Introduction
Glioblastoma multiforme (GBM), also known as
astrocytoma grade IV (1), is one of
the most common and fatal forms of malignant primary brain tumor.
In total, 14.6% of all brain tumors in the United States between
2012 and 2016 were GBM, with a 5-year survival rate of 6.8%
(2). Temozolomide (TMZ), an oral
alkylating agent, is the first-line chemotherapy drug for GBM, as
it can cross the blood-brain barrier (3). TMZ causes the methylation of the
O6 position of guanine in DNA, leading to a mismatch
between O6-methylguanine and thymine. Subsequently, the
cells undergo DNA replication and the mismatch repair promotes the
formation of DNA double-strand breaks (DSBs) (4), which may further trigger GBM cell death
(5–7).
Most patients show a significant initial response to
TMZ; however, the overall response to TMZ chemotherapy is still
poor due to the development of drug resistance. TMZ resistance may
involve multiple mechanisms, such as DNA methyltransferase (MGMT)
and DNA repair (8–10), and accelerating the repair of DSBs
can enhance the TMZ chemical resistance of GBM cells (11–13). The
molecular mechanism mediating TMZ resistance has not been fully
understood; therefore, an improved understanding of TMZ resistance
will assist with the development of new sensitizers to improve the
efficacy of TMZ treatment. The standard of care for patients with
GBM is maximum tumor resection, followed by radiotherapy and
adjuvant chemotherapy with TMZ; however, patients globally rarely
survive for >2 years after diagnosis (14,15).
Therefore, new treatment strategies are required to improve patient
survival.
RNA interference (RNAi) is a revolutionary technique
for studying the biological functions of a particular gene, by
silencing its gene expression (16,17). A
lentiviral short hairpin (sh)RNA subset was used in the present
study to identify genes that increase TMZ sensitivity in GBM cells.
Such screening methods have proven to be effective tools for
identifying key targets of drug sensitivity (18,19).
X-ray repair cross-complementing protein 5 (XRCC5),
also known as Ku80 or Ku86, is encoded by a gene located on human
chromosome 2q33. The non-homologous end joining (NHEJ) is the
predominant DSB repair mechanism in human cells (20). During the repair process, DSB is
first recognized by the heterodimer composed of XRCC6 (also denoted
as Ku70)/XRCC5 in NHEJ, then DNA-dependent protein kinase catalytic
subunit (DNA-PKcs) is subsequently recruited to repair the DSB
(21,22). Several studies have determined that
the increased protein expression level of XRCC5 has been associated
with treatment resistance and the development of numerous malignant
tumors, and if there is reduced XRCC5 protein expression, cancer
cells have reduced resistance to treatment and degree of malignancy
(23–29). However, whether XRCC5 affects TMZ
sensitivity in GBM is completely unknown.
Numerous large public databases provide complete
genetic information and clinical data, that enables bioinformatics
tools to analyze associations between gene expression levels and
clinical pathological features. These advances can assist in
quickly evaluating the differentially expressed genes associated
with progression, diagnosis and prognosis in different cancer
types, which is an important foundation for developing potential
therapeutic strategies (30–32). Bioinformatics data analysis was also
used to examine whether XRCC5 could be a clinical indicator for the
progression and prognosis in GBM in the present study. Acquired
drug resistance is a limiting factor in the clinical treatment of
GBM (33,34). The present study aimed to investigate
whether XRCC5 could be involved in TMZ resistance, which may
indicate a potential therapeutic target to improve the efficacy of
TMZ treatment.
Materials and methods
Cell culture
The human glioblastoma cell lines: U-87 MG (cat. no.
HTB-14; glioblastoma of unknown origin), M059K (cat. no. CRL-2365)
and DBTRG-05MG (cat. no. CRL-2020) were purchased from American
Type Culture Collection (ATCC). All the cells were cultured in
DMEM, supplemented with 10% FBS and 100 U/ml
penicillin/streptomycin (complete medium) at 37°C in a humidified
incubator with 5% CO2. The medium was refreshed every
2–3 days. After reaching 90% confluence, the cells were washed with
PBS and trypsinized with 0.05% trypsin-EDTA. The trypsinization
effect was neutralized with DMEM, supplemented with 10% FBS.
Subsequently, the cells were centrifuged at 350 × g for 5 min at
room temperature and the cell pellet was resuspended in the
complete medium. The cell suspension was diluted 1:1 (v/v) with
0.4% trypan blue and the viable cells (unstained) were counted with
a hemocytometer. DMEM, FBS, trypsin-EDTA and trypan blue solution
were purchased from Thermo Fisher Scientific, Inc. All the
experiments were performed within 1 year following purchasing the
cells from ATCC.
Lentivirus array-based shRNA library
screening
The vesicular stomatitis virus G protein
(VSV-G)-pseudotyped lentivirus-based subset was obtained from the
National RNAi Core Facility, Academia Sinica (Taipei, Taiwan). The
kinase and phosphatase (KP) gene subset was selected for screening.
The RNAi Consortium (TRC) designed multiple distinct shRNA clones
to target specific genes. A total of 428 shRNAs targeting 84
kinases or phosphatases were used for the functional screen in a
96-well format (one shRNA per well). Each viral clone, with a
unique target sequence, represented a kinase/phosphatase and each
infected cell would produce a gene-specific knockdown effect. In
brief, for a single shRNA clone, the U-87 MG cells were seeded
(3×103 cells/well) in 96-well plates, 24 h prior to
infection. The cells were then infected with KP subset lentiviruses
(multiplicity of infection, 3) in the presence of 8 µg/ml polybrene
(Sigma-Aldrich; Merck KGaA) at 37°C in a humidified incubator with
5% CO2. The effect of each gene knockdown on TMZ
sensitivity in the U-87MG cells was analyzed using an MTT assay as
later described.
Cell cytotoxicity
After U-87MG cells were transduced with
shRNA-expressing lentivirus for 48 h, the DMEM with FBS was removed
and replaced with fresh DMEM. Each shRNA clone infection was
performed in duplicate, in two independent 96-well plates. Then,
each replicate was treated with either vehicle (DMSO) or 500 µM TMZ
for 48 h at 37°C in a humidified incubator with 5% CO2.
A MTT assay was used to evaluate relative cell viability. Briefly,
the cells were plated at a density of 5×103 cells/per
well in 100 µl complete medium and in 96-well microplates. After
overnight incubation, the medium was replaced by serum-free medium,
containing TMZ concentration (0-1,000 µM). After incubation for 48
h, the MTT reagent (0.5 mg/ml) was added to each well, then the
cells were incubated for a further 4 h. After incubation, the
medium was removed and the purple formazan was solubilized with
dimethyl sulfoxide (DMSO). The absorbance was measured at 570 nm,
with background subtraction at 630 nm using a Microplate ELISA
Reader. The percentage of cell viability is shown relative to
untreated cells. The MTT, TMZ and DMSO reagents were purchased from
Sigma-Aldrich (Merck KGaA).
Public database analysis
XRCC5 mRNA expression level in human lower-grade
glioma and GBM tissues were analyzed through cBioPortal (https://www.cbioportal.org/). Two datasets in
cBioPortal were used: The Cancer Genome Atlas (TCGA) Pancancer
Atlas dataset (https://www.cell.com/pb-assets/consortium/pancanceratlas/pancani3/index.html)
(including lower grade glioma [oligoastrocytoma 25.3%, anaplastic
astrocytoma 24.5%, oligodendroglioma 22.6%, anaplastic
oligoastrocytoma 14.7%, astrocytoma 12.6% and diffuse glioma 0.2%)
provisional dataset (http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/LGG/20160128/)
(including lower grade glioma [astrocytoma 37.7%, oligodendroglioma
36.8%, oligoastrocytoma 25.3%, encapsulated glioma 0.2%, and
low-grade glioma (nos) 0.2%]). XRCC5 mRNA expression level in human
br ain cancer tissues and normal brain tissues were analyzed
through Oncomine (https://www.oncomine.org/). and the R2 Genomics
Analysis and Visualization Platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Median
and interquartile ranges are presented. In addition, the online
software R2 Genomics Analysis and Visualization Platform was used
to analyze the correlation between XRCC5 expression and clinical
prognosis. Kaplan-Meier curves were generated using the following
datasets: Tumor Brain-Madhavan-550-MAS5.0-u133p2; 208642_s_at.
(35), where a cut-off between high
expression and low expression groups.
Lentiviral systems for XRCC5
knockdown
The pLKO.1-puro-based lentiviral vectors (harboring
a specific shRNA encoding sequence, packaging plasmid pCMV-R8.91,
and pMD) were obtained from the National RNAi Core Facility at
Academia Sinica (Taipei, Taiwan). Recombinant lentiviruses were
packaged according to the manufacturer's instructions. Lentivirus
was produced by transfecting the 293T cells with the lentiviral
vector (4 µg) plus the packaging plasmids, pCMVΔR8.91 (4 µg) and
pMD (0.4 µg) using TurboFect reagent (Thermo Fisher Scientific,
Inc.). The lentiviral plasmids targeting XRCC5 were TRCN0000288701
(shXRCC5#1:
5′CCGGCGTGGGCTTTACCATGAGTAACTCGAGTTACTCATGGTAAAGCCCACGTTTTTG3′),
TRCN0000295856 (shXRCC5#2:
5′CCGGAGAGGAAGCCTCTGGAAGTTCCTCGAGGAACTTCCAGAGGCTTCCTCTTTTTTG3′) and
TRC2-pLKO_TRC005 (scrambled shControl). U-87MG cells were exposed
to lentiviral supernatants containing 8 µg/ml polybrene for 24 h,
the medium was replaced and then they were incubated for another 48
h. Puromycin (5 µg/ml) was added 48 h after transfection to select
stable cell lines. Stable cells were collected to determine
knockdown efficiency using western blot analysis, and the effect on
TMZ sensitivity was evaluated using an MTT assay and western blot
analysis.
Western blot analysis
The GBM cells were lysed with RIPA buffer (25 mM
Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate and
0.1% SDS) containing a protease inhibitor cocktail. The protein
concentration was determined using a Pierce BCA Protein Assay kit
(cat. no. 23235; Pierce; Thermo Fisher Scientific, Inc.) using
bovine serum albumin (BSA; cat. no. 23209; Pierce; Thermo Fisher
Scientific, Inc.) as a standard. An equal amount of total protein
(40 µg/lane) was resolved using an 8–15% SDS-PAGE and transferred
to PVDF membranes (EMD Millipore). The membrane was blocked with 5%
BSA (Thermo Fisher Scientific, Inc.), then probed with the
following primary antibodies at 4°C overnight: XRCC5 (1:5,000; cat.
no. 16389–1-AP; ProteinTech Group, Inc.), cleaved caspase-3
(1:1,000; cat. no. 9661; Cell Signaling Technology, Inc.), cleaved
PARP (1:1,000; cat. no. ab32064; Abcam) α-tubulin (1:10,000; cat.
no. 05-829; EMD Millipore). After washing with Tris-buffered saline
and 0.05% Tween-20, the membrane was subsequently incubated with
appropriate horseradish peroxidase-coupled secondary antibodies:
Goat Anti-Rabbit IgG (1:5,000; cat. no. 20202; Biotium, Inc.) and
goat anti-Mouse IgG (1:5,000; cat. no. 115-035-003; Jackson
ImmunoResearch Labs, Inc.) for 1 h at room temperature. Bound
antibodies were detected using enhanced chemiluminescence reagents
(Merck KGaA) and signals were visualized using X-ray film. Signal
intensities were quantified using the UN-SCAN-IT gel 6.1 software
(Silk Scientific, Inc.).
XRCC5 overexpression
According to the manufacturer's instructions, the
U-87MG and M059K cells were transfected with XRCC5 overexpression
plasmid (1 µg; pCMV3-XRCC5) or empty vector (1 µg; pCMV3) using
TurboFect transfection reagent (Thermo Fisher Scientific, Inc.) at
37°C in a humidified incubator with 5% CO2. All plasmids
were purchased from Sino Biological Inc. After 48 h of
transfection, cells were collected to determine the overexpression
efficiency using quantitative (q)PCR or western blot analysis, and
the effect on TMZ sensitivity was evaluated using an MTT assay.
Reverse transcription-qPCR
Total RNA was isolated using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). RT was conducted
using the PrimeScript RT reagent kit (Takara Bio Inc.) using the
following conditions: Incubation at 37°C for 30 min and heating to
85°C for 5 sec. qPCR was performed using the KAPA SYBR®
FAST qPCR Master Mix (2X) kit (Roche Diagnostics) and the
StepOnePlus sequence detection system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with the following thermal cycling
conditions: Initial denaturation at 95°C for 30 sec, denaturation
at 95°C for 2 min and annealing/extension at 60°C for 30 sec for 40
cycles. The following primers were used: XRCC5 forward,
5′-GACGTGGGCTTTACCATGAGT-3′ and reverse,
5′-TCAGTGCCATCTGTACCAAAC-3; and GAPDH forward
5′-ACATCCCCTCACCAATAACAAC-3′ and reverse,
5′-TAGCCAAATCATACTGCTCGTC-3′. All experiments were performed
according to the manufacturer's instructions. Relative gene
expression was calculated using the comparative Cq
(2−ΔΔCq) method (36) and
normalized to GAPDH.
Establishment of TMZ-resistant
cells
The TMZ-resistant cell lines, DBTRG-05MG-R and U-87
MG-R were established using a step-wise exposure of the parental
cells (DBTRG-05MG and U-87 MG cell lines, respectively) to
increasing concentrations of TMZ, ranging from 15.625 to 250 µM for
>6 months. TMZ-resistant and parent cells were collected to
analyze TMZ sensitivity using an MTT assay and western blot
analysis, and the levels of XRCC5 protein under TMZ treatment were
also assessed using western blot analysis.
Xenograft mouse model
A total of six female BALB/c nude mice (4-6 weeks
old, 15–20 g weight) were purchased from BioLASCO Co., Ltd. and
maintained in appropriate sterile filter capped cages at an animal
center certified by the Association for Assessment and
Accreditation of Laboratory Animal Care International at Chang Gung
Memorial Hospital (Chiayi, Taiwan). Mice were kept in an
environment with a temperature of 23–25°C, a relative humidity of
50–70% and a light-dark cycle of 12/12 h, with free access to food
and water. The experiment was conducted in 2019. The U87MG and
U87MG-R (5×106) cell lines were injected subcutaneously
into the right flanks of the mice (n=3/group). When the tumor
volumes (length × width2 ×0.5) had reached ~60
mm3, as measured by digital calipers, the mice were
administered with TMZ (10 mg/kg), once every 3 days for 15 days by
intraperitoneal injection. The experiment was terminated on the
15th day and mice were euthanized with excess CO2, with
a 10–30% volume displacement rate per minute. The death of the mice
was assessed by cardiac arrest and fixed/dilated pupils. All the
mice were handled following the Animal Care and Use Guidelines of
the Chang Gung Memorial Hospital (Chiayi, Taiwan) under a protocol
approved by the Institutional Animal Care and Use Committee.
Immunohistochemistry
The specimens from the mice were fixed with 3.7%
formaldehyde for 24 h at room temperature, then dehydrated in a
series of graded alcohol baths and embedded in paraffin. The
samples were cut into sections (4-µm) and heated at 65°C for 30
min. The sections were then de-paraffinized with xylene and
rehydrated with a descending alcohol series (100, 70, 50 and 30%)
followed by distilled water. Next, tissues treated with 5% hydrogen
peroxide at room temperature for 20 min, to inhibit endogenous
peroxidase activity, then incubated with 1% BSA (Thermo Fisher
Scientific, Inc.) for 1 h at room temperature to block non-specific
binding. The slides were incubated overnight at 4°C with a primary
antibody against XRCC5 (1:500; 16389-1-AP; ProteinTech Group,
Inc.). At room temperature, the slides were incubated with goat
anti-rabbit IgG (1:10,000; cat. no. AP132R; Sigma-Aldrich) The
DAB-substrate chromogen solution (1:50) was subsequently added for
3 min at room temperature and the slides were counterstained with
hematoxylin for 30 sec.
Statistical analysis
The data are presented as the mean ± SEM, from at
least 3 independent experiments. The box plots obtained by
cBioPortal analysis data were presented as median and interquartile
ranges. Statistical analysis for assessing significant differences
between two groups was performed using a paired Student's t-test.
For comparison among multiple groups, the data was analyzed using
one-way ANOVA followed by Tukey's post hoc test. An unpaired
Student's t-test was used for comparison of data between two groups
from Oncomine. Survival analyses were performed using the R2
database algorithm and statistical significance levels for multiple
testing were adjusted using Bonferroni's correction. P<0.05 was
considered to indicate a statistically significant difference.
Results
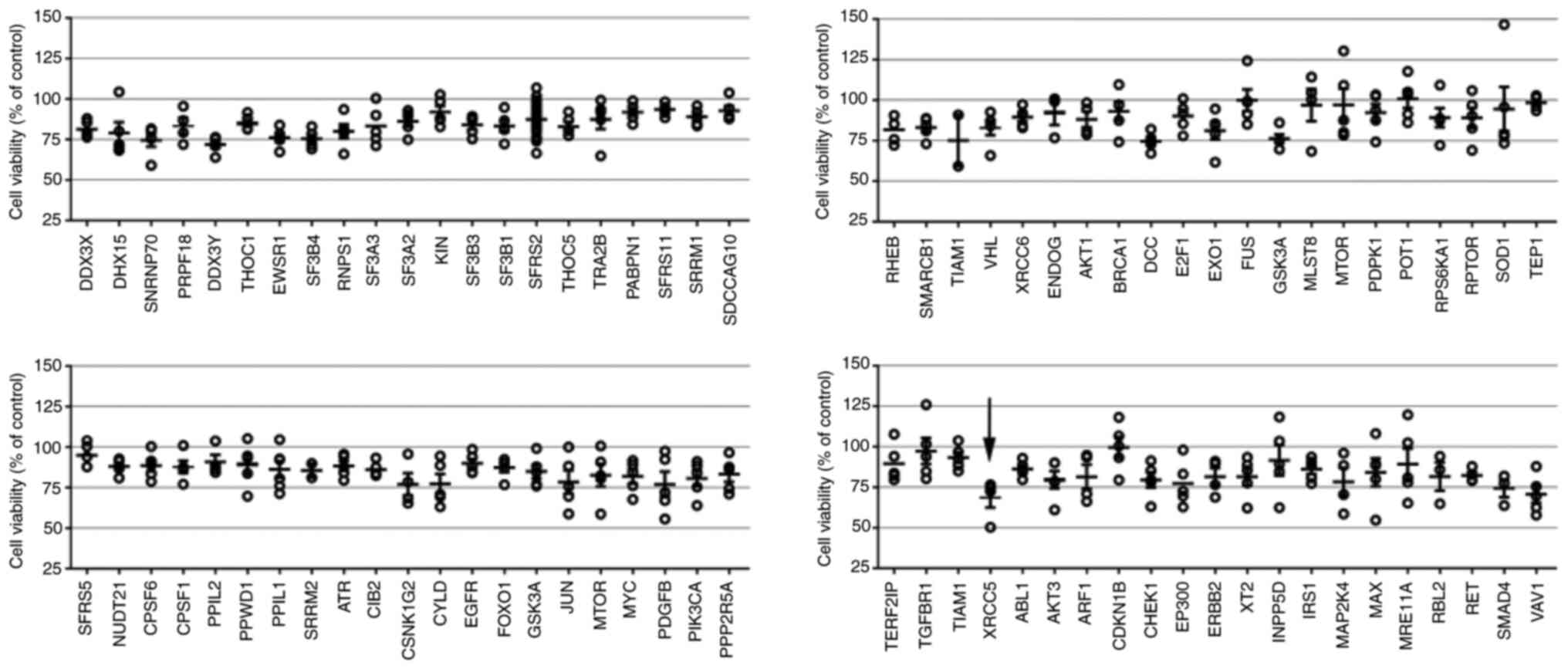
Array-based RNAi library for screening
a novel gene to enhance TMZ sensitivity
To identify genes that could enhance TMZ
cytotoxicity, a partial KP lentiviral shRNA library was screened in
the U-87 MG cell line. Preliminary screening showed that XRCC5 was
the most effective among the 84 genes and could increase TMZ
cytotoxicity. In cells with XRCC5 knockdown and treated with TMZ,
the average cell survival rate was 68.6±6.1%. Knockdown of the VAV1
gene was the second most effective in increasing cell cytotoxicity
with the average cell survival rate of 70.7±5.3% (Fig. 1).
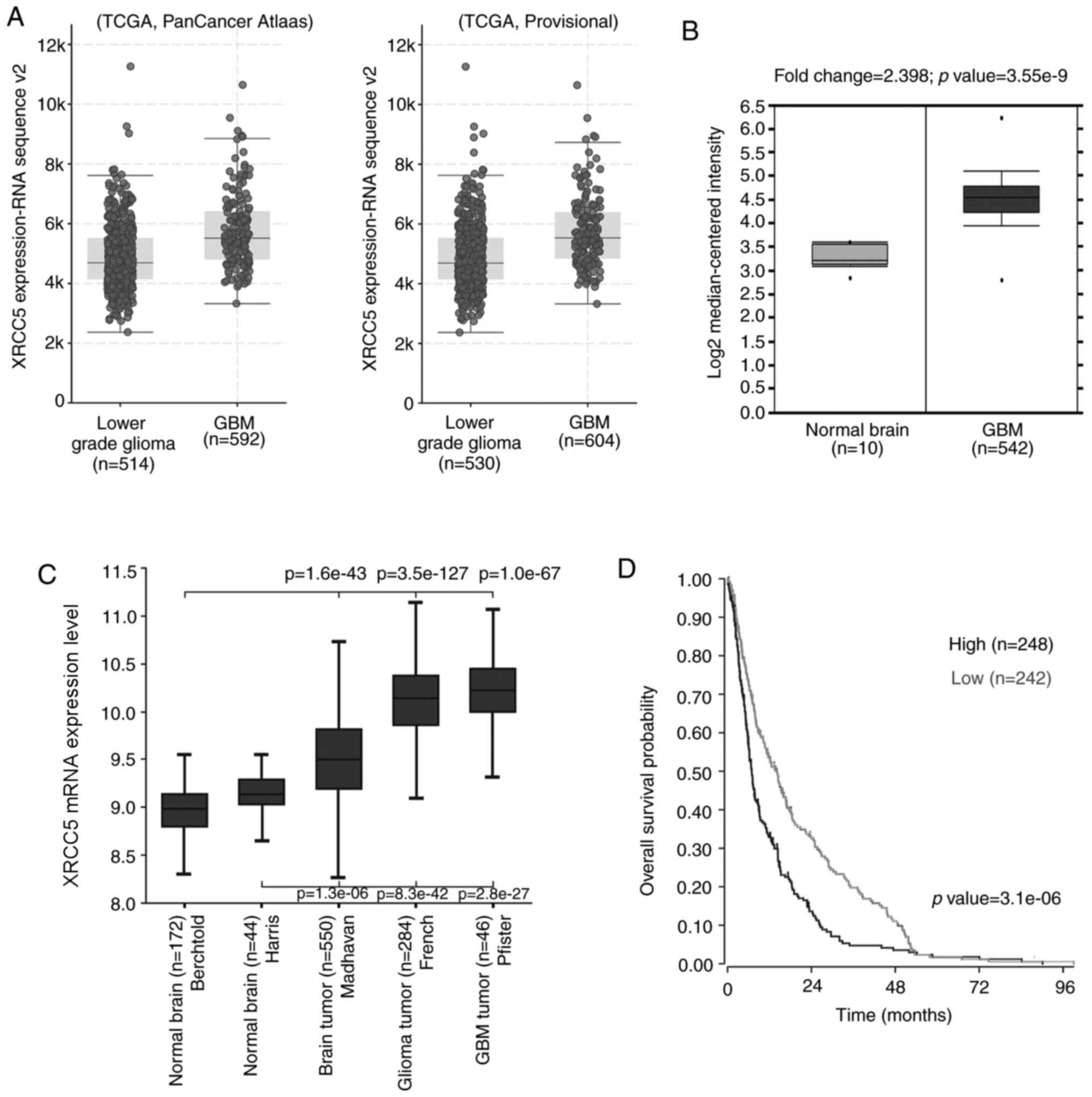
XRCC5 expression is upregulated in GBM
tissues and is associated with poor prognosis
To investigate the expression level of XRCC5 in
clinical tissues, analysis tools of three publicly available
databases were used. As shown in Fig.
2A, the cBioPortal online analysis tool to analyze two datasets
in TCGA, the data showed that the expression level of XRCC5 mRNA in
GBM was higher than that in low-grade gliomas. (TCGA, Pancancer
Atlas dataset: Lower grade glioma, n=514; GBM, n=592; TCGA,
Provisional dataset: Lower grade glioma, n=530; GBM, n=604). From
the Pancancer Atlas data, 37.7% of the patients with lower grade
glioma had astrocytoma, 36.8% had oligodendroglioma, 25.3% had
oligoastrocytoma and 0.2% had encapsulated glioma. From the
Provisional TCGA data, 25.3% of the patients with lower grade
glioma had oligoastrocytoma, 24.5% had anaplastic astrocytoma,
22.6% oligodendroglioma, 14.7% had anaplastic oligoastrocytoma,
12.6% had astrocytoma and 0.2% had diffuse glioma.
According to the Oncomine database, XRCC5 mRNA
expression was significantly increased (fold-change, 2.398) in 542
GBM tissues compared with that in 10 normal brain tissues
(P=3.55×10−9; Fig. 2B).
In addition, data was also downloaded from the R2 Genomics Analysis
and Visualization Platform database to compare the expression
levels of XRCC5 mRNA in normal brain and tumor brain tissues. As
shown in Fig. 2C, comparing brain
cancer tissues, from three different datasets (Madhavan, French and
Pfister) with normal brain tissues from two different datasets
(Madhavan compared with Berchtold (P=1.6×10−43), French
compared with Berchtold (P=3.5×10−127), Pfister compared
with Berchtold (P=1.0×10−67); Madhavan compared with
Haris (P=1.3×10−6), French compared with Haris
(P=8.3×10−42), Pfister compared with Haris
(P=2.8×10−27) it was found that XRCC5 was significantly
higher in cancerous tissues compared with that in normal tissues.
To investigate the association between XRCC5 expression level and
clinical prognosis, the R2 database online tool was used and
information was extracted from a tumor brain dataset (Madhavan-550
MAS5.0 u133p2) to analyze the potential effect of XRCC5 expression
on the overall survival time of patients with GBM. The Kaplan-Meier
curve in Fig. 2D showed that
patients whose tumors had high expression levels of XRCC5 had
poorer survival outcome (P=3.1×10−6).
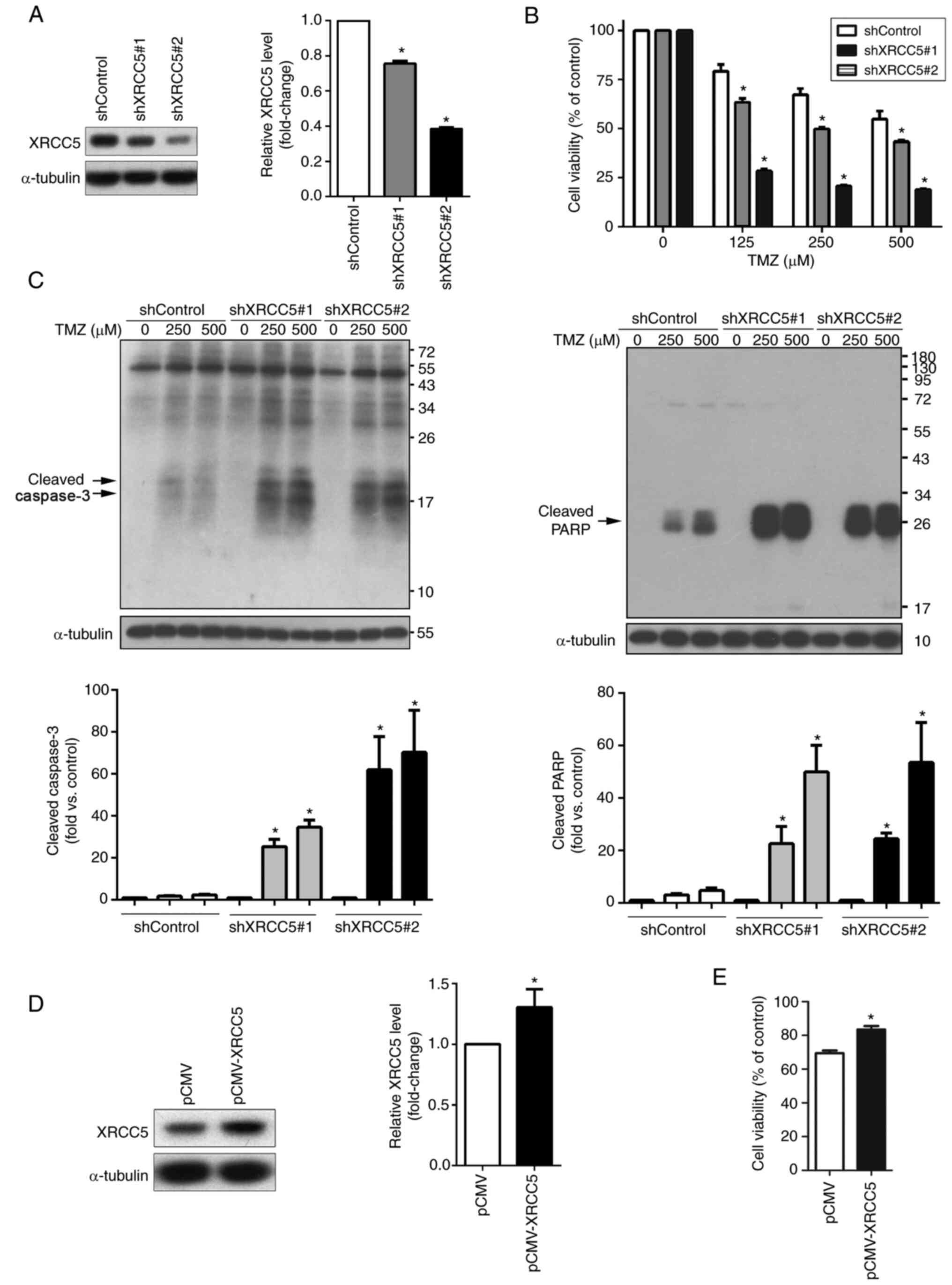
XRCC5 is involved in TMZ-induced
apoptosis in U-87MG cells
Next, the U-87MG cells were stably transfected with
the lentiviral XRCC5 shRNA vector to determine whether XRCC5
expression level was associated with sensitivity to TMZ. Knockdown
efficiency of XRCC5 was determined using western blot analysis
(Fig. 3A). As Fig. 3B shows, knockdown of XRCC5 mRNA
expression led to increased sensitivity to TMZ in a dose-dependent
manner. To further clarify that XRCC5 was associated with
TMZ-induced apoptosis, the cleaved forms of caspase-3 and PARP were
also analyzed using western blot analysis. Knockdown of XRCC5, as
shown in Fig. 3C, markedly increased
the expression level of cleaved-caspase-3 and -PARP. In addition,
XRCC5 overexpression was performed in the U-87MG cells to further
verify the role of XRCC5 in TMZ sensitivity. XRCC5 overexpression
was confirmed using western blot analysis (Fig. 3D) and overexpression of XRCC5 in the
U-87MG cells significantly increased resistance to TMZ, as
determined using a MTT assay (Fig.
3E).
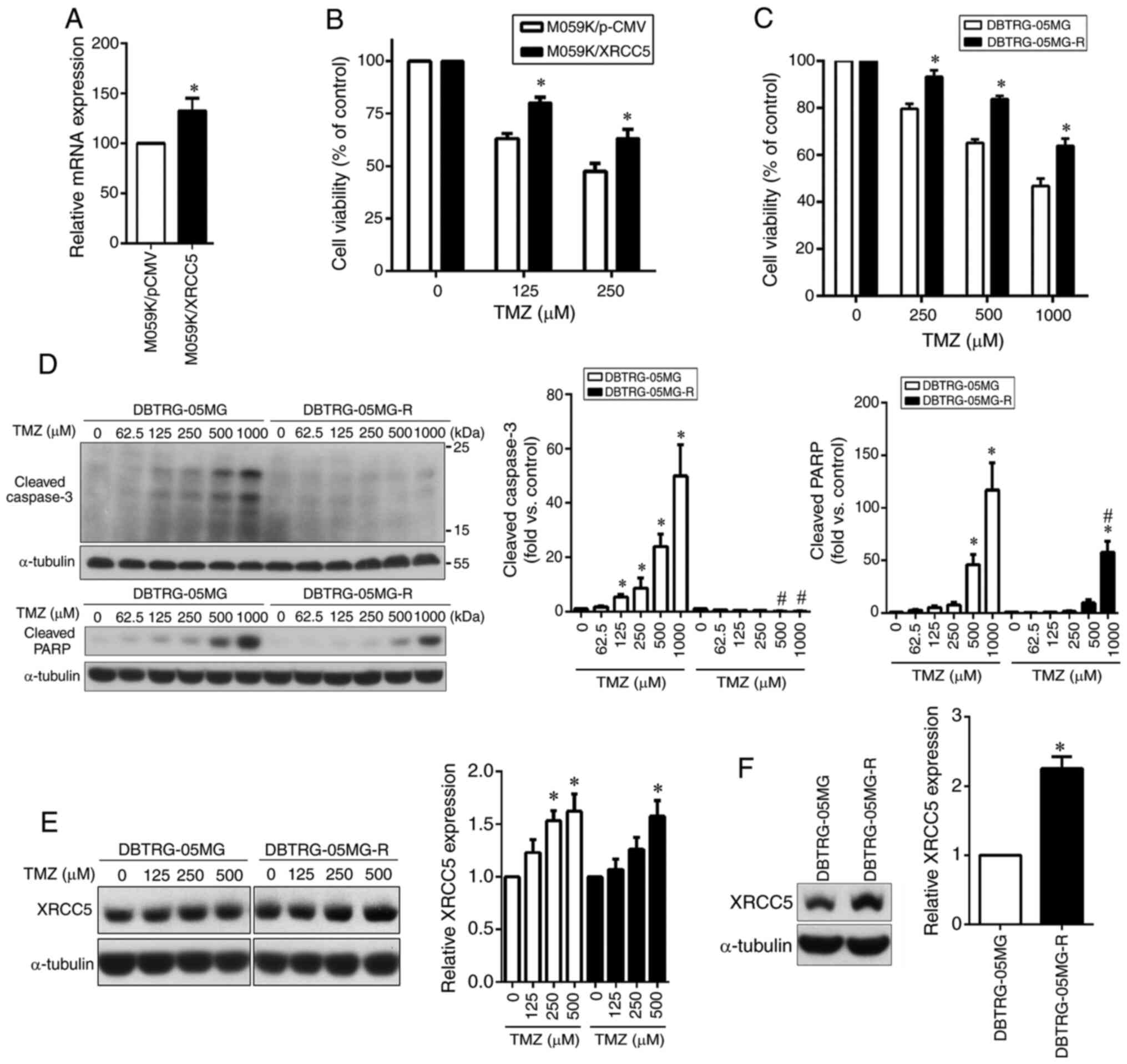
High expression level of XRCC5 in GBM
cells can promote TMZ resistance
Subsequently, the pCMV–XRCC5 plasmid was transfected
into the TMZ-sensitive M059K cell line to overexpress XRCC5 to
analyze whether it could increase TMZ drug resistance. The XRCC5
overexpression level was confirmed using qPCR (Fig. 4A). The results revealed that
overexpression of XRCC5 conferred increased resistance to TMZ
compared with that in the M059K/pCMV group, as determined using a
MTT assay (Fig. 4B). To further
confirm the effect of XRCC5 against TMZ cytotoxicity in the GBM
cell line, the TMZ-resistant DBTRG-05MG cell line was established.
As determined by a MTT assay, the DBTRG-05MG-R cells were more
resistant to TMZ compared with that in their parental cells
(Fig. 4C). The protein expression
levels of cleaved caspase-3 and cleaved PARP were also detected
using western blot analysis, showing that the two cleaved markers
of apoptosis were significantly increased in the TMZ-treated
DBTRG-05MG cells compared with that in the DBTRG-05MG-R cells
(Fig. 4D). Next, the dose-dependent
effect of TMZ on XRCC5 protein expression in the DBTRG-05MG and
DBTRG-05MG-R cells was investigated. As shown in Fig. 4E, the expression level of XRCC5 was
increased in both cell lines in a dose-dependent manner. However,
the basal expression level of XRCC5 in the TMZ-resistant cell line
(DBTRG-05MG-R) was higher compared with that in the parental cell
line (DBTRG-05MG) (Fig. 4F).
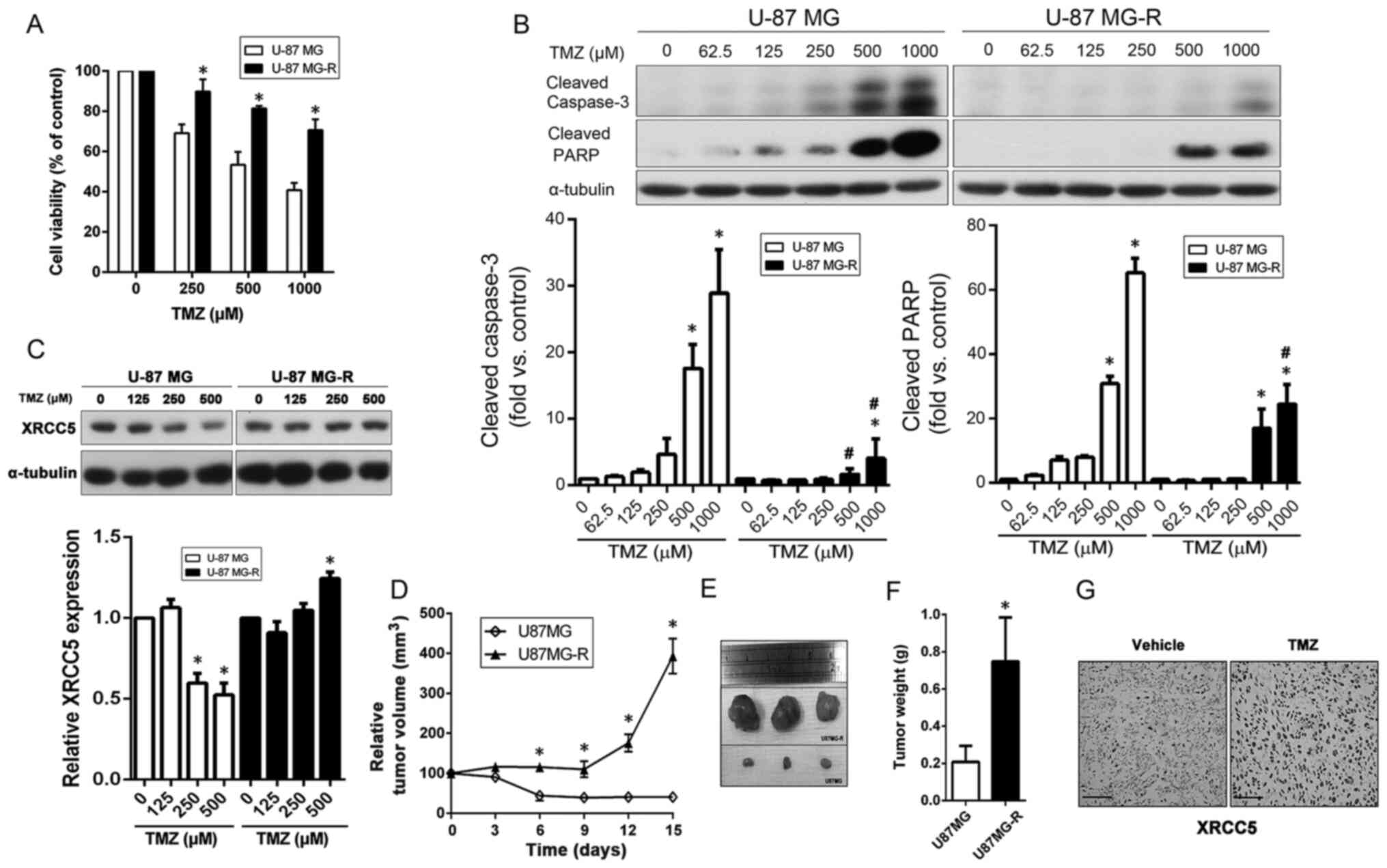
TMZ induces XRCC5 expression level in
TMZ-resistant U-87 MG cells to promote resistance to TMZ both in
vivo and in vitro
In addition to the DBTRG-05MG-R cell line, a
TMZ-resistant U-87 MG cell line was also established (U-87 MG-R).
As shown in Fig. 5A, the U-87 MG-R
cells were highly resistant to TMZ compared with that in the
parental cells. Following TMZ treatment, the protein expression
levels of cleaved caspase-3 and cleaved PARP were significantly
higher in the U-87 MG cells compared with that in the U-87 MG-R
cells, at high doses of TMZ (Fig.
5B). Next, the dose-dependent effects of TMZ on XRCC5 protein
expression level in the parental and TMZ-resistant U-87 MG cell
lines and the results showed that XRCC5 expression level, in the
TMZ-resistant cells, continued to be expressed under 250 and 500 µM
TMZ treatments, while the parental cell line exhibited a
significant decrease (Fig. 5C).
A nude mouse xenograft model was also used to
further validate the effects of TMZ on XRCC5 expression level. As
shown in Fig. 5D, TMZ effectively
inhibited the tumor growth of the U-87 MG cells, but not the U-87
MG-R cells. The size of the subcutaneous tumor was also compared at
the end of the experiment, as shown in Fig. 5E and it was found that the mean tumor
weight of tumors derived from the U-87 MG-R cells was significantly
higher compared with that in those derived from the U-87 MG cell
tumors (Fig. 5F). Subsequently, the
tumor sections were stained with XRCC5 and the results showed that
TMZ could markedly induce the expression of XRCC5 in the xenograft
tumors derived from U-87 MG-R cells (Fig. 5G). These results indicated the
protective role of XRCC5 against TMZ treatment and may cause GBM
cells to develop drug resistance.
Discussion
Collectively, functional screening using a shRNA
library showed that XRCC5 was effective at increasing the
cytotoxicity of GBM cells to TMZ. The association between XRCC5
mRNA expression level and clinical characteristics was subsequently
analyzed using bioinformatics, and it was found that the expression
level of XRCC5 in GMB specimens was higher compared with that in
low grade glioma and normal brain tissue. In addition, high XRCC5
expression was also associated with lower overall survival. To
verify the effect of XRCC5 on TMZ sensitivity, lentiviral shRNA was
used to silence XRCC5, while pCMV was used to overexpress it and
the effects on TMZ sensitivity were subsequently investigated. The
results revealed that XRCC5 could increase cancer cell resistance
to TMZ. In addition, TMZ-resistant cell lines were established and
used in in vitro and in vivo experiments to further
determine whether XRCC5 could be induced in the TMZ-resistant cells
following TMZ treatment. In the U-87 MG and U-87 MG-R cell lines,
the protein expression level of XRCC5 in the resistant cells was
increased following TMZ treatment (at 125 µM), while it was
decreased, from 250 µM, in the sensitive cells. Both the DBTRG-05MG
and DBTRG-05MG-R cells had a significant increase in XRCC5 protein
expression level following TMZ treatment. This could be the result
of DBTRG-05MG itself being a relatively resistant cell line
initially (37).
Eukaryotic cells depend on two major mechanisms for
DNA: Repair homologous recombination (HR) or non-homologous end
joining (NHEJ) (38). As DSB is
important in TMZ-induced cell death, repairing DSB by HR and NHEJ
could reduce the effect on the TMZ-triggered killing response. In
GBM, DNA repair is highly variable, which has strong effects on TMZ
resistance; therefore, DNA repair is an ideal choice for
personalized treatment (39).
Previous studies have shown that augmented HR repair is an
important mechanism underlying acquired TMZ resistance in GBM
(13). Once the components of HR,
such as RAD51 (40), XRCC3 (11), BRCA1 (41), BRCC3 (42) and BRCA2 (43), were suppressed, glioma cells would be
sensitive to TMZ. Similarly, numerous studies have also shown that
some components of NHEJ, including XLF, 53BP1 (44), XRCC4 (45), DNA-PKcs (46) and DNA ligase IV (47), if they were inhibited, could enhance
the therapeutic effect of TMZ.
XRCC5 is an important molecule in the NHEJ process
(48). A previous study has shown
that overexpression of XRCC5 in the NIH3T3 cell line derived from
mouse embryonic fibroblasts could protect cells against γ-ray
irradiation, leading to radioresistance (23). Similarly, in head and neck cancer,
the protein expression of XRCC5 was significantly increased in
radioresistant cells (24).
Radiotherapy combined with cisplatin chemotherapy is the primary
therapeutic strategy for cervical cancer and simultaneous
inhibition of XRCC5 could increase sensitization in cervical cancer
cells (27). In clinical lung
adenocarcinoma specimens, chemoresistant tumors exhibited higher
protein expression levels of XRCC5; therefore, XRCC5 could predict
the responsiveness and prognosis in patients with lung cancer
(25). Furthermore, XRCC5
overexpression was found to increase the resistance to chemotherapy
drugs, while XRCC5 knockdown augmented drug sensitivity in lung
cancer cells (26). In thyroid
carcinoma, XRCC5 protein expression levels in carcinoma tissues
were significantly higher compared with that in non-neoplastic
adjacent tissue and XRCC5 knockdown decreased malignancy in thyroid
cancer cells (29). In
chondrosarcoma, doxorubicin-resistant cells showed a dose-dependent
increase in XRCC5 protein expression level following doxorubicin
treatment, and silencing XRCC5 expression increased drug
sensitivity in resistant cells (28).
Currently; however, there has been no study on the
function of XRCC5 with respect to drug resistance in GBM, although
one report showed that the DNA methylation level of XRCC5 in blood
samples from patients with glioma was significantly higher compared
with that in healthy individuals. In addition, XRCC5 methylation
levels were significantly higher in patients treated with
radiotherapy and chemotherapy compared with that in patients who
were not treated (49). These
studies indicated that the methylation level of XRCC5 in blood may
be a potential indicator for evaluating the efficacy of
radiotherapy and chemotherapy in patients with glioma. In a Han
Chinese population, astrocytoma, another type of glioma, the XRCC5
genotype (SNP: rs9288516) was associated with increased risk of the
disease and poor prognosis (50).
In conclusion, to the best of our knowledge, this is
the first study to reveal the role of XRCC5 in TMZ resistance of
GBM. An array-based shRNA library was used to inactivate KP gene
activity and screen for genes, which leads to GBM cells becoming
more sensitive to TMZ treatment. Of these genes, the knockdown of
XRCC5 markedly sensitized GBM cells to TMZ-induced cell death. In
addition, the clinical relevance of XRCC5 was analyzed using the
cBioportal, Oncomine and R2 databases. The results showed that
XRCC5 mRNA expression level was increased in GBM tissues and was
associated with poor prognosis. In addition, in vitro and
in vivo analyses revealed that XRCC5 could play a role in
the protection against TMZ, suggesting that XRCC5 could be an
effective target for the development of novel chemotherapy for
treating drug resistant cancer cells.
Acknowledgements
Not applicable.
Funding
This study was supported by The Chang Gung Medical
Research Council (grant nos. CMRPG6F0351, CMRPG6F0352 and
CMRPG6F0353).
Avaliability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request. The datasets generated and/or analyzed during the current
study are available in the cBioPortal (https://www.cbioportal.org/), Oncomine (https://www.oncomine.org/) R2 Genomics Analysis and
Visualization Platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi).
Author's contributions
INL, JTY designed and supervised the experiments,
edited the manuscript and acquired funding. CH, HCH analyzed the
data. INL, YPW performed the experiments. JCC conceived, carried
out experimental work, data analysis, manuscript preparation and
editing. INL and JCC confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Cioffi G, Gittleman H, Patil N,
Waite K, Kruchko C and Barnholtz-Sloan JS: CBTRUS Statistical
Report: Primary brain and other central nervous system tumors
diagnosed in the United States in 2012–2016. Neuro Oncol. 21 (Suppl
5):v1–v100. 2019. View Article : Google Scholar
|
|
3
|
Yung WK, Albright RE, Olson J, Fredericks
R, Fink K, Prados MD, Brada M, Spence A, Hohl RJ, Shapiro W, et al:
A phase II study of temozolomide vs. procarbazine in patients with
glioblastoma multiforme at first relapse. Br J Cancer. 83:588–593.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hottinger AF, Ben Aissa A, Espeli V,
Squiban D, Dunkel N, Vargas MI, Hundsberger T, Mach N, Schaller K,
Weber DC, et al: Phase I study of sorafenib combined with radiation
therapy and temozolomide as first-line treatment of high-grade
glioma. Br J Cancer. 110:2655–2661. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fu D, Calvo JA and Samson LD: Balancing
repair and tolerance of DNA damage caused by alkylating agents. Nat
Rev Cancer. 12:104–120. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang J, Stevens MF and Bradshaw TD:
Temozolomide: Mechanisms of action, repair and resistance. Curr Mol
Pharmacol. 5:102–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Quiros S, Roos WP and Kaina B: Processing
of O6-methylguanine into DNA double-strand breaks requires two
rounds of replication whereas apoptosis is also induced in
subsequent cell cycles. Cell Cycle. 9:168–178. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bocangel DB, Finkelstein S, Schold SC,
Bhakat KK, Mitra S and Kokkinakis DM: Multifaceted resistance of
gliomas to temozolomide. Clin Cancer Res. 8:2725–2734.
2002.PubMed/NCBI
|
|
9
|
Fan CH, Liu WL, Cao H, Wen C, Chen L and
Jiang G: O6-methylguanine DNA methyltransferase as a promising
target for the treatment of temozolomide-resistant gliomas. Cell
Death Dis. 4:e8762013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Happold C, Roth P, Wick W, Schmidt N,
Florea AM, Silginer M, Reifenberger G and Weller M: Distinct
molecular mechanisms of acquired resistance to temozolomide in
glioblastoma cells. J Neurochem. 122:444–455. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roos WP, Frohnapfel L, Quiros S, Ringel F
and Kaina B: XRCC3 contributes to temozolomide resistance of
glioblastoma cells by promoting DNA double-strand break repair.
Cancer Lett. 424:119–126. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang B, Fu X, Hao J, Sun J, Li Z, Li H and
Xu H: PAXX Participates in Base Excision Repair via Interacting
with Pol β and Contributes to TMZ Resistance in Glioma Cells. J Mol
Neurosci. 66:214–221. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gil Del Alcazar CR, Todorova PK, Habib AA,
Mukherjee B and Burma S: Augmented HR repair mediates acquired
temozolomide resistance in glioblastoma. Mol Cancer Res.
14:928–940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hata N, Mizoguchi M, Kuga D, Hatae R,
Akagi Y, Sangatsuda Y, Amemiya T, Michiwaki Y, Fujioka Y, Takigawa
K, et al: First-line bevacizumab contributes to survival
improvement in glioblastoma patients complementary to temozolomide.
J Neurooncol. 146:451–458. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Caplen NJ, Parrish S, Imani F, Fire A and
Morgan RA: Specific inhibition of gene expression by small
double-stranded RNAs in invertebrate and vertebrate systems. Proc
Natl Acad Sci USA. 98:9742–9747. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agrawal N, Dasaradhi PV, Mohmmed A,
Malhotra P, Bhatnagar RK and Mukherjee SK: RNA interference:
Biology, mechanism, and applications. Microbiol Mol Biol Rev.
67:657–685. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salm F, Cwiek P, Ghosal A, Lucia
Buccarello A, Largey F, Wotzkow C, Höland K, Styp-Rekowska B,
Djonov V, Zlobec I, et al: RNA interference screening identifies a
novel role for autocrine fibroblast growth factor signaling in
neuroblastoma chemoresistance. Oncogene. 32:3944–3953. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ding Y, Huang D, Zhang Z, Smith J, Petillo
D, Looyenga BD, Feenstra K, Mackeigan JP, Furge KA and The BT:
Combined gene expression profiling and RNAi screening in clear cell
renal cell carcinoma identify PLK1 and other therapeutic kinase
targets. Cancer Res. 71:5225–5234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pannunzio NR, Watanabe G and Lieber MR:
Nonhomologous DNA end-joining for repair of DNA double-strand
breaks. J Biol Chem. 293:10512–10523. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meek K, Dang V and Lees-Miller SP: DNA-PK:
The means to justify the ends? Adv Immunol. 99:33–58. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lieber MR: The mechanism of double-strand
DNA break repair by the nonhomologous DNA end-joining pathway. Annu
Rev Biochem. 79:181–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chang IY, Youn CK, Kim HB, Kim MH, Cho HJ,
Yoon Y, Lee YS, Chung MH and You HJ: Oncogenic H-Ras up-regulates
expression of Ku80 to protect cells from gamma-ray irradiation in
NIH3T3 cells. Cancer Res. 65:6811–6819. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang HW, Kim SY, Yi SL, Son SH, Song DY,
Moon SY, Kim JH, Choi EK, Ahn SD, Shin SS, et al: Expression of
Ku80 correlates with sensitivities to radiation in cancer cell
lines of the head and neck. Oral Oncol. 42:979–986. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma Q, Li P, Xu M, Yin J, Su Z, Li W and
Zhang J: Ku80 is highly expressed in lung adenocarcinoma and
promotes cisplatin resistance. J Exp Clin Cancer Res. 31:992012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shang B, Jia Y, Chen G and Wang Z: Ku80
correlates with neoadjuvant chemotherapy resistance in human lung
adenocarcinoma, but reduces cisplatin/pemetrexed-induced apoptosis
in A549 cells. Respir Res. 18:562017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhuang L, Liu F, Peng P, Xiong H, Qiu H,
Fu X, Xiao Z and Huang X: Effect of Ku80 on the radiosensitization
of cisplatin in the cervical carcinoma cell line HeLa. Oncol Lett.
15:147–154. 2018.PubMed/NCBI
|
|
28
|
Hsieh MJ, Huang C, Lin CC, Tang CH, Lin
CY, Lee IN, Huang HC and Chen JC: Basic fibroblast growth factor
promotes doxorubicin resistance in chondrosarcoma cells by
affecting XRCC5 expression. Mol Carcinog. 59:293–303. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan Y, Li J, Wei W, Fang H, Duan Y, Li N,
Zhang Y, Yu J and Wang J: Ku80 gene knockdown by the CRISPR/Cas9
technique affects the biological functions of human thyroid
carcinoma cells. Oncol Rep. 42:2486–2498. 2019.PubMed/NCBI
|
|
30
|
Torcivia-Rodriguez J, Dingerdissen H,
Chang TC and Mazumder R: A primer for access to repositories of
cancer-related genomic big data. Methods Mol Biol. 1878:1–37. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saha SK, Islam SMR, Kwak KS, Rahman MS and
Cho SG: PROM1 and PROM2 expression differentially modulates
clinical prognosis of cancer: A multiomics analysis. Cancer Gene
Ther. 27:147–167. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang C, Chen E, Peng K, Wang H, Cheng X,
Wang Y, Yu S, Yu Y, Cui Y and Liu T: Mining the role of
angiopoietin-like protein family in gastric cancer and seeking
potential therapeutic targets by integrative bioinformatics
analysis. Cancer Med. 9:4850–4863. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Osuka S and Van Meir EG: Overcoming
therapeutic resistance in glioblastoma: The way forward. J Clin
Invest. 127:415–426. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shergalis A, Bankhead A III, Luesakul U,
Muangsin N and Neamati N: Current challenges and opportunities in
treating glioblastoma. Pharmacol Rev. 70:412–445. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gusev Y, Bhuvaneshwar K, Song L, Zenklusen
JC, Fine H and Madhavan S: The REMBRANDT study, a large collection
of genomic data from brain cancer patients. Sci Data. 5:1801582018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen JC, Lee IN, Huang C, Wu YP, Chung CY,
Lee MH, Lin MH and Yang JT: Valproic acid-induced amphiregulin
secretion confers resistance to temozolomide treatment in human
glioma cells. BMC Cancer. 19:7562019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chapman JR, Taylor MR and Boulton SJ:
Playing the end game: DNA double-strand break repair pathway
choice. Mol Cell. 47:497–510. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kaina B and Christmann M: DNA repair in
personalized brain cancer therapy with temozolomide and
nitrosoureas. DNA Repair (Amst). 78:128–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Quiros S, Roos WP and Kaina B: Rad51 and
BRCA2-New molecular targets for sensitizing glioma cells to
alkylating anticancer drugs. PLoS One. 6:e271832011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding J, Wu S, Zhang C, Garyali A,
Martinez-Ledesma E, Gao F, Pokkulandra A, Li X, Bristow C, Carugo
A, et al: BRCA1 identified as a modulator of temozolomide
resistance in P53 wild-type GBM using a high-throughput shRNA-based
synthetic lethality screening. Am J Cancer Res. 9:2428–2441.
2019.PubMed/NCBI
|
|
42
|
Chai KM, Wang CY, Liaw HJ, Fang KM, Yang
CS and Tzeng SF: Downregulation of BRCA1-BRCA2-containing complex
subunit 3 sensitizes glioma cells to temozolomide. Oncotarget.
5:10901–10915. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kondo N, Takahashi A, Mori E, Noda T,
Zdzienicka MZ, Thompson LH, Helleday T, Suzuki M, Kinashi Y,
Masunaga S, et al: FANCD1/BRCA2 plays predominant role in the
repair of DNA damage induced by ACNU or TMZ. PLoS One.
6:e196592011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang T, Chai J and Chi L: Induction Of
XLF And 53BP1 expression is associated with temozolomide resistance
in glioblastoma cells. Onco Targets Ther. 12:10139–10151. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zeng A, Wei Z, Yan W, Yin J, Huang X, Zhou
X, Li R, Shen F, Wu W, Wang X and You Y: Exosomal transfer of
miR-151a enhances chemosensitivity to temozolomide in
drug-resistant glioblastoma. Cancer Lett. 436:10–21. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Roos WP, Batista LF, Naumann SC, Wick W,
Weller M, Menck CF and Kaina B: Apoptosis in malignant glioma cells
triggered by the temozolomide-induced DNA lesion O6-methylguanine.
Oncogene. 26:186–197. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kondo N, Takahashi A, Mori E, Ohnishi K,
McKinnon PJ, Sakaki T, Nakase H and Ohnishi T: DNA ligase IV as a
new molecular target for temozolomide. Biochem Biophys Res Commun.
387:656–660. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Meek K, Gupta S, Ramsden DA and
Lees-Miller SP: The DNA-dependent protein kinase: The director at
the end. Immunol Rev. 200:132–141. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou C, Tang H, Yu J, Zhuang D and Zhang
H: Blood-based DNA methylation of DNA repair genes in the
non-homologous end-joining (NEHJ) pathway in patient with glioma.
Int J Clin Exp Pathol. 8:9463–9467. 2015.PubMed/NCBI
|
|
50
|
He X, Zhu X, Li L, Zhang J, Wu R, Zhang Y,
Kang L, Yuan D and Jin T: The relationship between polymorphisms of
XRCC5 genes with astrocytoma prognosis in the Han Chinese
population. Oncotarget. 7:85283–85290. 2016. View Article : Google Scholar : PubMed/NCBI
|