filtered using Ion Reporter Software (Thermo Fisher Scientific,
Inc.). The quality of all variants calls was manually confirmed by
IGV software (ver. 2.3.59; Broad Institute, Cambridge, MA,
USA).
Mutation detection by ddPCR
For the detection of the AKT1 c.49G>A
mutation, a specific primer/probe set was utilized (dHsaCP2000031;
Bio-Rad Laboratories). For PIK3CA and TP53 mutations,
specific assays were designed in-house and synthesized by
Integrated DNA Technologies (IDT); nucleotide sequences of primers
and probes are shown in Table SI).
ddPCR assays were performed as previously described (11). Reaction mixtures were made of ddPCR
Supermix for probes (no dUTP; Bio-Rad), primer, wild-type- and
mutant-specific probes, and template DNA in a total volume of 22
µl. Droplets were generated by mixing the reaction mixture with
Droplet Generation Oil (Bio-Rad Laboratories) in a QX200 droplet
generator (Bio-Rad Laboratories). PCR was performed on a Veriti
Thermal Cycler (Thermo Fisher Scientific, Inc.) using the following
cycling conditions: 10 min at 95°C, 40 cycles of 94°C for 30 sec
followed by 60°C for TP53 or 57°C for PIK3CA for one
minute, followed by 98°C for 10 min. Samples were transferred to
the QX200 Droplet Reader (Bio-Rad Laboratories) to measure the
fluorescence of 6-fluorescein amidite and hexachloro-fluorescein
probes. Droplets were scored as positive or negative based on their
fluorescence intensity, which was determined by gating thresholds
defined with positive and negative controls. Finally, absolute copy
number input in the reaction and the ratio of mutated fragments
were calculated by QuantaSoft ver 1.7 (Bio-Rad Laboratories) based
on a Poisson distribution. Samples were scored as positive for the
mutation when at least three mutation-containing droplets were
detected by the mutant-specific probe as reported previously
(12).
Immunohistochemical analysis
Immunohistochemistry (IHC) was performed using the
Envision™ HRP System (#K5361; Dako) using formalin-fixed sections,
as described previously (13). Two
samples with a low tumor cellularity were excluded. Heat-induced
antigen retrieval was performed for phospho-AKT (pAKT; #4060) and
phospho-S6RP (pS6RP; #4858; Cell Signaling Technology, Inc.)
antibodies in EDTA buffer at pH 9.0, and endogenous peroxidase
activity was inhibited according to the manufacturer's
instructions. The slides were then incubated at 4°C overnight with
anti-pAKT at 1/25 and anti-pS6RP at 1/100 [diluted in Dako Real
Antibody Diluent (Dako)], followed by incubation with an
HRP-conjugated secondary antibody and substrate.
Staining for pAKT and pS6RP was evaluated by
consensus by two observers blinded to the clinical data. For
scoring, the highest staining intensity in malignant cells was
classified as 0, no staining; 1, weak staining; 2, moderate
staining; or 3, strong staining (Fig.
S1).
Statistical analyses
Patients who were still alive at the most recent
follow-up were censored for survival on the date of that follow-up.
The Kaplan-Meier method was used to estimate overall survival (OS)
rates. Fisher's exact test was used to compare our data on the
frequency of gene mutations with that in the METABRIC database,
which is the largest global study of breast cancer tissue samples
ever performed (14). Pearson's
correlation coefficient was calculated to examine the relevance of
allele frequency obtained by targeted amplicon sequencing and
ddPCR. The Mann-Whitney U test was used to assess IHC score
differences between PIK3CA H1047R- and AKT1
E17K-mutant samples and wild-type samples. A two-sided P-value of
<0.05 was considered statistically significant. All statistical
analyses were two-sided and performed using R software (version
3.4.1; The R Foundation, Vienna, Austria) (15).
Results
Characteristics of early recurrent
TNBCs
The clinicopathological characteristics of the 36
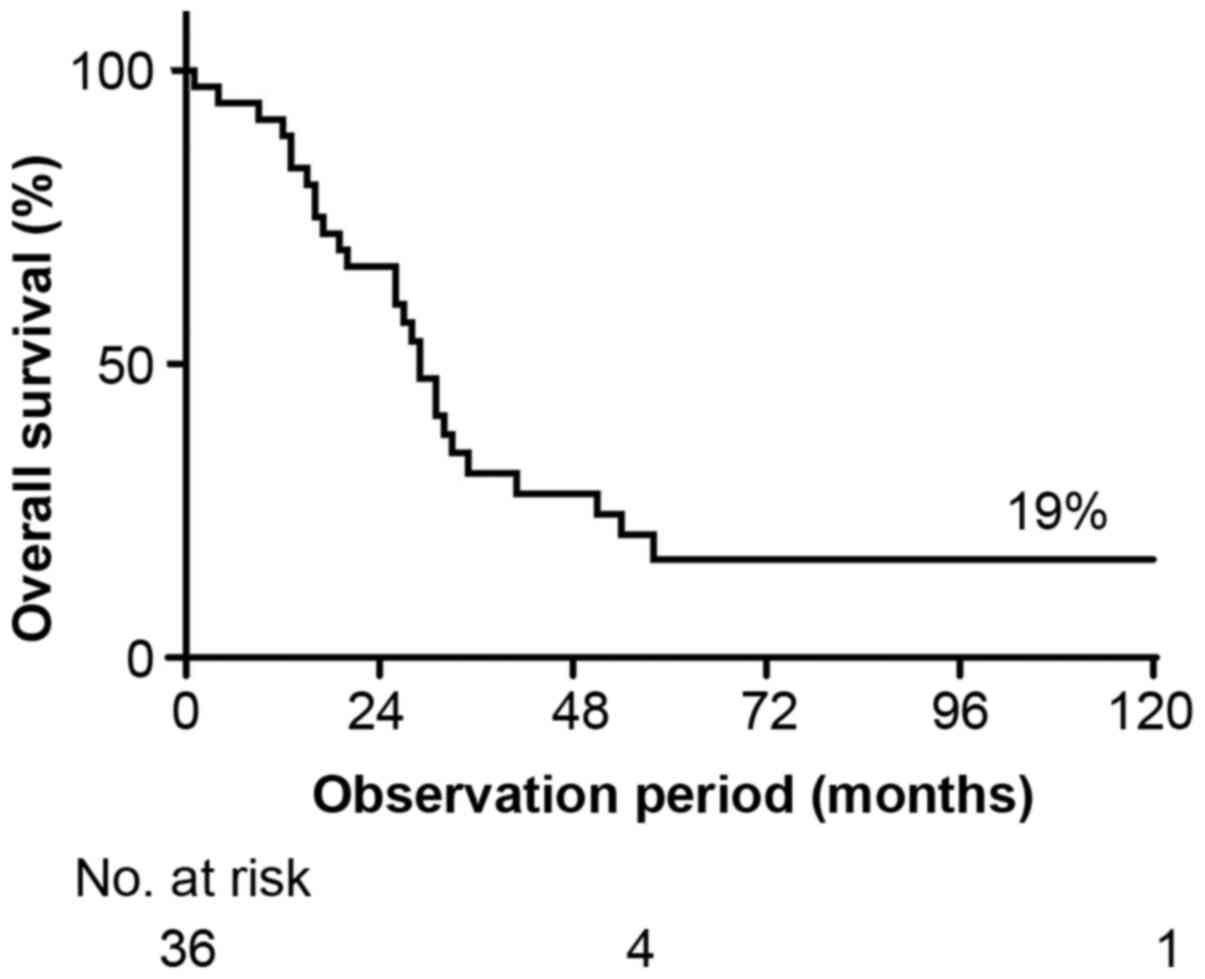
enrolled patients are summarized in Table I. The 5-year OS rate was 19%
(Fig. 1). The median age at surgery
was 56.6 years. Over half (63.9%) of the patients were
postmenopausal, and 89.0% had invasive ductal carcinoma histology.
Four patients (11.1%) were initially diagnosed with stage IV
cancer. Neoadjuvant and adjuvant treatments were administered to 35
of 36 patients (97.2%). Neoadjuvant chemotherapy was performed in
11 patients. Oral chemotherapy (tegafur-uracil) was administered to
four patients. One patient did not receive chemotherapy because of
liver cancer treatment. The median time to progression and median
OS from surgery were 12.6 months (range: 0–24 months) and 31.9
months (range: 1–175 months), respectively.
 | Table I.Demographic and clinicopathological
features of patients. |
Table I.
Demographic and clinicopathological
features of patients.
|
Characteristics | N | % |
|---|
| Total | 36 |
|
| Median age at
surgery, years (range) | 56.6 (30–81) |
|
| Menopausal
status |
|
|
|
Premenopausal | 13 | 36.1 |
|
Postmenopausal | 23 | 63.9 |
| T stage |
|
|
| T1
(≤2.0 cm) | 11 | 30.6 |
| T2
(2.1–5.0 cm) | 18 | 50 |
| T3
(>5.0 cm) | 5 | 13.9 |
| T4
(direct extension to the chest wall or skin) | 2 | 5.5 |
| No. of positive
lymph nodes |
|
|
| 0 | 16 | 44.4 |
|
1-3 | 10 | 27.8 |
|
4-9 | 7 | 19.5 |
|
≥10 | 2 | 5.5 |
|
Unknown | 1 | 2.8 |
| Stage |
|
|
| I | 7 | 19.5 |
| II | 15 | 41.6 |
|
III | 10 | 27.8 |
| IV | 4 | 11.1 |
| Histological
grade |
|
|
| 1 | 2 | 5.5 |
| 2 | 7 | 19.5 |
| 3 | 23 | 63.9 |
|
Unknown | 4 | 11.1 |
| Histological
type |
|
|
|
Invasive ductal | 32 | 89 |
|
Invasive lobular | 2 | 5.5 |
|
Others | 2 | 5.5 |
| Ki67 LI |
|
|
|
≤14 | 2 | 5.5 |
|
>14 | 22 | 61.2 |
|
Unknown | 12 | 33.3 |
| Chemotherapy
received |
|
|
|
Yes | 35 | 97.2 |
| No | 1 | 2.8 |
| Median PFS, months
(range) | 12.6 (0–24) |
|
| Median OS, months
(range) | 31.9
(1–175) |
|
Genetic alterations in early recurrent
TNBCs
Of the 36 cases, the tumor content was low in two
cases, so they were excluded from DNA analysis. There were 13
patients who had undergone resection of the recurrence, each with
eight lymph nodes, two lungs, two brains, and one pleura. Of 1,835
variants obtained from 47 samples, 72 variants were selected by
coverage, type of function, sequencing quality, filtering of
single-nucleotide polymorphisms (SNPs) using Variant Caller
software, and manual selection of false-positives (Fig. S2) (16). Forty-eight were single nucleotide
variants (66.7%) and 24 were small insertion-deletions (33.3%). Of
these variants, 54 and seven were unique to the primary and
metastatic lesions, respectively, and 11 were found in both.
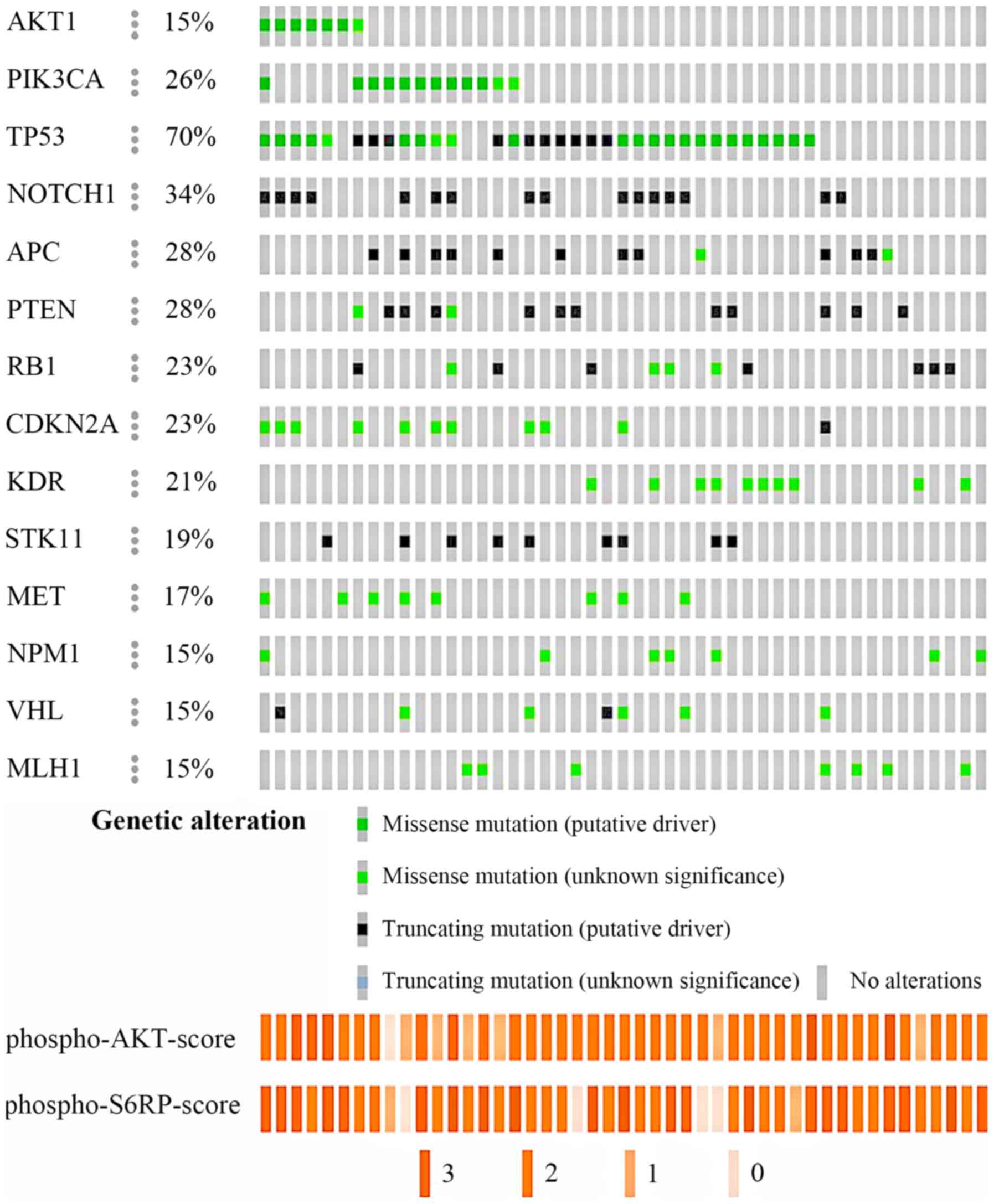
Thirty-three of 34 tumors (97.1%) obtained from primary tumors had
the following mutations (average 5.2, range 1–12 mutations):
TP53 (27, 79.4%); NOTCH1 (16, 47.1%); PTEN
(13, 38.2%); PIK3CA (11, 32.3%); CDKN2A (11, 32.3%);
APC (10, 29.4%); VHL (8, 23.5%); and AKT1 (7,
20.6%) (Fig. 2). All 13 recurrent
samples harbored mutations (average 2.5, range: 1–5 mutations), and
mutation in KDR was the most frequent (8, 61.5%). The
comparison of genetic mutations in 13 metastatic tumors and
corresponding primary tumors were shown in Fig. S3. The left side column shows the
data of the primary tumor and the right column shows the data of
the metastatic tumor. AKT1 mutations were not found in these
samples. For PIK3CA mutation, samples from 4 patients were
detected including from 3 primary tumors. The sites and frequencies
of mutations in TP53, AKT1, and PIK3CA are shown in
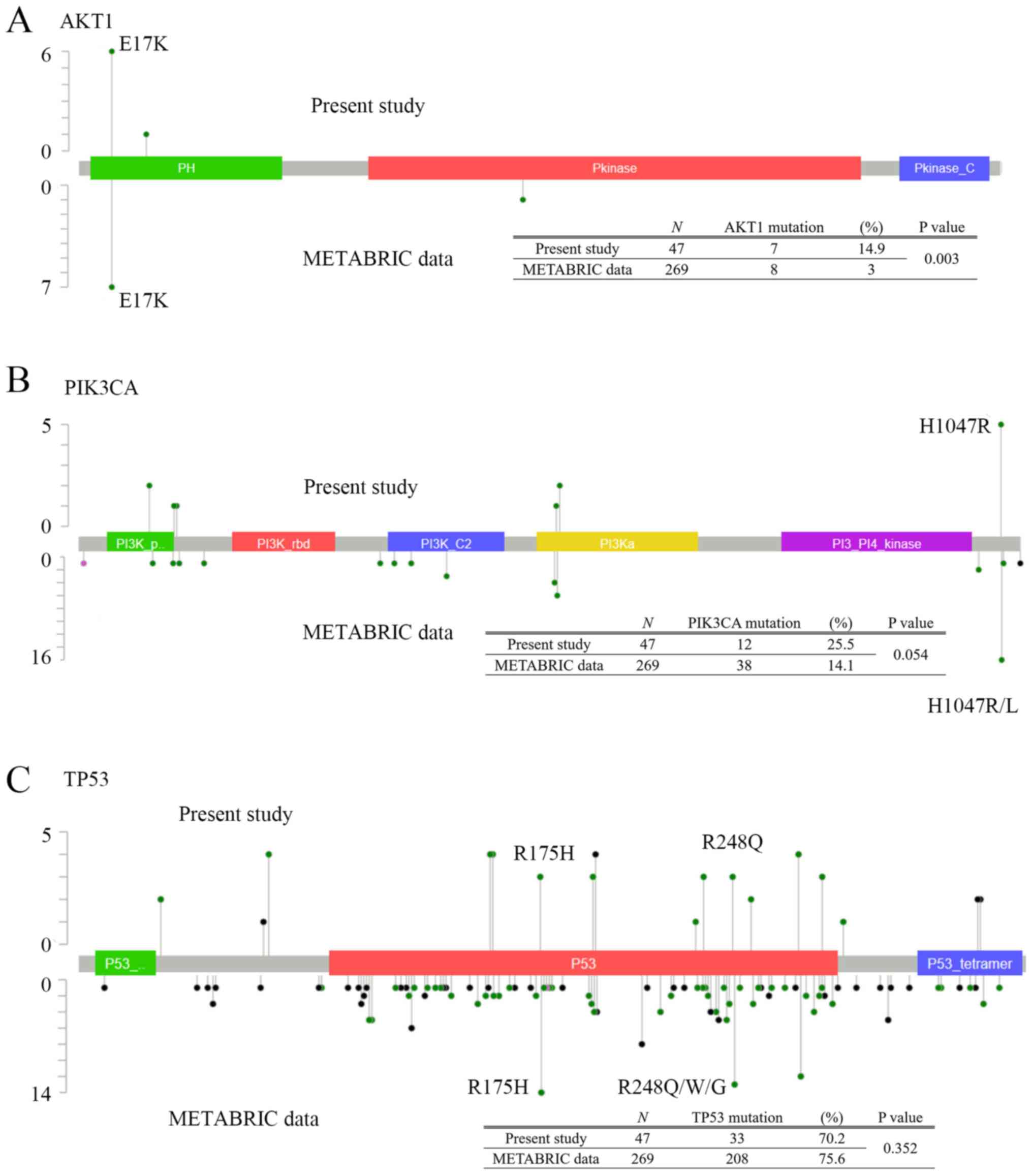
Fig. 3. AKT1 E17K was the most
common mutation in our study (7/47, 14.9%) (Fig. 3A). For PIK3CA gene (Fig. 3B), PIK3CA H1047R was the most
frequent mutation (5/47, 10.6%). In contrast, for mutations in
TP53 gene, there were various mutation sites, but no
characteristic mutations were found (Fig. 3C).
Patterns of expression of phospho-AKT
and phospho-S6RP
AKT1 and PIK3CA were reported as
driver oncogenes whose activity was sufficient to transform a
normal cell into a cancerous one. PI3K/AKT signaling pathway
activity can be detected by the phosphorylation of AKT and S6RP
proteins. Therefore, we investigated the correlation between
genetic alterations and protein phosphorylation in tumor samples by
IHC (Fig. 2). Representative images
of tumor cells with strong, moderate, weak, and no staining for
pAKT1 and pS6RP are shown in Fig.
S1. Tumors with PIK3CA H1047R or AKT1 E17K
mutations had higher expression level of phospho-AKT (pAKT) and
phospho-S6RP (pS6RP) than PIK3CA and AKT1 wild-type
tumors (P=0.044 and P=0.105, respectively; Table SII), suggesting that genetic
alteration of AKT1 or PIK3CA leads to induction of
abnormal activation of survival pathways in cancer.
Mutation allele frequencies of FFPE
samples
We sought to develop a highly sensitive blood-based
method for detection of genetic alterations that can be used as a
clinical application for serial monitoring. We designed or
purchased allele-specific probes for ddPCR that yielded 0.01%
detection sensitivity between mutant and wild-type alleles
(Table SI; Fig. S4; Appendix S1). Genomic DNA extracted from
HCT116, SKBR-3 or HCC70 cells was used as a ddPCR mutation control
for PIK3CA H1047R, TP53 R175H or TP53 R248Q, respectively, and
genomic DNA extracted from HCC38 cells was used as a wild-type
control for PIK3CA or TP53 (Table
SIII).
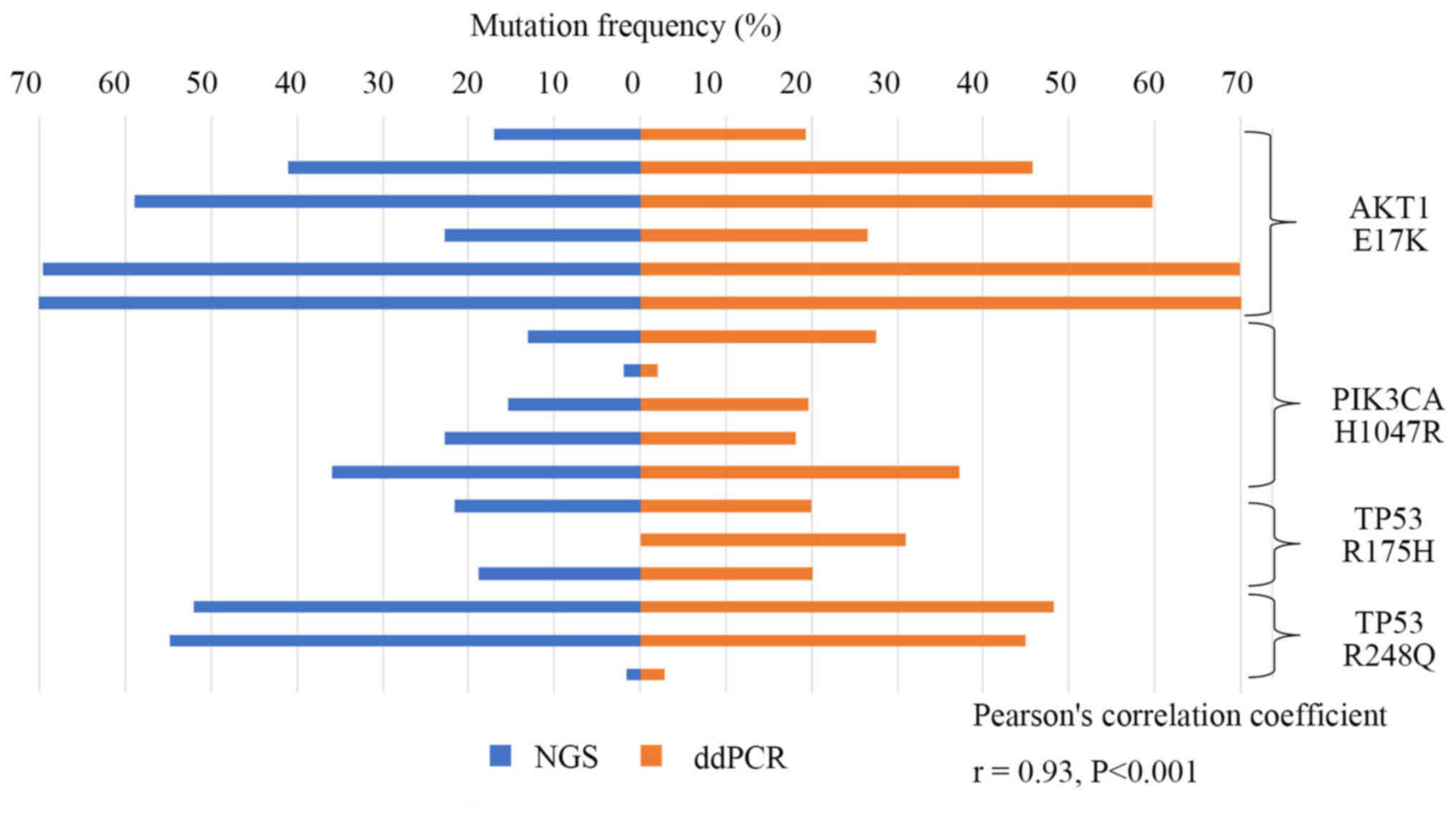
Then the frequencies of mutations identified by
ddPCR were compared to the ones previously determined by NGS using
DNA derived from the FFPE tissue samples containing 47 specimens of
early TNBC recurrence. As shown in Fig.
4, AKT1 E17K, PIK3CA H1047R, and TP53
R175H and R248Q mutations detected by NGS significantly correlated
with frequencies in ddPCR (r=0.93, P<0.001).
Results of digital PCR detection of
clinical samples
In our early recurrent TNBCs patients, 4 patients
out of 36 patients still survived. Among them, consent was obtained
in three cases, and plasma was collected and analyzed. Patients
nos. 1422 and 2443 were harboring PIK3CA H1047R mutation and
patient no. 2152 was harboring AKT1 E17K mutation in primary
tumor, however in patient no. 1422, PIK3CA H1047R mutation
was not detected in lymph node metastasis site nor cfDNA due to the
adjuvant chemotherapy after surgery (Fig. S5A).
In other TNBC cohort including 3 cases with early
recurrence within 2 years after surgery, 2 cases of late
recurrence, 4 cases without metastasis were analyzed for cfDNA.
Within this cohort, 2 patients (patient nos. 2667 and 3019) were
harboring PIK3CA H1047R (Fig.
S5B and C) or a patient (patient no. 996) was harboring
AKT1 E17K mutation (Fig.
S5D) and all corresponding cfDNA from plasma was detected in
the same mutations as primary lesion.
Collectively, 6 patients with metastatic TNBC
patients harboring PIK3CA H1047R or AKT1 E17K
mutation, 5 patients (83.3%) were detected corresponding mutation
by digital PCR methods.
Clinical course of a PIK3CA
H1047R-positive TNBC patient
A 58-year-old woman (patient no. 2443) with a
diagnosis of TNBC (T3N0M0, Stage IIb) received neoadjuvant
chemotherapy with 5-fluorouracil/epirubicin/cyclophosphamide
followed by docetaxel. After neoadjuvant chemotherapy, she
underwent mastectomy and sentinel lymph node biopsy. Histological
examination of the resected specimens revealed pathological
complete response. Needle biopsy revealed invasive lobular
carcinoma with high proliferative rate (MIB-1 index 28%). Thirteen
months after the surgery, multiple liver metastases were found.
Furthermore, 39 months after surgery, a liquid biopsy was performed
(and the cfDNA was extracted, concentration: 20.6 ng/ml of plasma).
Genetic mutation analysis of cfDNA indicated the presence of
PIK3CA c.3140A>G mutation (H1047R), as did genetic
analysis of the primary site, and the allele frequency was 0.4% as
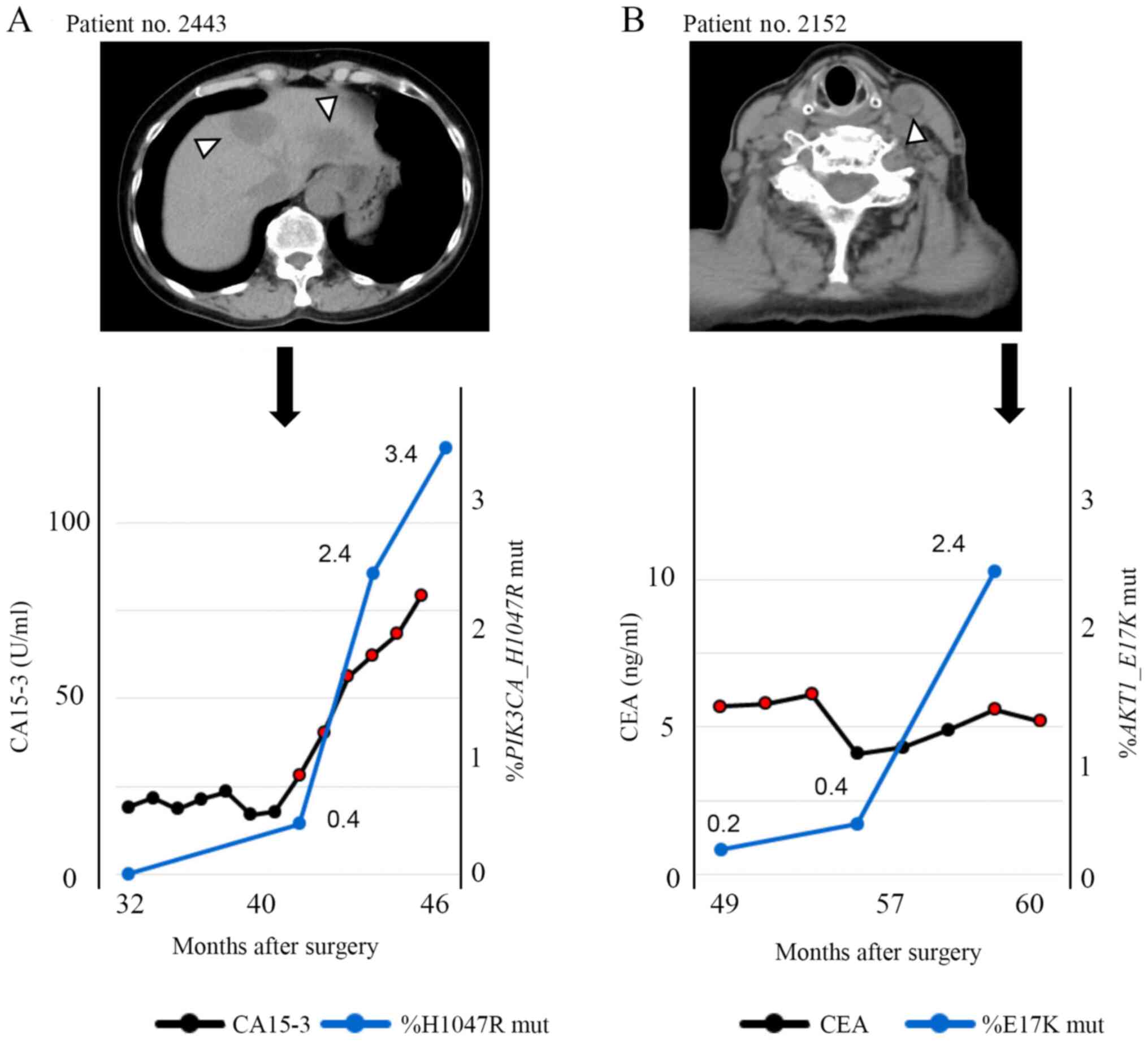
defined by the liquid biopsy. One month later, progression of liver
metastases was detected by CT (Fig.
5A). Liquid biopsies were performed every three months
thereafter, and the increase of mutant allele frequency was
correlated to increase of CA-15-3. As shown in Fig. S5B and C, mutations were detectable
by liquid biopsy in two other patients, and one of them had brain
metastasis while a tumor marker was negative.
Clinical course of an AKT1
E17K-positive TNBC patient
A 77-year-old woman (patient no. 2152) underwent
left breast resection and axillary lymph node dissection. She was
diagnosed with Stage IIIB TNBC with direct extension to the skin
and lymph node metastasis. Examination of the biopsy sample
revealed invasive ductal carcinoma with high proliferative rate
(MIB-1 index 38%). Eleven months after surgery, CT confirmed lymph
node progression. Fifty-two months after surgery, a liquid biopsy
was performed and the concentration of extracted cfDNA was 16.6
ng/ml of plasma. Seven months later, a second liquid biopsy was
performed and the concentration of extracted cfDNA was 18.8 ng/ml
of plasma. Ten months later, a third liquid biopsy was performed
and the concentration of cfDNA was 13.5 ng/ml of plasma. Genetic
mutation analysis of cfDNA indicated the presence of AKT1
c.49G>A (E17K) as in the primary site, and the mutation allele
frequency was 0.2% in the first biopsy and 0.4% in the second
biopsy (Fig. 5B). In the second
liquid biopsy, although tumor markers were undetectable, the AKT1
c.49G>A (E17K) mutation could be detected. In the third liquid
biopsy, the allele frequency rose to 2.4%, and one month later,
metastasis of left the cervical lymph node was detected by CT. As
shown in Fig. S5D, mutation was
detectable by liquid biopsy in another patient.
Discussion
We studied a high-sensitivity detection system based
on ddPCR that enabled the detection of low-level copy mutations and
the identification of mutations in plasma samples of patients with
metastatic TNBC. ddPCR is a relatively inexpensive method and has
rapid turnover. The ddPCR assay is especially useful when there is
insufficient genomic DNA for NGS analysis. Circulating DNA in the
cell-free plasma fraction originates from many different cells,
including lymphocytes and neoplastic cells (17). Their DNA is released into the
circulation in the process of cellular destruction (18). Therefore, circulating-tumor DNA
(ctDNA) in cell-free plasma represents a very low fraction of the
total amount of circulating DNA, but there are several reports in
which ctDNA in TNBC patients was analyzed. Chen et al
(7) reported that ctDNA was detected
in four out of the 33 early-stage TNBC patients after neoadjuvant
chemotherapy by NGS analysis. All four cases relapsed early after
surgery, and disease-free survival (DFS) was significantly inferior
compared to cases in which ctDNA could not be detected. On the
other hand, ctDNA could not be detected in nine of the recurrent
cases, and the sensitivity remained at 31%. Riva et al
(8) reported that in 27 out of 36
early TNBC patients before treatment, it was possible to detect
ctDNA, and in patients, for whom it was possible to detect ctDNA
even after 1 cycle of neoadjuvant chemotherapy, DFS and OS were
significantly lower. These findings suggest that the detection and
quantification of ctDNA is a very promising tool for assessing a
response to neoadjuvant chemotherapy. Our detection system using
ddPCR can be used for liquid biopsy because of its high
sensitivity. Importantly, using this protocol, we were able to
understand the disease progression during chemotherapy in patients
with metastatic TNBC. In patient no. 2152 in particular, an
increase in the mutation copy ratio was observed reflecting on the
disease state progression more sensitively than the tumor marker
did. In solid tumors other than breast cancer, biomarkers have been
used as a tool to detect recurrence. These biomarkers include
prostate-specific antigen for prostate cancer, carcinoembryonic
antigen (CEA) for colon cancer, and cancer antigen 125 for ovarian
cancer (19). On the other hand, in
breast cancer or non-small cell lung cancer, Clinical Practice
Guidelines does not recommend the surveillance tumor marker testing
for the patients treated with curative intent. It because
biomarkers measured postoperatively were not sensitive nor specific
for cancer relapse (20,21).
In our previous study, the usefulness of cfDNA
measurements rather than tumor markers in NSCLC patients was
indicated (11). Ideally, if we
could design digital PCR probes/primers for all the detected
genetic mutations by NGS, all patients could be eligible for
detection from cfDNA. However, NGS was not performed for the
purpose of screening of functioning genetic mutations in TNBCs, but
rather for genetic mutations that should be searched in cfDNA. In
the present study, we identified frequently mutated and associated
with molecularly targeted therapies using NGS based genetic
alteration screening and we focused on AKT1 and
PIK3CA mutations and showed that analysis of cfDNA over time
could be useful for determining recurrence.
The prognosis is poor for metastatic TNBC and TNBC
that recurs within 2–3 years (1).
Although clinical trials have examined various treatments (22–24), no
cure has been established. In this study, we found that PI3K/AKT
signaling, and in particular PIK3CA H1047R or AKT1
E17K mutations, were significantly associated with high incidence
of metastasis as compared to the METABRIC cohort. The METABRIC
database contains the details of genetic mutations for 269 TNBC
cases, which we compared to our results (Fig. 3). The frequency of AKT1
mutation was higher in our study than in METABRIC. AKT1 E17K was
the most common mutation in both groups (Fig. 3A). For PIK3CA gene, the
mutation sites were generally consistent between our data and those
in METABRIC; however, the mutational frequency was not
statistically significant, although there was a tendency for higher
mutational frequency in our study compared with that shown in
METABRIC (Fig. 3B). In both groups,
PIK3CA H1047R was the most frequent mutation (our data:
5/47, 10.6% and METABRIC: 15/269, 5.6%). While Takeshita et
al (5) reported that the
presence of PIK3CA H1047R or E542K in cfDNA was
associated with favorable prognosis, in our cohort, patients with
recurrence within two years after surgery were corrected in this
study, therefore we could not verify whether PIK3CA H1047R
was prognosis marker or not. In contrast, the mutations in
TP53 were generally consistent, but no statistically
significant difference in its frequency was observed (Fig. 3C). The frequencies of TP53
R175H and R248Q mutations were higher in METABRIC (13/269, 4.8% and
7/269, 2.6%, respectively), but these mutations were also common in
our study (3/47, 6.4%). A previous report suggested that Asian TNBC
patients were more likely to harbor PI3K/AKT mutations than
patients of other ethnicities (25).
AKT1 E17K is considered a driver oncogene (26–28), and
AKT inhibitors have been assessed in preclinical and clinical
trials in multiple cancer types known to harbor this mutation,
suggesting that AKT-targeting therapy may be effective for patients
with activated AKT signaling (29).
Based on the above, Asian TNBC patients have a higher frequency of
this mutation, and AKT inhibitors may be an effective treatment
option for this population.
There are limitations in this study. First, the
number of patient samples was small; however, we analyzed 36
patients with TNBC who relapsed within 2 years after surgical
resection, the frequency of AKT1 E17K was higher in this
cohort, and there may be clinical benefits in monitoring this
mutation, as it is a marker of recurrence. A long-term surveillance
study in a large cohort of patients is warranted to confirm these
important findings. Second, the liquid biopsy was performed on
patients whose tumor has already metastasized. For early detection,
it is necessary to confirm similar events in patients whose tumor
has not yet metastasized.
Nevertheless, we believe that molecular target
therapy for druggable driver oncogene will be applicable for the
treatment of TNBC in the near future. Intervention with kinase
inhibitors could improve prognosis as seen in other types of
cancers. So, it is important to know what kinds of driver oncogene
are present in TNBC patients with poor prognosis that can be
detected in cfDNA mutation analysis by cost-effective and highly
sensitive way.
In this study, considering the possibility for AKT
inhibitor treatment as a therapy for TNBC, liquid biopsy was
performed on patients with AKT1 and PIK3CA mutations.
Based on the results obtained, detecting single mutation detection
like PIK3CA and AKT for surveillance of TNBC which
comprised ~32% of TNBC was not sufficient for the early detection
of TNBC recurrence; however, we believe that using predefined
genetic alteration, multiple targets in ddPCR, e.g., TP53
R175H/R248Q or high-sensitive NGS-based liquid biopsy could be
employed to yield higher detection rates.
In conclusion, in our TNBC early recurrent patient
cohort, AKT1 E17K (14.9%) and PIK3CA H1047R (10.6%)
were common in surgically resected samples detected by target
amplicon sequencing. The PI3K/AKT pathway was active in patients
harboring AKT1 E17K and PIK3CA H1047R mutations,
suggesting an inhibitor of this pathway needed to be explored as a
possible therapeutic agent. Finally, we successfully detected
predefined genetic mutations from circulating free DNA using ddPCR
method; however, for early recurrence detection or treatment
monitoring, we suggest using multiple target detection strategy
with high detection sensitivity.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Munehiko Ogata
and Ms. Mayumi Suzuki (Sapporo Higashi Tokushukai Hospital) for
technical support during genetic analyses.
Funding
This work was supported by JSPS KAKENHI [grant no.
18K15262 (to RY)]
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to their containing
genetic information that could compromise the privacy of research
participants but are available from the corresponding author on
reasonable request.
Authors' contributions
Conception and design: SaO and TS. Provision of
study materials, and acquisition, analysis and interpretation of
data: SaO, TS, SY, MA, NY, RY, KI, YoM, ShO, SC, HT, RH, TN, HK,
AS, YOn, YuM, MK and YOh. All authors wrote, read and approved the
final manuscript.
Ethics approval and consent to
participate
This study protocol, including tumor sample
collection and genetic analysis, was approved by the Asahikawa
Medical University Research Ethics Committee (approval nos. 17043
and 18118). For the prospective study (approval nos. 17043),
written informed consent was obtained from all patients before
their enrollment for genetic analysis. For the retrospective study
for survival analysis of 36 patients with TNBC (approval no.
18118), informed consent was waived by the Asahikawa Medical
University Research Ethics Committee and an opportunity for opt-out
regarding the study was provided through the institutional
website.
Patient consent for publication
Written informed consent for publication of clinical
details and /or clinical images was obtained from the patients.
Competing interests
YMiz and YOn receive funding from Hitachi High
Technologies, Inc. for the experimental equipment. No potential
conflicts of interest were disclosed by the other authors.
References
|
1
|
Massihnia D, Galvano A, Fanale D, Perez A,
Castiglia M, Incorvaia L, Listì A, Rizzo S, Cicero G, Bazan V, et
al: Triple negative breast cancer: Shedding light onto the role of
pi3k/akt/mtor pathway. Oncotarget. 7:60712–60722. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cossu-Rocca P, Orrù S, Muroni MR, Sanges
F, Sotgiu G, Ena S, Pira G, Murgia L, Manca A, Uras MG, et al:
Analysis of PIK3CA mutations and activation pathways in triple
negative breast cancer. PLoS One. 10:e01417632015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oxnard GR, Paweletz CP, Kuang Y, Mach SL,
O'Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P,
Jackman DM, et al: Noninvasive detection of response and resistance
in EGFR-mutant lung cancer using quantitative next-generation
genotyping of cell-free plasma DNA. Clin Cancer Res. 20:1698–1705.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takeshita T, Yamamoto Y, Yamamoto-Ibusuki
M, Tomiguchi M, Sueta A, Murakami K and Iwase H: Clinical
significance of plasma cell-free DNA mutations in PIK3CA, AKT1, and
ESR1 gene according to treatment lines in ER-positive breast
cancer. Mol Cancer. 17:672018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takeshita T, Yamamoto Y, Yamamoto-Ibusuki
M, Tomiguchi M, Sueta A, Murakami K, Omoto Y and Iwase H: Analysis
of ESR1 and PIK3CA mutations in plasma cell-free DNA from
ER-positive breast cancer patients. Oncotarget. 8:52142–52155.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen YH, Hancock BA, Solzak JP, Brinza D,
Scafe C, Miller KD and Radovich M: Next-generation sequencing of
circulating tumor DNA to predict recurrence in triple-negative
breast cancer patients with residual disease after neoadjuvant
chemotherapy. NPJ Breast Cancer. 3:242017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Riva F, Bidard FC, Houy A, Saliou A, Madic
J, Rampanou A, Hego C, Milder M, Cottu P, Sablin MP, et al:
Patient-Specific Circulating Tumor DNA Detection during Neoadjuvant
Chemotherapy in Triple-Negative Breast Cancer. Clin Chem.
63:691–699. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan pH
and van de Vijver MJ: WHO Classification of Tumours of the Breast.
4:(4th edition). IARC WHO Classification of Tumours. 2012.
|
|
10
|
Yoshida R, Sasaki T and Ohsaki Y: EGFR and
KRAS mutations in triple-mutated lung cancer. J Thorac Oncol.
12:e92–e93. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoshida R, Sasaki T, Umekage Y, Tanno S,
Ono Y, Ogata M, Chiba S, Mizukami Y and Ohsaki Y: Highly sensitive
detection of ALK resistance mutations in plasma using droplet
digital PCR. BMC Cancer. 18:11362018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ono Y, Sugitani A, Karasaki H, Ogata M,
Nozaki R, Sasajima J, Yokochi T, Asahara S, Koizumi K, Ando K, et
al: An improved digital polymerase chain reaction protocol to
capture low-copy KRAS mutations in plasma cell-free DNA by
resolving ‘subsampling’ issues. Mol Oncol. 11:1448–1458. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishibashi K, Kumai T, Ohkuri T, Kosaka A,
Nagato T, Hirata Y, Ohara K, Oikawa K, Aoki N, Akiyama N, et al:
Epigenetic modification augments the immunogenicity of human
leukocyte antigen G serving as a tumor antigen for T cell-based
immunotherapy. OncoImmunology. 5:e11693562016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers
refines their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McCall CM, Mosier S, Thiess M, Debeljak M,
Pallavajjala A, Beierl K, Deak KL, Datto MB, Gocke CD, Lin MT, et
al: False positives in multiplex PCR-based next-generation
sequencing have unique signatures. J Mol Diagn. 16:541–549. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barak V, Holdenrieder S, Nisman B and
Stieber P: Relevance of circulating biomarkers for the therapy
monitoring and follow-up investigations in patients with non-small
cell lung cancer. Cancer Biomark. 6:191–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Colt HG, Murgu SD, Korst RJ, Slatore CG,
Unger M and Quadrelli S: Follow-up and surveillance of the patient
with lung cancer after curative-intent therapy: Diagnosis and
Management of Lung Cancer, 3rd ed: American College of Chest
Physicians Evidence-Based Clinical Practice Guidelines. Chest. 143
(Suppl 5):e437S–e454S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Emens LA and Davidson NE: The follow-up of
breast cancer. Semin Oncol. 30:338–348. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Albain KS, Nag SM, Calderillo-Ruiz G,
Jordaan JP, Llombart AC, Pluzanska A, Rolski J, Melemed AS,
Reyes-Vidal JM, Sekhon JS, et al: Gemcitabine plus Paclitaxel
versus Paclitaxel monotherapy in patients with metastatic breast
cancer and prior anthracycline treatment. J Clin Oncol.
26:3950–3957. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Perez EA, Hillman DW, Mailliard JA, Ingle
JN, Ryan JM, Fitch TR, Rowland KM, Kardinal CG, Krook JE, Kugler
JW, et al: Randomized phase II study of two irinotecan schedules
for patients with metastatic breast cancer refractory to an
anthracycline, a taxane, or both. J Clin Oncol. 22:2849–2855. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Talbot DC, Moiseyenko V, Van Belle S,
O'Reilly SM, Alba Conejo E, Ackland S, Eisenberg P, Melnychuk D,
Pienkowski T, Burger HU, et al: Randomised, phase II trial
comparing oral capecitabine (Xeloda) with paclitaxel in patients
with metastatic/advanced breast cancer pretreated with
anthracyclines. Br J Cancer. 86:1367–1372. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X,
Xiao Y, Yu KD, Liu YR, Yu Y, et al: Genomic and transcriptomic
landscape of triple-negative breast cancers: Subtypes and treatment
strategies. Cancer Cell. 35:428–440.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Bruin EC, Whiteley JL, Corcoran C, Kirk
PM, Fox JC, Armisen J, Lindemann JPO, Schiavon G, Ambrose HJ and
Kohlmann A: Accurate detection of low prevalence AKT1 E17K mutation
in tissue or plasma from advanced cancer patients. PLoS One.
12:e01757792017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim MS, Jeong EG, Yoo NJ and Lee SH:
Mutational analysis of oncogenic AKT E17K mutation in common solid
cancers and acute leukaemias. Br J Cancer. 98:1533–1535. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rudolph M, Anzeneder T, Schulz A, Beckmann
G, Byrne AT, Jeffers M, Pena C, Politz O, Köchert K, Vonk R, et al:
AKT1 (E17K) mutation profiling in breast cancer: Prevalence,
concurrent oncogenic alterations, and blood-based detection. BMC
Cancer. 16:6222016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hyman DM, Smyth LM, Donoghue MTA, Westin
SN, Bedard PL, Dean EJ, Bando H, El-Khoueiry AB, Pérez-Fidalgo JA,
Mita A, et al: AKT inhibition in solid tumors with AKT1 mutations.
J Clin Oncol. 35:2251–2259. 2017. View Article : Google Scholar : PubMed/NCBI
|