Introduction
According to the global cancer statistics of 2018,
gastric cancer (GC) is the fifth most common type of cancer
(1,033,701 new cases) and the third leading cause of
cancer-associated mortality (782,685 deaths) worldwide (1). The incidence rate of GC has been
declining in North America and most Western European countries;
however, the burden of GC remains high in Asia, Latin America, and
Central and Eastern Europe (2).
Notably, half of all cases of GC worldwide occur in East Asia
(3). Despite the progress in the
development of novel therapeutic approaches, the prognosis of GC
remains poor, as the traditional histological subtypes cannot be
used to determine the different prognoses and chemotherapeutic
responses (4–6). Emerging data have suggested that GC is
a complex and heterogenic disease. The underlying molecular and
histopathological tumor features are responsible for the diverse
outcomes and chemotherapeutic responses of patients (7–9). The
genomic and molecular classification of GC have been addressed by
The Cancer Genome Atlas and the Asian Cancer Research Group, both
of which have indicated that microsatellite instability (MSI) GC is
a separate subgroup of GC (10,11).
MSI is a type of genetic instability, characterized
by length alteration in the tandem repeat sequence, which is
referred to as a microsatellite (MS) (12), is known as a hypermutable phenotype
and occurs due to a defective DNA mismatch repair (MMR) system,
with key MMR gene inactivations, such as germline mutations in mutS
homolog 2 (MSH2) or mutL homolog 1 (MLH1) (13). At present, MSI detection is widely
used in the screening of Lynch syndrome and the prognosis
evaluation of colorectal cancer (CRC) using the Bethesda (B5) panel
recommended by the American National Cancer Institute (14). However, Hause et al (15) identified MSI-positive tumors in 14
out of 18 cancer types, using 5,930 cancer exomes and the
mutation-prone MS loci of different tumors were largely
distinctive, indicating that loci that are stable in one type of
cancer may be frequently altered in another. In GC, the association
between MSI and clinical features remains ambiguous, which may be
due to the arbitrary use of MS loci, which is mostly based on CRC
studies to define MSI status in different studies, and these loci
are not likely to be sufficient for making clinical decisions.
Therefore, there is an urgent requirement to identify novel MS loci
for the clinical guidance of GC (11,16,17).
It is well-known that the alteration of
tumor-related genes, such as the inactivation of tumor suppressor
(TS) genes and the amplification of oncogenes, is a main factor in
GC development (18,19). On the other hand, MSs are frequently
used as markers for the detection of loss of heterozygosity (LOH),
which has been associated with GC progression (20). MSI and LOH status have been
considered to be valuable and independent prognostic markers in
patients with CRC (21).
The present study first focused on a wide range of
MS loci located in GC-related genes, which were likely to be
associated with GC development, and aimed to identify a panel of MS
loci, which could be potential predictive biomarkers for clinical
features and outcomes of GC.
Materials and methods
Enrolled patients
The present retrospective study comprised of
patients with GC (226 specimens), who underwent surgical resection
between March 2008 and November 2014, and in which normal adjacent
mucosa and tumor tissues were collected. In addition, clinical
information from the Clinical Data and Biobank Resource of Beijing
Friendship Hospital (Beijing, China), was also collected for these
patients. This has also been assessed and certified to meet the
requirements of the China Human Genetic Resources Management Office
and ISO certification 9001:2015. The inclusion criteria for
patients were: i) Presence of a paired tissue sample (paired normal
adjacent mucosa and tumor tissues); and ii) Complete or partial
clinical information collected in the Clinical Data and Biobank
Resource of Beijing Friendship Hospital (Beijing, China). The
following exclusion criteria were used for patient recruitment: i)
Does not have frozen tumor tissues or paired normal adjacent
mucosa; ii) Does not have any clinical information; iii) Was not
admitted between March 2008 and November 2014; iv) And the initial
surgical treatment was not performed at Beijing Friendship Hospital
(Beijing, China). The tumor and normal adjacent mucosa tissues were
collected for research. To distinguish the normal adjacent mucosa
from the tumor tissues, the normal tissues were first defined by
its macroscopical character and then harvested by keeping a minimum
distance of 5 cm from the tumor tissue. Histological classification
was defined according to the World Health Organization guidelines
(22). Detailed information of the
patients is shown in Table I. As
aforementioned, the samples and the clinical information were
collected directly from the Clinical Data and Biobank Resource of
Beijing Friendship Hospital, and certain clinical information of a
few patients were missing, due to the lack of routine detection,
such as HP infection or other unknown reason. The Ethics Committee
of Beijing Friendship Hospital (Beijing, China) approved the study
proposal (approval. no. 2017-P2-013-03) and all patients involved
in the study provided written informed consent. A total of 226 GC
samples were defined as the validation set and a sub-group of 90
samples among the 226 GCs was defined as the training set.
 | Table I.Association between MSI status
detected using the Bethesda panel and clinicopathological
characteristics in patients with GC. |
Table I.
Association between MSI status
detected using the Bethesda panel and clinicopathological
characteristics in patients with GC.
| Clinicopathological
characteristic | Number (%) | MSI-H | MSI-L/MSS | P-value |
|---|
| Mean age,
years | 226 (100) | 65.9 | 62.4 | 0.074 |
| Sex |
|
Male | 166 (73.45) | 10 | 156 | 0.156 |
|
Female | 60 (26.55) | 7 | 53 |
|
| Smoking status |
|
Yes | 112 (51.38) | 6 | 106 | 0.167 |
| No | 106 (48.62) | 11 | 95 |
|
| Drinking
statusa |
|
Yes | 66 (30.28) | 1 | 65 | 0.026b |
| No | 152 (69.72) | 16 | 136 |
|
| TNM
stagea |
| I | 21 (9.81) | 2 | 19 | 0.947 |
| II | 42 (19.63) | 3 | 39 |
|
|
III | 130 (60.75) | 10 | 120 |
|
| IV | 21 (9.81) | 1 | 20 |
|
| Depth of tumor
invasiona |
|
pT1 | 11 (5.19) | 1 | 10 | 0.761 |
|
pT2 | 24 (11.32) | 3 | 21 |
|
|
pT3 | 79 (37.26) | 6 | 73 |
|
|
pT4 | 98 (46.23) | 6 | 92 |
|
| Lymph node
involvementa |
|
pN0 | 45 (21.33) | 8 | 37 | 0.0036b |
|
pN1-pN3a | 166 (78.67) | 8 | 158 |
|
| Presence of
metastasisa |
| M0 | 193 (90.19) | 15 | 178 | >0.999 |
| M1 | 21 (9.81) | 1 | 20 |
|
| HP
infectiona |
|
Negative | 29 (56.86) | 3 | 26 | >0.999 |
|
Positive | 22 (43.14) | 2 | 20 |
|
| Pathological
typea |
|
Adenocarcinoma | 128 (62.14) | 9 | 119 | 0.581 |
|
Mucinous carcinoma | 78 (37.86) | 7 | 71 |
|
| Histological
gradea |
|
Well-differentiated | 11 (6.59) | 1 | 10 | 0.571 |
|
Moderately/poorly | 156 (93.41) | 11 | 145 |
|
DNA isolation
Total genomic DNA was isolated and purified from 226
tumor and normal adjacent tissues using a standard
phenol-chloroform extraction and ethanol precipitation method, as
previously described by Du et al (23). In addition, the quality of DNA was
evaluated by measuring the A260/A280 ratio with a micro-volume
spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific, Inc.)
and further assessed using 1.5% agarose gel electrophoresis.
MS analysis
In total, 20 genes with mononucleotide (mono-),
dinucleotide (di-), trinucleotide (tri-), tetranucleotide (tetra-),
pentanucleotide (penta-) or compound (more than one type of repeat
motif) MSs were selected for screening. Cancer-associated genes,
including ARID1A, APC, BAX, BRAF, CDKN1A, CDX2, CTNNB1, ERBB2,
E2F4, MCC, PTN, PTEN, PRR11, RUNX3, TGFBRII and TP53,
which have been reported to be associated with GC in the
literature, and MMR genes, including MSH2, MLH1, MGMT and
PMS2, were included in the present study (24–31). In
total, 91 MS loci were selected from the 5′ untranslated region
(UTR), intron, exon or 3′UTR of these 20 genes using the SSRHunter
software (v1.3) (Li Qiang, Nanjing Agricultural University) and
manual searching (Table SI), then
primers for each selected MS were designed based on the flanking
sequence using Primer Blast in the Pubmed website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).
Subsequently, the suitability of the designed primers was
determined using PCR and the conditions were optimized. The B5
panel (BAT25, BAT26, D2S123, D5S346 and D17S250) which was used as
the control to compare with the loci in tumor-related genes was
also included in the present study. PCR was used to amplify these
loci and short tandem repeat (STR) scanning (fluorescence capillary
electrophoresis) was used for the detection of MSI, which were both
performed on an ABI-3730XL DNA Analyzer system (Applied Biosystems;
Thermo Fisher Scientific, Inc.), as previously described by Huo
et al (32). Briefly, the PCR
(20 µl) was performed using a Taq DNA polymerase kit (cat. no.
R001AM; Takara Bio, Inc.,) and included 2 µl 10X buffer, 0.5 µmol/l
of each primer (synthesized by Sangon Biotech Co., Ltd.), 125
µmol/l dNTP (4X), 1.0 U Taq DNA polymerase, 1.5–2.5 mmol/l
MgCl2 and 100 ng template DNA. The following
thermocycling conditions were used: Initial denaturation at 94°C
for 5 min; followed by 35 cycles of denaturation at 94°C for 30
sec, annealing at gradient temperatures for each MS for 30 sec and
extension at 72°C for 30 sec; and a final extension at 72°C for 5
min.
A result was determined to be a MSI when the PCR
product generated for the tumor sample exhibited at least one new
peak compared with that in the product from the adjacent normal
tissue using GeneMarker v1.75 (SoftGenetics, LLC). For the
heterozygous locus, a sample was determined to exhibit LOH when one
of the alleles was completely or partially lost. More specifically,
the ratio of the peak height between the two alleles in the
adjacent normal tissue compared with that in the tumor tissue was
<0.67 or >1.5. The peak height was determined using
GeneMarker v1.75 (SoftGenetics, LLC).
The mutation profiles of 91 MS loci in tumor-related
genes and 5 B5 loci in the training group (n=90) were first
examined and then validated using a sub-set of the 91 loci (n=27)
and 5 B5 loci in the second set of samples (n=136). The present
study evaluated the efficiency of the loci using the Efficiency
Score (ES). The ES was calculated as follows: ES =
RankSensitivity + RankSpecificity. The lower
the ES was, the more efficient the locus was. Sensitivity equals
the number of detected MSI/the total. Specificity equals the number
of detected MSS/the total number.
DNA sequencing
To validate the STR scanning results, PCR fragments
were cloned into the PMD19-TVector (TA cloning Vector) (Takara Bio,
Inc.), and sequence analysis was performed using an ABI 3730XL DNA
Sequencer and Sanger sequencing, and was commissioned to be
completed by Annoroad Gene Technology Co., Ltd. To define the
occurrence of MSI, the sequencing results of the examined loci in
MSI-positive GC tissues were compared with those in the adjacent
normal tissues.
Statistical analysis
Descriptive statistics were used to summarize the
relevant clinicopathological information. The comparation of
MSI/LOH frequency, core sequence length and repeat unit of mono-,
di-, tri-, tetra- and penta- was analyzed using one-way ANOVA and a
Tukey's post hoc test. The comparation of age in different groups
was analyzed using an unpaired t-test and the data is presented as
mean ± SD.; sex, smoking, drinking, lymph node involvement, TNM
stage, depth of tumor invasion, metastasis, HP infection,
pathological type, and histological grade, were analyzed using a
Pearson's χ2 test or a Fisher's exact test, and
Spearman's correlation analysis was used to investigate the
correlation between intrinsic features (length and repeat number of
the MS core sequence) and the MSI/LOH frequency. Overall survival
[OS; the time between the start of randomization (which means the
patients received different treatments after surgical resection)
and death] and disease-free survival (DFS; the time between the
start of randomization and recurrence of the disease) analyses were
determined using the Kaplan-Meier method, and statistically
analyzed using a log-rank test. Data analysis was performed using
SPSS Statistics v16.0 software (SPSS, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
MSI/LOH analysis in tumor-related
genes
A total of 586 variations, including MSI and LOH
events, were identified when analyzing 91 MS markers in 20
tumor-related genes, using STR scanning in the first set of
patients with GC (n=90). It was revealed that 35 out of the 90 GC
samples (38.9%) did not exhibit any MSI or LOH. Among the 91 loci,
91.2% were mutated in ≥1 patient (Fig.
S1; Table SII). To validate the
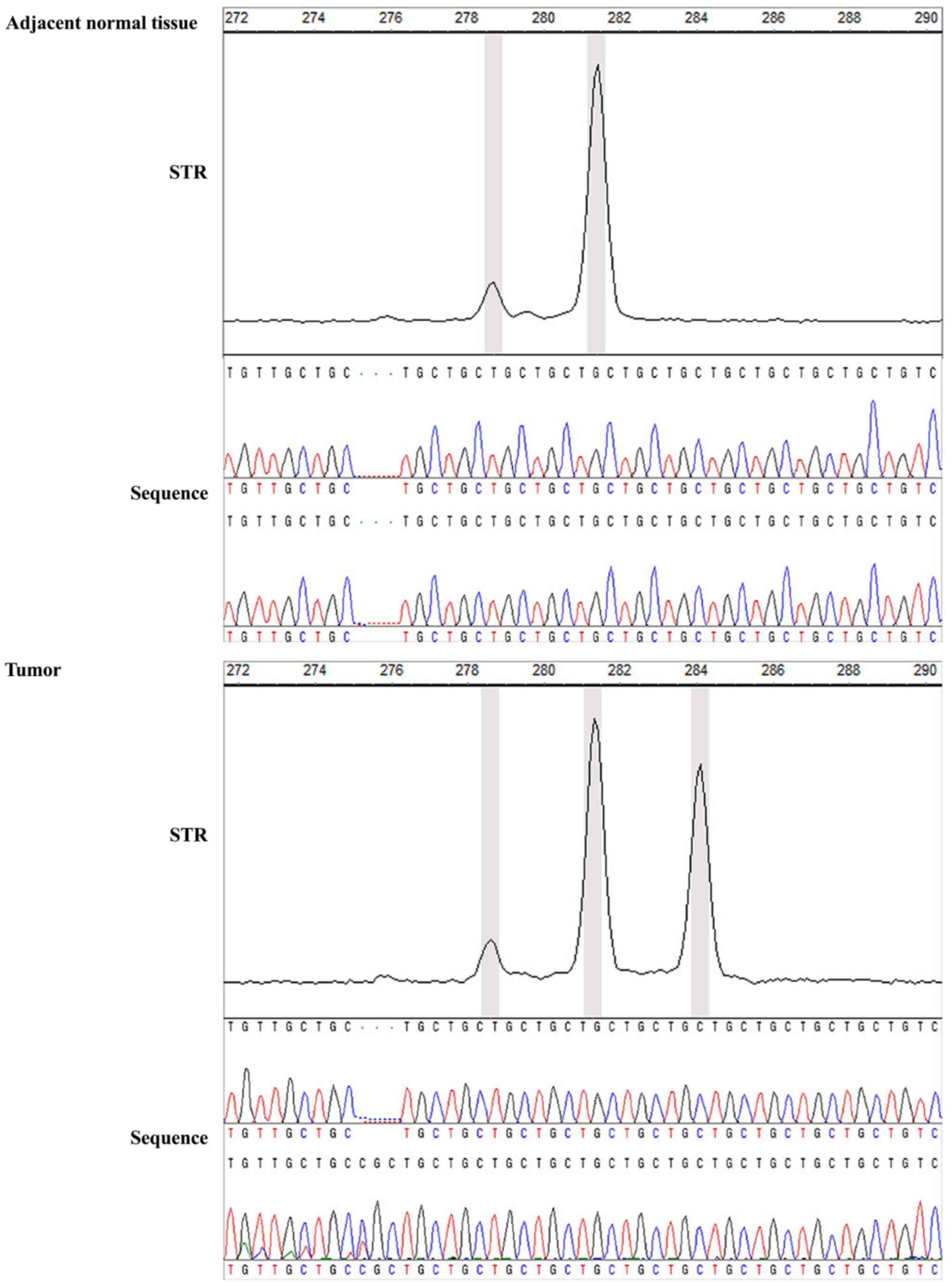
scanning results, one of the MSI-positive GC sample of locus E2F4
and its adjacent normal tissue were selected for TA clone
sequencing. The STR scanning result in adjacent normal tissue was
281/281 bp and was mutated to 281/284 bp (+3 bp) in the matched
tumor sample. In addition, there was a TGC (3 bp) insertion in the
sequencing result of the tumor tissue (Fig. 1). The MSI/LOH frequency of the
individual MSs not only varied markedly from locus to locus, but
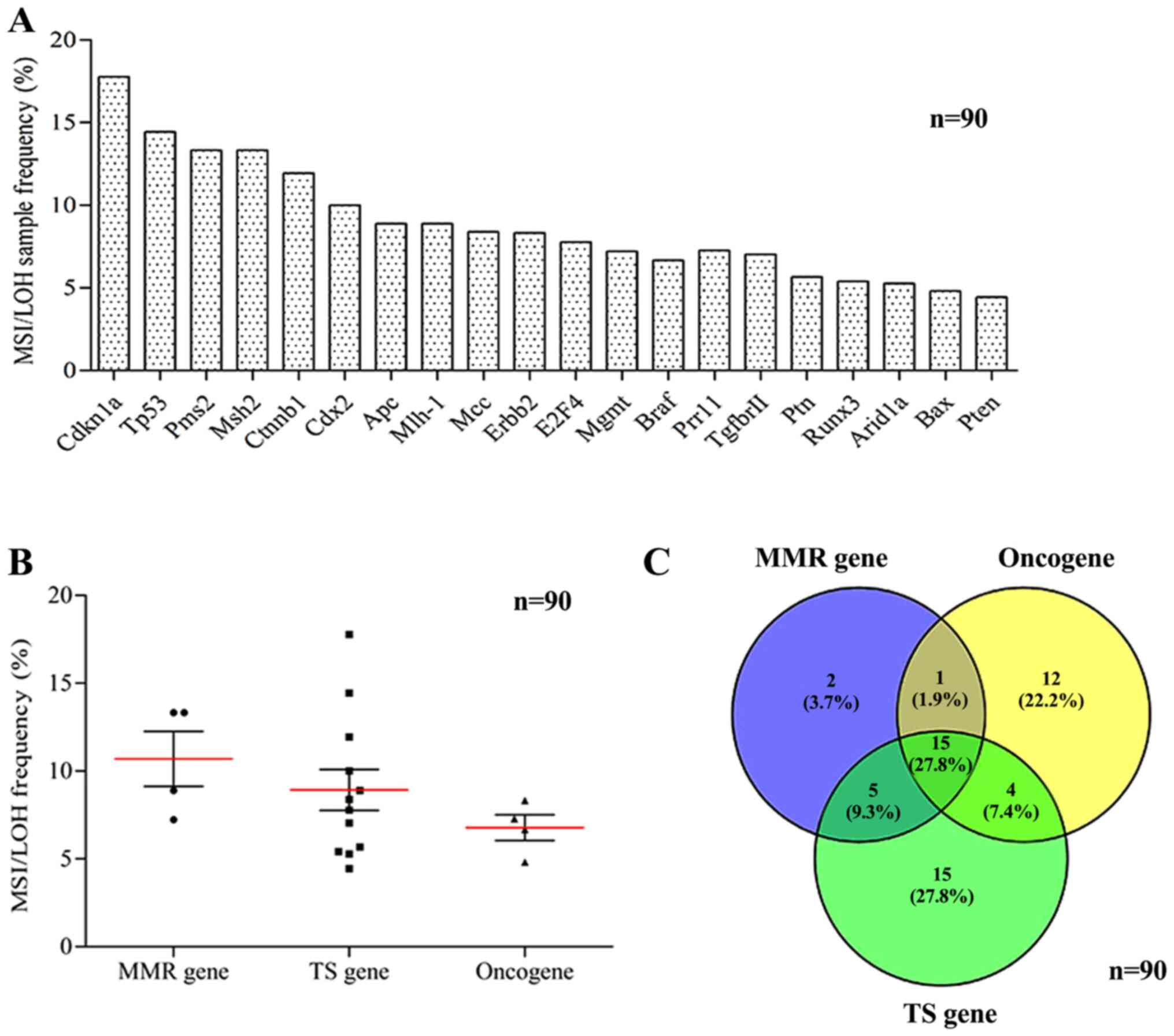
also from gene to gene (Fig. 2A).
There was no significant difference in MS variation among MMR
genes, oncogenes or TS genes (P=0.332; Fig. 2B). However, most MS variations in the
MMR genes (16/23; 69.6%) exhibited consistency compared with that
in oncogenes, and similarly, 87.0% (20/23) of the mutations in the
MMR genes were consistent compared with that in the TS genes. In
addition, the consistency between MS variations in oncogenes
(19/32, 59.4%) and TS genes (19/39, 48.7%) were much lower
(Fig. 2C). The MS variation
frequency varied markedly between the different loci; however, the
differences between the MSI/LOH profiles and the MMR genes,
oncogenes and TS genes suggested that the mutations (MSI/LOH)
occurring in the genes were not completely random, but followed a
certain regularity. Among the mutations that occurred in MMR genes,
only 8.70% (2/23) of MSI/LOH events occurred in only the MMR genes,
while 91.30% (21/23) of MSI/LOH events simultaneously occurred in
the MMR genes and oncogenes or TS genes (Fig. 2C).
Effect of the intrinsic features of MS
on mutations
In cancer, the causes of MSI/LOH are not completely
understood (33). To investigate the
intrinsic features, which may contribute to MSI/LOH frequency in
tumors, the present study analyzed the repeat units, length and
repeated motif of the 91 MS loci. The MS locus number of the mono-,
di-, tri-, tetra-, penta-MS and compounds was 42, 22, 11, 9, 2 and
5, respectively (Fig. 3A).
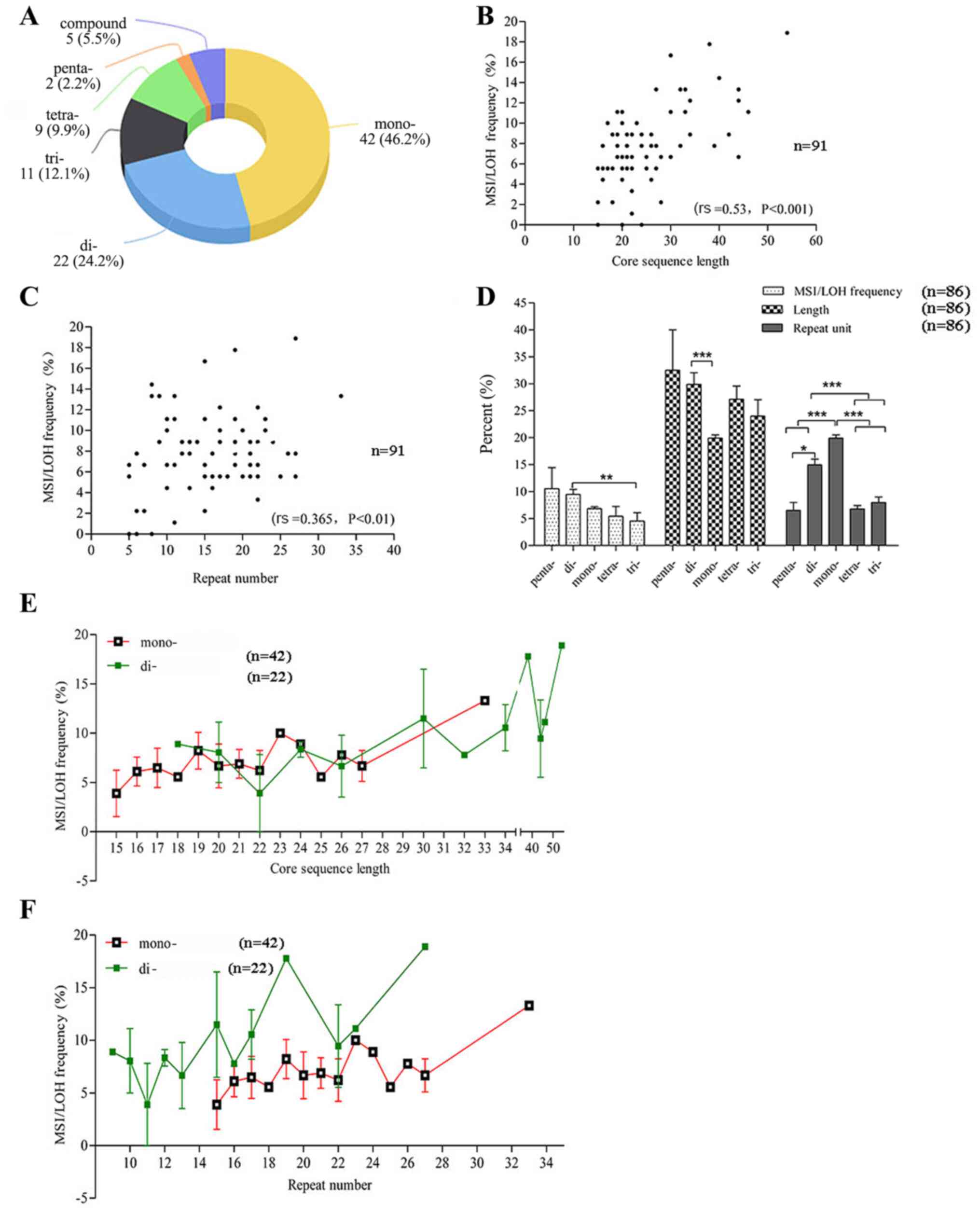 | Figure 3.Effect of the intrinsic features of
MS on MSI/LOH occurrence. (A) Repeat motif composition of the 91 MS
loci. Correlation between (B) core sequence length and (C) repeat
number with MSI/LOH frequency. P-values were determined using a
Spearman's correlation. (D) Mean MSI/LOH frequency, length and
repeat number of different motif groups. P-values were determined
using one-way ANOVA followed by Tukey's post hoc test. (E) MSI
frequency of different sequence lengths in the mono- and di-MS
groups. (F) MSI/LOH frequency of different repeat numbers in the
mono- and di-MS groups. *P<0.05, **P<0.01 and ***P<0.001.
MS, microsatellite; MSI, microsatellite instability; LOH, loss of
heterozygosity; mono-, mononucleotide; di-, dinucleotide; tetra-,
tetranucleotide; tri-, trinucleotide; penta-, pentanucleotide. |
It was first revealed that the length and repeat
number of the MS core sequence were positively correlated with MSI
and LOH incidence (rs=0.53; P<0.001 and
rs=0.365; P<0.01; Fig. 3B
and C, respectively).
The MSI/LOH frequency of the di-MS group was
markedly higher compared with that in the tri- MS groups
(P<0.01); however, it was unclear if this was due to the
different repeat motifs or the different length/repeat number of
the core sequence. The core sequence length of di-MS was
significantly longer compared with mono-MS (P<0.001) and the
repeat number of mono-MS was the highest among all the type of MS
followed by di-MS (5 compound MSs were excluded, so the loci number
was 91–5=86) (Fig. 3D). Due to the
limited number of tri-, tetra- and penta-MS loci, only the mono-
and di-MS loci were selected for the next step. To investigate
whether the diversity in length and repeat number of the core
sequence contributed to the different MSI/LOH frequency, the
MSI/LOH frequency was further compared between the di- and mono-MS
groups, when the same length or repeat number was noted. Based on
the trend of the curves, it seemed that for the same length, the
MSI/LOH frequency was not very different, since crossover was
observed between the two curves (Fig.
3E). However, for the same repeat number, the MSI/LOH frequency
of the di-MS loci was higher compared with that in the mono-MS loci
(Fig. 3F). Therefore, it was
hypothesized that the difference in the MSI/LOH rate between mono-
and di-MS was mainly due to the different lengths rather than the
repeat motif or repeat number only.
MSI/LOH of the validation loci in 226
GC samples
Based on the frequency and sample coverage of the
MSI/LOH within the tumor-associated genes, in the first set of
samples (n=90), a total of 27 loci in 15 genes were optimized.
Subsequently, the same method was used to detect the variation of
the 27 selected loci and 5 B5 panel loci in the second set of 136
samples (226–90=136). The results are shown in Fig. S2.
The results of the MSI/LOH in the 32 loci in the
first set (n=90) and second set (n=136) of patients with GC were
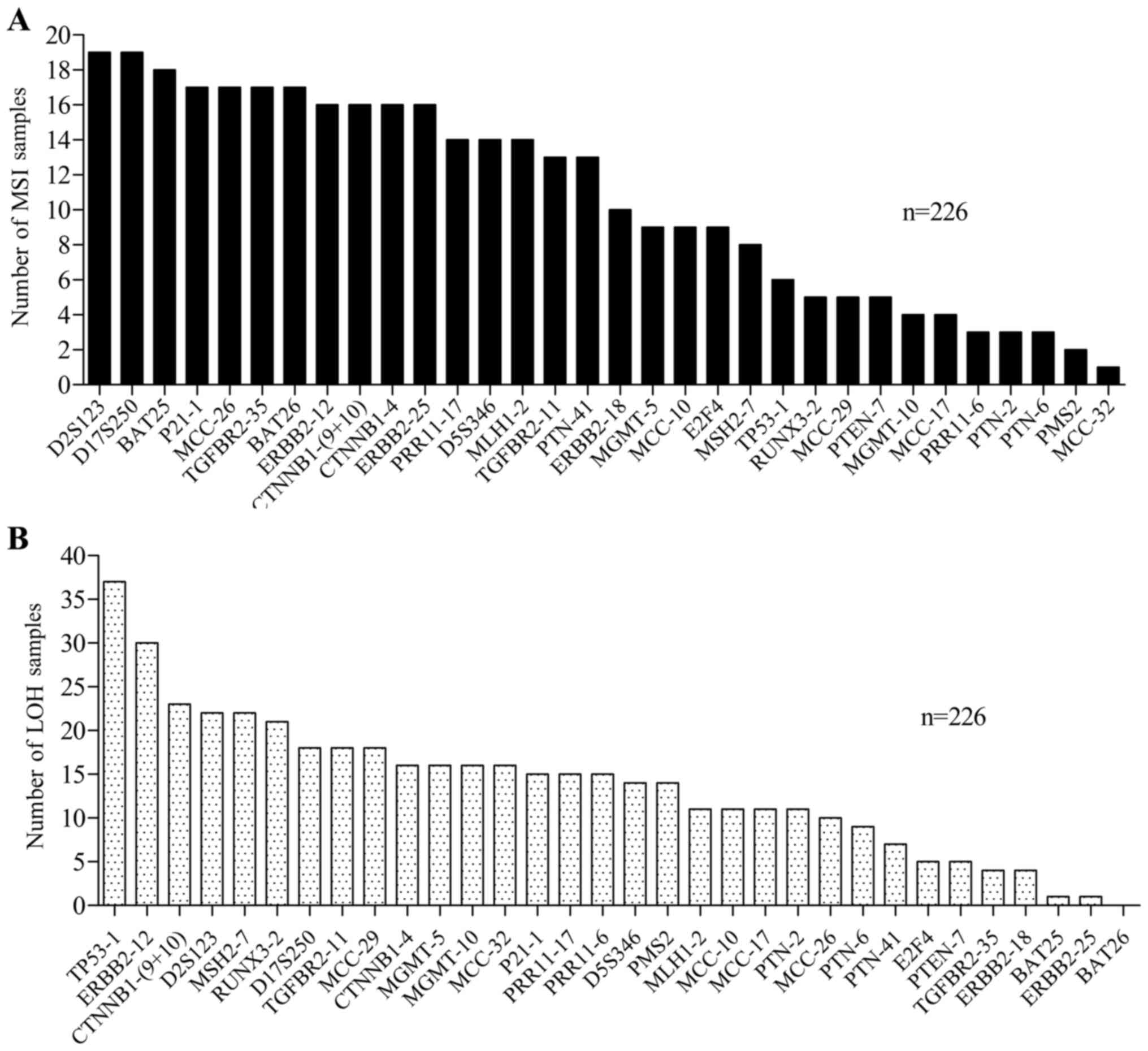
integrated in the validation group (n=90+136=226) (Fig. 4). The MSI frequencies of the 32 loci
in the 226 samples are shown in Fig.
4A. The MSI frequencies of the most frequent MS loci were:
D2S123 (19/226, 8.41%), D17S250 (19/226, 8.41%), BAT25 (18/226,
7.96%), P21-1 (17/226, 7.52%), MCC26 (17/226, 7.52%), TGFBR2-35
(17/226, 7.52%), BAT26 (17/226, 7.52%), ERBB2-12 (16/226, 7.08%),
CTNNB1-(9+10) (16/226, 7.08%), CTNNB1-4 (16/226, 7.08%) and
ERBB2-25 (16/226, 7.08%). Furthermore, the highest LOH occurrence
rate was observed for TP53-1 (37/226, 16.37%), followed by ERBB2-12
(30/226, 13.37%), CTNNB1-(9+10) (23/226, 10.18%), D2S123 (22/226,
9.73%), MSH2-7 (22/226, 9.73%) and RUNX3-2 (21/226, 9.29%)
(Fig. 4B).
MS panel for MSI-high (MSI-H)
detection
In previous study, MSI-H was defined as MSI
occurrence at ≥40% of the loci of the B5 panel, while occurrence at
<40% of the loci has been defined as MSI-low (MSI-L) and no MSI
occurrence at all detected loci was defined as microsatellite
stable (MSS) (14). In the present
study, the B5 panel analysis revealed that among the 226 samples,
17 (7.5%) were MSI-H, 9 (4.0%) were MSI-L and 200 (88.5%) were MSS.
The detected loci exhibited different sensitivity and specificity
(Table II). The optimal locus to
define MSI-H samples was BAT26, which had a sensitivity and
specificity of 100%. This was followed by BAT25 with a sensitivity
of 100% and a specificity of 99.5%. Therefore, BAT26 and BAT25 were
the most sensitive loci to identify MSI-H status in GC. The top 5
most efficient loci were BAT26, BAT25, CTNNB1-(9+10), ERBB2-25 and
TGFBR2-35. However, the B5 panel loci, D2S123, D5S346 and D17S250
had a higher ES, which was associated with a relatively low
efficiency for the definition of MSI-H status. The aim of this part
was to compare the detection efficiency of the loci in
tumor-related genes and B5 loci and find the top 5 loci, so we just
listed the loci which score whose score was >10 (Table II).
 | Table II.Efficiency of MSI-H detection of the
MS loci. |
Table II.
Efficiency of MSI-H detection of the
MS loci.
|
| Sensitivity | Specificity |
|
|---|
|
|
|
|
|
|---|
| Loci | Number of MSI | Rate (%) | Rank | Number of MSS | Rate (%) | Rank | ES |
|---|
| BAT26 | 17 | 100 | 1 | 209 | 100 | 1 | 2 |
| BAT25 | 17 | 100 | 1 | 208 | 99.5 | 2 | 3 |
| CTNNB1-(9+10) | 16 | 94.1 | 2 | 209 | 100 | 1 | 3 |
| ERBB2-25 | 16 | 94.1 | 2 | 209 | 100 | 1 | 3 |
| TGFBR2-35 | 16 | 94.1 | 2 | 208 | 99.5 | 2 | 4 |
| ERBB2-12 | 15 | 88.2 | 3 | 208 | 99.5 | 2 | 5 |
| D2S123 | 16 | 94.1 | 2 | 206 | 98.6 | 4 | 6 |
| P21-1 | 15 | 88.2 | 3 | 207 | 99 | 3 | 6 |
| D5S346 | 13 | 76.5 | 5 | 208 | 99.5 | 2 | 7 |
| D17S250 | 15 | 88.2 | 3 | 205 | 98.1 | 5 | 8 |
| MLH1-2 | 14 | 82.4 | 4 | 209 | 100 | 1 | 8 |
| PRR11-17 | 14 | 82.4 | 4 | 208 | 99.5 | 2 | 8 |
| TGFBR2-11 | 12 | 70.6 | 6 | 208 | 99.5 | 2 | 8 |
| PTN-41 | 12 | 70.6 | 6 | 208 | 99.5 | 2 | 8 |
| CTNNB1-4 | 13 | 76.5 | 5 | 206 | 98.6 | 4 | 9 |
| MCC-26 | 13 | 76.5 | 5 | 205 | 98.1 | 5 | 10 |
MSI/LOH status and clinical
features
Numerous studies have demonstrated that the MSI-H
status was associated with the clinicopathological features of
patients with GC; however, the results remain ambiguous (34,35). The
present study investigated the association between MSI-H status
detected using the B5 panel and clinicopathological
characteristics. The results indicated that MSI-H status was
associated with drinking (P=0.026) and the absence of lymph node
involvement (P=0.0036) In addition, MSI-H status was slightly more
common in elderly individuals (P=0.074; Table I). However, there was no association
between MSI-H status and the other clinicopathological
characteristics (Table I).
The association between the MSI/LOH profile at each
locus (n=27) in the tumor-related genes/the B5 panel (n=5) and 5
important clinical features (lymph node involvement, tumor invasion
depth, TNM Stage, pathological type and the presence of metastasis)
was further analyzed in 226 GC samples (Table III).
 | Table III.Association between the MSI/LOH
profile in the tumor-related genes and clinicopathological
characteristics. |
Table III.
Association between the MSI/LOH
profile in the tumor-related genes and clinicopathological
characteristics.
|
| MSI loci | LOH loci |
|---|
|
|
|
|
|---|
| Clinicopathological
characteristic | Name | P-value | Name | P-value |
|---|
| Lymph node
involvement |
|
|
|
|
|
pN0 | BAT25 | 0.008 | – | – |
|
| BAT26 | 0.008 |
|
|
|
| D2S123 | 0.049 |
|
|
|
| D17S250 | 0.012 |
|
|
|
| MLH1-2 | 0.008 |
|
|
|
| MGMT-5 | 0.012 |
|
|
|
| CTNNB1-(9+10) | 0.005 |
|
|
|
| ERBB2-12 | 0.021 |
|
|
|
| ERBB2-18 | 0.007 |
|
|
|
| ERBB2-25 | 0.005 |
|
|
|
| TGFβ-11 | 0.008 |
|
|
|
| TGFβ-35 | 0.002 |
|
|
|
pN1-pN3a | – | – | – | – |
| Tumor invasion
depth |
|
|
|
|
|
pT1T2 | TP53-1 | 0.003 | MGMT-10 | 0.01 |
|
pT3T4 | – | – | – | – |
| TNM stage |
|
|
|
|
|
I/II | – | – | MGMT-10 | 0.0003 |
|
|
|
| PTN-2 | 0.017 |
|
III/IV | – | – | MCC-17 | 0.036 |
| Pathological
type |
|
|
|
|
|
Adenocarcinoma | – | – | TP53-1 | 0.001 |
|
|
|
| ERBB2-12 | 0.025 |
|
Mucinous carcinoma | – | – | – | – |
The results demonstrated that the MSI status of 12
loci, including BAT25 (P=0.008), BAT26 (P=0.008), D2S123 (P=0.049),
D17S250 (P=0.012), MLH1-2 (P=0.008), MGMT-5 (P=0.012),
CTNNB1-(9+10) (P=0.005), ERBB2-12 (P=0.021), ERBB2-18 (P=0.007),
ERBB2-25 (P=0.005), TGFβ-11 (P=0.008) and TGFβ-35 (P=0.002) was
significantly associated with the absence of lymph node involvement
(Table III). The MSI in TP53-1
(P=0.003) or the LOH in MGMT-10 (P=0.01) were also significantly
associated with T1/T2 stage of depth of tumor invasion. In
addition, the LOH status of MGMT-10 (P=0.0003) and PTN-2 (P=0.017)
was associated with TNM stage I/II, while LOH in MCC-17 (P=0.036)
was significantly associated with TNM stage III/IV. Finally, the
LOH status of TP53-1 (P=0.001) and ERBB2-12 (P=0.025) was
significantly associated with adenocarcinoma compared with that in
mucinous carcinoma (Table III).
However, the MSI/LOH of the loci in tumor-related genes were not
significantly associated with the presence of metastasis.
MSI/LOH status and prognosis
At present, studies analyzing the prognostic value
of MSI remain controversial (17).
The present study investigated the association between MSI status
of the B5 panel and prognosis (OS, n=176; DFS, n=177; the low
number of samples for OS and DFS is because the follow-up
information of some patients were lacking). The results revealed
that there was no association between MSI-H status and OS
(P=0.9521) or DFS (P=0.8904; Fig. 5A and
B).
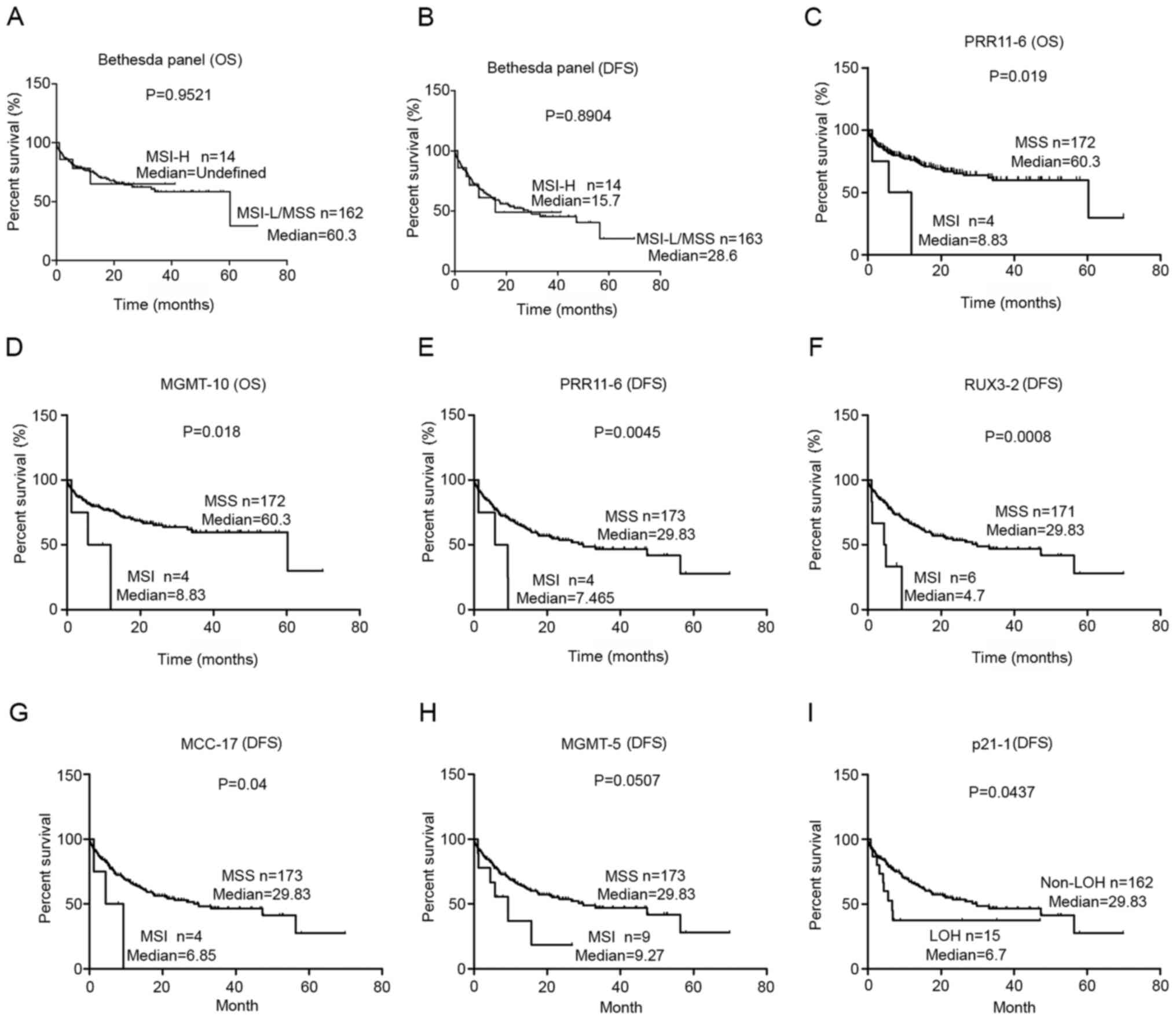 | Figure 5.Association between MSI/LOH and the
prognosis of gastric cancer. Association between MSI-high status of
the Bethesda panel and (A) OS or (B) DFS. Association between MSI
in (C) PRR11-6 and (D) MGMT-10 with OS, and MSI in (E) PRR11-6, (F)
RUX3-2, (G) MCC-17 and (H) MGMT-5 with DFS. (I) Association between
LOH of p21-1 and DFS. P-values were determined using Kaplan-Meier
analysis and the log-rank test. MSI, microsatellite instability;
LOH, loss of heterozygosity; OS, overall survival; DFS,
disease-free survival; MSS, microsatellite stable; H, high; L,
low. |
The present study further analyzed the association
between MSI/LOH status in the tumor-related genes and survival. The
results demonstrated that the MSI or LOH in 6 loci in the
tumor-related genes were significantly associated with prognosis
(Fig. 5). The MSI in PRR11-6
(P=0.019) and MGMT-10 (P=0.018) was associated with poor OS
(Fig. 5C and D). Furthermore, the
MSI in PRR11-6 (P=0.0045), RUNX3-2 (P=0.0008), MCC-17 (P=0.04) and
MGMT-5 (P=0.0507) were significantly associated with poor DFS
(Fig. 5E-H). In addition, the LOH in
P21-1 (P=0.0437) was significantly associated with worse DFS
(Fig. 5I). Notably, all six loci
predicted a poor prognosis, and none of these loci were
mononucleotide loci.
Discussion
Increasing evidence suggests that MSI is one of the
most robust subgroups of GC with specific molecular features
(10,11). In recent years, MSI has also been
developed as a predictive biomarker for the response to immune
checkpoint inhibitors (programmed cell death protein 1/programmed
death-ligand 1 blockade) in GC. For example, it was proved that
PD-L1 positive tumors had the best outcome observed for patients
with MSI-high/PD-L1 positive tumors and those with MSS/PD-L1
negative tumors had the worst outcome (36). However, one of the most important
problems, which seriously hinders the practical application of MSI
in GC, is that a wide variety of different MS markers have been
used to investigate MSI status (37). This includes the MS panel used in the
study by Angell et al (36),
containing five mononucleotide repeat markers (NR27, NR21, NR24,
BAT25 and BAT26). This was different from numerous other studies,
which used a different panel of loci, for example: A combination of
mono- and dinucleotide markers (i.e., BAT25, BAT26, D1S104, D2S123,
D3S1611, D5S107, D17S261 and D18S342) (38), only dinucleotide markers (i.e.,
D2S123, D3S966, D3S1076, D5S82, DP1, D10S197, D11S904, D13S175 and
NM23) (39), only one mononucleotide
marker (i.e., BAT26) (40) and the
present study (B5 panel and markers in tumor-related genes).
Therefore, it is important to identify more efficacious and
specific loci for GC MSI detection.
The present study examined the MSI of 91 loci, all
of which were located in GC-related genes in 90 GC samples. To
further characterize the genetic alterations found in the GC
samples, the present study also evaluated LOH, using the same panel
of loci. As expected, the majority of the loci (91.2%) exhibited
MSI/LOH mutations. The results revealed that, in a patient who
possessed a mutation in the oncogenes or TS genes, this was usually
accompanied by a mutation in the MMR genes; however, there was no
significant association between the mutation profile of oncogenes
and TS genes. This suggested that instability in the MMR genes
served an important role in the mutation status of the oncogenes
and TS genes. This is in accordance with previous studies, which
indicated that the loss-of-function of the MMR genes was associated
with a highly mutated phenotype in key oncogenes and TS genes
(41,42). Human MS evolution research has
illustrated that the fragility is primarily determined by the base
composition (33,43). According to the present data, the MS
mutation rate was more likely determined by the length of the core
sequence, but not by the type of repeat motif or the repeat number
only.
To identify the effective loci which have a
potential role in the prediction of clinical features or the
prognosis of GC, the present study further analyzed 27 potential
loci among the 91 loci in the tumor-related genes. In addition to
the B5 panel, 27 loci were detected in the second set of GC
samples. Subsequently, the mutations at these sites, in the two
sets of patients were integrated. The results demonstrated that the
B5 loci, D2S123 (8.41%), D17S250 (8.41%) and BAT25 presented a
relatively higher potential of variation, whereas the locus D5S346
did not exhibit a high rate of MSI or LOH in GC. The results
indicated that, although recommended as classic markers in CRC, not
all of the five loci in the B5 panel were sensitive to MSI in GC.
In addition, TP53-1 was considered to be the most LOH-prone locus,
which was in line with a previous study, which suggested that TP53
was the most mutated gene (18).
Currently, the loci used in GC research,
particularly the B5 panel recommended by the National Cancer
Institute, are mainly based on hereditary non-polyposis CRC
research (44). In the present
study, as defined by the B5 panel, MSI-H GC accounted for 7.5%,
which was consistent with previous studies, revealing that the
MSI-H incidence of GC ranges between 5.6 and 33.3% (45,46). A
combination of the sensitivity and specificity for the detection of
MSI was used to evaluate the efficiency of the loci. Notably, the
top 5 effective loci (BAT26, BAT25, CTNNB1-(9+10), ERBB2-25 and
TGFBR2-35) were mononucleotide MSs, which was similar to the
results of a study, which indicated that mononucleotide markers are
more effective for the identification of MSI status compared with
that in dinucleotide markers (47).
The predictive and prognostic role of MSI status has been well
established in CRC; however, there is still a large heterogeneity
in the results regarding the association between MSI and
clinicopathological features across different GC studies (34,35). To
the best of our knowledge, few studies have explored the
association between drinking and MSI (48,49). The
present study demonstrated that the MSI-H status of the B5 panel
was significantly associated with drinking (P=0.026). In addition,
the results revealed that the MSI-H status of the B5 panel was also
significantly associated with the absence of lymph node involvement
(P=0.0036) and slightly more common in elderly people (P=0.074).
This was similar to the results reported in a previous
meta-analysis (50).
The present study subsequently evaluated the
association between the MSI/LOH in the loci of tumor-related genes
and clinicopathological characteristics, which are likely to affect
GC treatment, including lymph node involvement, the depth of tumor
invasion, presence of metastasis, TNM stage and pathological
type.
It remains a challenge to predict the presence of
lymph node metastasis in clinical settings, even following surgery,
due to the preference of limited resections (51). Due to its significance in the
prognosis of GC, a previous study investigated lymph node
metastasis according to MSI status and the result indicated that
MSI-H phenotype was significantly associated with the presence of
lymphatic invasion (P=0.036) (52).
In the present study, the MSI status of 12 single loci, including 4
from the B5 panel and 8 from the tumor-related genes, was
significantly associated with lymph node metastasis as previously
reported. In addition, it was revealed that patients with a MSI in
TP53-1 or a LOH in MGMT-10 were more prone to having earlier stages
of depth of tumor invasion. Patients presenting with LOH in MGMT-10
or PTN-2 were more likely to be in earlier TNM stages, while
patients with LOH in MCC-17 were likely to present with advanced
TNM stage. The LOH in TP53-1 or ERBB2-12 was higher in
adenocarcinoma compared with that in mucinous carcinoma. These
findings indicated that the MSI/LOH at these loci may serve an
important role in lymph node metastasis, tumor invasion, TNM or
pathological type, and may be useful and applicable for the
prediction of clinical features in patients with GC.
The association between the survival of patients
with GC and MSI has yet to be determined. As evaluated by the B5
panel, patients with GC and MSI-H or MSI-L/MSS status exhibited
similar outcomes with respect to OS and DFS. The results from the
present study are inconsistent with a previous study, which
demonstrated that the prognosis of patients in the MSI group was
improved compared with that in patients with MSS GC (50). This could be due to the different
loci used for the determination of MSI. However, the present study
found that mutations of 6single loci in tumor-related genes,
including MSI in five loci and LOH in 1 loci, were associated with
poorer outcomes with respect to OS or DFS. Among these valuable
loci (Fig. 5), only the MSI status
of MGMT-5 was significantly associated with lymph node involvement
(Table III), which indicates that
lymph node involvement maybe an important factor in the association
between the MSI status of MGMT-5 and survival. In the present study
it was found that, with the exception of the MSI status in MGMT-5,
all the other MSI/LOH events, which were associated with prognosis,
were not associated with the clinical features, indicating that the
MSI/LOH status of these loci maybe a prognostic factor without the
affect from other clinical features. However, the predictive impact
of MSI or MMR status and survival remains a contentious issue
(50). The possible reasons may be
due to i) Numerous other important clinical features not being
analyzed and ii) Not enough patients with GC were included in the
present study.
There are also additional limitations to the present
study. The 91 MS loci selected from the 20 genes were predetermined
based on the PCR amplification efficacy, and not all MS loci in
these tumor-related genes were included, potentially missing other
important MSI loci in these genes. However, it was identified that
the same repeat motif and similar repeat unit locus in the same
gene exhibited highly consistent distribution and frequency of MSI
events (data not shown). An important challenge for the future is
to standardize the laboratory methodology for MSI detection in
GC.
In conclusion, the present study provided several
valuable MS biomarkers for the prediction of clinical features and
prognosis, and has important implications for MSI detection in
clinical applications and practice.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Science Foundation of China (grant no. 31772545) and Support
Project of High-Level Teachers in Beijing Municipal Universities in
the Period of 13th five-year Plan (grant no. IDHT 20170516).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZC and ZB designed the study. XX performed the
experiments. XH analyzed the data, wrote and revised the
manuscript, and completed the submission. SZ, XD and CL
participated in data analysis including offering advice and answers
to some of the questions from the reviewers and editors. ZC, XD and
XH confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Beijing Friendship Hospital
(Beijing, China) approved the study proposal (approval number,
2017-P2-013-03) and all patients involved in the study provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferro A, Peleteiro B, Malvezzi M, Bosetti
C, Bertuccio P, Levi F, Negri E, La Vecchia C and Lunet N:
Worldwide trends in gastric cancer mortality (1980–2011), with
predictions to 2015, and incidence by subtype. Eur J Cancer.
50:1330–1344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bang YJ, Van Cutsem E, Feyereislova A,
Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T,
et al: Trastuzumab in combination with chemotherapy versus
chemotherapy alone for treatment of HER2-positive advanced gastric
or gastro-oesophageal junction cancer (ToGA): A phase 3,
open-label, randomised controlled trial. Lancet. 376:687–697. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smyth EC and Moehler M: Pembrolizumab in
first-line gastric cancer: Win, lose, or draw? JAMA Oncol.
1:1539–1541. 2020. View Article : Google Scholar
|
|
6
|
Bükakkaramikli NC, Blommestein HM, Riemsma
R, Armstrong N, Clay FJ, Ross J, Worthy G, Severens J, Kleijnen J
and Al MJ: Ramucirumab for treating advanced gastric cancer or
gastro-oesophageal junction adenocarcinoma previously treated with
chemotherapy: An evidence review group perspective of a NICE single
technology appraisal. Pharmacoeconomics. 35:1211–1221. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilke H, Muro K, Van Cutsem E, Oh SC,
Bodoky G, Shimada Y, Hironaka S, Sugimoto N, Lipatov O, Kim TY, et
al: Ramucirumab plus paclitaxel versus placebo plus paclitaxel in
patients with previously treated advanced gastric or
gastro-oesophageal junction adenocarcinoma (RAINBOW): A
double-blind, randomised phase 3 trial. Lancet Oncol. 15:1224–1235.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheong JH, Yang HK, Kim H, Kim WH, Kim YW,
Kook MC, Park YK, Kim HH, Lee HS, Lee KH, et al: Predictive test
for chemotherapy response in resectable gastric cancer: A
multi-cohort, retrospective analysis. Lancet Oncol. 19:629–638.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roh CK, Choi YY, Choi S, Seo WJ, Cho M,
Jang E, Son T, Kim HI, Kim H, Hyung WJ, et al: Single patient
classifier assay, microsatellite instability, and epstein-barr
virus status predict clinical outcomes in stage II/III gastric
cancer: Results from CLASSIC trial. Yonsei Med J. 60:132–139. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choi YY, Jang E, Seo WJ, Son T, Kim HI,
Kim HI, Hyung WJ, Huh YM, Noh SH and Cheong JH: Modification of the
TNM staging system for stage II/III gastric cancer based on a
prognostic single patient classifier algorithm. J Gastric Cancer.
18:142–151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mizoshita T, Tsukamoto T, Cao X, Otsuka T,
Ito S, Takahashi E, Nakamura S, Nakamura T, Yamamura Y and
Tatematsu M: Microsatellite instability is linked to loss of hMLH1
expression in advanced gastric cancers: Lack of a relationship with
the histological type and phenotype. Gastric Cancer. 8:164–172.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ionov Y, Peinado MA, Malkhosyan S, Shibata
D and Perucho M: Ubiquitous somatic mutations in simple repeated
sequences reveal a new mechanism for colonic carcinogenesis.
Nature. 363:558–561. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boland CR, Thibodeau SN, Hamilton SR,
Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA,
Fodde R, Ranzani GN and Srivastava S: A national cancer institute
workshop on microsatellite instability for cancer detection and
familial predisposition: Development of international criteria for
the determination of microsatellite instability in colorectal
cancer. Cancer Res. 58:5248–5257. 1998.PubMed/NCBI
|
|
16
|
Hause RJ, Pritchard CC, Shendure J and
Salipante SJ: Classification and characterization of microsatellite
instability across 18 cancer types. Nat Med. 22:1342–1350. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang WL, Chen MH, Huang KH, Chang SC, Lin
CH, Chao Y, Lo SS, Li AF, Wu CW and Shyr YM: The
clinicopathological features and genetic mutations in gastric
cancer patients according to EMAST and MSI status. Cancers (Basel).
27:5512020. View Article : Google Scholar
|
|
18
|
An JY, Kim H, Cheong JH, Hyung WJ, Kim H
and Noh SH: Microsatellite instability in sporadic gastric cancer:
Its prognostic role and guidance for 5-FU based chemotherapy after
R0 resection. Int J Cancer. 131:505–511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gurzu S, Jung I, Sugimura H, Stefan-van
Staden RI, Yamada H, Natsume H, Iwashita Y, Szodorai R and
Szederjesi J: Maspin subcellular expression in wild-type and mutant
TP53 gastric cancers. World J Gastrointest Oncol. 15:741–755. 2020.
View Article : Google Scholar
|
|
20
|
Kanayama K, Imai H, Usugi E, Matsuda C,
Ichishi M, Hirokawa YS and Watanabe M: Cancer-Related gene
mutations and intratumoral genetic heterogeneity in human epidermal
growth factor receptor 2 heterogeneous gastric cancer. Pathol Int.
70:865–870. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grundei T, Vogelsang H, Ott K, Mueller J,
Scholz M, Becker K, Fink U, Siewert JR, Höfler H and Keller G: Loss
of heterozygosity and microsatellite instability as predictive
markers for neoadjuvant treatment in gastric carcinoma. Clin Cancer
Res. 6:4782–4788. 2000.PubMed/NCBI
|
|
22
|
Chang SC, Lin JK, Lin TC and Liang WY:
Loss of heterozygosity: An independent prognostic factor of
colorectal cancer. World J Gastroenterol. 11:7787842005. View Article : Google Scholar
|
|
23
|
Lin X, Zhao Y, Song WM and Zhang B:
Molecular classification and prediction in gastric cancer. Comput
Struct Biotechnol J. 13:448–458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du X, Chen Z, Li W, Tan Y, Lu J, Zhu X,
Zhao T, Dong G and Zeng L: Development of novel microsatellite DNA
markers by cross-amplification and analysis of genetic variation in
gerbils. J Hered. 101:710–716. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Plevová P: An update on inherited colon
cancer and gastrointestinal polyposis. Klin Onkol. 32:97–108. 2019.
View Article : Google Scholar
|
|
26
|
Yamamoto H and Imai K: Microsatellite
instability: An update. Arch Toxicol. 89:899–921. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu WK, Cho CH, Lee CW, Fan D, Wu K, Yu J
and Sung JJ: Dysregulation of cellular signaling in gastric cancer.
Cancer Lett. 28:144–153. 2010. View Article : Google Scholar
|
|
28
|
Yan LH, Wei WY, Xie YB and Xiao Q: New
insights into the functions and localization of the homeotic gene
CDX2 in gastric cancer. World J Gastroenterol. 14:3960–3966. 2014.
View Article : Google Scholar
|
|
29
|
Xanthoulis A and Tiniakos DG: E2F
transcription factors and digestive system malignancies: How much
do we know? World J Gastroenterol. 19:3189–3198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu H, Li C, Cai S, Zhu C, Tian Y, Zheng J,
Hu J, Chen C and Liu W: Increased expression of pleiotrophin is a
prognostic marker for patients with gastric cancer.
Hepatogastroenterology. 61:1478–1482. 2014.PubMed/NCBI
|
|
31
|
Hu H, Song Z, Yao Q, Geng X, Jiang L, Guo
C and Li H: Proline-Rich protein 11 regulates self-renewal and
tumorigenicity of gastric cancer stem cells. Cell Physiol Biochem.
47:1721–1728. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lotem J, Levanon D, Negreanu V, Bauer O,
Hantisteanu S, Dicken J and Groner Y: Runx3 in immunity,
inflammation and cancer. Adv Exp Med Biol. 962:369–393. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huo X, Zhang S, Li Z, Gao J, Wang C, Li C,
Guo M, Du X and Chen Z: Analysis ofthe relationship between
microsatellite instability and thymic lymphoma induced by
methyl-N-nitrosourea in C57BL/6J mice. Mut Res. 771:21–28. 2015.
View Article : Google Scholar
|
|
34
|
Fujimoto A, Fujita M, Hasegawa T, Wong JH,
Maejima K, Oku-Sasaki A, Nakano K, Shiraishi Y, Miyano S, Yamamoto
G, et al: Comprehensive analysis of indels in whole-genome
microsatellite regions and microsatellite instability across 21
cancer types. Genome Res. 24:334–346. 2020. View Article : Google Scholar
|
|
35
|
Huang KH, Chen MH, Fang WL, Lin CH, Chao
Y, Lo SS, Li AF, Wu CW and Shyr YM: The clinicopathological
characteristics and genetic alterations of signet-ring cell
carcinoma in gastric cancer. Cancers (Basel). 17:23182020.
View Article : Google Scholar
|
|
36
|
Marrelli D, Polom K, Pascale V, Vindigni
C, Piagnerelli R, De Franco L, Ferrara F, Roviello G, Garosi L,
Petrioli R and Roviello F: Strong prognostic value of
microsatellite instability in intestinal type non-cardia gastric
cancer. Ann Surg Oncol. 23:943–950. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Angell HK, Lee J, Kim KM, Kim K, Kim ST,
Park SH, Kang WK, Sharpe A, Ogden J and Davenport A: PD-L1 and
immune infiltrates are differentially expressed in distinct
subgroups of gastric cancer. Oncoimmunology. 8:e15444422018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Choi YY, Bae JM, An JY, Kwon IG, Cho I,
Shin HB, Eiji T, Aburahmah M, Kim HI, Cheong JH, et al: Is
microsatellite instability a prognostic marker in gastric cancer? A
systematic review with meta-analysis. J Surg Oncol. 110:129–135.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Falchetti M, Saieva C, Lupi R, Masala G,
Rizzolo P, Zanna I, Ceccarelli K, Sera F, Mariani-Costantini R,
Nesi G, et al: Gastric cancer with high-level microsatellite
instability: Target gene mutations, clinicopathologic features, and
long-term survival. Hum Pathol. 39:925–932. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hayden JD, Cawkwell L, Quirke P, Dixon MF,
Goldstone AR, Sue-Ling H, Johnston D and Martin IG: Prognostic
significance of microsatellite instability in patients with gastric
carcinoma. Eur J Cancer. 33:2342–2346. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu MS, Lee CW, Sheu JC, Shun CT, Wang HP,
Hong RL, Lee WJ and Lin JT: Alterations of BAT-26 identify a subset
of gastric cancer with distinct clinicopathologic features and
better postoperative prognosis. Hepatogastroenterology. 49:285–289.
2002.PubMed/NCBI
|
|
42
|
Corso G, Velho S, Paredes J, Pedrazzani C,
Martins D, Milanezi F, Pascale V, Vindigni C, Pinheiro H, Leite M,
et al: Oncogenic mutations in gastric cancer with microsatellite
instability. Eur J Cancer. 47:443–451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Leite M, Corso G, Sousa S, Milanezi F,
Afonso LP, Henrique R, Soares JM, Castedo S, Carneiro F, Roviello
F, et al: MSI phenotype and MMR alterations in familial and
sporadic gastric cancer. Int J Cancer. 128:1606–1613. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Maruvka YE, Mouw KW, Karlic R, Parasuraman
P, Kamburov A, Polak P, Haradhvala NJ, Hess JM, Rheinbay E, Brody
Y, et al: Analysis of somatic microsatellite indels identifies
driver events in human tumors. Nat Biotechnol. 35:951–959. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim DG, An JY, Kim H, Shin SJ, Choi S, Seo
WJ, Roh CK, Cho M, Son T, Kim HI, et al: Clinical implications of
microsatellite instability in early gastric cancer. J Gastric
Cancer. 19:427–437. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim JY, Shin NR, Kim A, Lee HJ, Park WY,
Kim JY, Lee CH, Huh GY and Park DY: Microsatellite instability
status in gastric cancer: A reappraisal of its clinical
significance and relationship with mucin phenotypes. Korean J
Pathol. 47:28–35. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin JT, Wu MS, Shun CT, Lee WJ, Wang JT,
Wang TH and Sheu JC: Microsatellite instability in gastric
carcinoma with special references to histopathology and cancer
stages. Eur J Cancer. 31A:1879–1882. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Suraweera N, Duval A, Reperant M, Vaury C,
Furlan D, Leroy K, Seruca R, Iacopetta B and Hamelin R: Evaluation
of tumor microsatellite instability using five quasimonomorphic
mononucleotide repeats and pentaplex PCR. Gastroenterology.
123:1804–1811. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huo X, Xiao X, Zhang S, Zhou D and Chen Z:
Association of intron microsatellite instability and exon
mutational profile of TP53 in human gastric cancers. Anticancer
Res. 37:4507–4514. 2017.PubMed/NCBI
|
|
50
|
Liu X, Feng D, Huo X, Xiao X and Chen Z:
Association of intron microsatellite status and exon mutational
profiles of TP53 in human colorectal cancer. Exp Ther Med.
18:4287–4294. 2019.PubMed/NCBI
|
|
51
|
Polom K, Marano L, Marrelli D, De Luca R,
Roviello G, Savelli V, Tan P and Roviello F: Meta-Analysis of
microsatellite instability in relation to clinicopathological
characteristics and overall survival in gastric cancer. Br J Surg.
105:159–167. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
An JY, Min JS, Lee YJ, Jeong SH, Hur H,
Han SU, Hyung WJ, Cho GS, Jeong GA, Jeong O, et al: Safety of
laparoscopic sentinel basin dissection in patients with gastric
cancer: An analysis from the SENORITA prospective multicenter
quality control trial. J Gastric Cancer. 18:30–36. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Choi J, Nam SK, Park DJ, Kim HW, Kim HH,
Kim WH and Lee HS: Correlation between microsatellite
instability-high phenotype and occult lymph node metastasis in
gastric carcinoma. APMIS. 123:215–222. 2015. View Article : Google Scholar : PubMed/NCBI
|