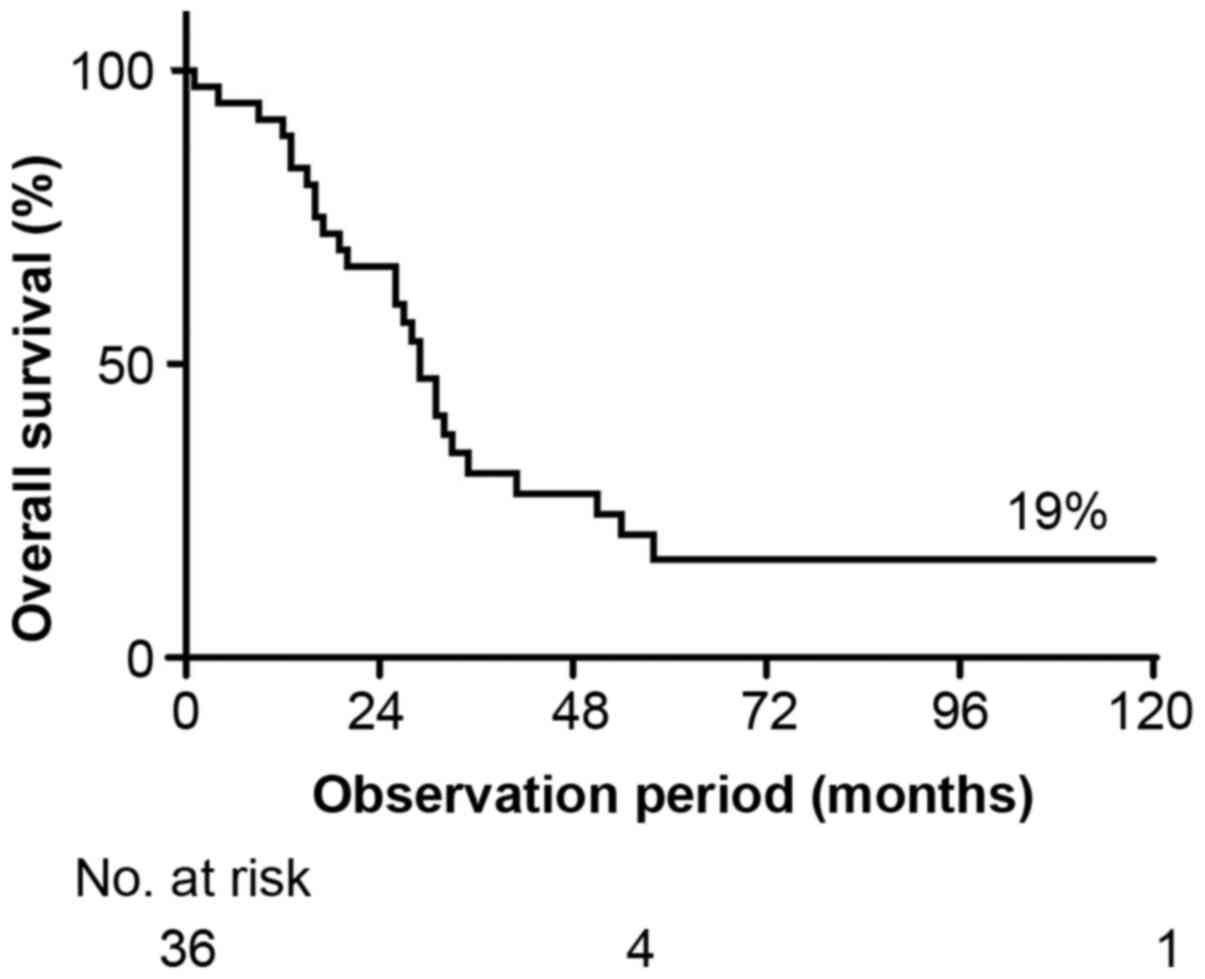
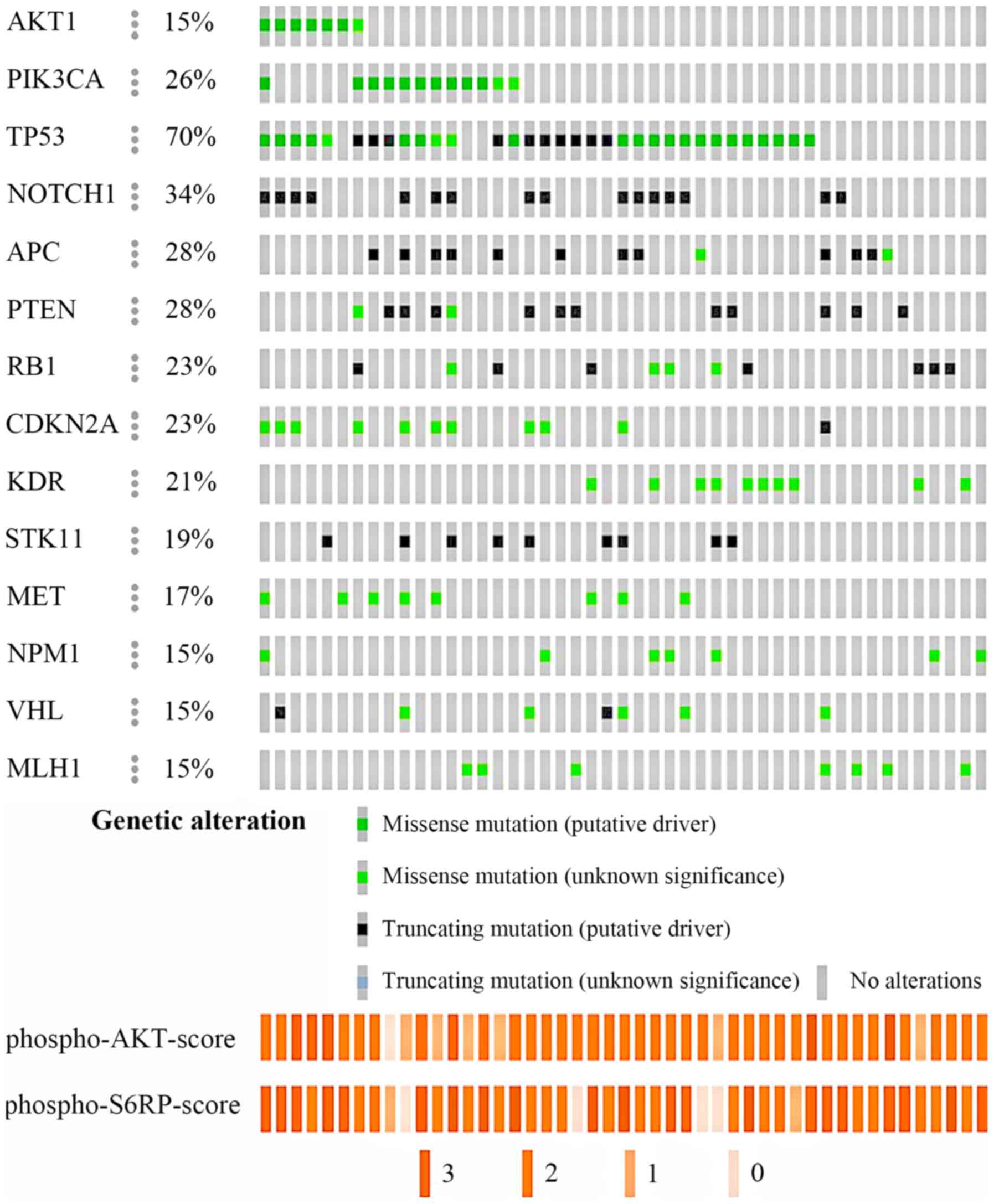
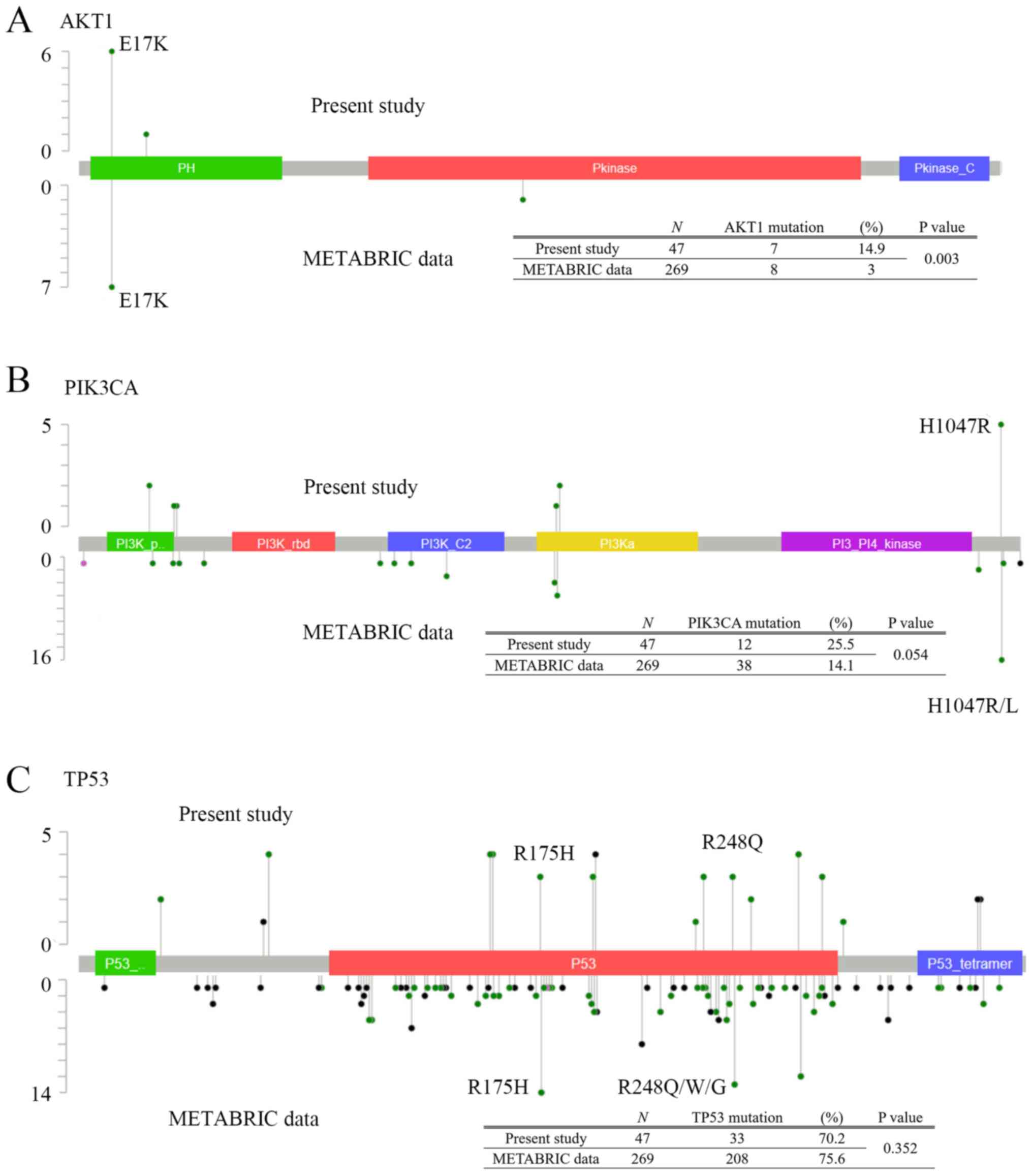
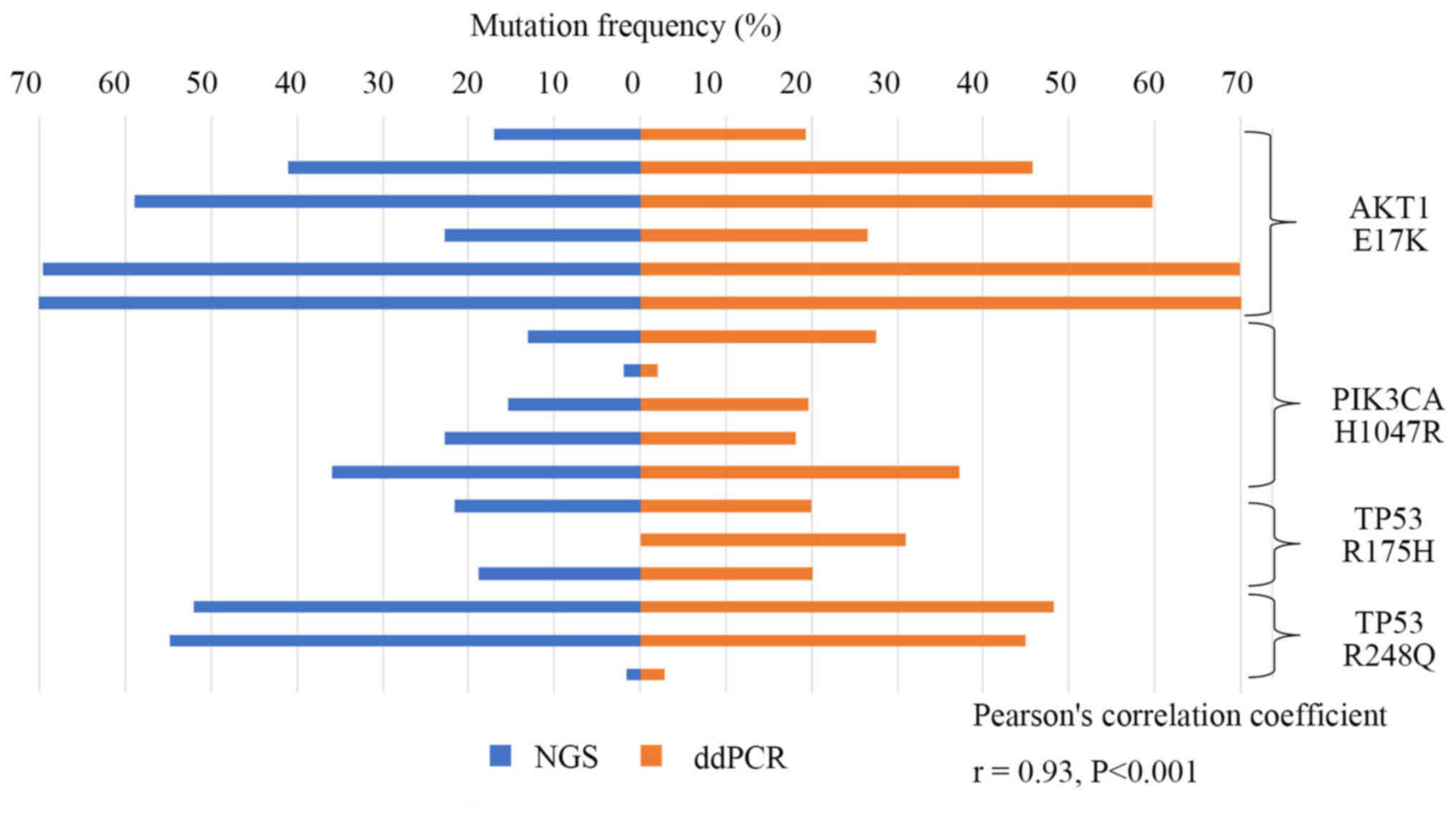
|
1
|
Massihnia D, Galvano A, Fanale D, Perez A,
Castiglia M, Incorvaia L, Listì A, Rizzo S, Cicero G, Bazan V, et
al: Triple negative breast cancer: Shedding light onto the role of
pi3k/akt/mtor pathway. Oncotarget. 7:60712–60722. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cossu-Rocca P, Orrù S, Muroni MR, Sanges
F, Sotgiu G, Ena S, Pira G, Murgia L, Manca A, Uras MG, et al:
Analysis of PIK3CA mutations and activation pathways in triple
negative breast cancer. PLoS One. 10:e01417632015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oxnard GR, Paweletz CP, Kuang Y, Mach SL,
O'Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P,
Jackman DM, et al: Noninvasive detection of response and resistance
in EGFR-mutant lung cancer using quantitative next-generation
genotyping of cell-free plasma DNA. Clin Cancer Res. 20:1698–1705.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takeshita T, Yamamoto Y, Yamamoto-Ibusuki
M, Tomiguchi M, Sueta A, Murakami K and Iwase H: Clinical
significance of plasma cell-free DNA mutations in PIK3CA, AKT1, and
ESR1 gene according to treatment lines in ER-positive breast
cancer. Mol Cancer. 17:672018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takeshita T, Yamamoto Y, Yamamoto-Ibusuki
M, Tomiguchi M, Sueta A, Murakami K, Omoto Y and Iwase H: Analysis
of ESR1 and PIK3CA mutations in plasma cell-free DNA from
ER-positive breast cancer patients. Oncotarget. 8:52142–52155.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen YH, Hancock BA, Solzak JP, Brinza D,
Scafe C, Miller KD and Radovich M: Next-generation sequencing of
circulating tumor DNA to predict recurrence in triple-negative
breast cancer patients with residual disease after neoadjuvant
chemotherapy. NPJ Breast Cancer. 3:242017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Riva F, Bidard FC, Houy A, Saliou A, Madic
J, Rampanou A, Hego C, Milder M, Cottu P, Sablin MP, et al:
Patient-Specific Circulating Tumor DNA Detection during Neoadjuvant
Chemotherapy in Triple-Negative Breast Cancer. Clin Chem.
63:691–699. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan pH
and van de Vijver MJ: WHO Classification of Tumours of the Breast.
4:(4th edition). IARC WHO Classification of Tumours. 2012.
|
|
10
|
Yoshida R, Sasaki T and Ohsaki Y: EGFR and
KRAS mutations in triple-mutated lung cancer. J Thorac Oncol.
12:e92–e93. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoshida R, Sasaki T, Umekage Y, Tanno S,
Ono Y, Ogata M, Chiba S, Mizukami Y and Ohsaki Y: Highly sensitive
detection of ALK resistance mutations in plasma using droplet
digital PCR. BMC Cancer. 18:11362018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ono Y, Sugitani A, Karasaki H, Ogata M,
Nozaki R, Sasajima J, Yokochi T, Asahara S, Koizumi K, Ando K, et
al: An improved digital polymerase chain reaction protocol to
capture low-copy KRAS mutations in plasma cell-free DNA by
resolving ‘subsampling’ issues. Mol Oncol. 11:1448–1458. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishibashi K, Kumai T, Ohkuri T, Kosaka A,
Nagato T, Hirata Y, Ohara K, Oikawa K, Aoki N, Akiyama N, et al:
Epigenetic modification augments the immunogenicity of human
leukocyte antigen G serving as a tumor antigen for T cell-based
immunotherapy. OncoImmunology. 5:e11693562016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers
refines their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McCall CM, Mosier S, Thiess M, Debeljak M,
Pallavajjala A, Beierl K, Deak KL, Datto MB, Gocke CD, Lin MT, et
al: False positives in multiplex PCR-based next-generation
sequencing have unique signatures. J Mol Diagn. 16:541–549. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barak V, Holdenrieder S, Nisman B and
Stieber P: Relevance of circulating biomarkers for the therapy
monitoring and follow-up investigations in patients with non-small
cell lung cancer. Cancer Biomark. 6:191–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Colt HG, Murgu SD, Korst RJ, Slatore CG,
Unger M and Quadrelli S: Follow-up and surveillance of the patient
with lung cancer after curative-intent therapy: Diagnosis and
Management of Lung Cancer, 3rd ed: American College of Chest
Physicians Evidence-Based Clinical Practice Guidelines. Chest. 143
(Suppl 5):e437S–e454S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Emens LA and Davidson NE: The follow-up of
breast cancer. Semin Oncol. 30:338–348. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Albain KS, Nag SM, Calderillo-Ruiz G,
Jordaan JP, Llombart AC, Pluzanska A, Rolski J, Melemed AS,
Reyes-Vidal JM, Sekhon JS, et al: Gemcitabine plus Paclitaxel
versus Paclitaxel monotherapy in patients with metastatic breast
cancer and prior anthracycline treatment. J Clin Oncol.
26:3950–3957. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Perez EA, Hillman DW, Mailliard JA, Ingle
JN, Ryan JM, Fitch TR, Rowland KM, Kardinal CG, Krook JE, Kugler
JW, et al: Randomized phase II study of two irinotecan schedules
for patients with metastatic breast cancer refractory to an
anthracycline, a taxane, or both. J Clin Oncol. 22:2849–2855. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Talbot DC, Moiseyenko V, Van Belle S,
O'Reilly SM, Alba Conejo E, Ackland S, Eisenberg P, Melnychuk D,
Pienkowski T, Burger HU, et al: Randomised, phase II trial
comparing oral capecitabine (Xeloda) with paclitaxel in patients
with metastatic/advanced breast cancer pretreated with
anthracyclines. Br J Cancer. 86:1367–1372. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X,
Xiao Y, Yu KD, Liu YR, Yu Y, et al: Genomic and transcriptomic
landscape of triple-negative breast cancers: Subtypes and treatment
strategies. Cancer Cell. 35:428–440.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Bruin EC, Whiteley JL, Corcoran C, Kirk
PM, Fox JC, Armisen J, Lindemann JPO, Schiavon G, Ambrose HJ and
Kohlmann A: Accurate detection of low prevalence AKT1 E17K mutation
in tissue or plasma from advanced cancer patients. PLoS One.
12:e01757792017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim MS, Jeong EG, Yoo NJ and Lee SH:
Mutational analysis of oncogenic AKT E17K mutation in common solid
cancers and acute leukaemias. Br J Cancer. 98:1533–1535. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rudolph M, Anzeneder T, Schulz A, Beckmann
G, Byrne AT, Jeffers M, Pena C, Politz O, Köchert K, Vonk R, et al:
AKT1 (E17K) mutation profiling in breast cancer: Prevalence,
concurrent oncogenic alterations, and blood-based detection. BMC
Cancer. 16:6222016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hyman DM, Smyth LM, Donoghue MTA, Westin
SN, Bedard PL, Dean EJ, Bando H, El-Khoueiry AB, Pérez-Fidalgo JA,
Mita A, et al: AKT inhibition in solid tumors with AKT1 mutations.
J Clin Oncol. 35:2251–2259. 2017. View Article : Google Scholar : PubMed/NCBI
|