Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide (1). In most countries, for lung cancer
patients diagnosed during 2005–2009, the 5-year survival rate
ranges between 10 and 20% (1), which
highlights the inadequacy of the current treatment strategies for
lung cancer (1). Lung cancer is
classified into two main types, small cell lung cancer and
non-small cell lung cancer (NSCLC), and NSCLC accounts for ~90% of
all cases (2). In the majority of
patients with NSCLC, local or advanced metastasis occurs after
diagnosis (3). Therefore, the
mechanisms underlying NSCLC development have become a focus of
research in recent years.
MicroRNA (miRNA/miR)-199a-5p expression is generally
downregulated in tumors and can inhibit the progression of various
tumor types, including prostate cancer, breast cancer,
hepatocellular carcinoma, ovarian cancer, colorectal cancer and
lung cancer, by downregulating different downstream genes (4–9). For
example, Hu et al (10)
reported that miR-199a-5p could inhibit the migration and invasion
of colorectal cancer by targeting discoidin domain receptor-1. In
liver cancer, miR-199a-5p inhibits glucose consumption and lactic
acid generation by targeting hexokinase 2, thus attenuating the
proliferation and tumorigenesis of cancer cells (11). In addition, Li et al (12) revealed that miR-199a-5p-mediated
silencing of ETS proto-oncogene 1, transcription factor represses
the invasion of breast cancer cells by decreasing the levels of
integrin 1 (12). Additionally,
miR-199a-5p has been reported to be a tumor suppressor in NSCLC
(9,13). However, the mechanisms of the
downregulation of miR-199a-5p expression and the downstream
effectors targeted by miR-199a-5p in NSCLC are not fully
understood.
A-kinase anchoring protein 1 (AKAP1) is a scaffold
protein, which recruits protein kinase A, other signaling proteins
and RNA to the outer mitochondrial membrane (14). In hepatocellular carcinoma, high
expression levels of AKAP1 are associated with a poor prognosis,
suggesting that AKAP1 may serve as a biomarker (15). Another previous study reported that
knockdown of AKAP1 sensitizes gastric cancer cells to cisplatin
treatment (16). Furthermore, AKAP1
is expressed at high levels in lung cancer cells compared with
non-tumoral cells, and is associated with the decreased survival
rate of patients with lung cancer (17). However, to the best of our knowledge,
the mechanisms underlying the aberrant expression of AKAP1 in NSCLC
and its cellular function remain unknown.
The present study demonstrated that the
hypermethylation of the promoter of miR-199a led to downregulation
of miR-199a-5p in NSCLC cells. Exogenous miR-199a-5p suppressed the
proliferation and tumorigenicity of NSCLC cells by decreasing AKAP1
expression, which was achieved via direct targeting of the 3′
untranslated region (UTR) of AKAP1 mRNA. These findings indicated a
novel regulatory mechanism of AKAP1, as well as its role in NSCLC
proliferation.
Materials and methods
Collection of NSCLC tissues
All patients with NSCLC (n=41; median age, 61 years;
age range, 35–84 years; 14 female and 27 male patients) were
diagnosed according to the International Association for Lung
Cancer Research 2015 version guidelines (18). NSCLC and paired para-carcinoma
tissues (2 cm away from the tumor edge) were collected between
January 2019 and June 2020 at The First Affiliated Hospital of
Wenzhou Medical University (Wenzhou, China). Patients with NSCLC
were included that had been diagnosed on the basis of pathological
examinations. Histological classification was performed according
to WHO Classification of Tumors of the Lung (2015) (19) and tumor staging was performed
according to the staging criteria of International Union Against
Cancer (UICC, 2017) (20). Patients
with NSCLC without radiotherapy or chemotherapy prior to the
surgical lung resection were included. Pregnant patients, patients
with mental or cognitive impairment and patients with another lung
disease were excluded from the present study. Written informed
consent was obtained from all patients, and the present study was
approved by the Ethics Committee in Clinical Research of The First
Affiliated Hospital of Wenzhou Medical University (issuing no.
2019-128; Wenzhou, China).
Online analysis of the cancer genome
atlas (TCGA) data
Kaplan-Meier analysis of TCGA database (https://cancergenome.nih.gov/abouttcga/overview) was
performed by OncoLnc (version 2016: http://www.oncolnc.org/) using the lung adenocarcinoma
(LUAD) dataset (21). Patients were
divided into high miR-199a-5p expression (40% upper percentile) and
low miR-199a-5p expression (40% lower percentile) groups.
Cell culture
The BEAS-2B normal human lung epithelial cell line
and the H1299 and H460 NSCLC cell lines were purchased from
American Type Culture Collection. Cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 mg/ml
streptomycin and 100 IU/ml penicillin in a 5% CO2
atmosphere at 37°C. Cell passage was performed every 2 days.
5-Aza-dC (2 µM) purchased from Sigma-Aldrich; Merck KGaA was used
to treat H1299 cells for 48 h at 37°C.
Plasmids and mimic RNAs
miR-199a-5p or negative control (NC) mimic RNAs
(miR-199a-5p mimics, 5′-CCCAGUGUUCAGACUACCUGUUC-3′; NC mimics,
5′-GGUUCGUACGUACACUGUUCA-3′, both 20 µM) were purchased from
Shanghai GenePharma Co., Ltd. miR-199a-5p and AKAP1 overexpression
plasmids (ov-miR-199a-5p and ov-AKAP1) were constructed using the
pcDNA3.1 vector (both 500 ng/µl, Invitrogen; Thermo Fisher
Scientific, Inc.). The 3′UTR of the AKAP1 gene was amplified based
on cDNA and then cloned into the pGL3 luciferase reporter vector
(500 ng/µl; Promega Corporation) to obtain the AKAP1-3′UTR reporter
plasmid (pGL3-AKAP1-3′UTR). A mutation reporter vector
(pGL3-AKAP1-3′UTR-mut) was constructed via subcloning (the sequence
ACU at the miR-199a-5p binding site was mutated to UGA).
Transfection was performed using Lipofectamine® 3000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols at 37°C for 24 h. At 48 h
after transfection, subsequent experiments were performed or G418
(Sigma-Aldrich; Merck KGaA) was used to screen H1299 cells with
stable transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells and tissues using
an RNAiso Plus kit (Takara Bio, Inc.). The RNA concentration was
measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.).
For mRNA, cDNA was reverse transcribed from 0.5 µg
mRNA using a PrimeScript RT reagent kit (Takara Bio, Inc.), and the
following conditions were used for RT: 37°C for 30 min, 85°C for 15
sec. qPCR for AKAP1 (forward, 5′-TCCGTGGATAGCTGTTGCAG-3′; reverse,
5′-CTGCTTGCCAATTAGCCGAC-3′) was performed using SYBR Green PCR
Master Mix (Takara Bio, Inc.). The following thermocycling
conditions were used: Initial denaturation at 95°C for 5 min,
followed by 40 cycles of 95°C for 45 sec, annealing at 55°C for 45
sec and extension at 72°C for 1 min. β-actin (forward,
5′-CTCCATCCTGGCCTCGCTGT-3′; reverse, 5′-GCTGTCACCTTCACCGTTCC-3′)
was used as an internal control to normalize mRNA levels. The
relative expression levels were calculated using the
2−ΔΔCq method (22).
For miRNA, a TaqMan microRNA RT kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used for reverse
transcription, and the following conditions for RT were used: 16°C
for 30 min, 42°C for 30 min and 85°C for 5 min. A TaqMan miRNA
assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used
to measure the expression levels of mir-199a-5p (forward,
5′-AACCATGCCCAGTGTTCAGACTA-3′; reverse, 5′-CAGTGCAGGGTCCGAGGT-3′).
The following thermocycling conditions were used: Initial
denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec, annealing and extension at 60°C for 1 min. U6 (forward,
5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′) was
used as an internal control to normalize miRNA levels. The relative
expression levels were calculated using the 2−ΔΔCq
method (22).
DNA sodium bisulfite conversion
Genomic DNA was extracted from NSCLC and
para-carcinoma tissues using the phenol-chloroform technique.
Bisulfite conversion was performed using an EpiJET Bisulfite
Conversion Kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols (incubation conditions: 98°C for 10 min,
60°C for 150 min). Specific primers [Forward,
5′-TATATTTGGAATTGTTTATAGT-3′; reverse, 5′-AAAAAAATATCTAACTCTTTAA-3′
(23)] for the converted promoter
region were used to generate the PCR product using Platinum™ Taq
Green Hot Start DNA Polymerase (Invitrogen; Thermo Fisher
Scientific, Inc.). Amplification was conducted by performing
initial denaturation at 94°C for 5 min, followed by 35 cycles of
94°C for 2 min, 94°C for 30 sec, 60°C for 30 sec and 72°C for 1
min. PCR products were separated by 2% agarose gel and cloned into
pGEM-Teasy (Promega Corporation) followed by sequencing with Sp6
primer. The dideoxy chain-termination method (Sanger method) was
used to sequence the inserted fragments. The lollipop diagram from
the sequencing data was generated by BiQ Analyzer v.2.0
(Max-Planck-Institut Informatik). Filled (black) circles
corresponded to methylated Cs, and unfilled (white) circles
corresponded to unmethylated Cs.
Western blotting
Whole cell protein was obtained using cold cell
lysis RIPA buffer (cat. no. 20-188; EMD Millipore), and the total
protein concentration was measured using a Bradford protein assay
(cat. no. 23236; Bio-Rad Laboratories, Inc.). Equivalent amounts of
protein (30 µg/lane) were separated via 10% SDS-PAGE and
transferred to a nitrocellulose membrane (Whatman plc; Cytiva). The
membrane was blocked with 5% milk for 2 h at room temperature, and
incubated at 4°C overnight with the following primary antibodies:
AKAP1 (dilution, 1:1,000; cat. no. 5203; Cell Signaling Technology,
Inc.) and GAPDH (dilution, 1:2,000; cat. no. sc-47724; Santa Cruz
Biotechnology, Inc.). Subsequently, the membrane was incubated with
an appropriate fluorescent secondary antibody (IRDye®
800CW- or IRDye® 680RD-conjugated antibodies (dilution,
1:10,000; cat. nos. 926-32211 and 926-68070; LI-COR Biosciences) at
room temperature for 1 h. Subsequently, protein bands were detected
using an Odyssey® infrared imaging system (LI-COR
Biosciences). The obtained signals were converted into grayscale
images using Application Software v.2.1.12 (LI-COR
Biosciences).
Prediction of miR-199a-5p targets
TargetScan (TargetScan Release 3.1; http://www.targetscan.org) online prediction tools
were used to predict miR-199a-5p target genes. Briefly, the default
settings were adopted when setting up the run parameters. The gene
was considered as a potential target gene of miR-199a-5p when its
total context++ score was <-0.6.
Luciferase reporter assay
H1299 or H460 cells (1×105 cells/well)
were plated onto a 24-well plate. The following day, the cells in
each well were co-transfected with 0.5 µg firefly luciferase report
plasmid (pGL3 with AKAP1 3′UTR) and 0.01 µg Renilla
luciferase report plasmid (pRL-SV40; Promega Corporation). The
pRL-SV40 plasmid was used to standardize the transfection
efficiency. Meanwhile, the cells in each well were also transfected
with 40 pmol miR-199a-5p or negative control (NC) mimic RNAs
(miR-199a-5p mimics, 5′-CCCAGUGUUCAGACUACCUGUUC-3′; NC mimics,
5′-GGUUCGUACGUACACUGUUCA-3′, both 20 µM, Shanghai GenePharma Co.,
Ltd.). Transfection was performed using Lipofectamine®
3000 transfection reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols at 37°C. The cells
were lysed using 100 µl passive lysis buffer (Promega Corporation)
at 2 days after transfection. The reporter activity was measured
using the dual luciferase reporter assay system (cat. no. E1910;
Promega Corporation) according to the manufacturer's protocols, and
a luminometer (model no. LB9507; Titertek-Berthold). Briefly, 20 µl
cell lysate was mixed with 100 µl LAR II in a luminometer tube and
then the tube was placed in the luminometer for the first
measurement (firefly luciferase activity). Subsequently, 100 µl
Stop & Glo® reagent was added and followed by gentle
mixing. The tube was then placed in the luminometer again for the
second measurement (Renilla luciferase activity). Relative
luciferase activity was calculated by dividing the first
measurement by the second.
Cell proliferation assay
Cell proliferation was analyzed using a Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay.
Cells (3×103) were added into each well of a 96-well
plate. After the indicated time (12, 24, 36, 48 and 60 h), 10%
CCK-8 reagent was added to each well and the cells were incubated
for 2 h. Measurements were obtained at a wavelength of 450 nm using
a spectrophotometer (Bio-Rad Laboratories, Inc.).
EdU incorporation assay
The EdU (5-ethynyl-2-deoxyuridine) incorporation
assay was used to represent DNA synthesis in cells. H1299 and H460
cells (5×104/well) were transfected with miR-199a-5p
mimics or AKAP1 expression plasmids for 48 h at 37°C. Next, cells
were washed 3 times with PBS, and then incubated in serum-free RPMI
1640 with 10 mM EdU (Sigma-Aldrich; Merck KGaA) for 2 h at 37°C.
After extensive washing with PBS, cells were blocked with 10% FBS
in PBS for 30 min at 37°C. Incorporated EdU was detected by the
fluorescent azide coupling reaction (Invitrogen; Thermo Fisher
Scientific, Inc.). Images of the cells were captured with a
fluorescence microscope (Nikon Ti-E) (magnification, ×40) and
analyzed by Image J v.1.4.3.67 (National Institutes of Health).
Colony formation assay
H1299 cells (5×103) were suspended in 2.5
ml of 0.3% agar and then placed in each well of a 6-well plate,
which was pre-coated with 1.0 ml 0.6% agar/well. Cells were
cultured in an incubator at 37°C with 5% CO2 and 95%
air. The RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
was replaced with fresh medium every 3 days. After 21 days, the
colonies were captured by a light microscope (Eclipse Ti), and at
least 5 individual fields at magnification, ×100 were chosen. Then
colonies were counted using ImageJ software (version 1.4.3.67;
National Institutes of Health).
Statistical analysis
SPSS v13.0 (SPSS, Inc.) and GraphPad Prism 5
(GraphPad Software, Inc.) were used for statistical analysis.
Experimental data were presented as mean ± SD from at least 3
independent experiments. An unpaired Student's t-test was used for
comparisons between two independent groups. A paired t-test was
used for comparisons between two paired groups. ANOVA (data with
univariate change were analyzed using one-way with Tukey's post hoc
test; data with multivariate change were analyzed using two-way
with Sidak's post hoc test) was used for multiple group
comparisons. A Pearson test was used for correlation analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
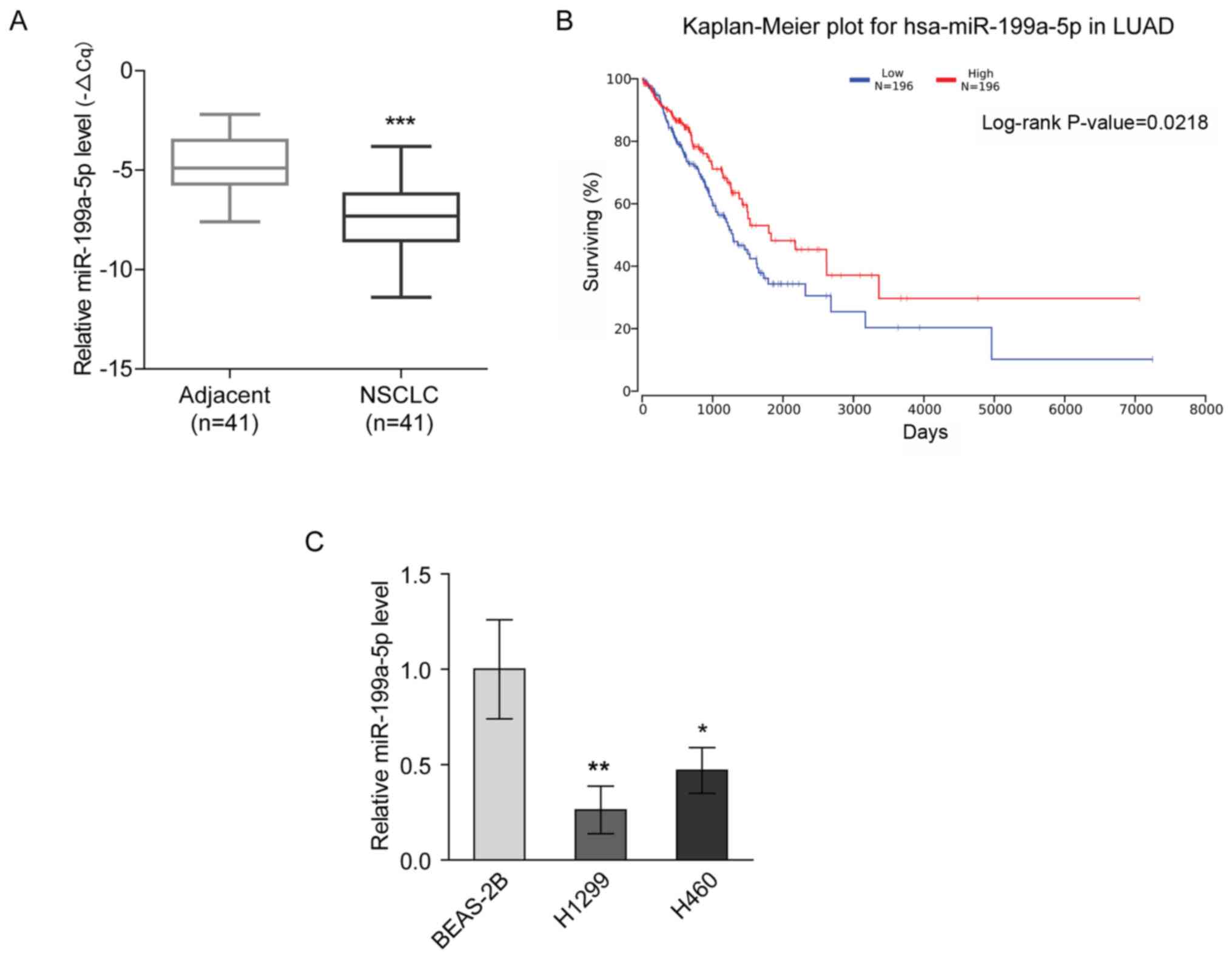
miR-199a-5p expression is decreased in
NSCLC
miR-199a-5p has been reported to be a tumor
suppressor in lung cancer (9,13). The
present study revealed that miR-199a-5p expression was
downregulated in NSCLC tissues compared with para-carcinoma tissues
(Fig. 1A). Furthermore, Kaplan-Meier
analysis of a TCGA dataset using OncoLnc demonstrated that the
overall survival rate of the patients with LUAD with high
expression levels of miR-199a-5p was significantly increased
compared with that of the patients with low miR-199a-5p expression
(Fig. 1B). Consistently, the
expression levels of miR-199a-5p were significantly lower in NSCLC
cell lines (H1299 and H460) compared with in the BEAS-2B normal
lung epithelial cell line (Fig. 1C).
These findings suggested that the downregulation of miR-199a-5p
expression was associated with NSCLC.
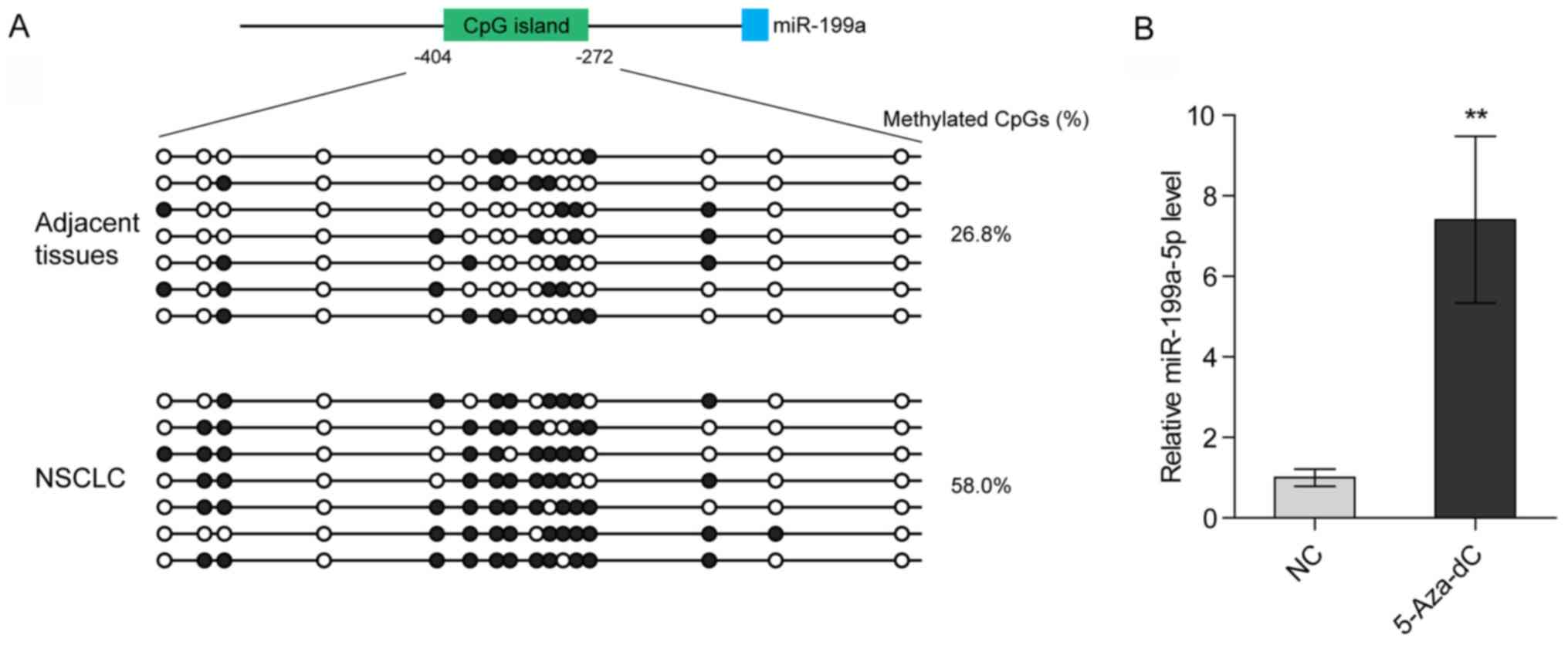
Hypermethylation of the promoter of
the miR-199a gene contributes to the low expression levels of
miR-199a-5p in NSCLC
Deng et al (23) reported that promoter methylation of
the miR-199a gene suppresses miR-199a-3p in ovarian cancer. To
determine whether the loss of miR-199a-5p in NSCLC also arises from
promoter hypermethylation, bisulfite genomic sequencing was
performed to analyze the methylation levels of the CpG island
(−404/-272) located at the miR-199a promoter. It was revealed that
the methylation levels of the CpG island were markedly higher in
NSCLC tissues compared with in para-carcinoma tissues (Fig. 2A; Table
SI). Additionally, treatment with a selective inhibitor of DNA
methyltransferases [5-Aza-2′-deoxycytidine (5-Aza-dC)]
significantly increased the expression levels of miR-199a-5p in
H1299 cells (Fig. 2B). These data
indicated that promoter hypermethylation may lead to the loss of
miR-199a-5p in NSCLC.
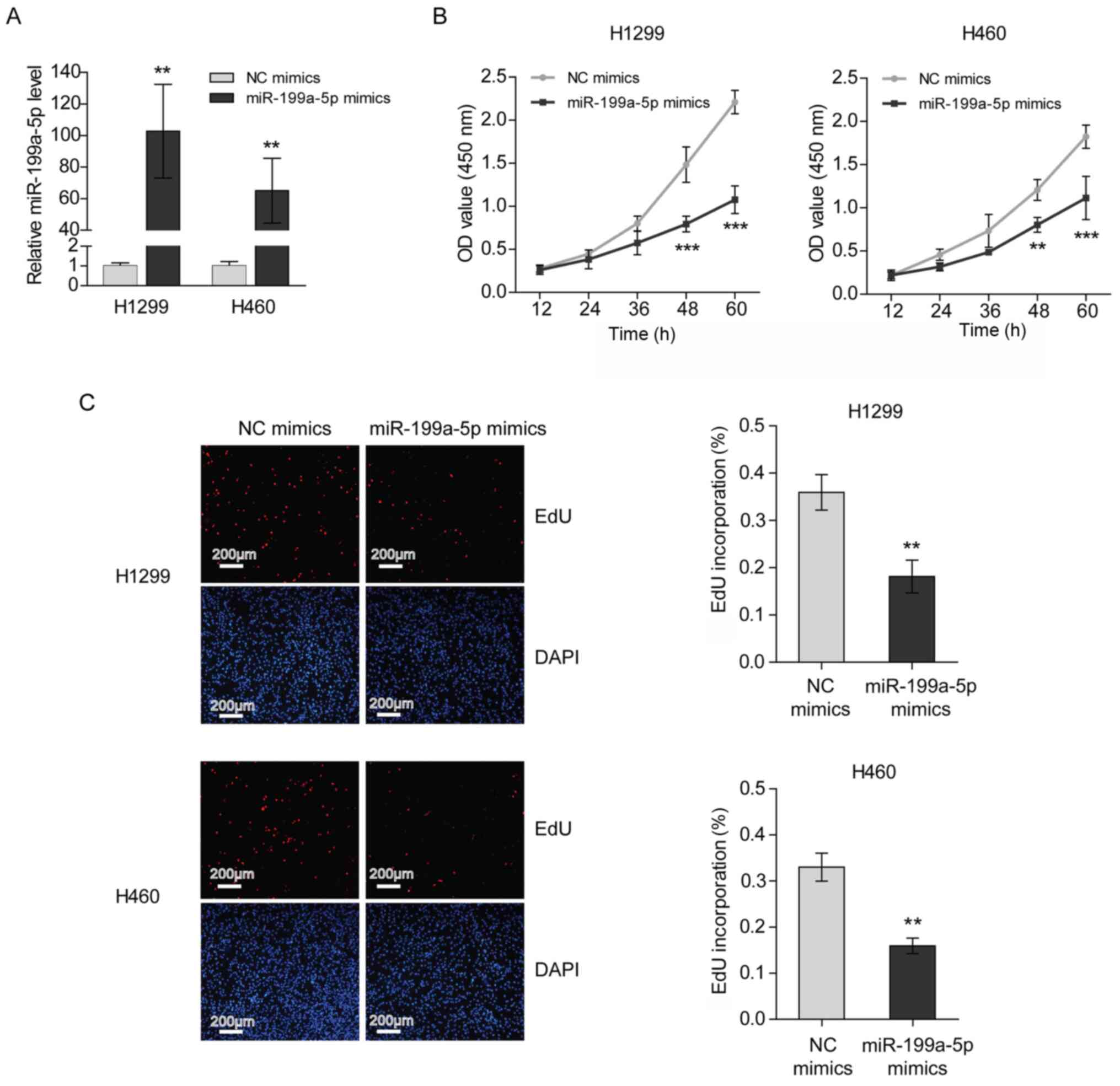
Transfection with miR-199a-5p mimics
suppresses the proliferation of NSCLC cells
To investigate the role of miR-199a-5p in NSCLC
development, the proliferation of H1299 and H460 cells transfected
with miR-199a-5p mimics was measured. The transfection efficiency
was verified by RT-qPCR (Fig. 3A).
The results of the CCK-8 assays demonstrated that the proliferation
rates of H1299 and H460 cells were decreased following miR-199a-5p
mimics transfection (Fig. 3B).
Furthermore, it was revealed that the transfection of miR-199a-5p
mimics interrupted 5-ethynyl-2′-deoxyuridine (EdU) incorporation,
indicating that miR-199a-5p inhibited the DNA synthesis of NSCLC
cells (Fig. 3C).
miR-199a-5p directly targets AKAP1
mRNA in NSCLC
To understand the mechanism underlying the antitumor
function of miR-199a-5p in NSCLC, TargetScan was used to predict
the potential transcripts targeted by miR-199a-5p. The seed
sequence of miR-199a-5p was complementary to the 3′UTR of AKAP1
mRNA and is highly conserved among different species (Fig. 4A). Recently, the unique involvement
of AKAP1 in tumor growth has been reviewed (14). In the present study, the luciferase
assay results demonstrated that miR-199a-5p inhibited the activity
of the reporter with wild-type 3′UTR of AKAP1 mRNA, but was not
able to change the activity of the reporter when the miR-199a-5p
binding site was mutated (Fig. 4B).
Consistently, the mRNA and protein expression levels of AKAP1 were
decreased following miR-199a-5p mimic transfection in H1299 and
H460 cells (Fig. 4C). Since
miR-199a-5p expression was induced by 5-Aza-dC treatment, the
expression levels of AKAP1 were measured after 5-Aza-dC treatment.
The results indicated that 5-Aza-dC decreased the mRNA and protein
expression levels of AKAP1 (Fig.
S1). In contrast to miR-199a-5p, clinical analysis demonstrated
that the mRNA expression levels of AKAP1 were significantly higher
in NSCLC tissues compared with in para-carcinoma tissues (Fig. 4D). Furthermore, the mRNA expression
levels of AKAP1 were negatively correlated with miR-199a-5p
expression in NSCLC tissues (Fig.
4E). Collectively, these findings suggested that miR-199a-5p
may downregulate AKAP1 in NSCLC.
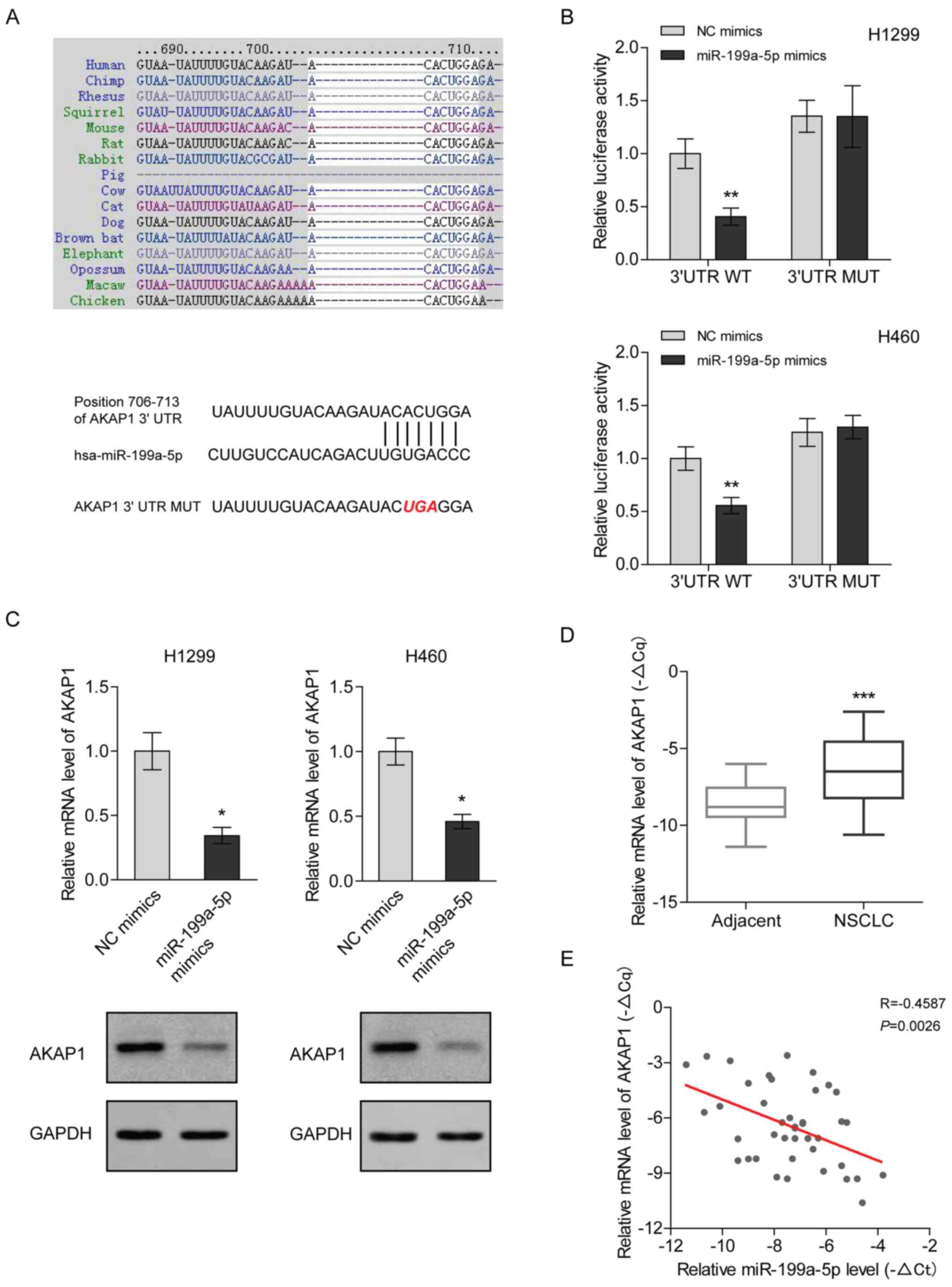 | Figure 4.miR-199a-5p directly targets the mRNA
of AKAP1 in NSCLC. (A) miR-199a-5p seed sequence complementary to
the 3′UTR of AKAP1 mRNA was analyzed using TargetScan Human 7.2.
(B) Luciferase activity was measured in H1299 and H460 cells
co-transfected with the luciferase constructs containing the WT or
MUT AKAP1 3′UTR, as well as miR-199a-5p mimics or NC mimics, and
was normalized to the WT/NC mimics group. Two-way ANOVA with
Sidak's post hoc test was used for analysis. **P<0.01 vs. WT/NC
mimics group. (C) mRNA and protein expression levels of AKAP1 were
analyzed using RT-qPCR and western blotting in H1299 and H460 cells
transfected with miR-199a-5p mimics. mRNA expression levels were
normalized to those of the NC mimics group. An unpaired t-test was
used for analysis. *P<0.05 vs. NC mimics group. (D) mRNA
expression levels of AKAP1 in 41 paired NSCLC and adjacent tissues
were quantified via RT-qPCR, using β-actin as an internal control.
-ΔCq [ΔCq: Cq (AKAP1)-Cq (β-actin)] represented the relative
expression level. A paired t-test was used for analysis.
***P<0.001 vs. adjacent tissue. (E) Correlation between the mRNA
expression levels of AKAP1 and miR-199a-5p in 41 NSCLC tissues. A
Pearson test was used for analysis. AKAP1, A-kinase anchoring
protein 1; miR, microRNA; MUT, mutant; NC, negative control; NSCLC,
non-small cell lung cancer; RT-qPCR, reverse
transcription-quantitative PCR; UTR, untranslated region; WT,
wild-type. |
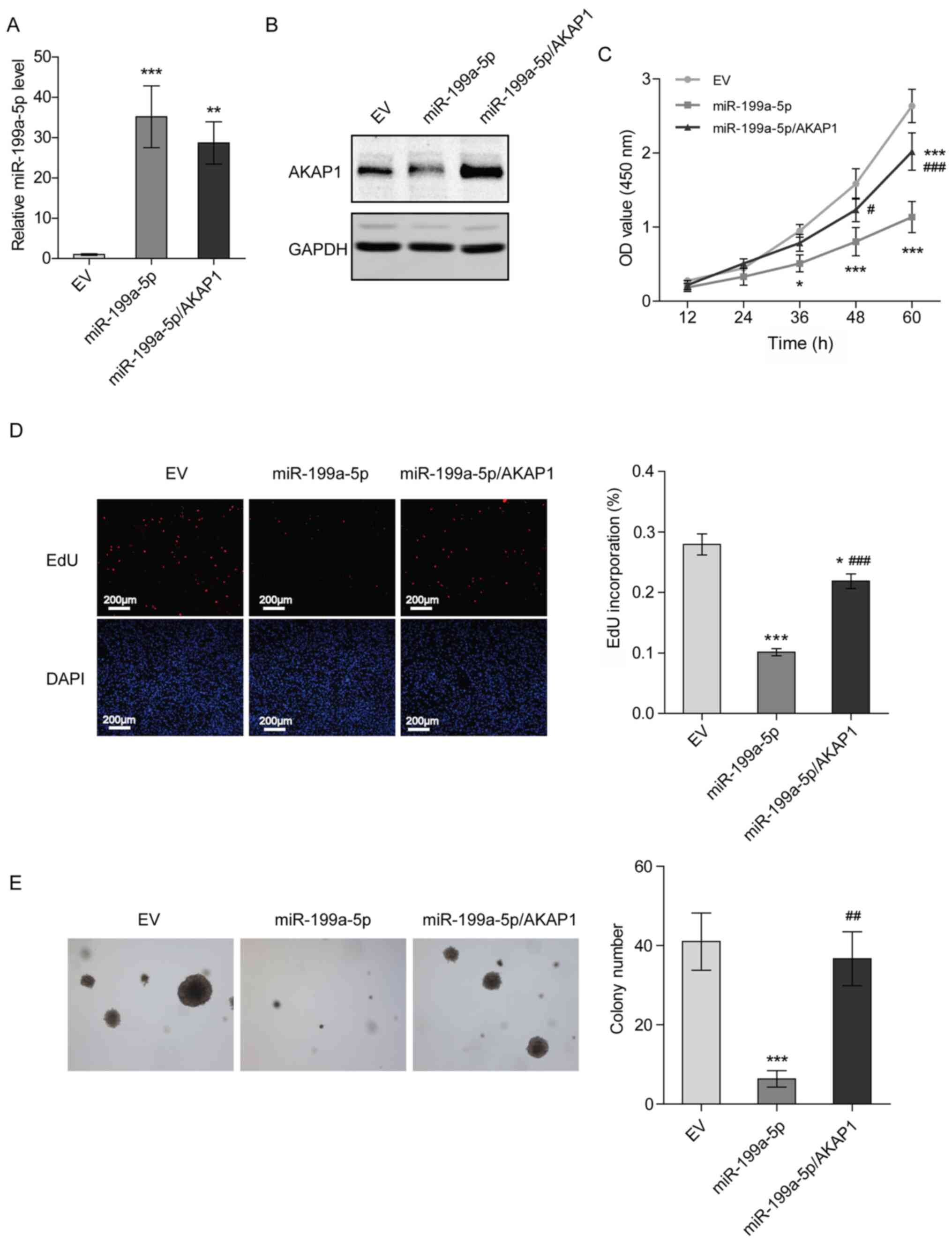
Overexpression of AKAP1 restores the
proliferation of NSCLC cells transfected with miR-199a-5p
mimics
To verify whether miR-199a-5p represses the
proliferation of H1299 cells by downregulating AKAP1, H1299 cells
stably overexpressing miR-199a-5p and AKAP1 were constructed
(Figs. 5A, B and S2). Compared with the control group (EV),
a suppression of cell proliferation and DNA synthesis was observed
in H1299 cells stably overexpressing miR-199a-5p after 36 h of
culturing, while overexpression of AKAP1 partially rescued cell
proliferation and DNA synthesis (Fig. 5C
and D). Additionally, overexpression of miR-199a-5p decreased
the colony formation ability of H1299 cells, and overexpression of
AKAP1, to a certain extent, recovered this attenuated colony
formation (Fig. 5E). These results
suggested that miR-199a-5p inhibited the proliferation of NSCLC
cells by targeting AKAP1.
Discussion
The present study demonstrated that promoter
hypermethylation inhibited the transcription of miR-199a-5p, which
decreased the expression levels of AKAP1 by targeting AKAP1 mRNA.
Furthermore, increased AKAP1 expression promoted the proliferation
and colony formation of NSCLC cells.
Abnormal regulation of miRNAs serves an important
role in the occurrence and development of lung cancer (24). The inhibition of miRNA biosynthesis
has been observed in lung cancer (24). For example, Karube et al
(25) reported that the expression
levels of Dicer, a key endoribonuclease in miRNA-mediated gene
silencing was decreased in NSCLC and low Dicer expression was
associated with poor prognosis. Additionally, the results of animal
experiments have demonstrated that conditional knockout of Dicer
promotes tumor development in K-RAS-induced lung cancer (26). In rats, the tobacco carcinogen
nicotine-derived nitrosamine ketone induces the dysregulation of
various miRNAs (27). In addition,
changes in the miRNA expression profile have been detected in
precancerous lesions of the bronchial epithelium (28). However, it is difficult to identify
specific miRNA markers for NSCLC due to tumor heterogeneity and
different analysis methods. Wang et al (29) retrospectively analyzed four studies
comparing the miRNA expression profiles between NSCLC and
corresponding para-cancerous tissues; however, the differentially
expressed miRNAs were different in each study.
The present study demonstrated that low expression
levels of miR-199a-5p in NSCLC were associated with a poor
survival, suggesting that miR-199a-5p may be a potential miRNA
marker for the prediction of the prognosis of patients with NSCLC.
Previously, Sanfiorenzo et al (30) reported that low expression levels of
miR-199a-5p in the plasma are associated with poor survival of
patients with NSCLC. Recently, it was revealed that miR-199a-5p
could increase the doxorubicin sensitivity of NSCLC (31). Therefore, miR-199a-5p may function as
a crucial tumor suppressor miRNA for NSCLC.
Several studies have revealed that long non-coding
RNAs promote the proliferation of NSCLC via sponging of miR-199a-5p
(32,33). In addition, hypermethylation of the
CpG island has been identified to be closely associated with the
abnormal miRNA expression profiles in tumors (34). In total ~1/2 of all miRNAs are
regulated by the CpG island (35).
Previous studies have reported that the altered status of DNA
methylation leads to the abnormal regulation of miRNA expression in
NSCLC (36–38). The miR-199a gene is located at two
different chromosomes (miR-199a1 is located at chromosome 19 and
miR-199a2 is located at chromosome 1) (23). Therefore, the present study first
analyzed the distribution of CpG islands at the promoter regions of
the two gene loci using bioinformatics software, and identified
that there was a CG-rich region (−404/-272) upstream of the
transcription start site of miR-199a1, while no potential CpG
island was found at the miR-199a2 promoter, which was consistent
with a previous study (23).
Therefore, the present study only analyzed the transcriptional
regulation of the miR-199a1 gene. Bisulfite sequencing demonstrated
that the methylation levels of the CpG island at the miR-199a
promoter were increased in NSCLC compared with in para-cancerous
tissues. Furthermore, treatment with a DNA methyltransferase
inhibitor increased the expression levels of miR-199a-5p in NSCLC
cells. These findings suggested that promoter hypermethylation
suppressed miR-199a-5p expression in NSCLC.
Deng et al (23) revealed that knockdown of DNA
methyltransferase 3α (DNMT3A) reverses the hypermethylation of the
miR-199a gene in ovarian cancer. Furthermore, DNMT3A is highly
expressed in various cancer types, including NSCLC, and can mediate
the epigenetic silencing of tumor suppressor genes (39). Therefore, it was hypothesized that
the promoter hypermethylation of the miR-199a gene may also be
induced by aberrantly expressed DNMT3A in NSCLC.
The present study demonstrated that miR-199a-5p
inhibited the proliferation of NSCLC cells by targeting AKAP1.
Rinaldi et al (17) reported
that AKAP1 promotes glioblastoma (GBM) growth by enhancing the mTOR
signaling pathway. In addition, these authors observed that
knockdown of AKAP1 severely impaired the oxidative metabolism of
GBM cells. In general, high oxidative phosphorylation (OXPHOS) is
adopted to promote tumor growth (40,41), and
AKAP1 stimulates mitochondrial respiration (42). Therefore, deregulated AKAP1 may
enhance OXPHOS to induce the proliferation of NSCLC cells.
In conclusion, the present study indicated that
promoter hypermethylation caused the downregulation of miR-199a-5p
expression in NSCLC, and miR-199a-5p was identified as a novel
regulator of AKAP1 expression influencing NSCLC proliferation.
Therefore, the present findings may provide a prognostic miRNA
marker for patients with NSCLC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Program of
Wenzhou Science and Technology Bureau (grant no. Y20190113).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
NY and LJ conceived and designed the study. NY, YFL,
TZ and YXL performed the experiments and wrote the manuscript. NY,
ZC and XZ analyzed and interpreted the data. NY, XZ and LJ
confirmed the authenticity of all the raw data. NY, XZ and LJ
drafted the manuscript. All other authors helped to draft the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Experiments using human tissues were approved by the
Ethics Committee in Clinical Research of the First Affiliated
Hospital of Wenzhou Medical University (issuing no. 2019-128;
Wenzhou, China) and written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al: Global surveillance of cancer survival 1995–2009: Analysis
of individual data for 25,676,887 patients from 279
population-based registries in 67 countries (CONCORD-2). Lancet.
385:977–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Riaz SP, Lüchtenborg M, Coupland VH,
Spicer J, Peake MD and Møller H: Trends in incidence of small cell
lung cancer and all lung cancer. Lung Cancer. 75:280–284. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Toschi L, Cappuzzo F and Jänne PA:
Evolution and future perspectives in the treatment of locally
advanced non-small cell lung cancer. Ann Oncol. 18 (Suppl
9):ix150–ix155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhong J, Huang R, Su Z, Zhang M, Xu M,
Gong J, Chen N, Zeng H, Chen X and Zhou Q: Downregulation of
miR-199a-5p promotes prostate adeno-carcinoma progression through
loss of its inhibition of HIF-1α. Oncotarget. 8:83523–83538. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Turashvili G, Lightbody ED, Tyryshkin K,
SenGupta SK, Elliott BE, Madarnas Y, Ghaffari A, Day A and Nicol
CJB: Novel prognostic and predictive microRNA targets for
triple-negative breast cancer. FASEB J. May 29–2018.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ghosh A, Dasgupta D, Ghosh A, Roychoudhury
S, Kumar D, Gorain M, Butti R, Datta S, Agarwal S, Gupta S, et al:
MiRNA199a-3p suppresses tumor growth, migration, invasion and
angiogenesis in hepatocellular carcinoma by targeting VEGFA,
VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 8:e27062017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gadducci A, Sergiampietri C, Lanfredini N
and Guiggi I: Micro-RNAs and ovarian cancer: The state of art and
perspectives of clinical research. Gynecol Endocrinol. 30:266–271.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye H, Pang L, Wu Q, Zhu Y, Guo C, Deng Y
and Zheng X: A critical role of mir-199a in the cell biological
behaviors of colorectal cancer. Diagn Pathol. 10:652015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ahmadi A, Khansarinejad B, Hosseinkhani S,
Ghanei M and Mowla SJ: miR-199a-5p and miR-495 target GRP78 within
UPR pathway of lung cancer. Gene. 620:15–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu Y, Liu J, Jiang B, Chen J, Fu Z, Bai F,
Jiang J and Tang Z: MiR-199a-5p loss up-regulated DDR1 aggravated
colorectal cancer by activating epithelial-to-mesenchymal
transition related signaling. Dig Dis Sci. 59:2163–2172. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen
T, Chen Z, Huang S, Gu J, Li J, et al: miR-199a-5p is negatively
associated with malignancies and regulates glycolysis and lactate
production by targeting hexokinase 2 in liver cancer. Hepatology.
62:1132–1144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li W, Wang H, Zhang J, Zhai L, Chen W and
Zhao C: miR-199a-5p regulates β1 integrin through Ets-1 to suppress
invasion in breast cancer. Cancer Sci. 107:916–923. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Wang D, Li X, Shao Y, He Y, Yu H and
Ma Z: MiR-199a-5p suppresses non-small cell lung cancer via
targeting MAP3K11. J Cancer. 10:2472–2479. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, Merrill RA and Strack S: A-kinase
anchoring protein 1: Emerging roles in regulating mitochondrial
form and function in health and disease. Cells. 9:2982020.
View Article : Google Scholar
|
|
15
|
Yu J, Zhang Y, Zhou D, Wu J and Luo R:
Higher expression of A-kinase anchoring-protein 1 predicts poor
prognosis in human hepatocellular carcinoma. Oncol Lett.
16:131–136. 2018.PubMed/NCBI
|
|
16
|
Li B, Wang W, Li Z, Chen Z, Zhi X, Xu J,
Li Q, Wang L, Huang X, Wang L, et al: MicroRNA-148a-3p enhances
cisplatin cytotoxicity in gastric cancer through mitochondrial
fission induction and cyto-protective autophagy suppression. Cancer
Lett. 410:212–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rinaldi L, Sepe M, Delle Donne R, Conte K,
Arcella A, Borzacchiello D, Amente S, De Vita F, Porpora M, Garbi
C, et al: Mitochondrial AKAP1 supports mTOR pathway and tumor
growth. Cell Death Dis. 8:e28422017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rami-Porta R, Bolejack V, Crowley J, Ball
D, Kim J, Lyons G, Rice T, Suzuki K, Thomas CF Jr, Travis WD, et
al: The IASLC Lung cancer staging project: Proposals for the
revisions of the T descriptors in the forthcoming eighth edition of
the TNM classification for lung cancer. J Thorac Oncol.
10:990–1003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Travis WD: The 2015 WHO classification of
lung tumors. Pathologe. 35 (Suppl 2):1882014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen K, Chen H, Yang F, Sui X, Li X and
Wang J: Validation of the eighth edition of the TNM staging system
for lung cancer in 2043 surgically treated patients with
non-small-cell lung cancer. Clin Lung Cancer. 18:e457–e466. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng Y, Zhao F, Hui L, Li X, Zhang D, Lin
W, Chen Z and Ning Y: Suppressing miR-199a-3p by promoter
methylation contributes to tumor aggressiveness and cisplatin
resistance of ovarian cancer through promoting DDR1 expression. J
Ovarian Res. 10:502017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Iqbal MA, Arora S, Prakasam G, Calin GA
and Syed MA: MicroRNA in lung cancer: Role, mechanisms, pathways
and therapeutic relevance. Mol Aspects Med. 70:3–20. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karube Y, Tanaka H, Osada H, Tomida S,
Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S,
Mitsudomi T and Takahashi T: Reduced expression of Dicer associated
with poor prognosis in lung cancer patients. Cancer Sci.
96:111–115. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kalscheuer S, Zhang X, Zeng Y and
Upadhyaya P: Differential expression of microRNAs in early-stage
neoplastic transformation in the lungs of F344 rats chronically
treated with the tobacco carcinogen
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis.
29:2394–2399. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mascaux C, Laes JF, Anthoine G, Haller A,
Ninane V, Burny A and Sculier JP: Evolution of microRNA expression
during human bronchial squamous carcinogenesis. Eur Respir J.
33:352–359. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang QZ, Xu W, Habib N and Xu R: Potential
uses of microRNA in lung cancer diagnosis, prognosis, and therapy.
Curr Cancer Drug Targets. 9:572–594. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sanfiorenzo C, Ilie MI, Belaid A, Barlési
F, Mouroux J, Marquette CH, Brest P and Hofman P: Two panels of
plasma microRNAs as non-invasive biomarkers for prediction of
recurrence in resectable NSCLC. PLoS One. 8:e545962013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jin Y, Wang H, Zhu Y, Feng H, Wang G and
Wang S: miR-199a-5p is involved in doxorubicin resistance of
non-small cell lung cancer (NSCLC) cells. Eur J Pharmacol.
878:1731052020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hua Q, Jin M, Mi B, Xu F, Li T, Zhao L,
Liu J and Huang G: LINC01123, a c-Myc-activated long non-coding
RNA, promotes proliferation and aerobic glycolysis of non-small
cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol.
12:912019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang C, Han C, Zhang Y and Liu F: LncRNA
PVT1 regulate expression of HIF1α via functioning as ceRNA for
miR-199a-5p in non-small cell lung cancer under hypoxia. Mol Med
Rep. 17:1105–1110. 2018.PubMed/NCBI
|
|
34
|
Suzuki H, Maruyama R, Yamamoto E and Kai
M: DNA methylation and microRNA dysregulation in cancer. Mol Oncol.
6:567–578. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Weber B, Stresemann C, Brueckner B and
Lyko F: Methylation of human microRNA genes in normal and
neoplastic cells. Cell Cycle. 6:1001–1005. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang J, Fu J, Pan Y, Zhang X and Shen L:
Silencing of miR-1247 by DNA methylation promoted non-small-cell
lung cancer cell invasion and migration by effects of STMN1. Onco
Targets Ther. 9:7297–7307. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu C, Lv D, Li M, Zhang X, Sun G, Bai Y
and Chang D: Hypermethylation of miRNA-589 promoter leads to
upregulation of HDAC5 which promotes malignancy in non-small cell
lung cancer. Int J Oncol. 50:2079–2090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia W, Chen Q, Wang J, Mao Q, Dong G, Shi
R, Zheng Y, Xu L and Jiang F: DNA methylation mediated silencing of
microRNA-145 is a potential prognostic marker in patients with lung
adenocarcinoma. Sci Rep. 5:169012015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT
and Wang YC: Alteration of DNA methyltransferases contributes to
5′CpG methylation and poor prognosis in lung cancer. Lung Cancer.
55:205–213. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Martinez-Outschoorn UE, Pestell RG, Howell
A, Tykocinski ML, Nagajyothi F, Machado FS, Tanowitz HB, Sotgia F
and Lisanti MP: Energy transfer in ‘parasitic’ cancer metabolism:
Mitochondria are the powerhouse and Achilles' heel of tumor cells.
Cell Cycle. 10:4208–4216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sotgia F, Martinez-Outschoorn UE and
Lisanti MP: Cancer metabolism: New validated targets for drug
discovery. Oncotarget. 4:1309–1316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Livigni A, Scorziello A, Agnese S,
Adornetto A, Carlucci A, Garbi C, Castaldo I, Annunziato L,
Avvedimento EV and Feliciello A: Mitochondrial AKAP121 links cAMP
and src signaling to oxidative metabolism. Mol Biol Cell.
17:263–271. 2006. View Article : Google Scholar : PubMed/NCBI
|