Introduction
Lung cancer (LC) is one of the most common causes of
death in the world and is the cause of 20% of cancer-associated
deaths (1). The incidence of LC in
Europe is >410,000 cases/year (1–3).
Occurrence of LC is in second place (>200,000 cases/year in
Europe) in men after prostate cancer and third in women (~100,000
cases/year) after breast and colorectal cancer (2). Non-small cell LC (NSCLC) is the most
frequent histopathological type of LC (~85% of all LC cases)
(3). At present, the most effective
therapeutic option for NSCLC seems to be immunotherapy targeting
immune checkpoints (4). One of these
checkpoints involves programmed death-ligand 1 (PD-L1); PD-L1
expression on neoplastic cells allows them to escape from immune
surveillance by interacting with programmed death-1 (PD-1) located
on T cells (4,5). Immunotherapies using anti-PD-1 or
anti-PD-L1 monoclonal antibodies block the PD-1/PD-L1 pathway and
make cancer cells visible to the immune system, which can now
recognize and destroy them (4,5).
Suitability for immunotherapy is based on the evaluation of protein
expression on tumor cells using immunohistochemical methods
(4–7).
Examination of PD-L1 expression using
immunohistochemistry (IHC) serves an important role in the
qualification of patients with NSCLC for first line immunotherapy
(6–10). Patients with PD-L1 expression on ≥50%
of tumor cells may be treated with first line pembrolizumab therapy
(6,7). Currently, monoclonal antibodies clone
22C3 (DAKO) or clone SP263 (Ventana) are recommended for assessing
PD-L1 expression on tumor cells (TCs) (8). However, five different antibody clones
(SP142, SP263, 22C3, 28-8, 73-10) and IHC tests were used for
assessment of PD-L1 expression in clinical trials with different
anti-PD-1 or anti-PD-L1 immune-check points inhibitors (such as
nivolumab, pembrolizumab and durvalumab) (9–12). These
resulted in divergent evaluation of PD-L1 expression on tumor and
immune cells (9,10). From a technical point of view, the
differences in IHC tests results are due to the different affinity
strength of particular antibodies and due to their different
binding site in PD-L1 molecule (extracellular or intracellular
domain of PD-L1 protein) (11,12). The
expression of PD-L1 protein on cancer cells may also depend on
PD-L1 expression, single nucleotide polymorphisms (SNPs) or
the number of PD-L1 copies [copy number variation
(CNV)].
There are several SNPs of PD-L1 described in
literature, such as rs2297136, rs4143815, rs4143815, rs822336 or
rs822337 (13–22). They have different locations in the
gene (introns, exons, promoters), however majority of them are
located in 3′-untranslated region (3-UTR) and are closely related
to posttranscriptional regulation of gene expression (related to
microRNA activity) (15–22). There is some evidence that
PD-L1 polymorphisms may be independent predictive biomarkers
in patients with NSCLC receiving first-line chemotherapy with
paclitaxel and cisplatin (15).
Studies on PD-L1 polymorphisms are conducted mainly in Asian
populations (15,16). The promoter region is involved in the
initial regulation of gene expression (transcription). Specific
transcription factors, recognizing the promoter sequence, attach
and recruit other proteins, including polymerases involved in
transcription initiation. Therefore, the sequence of promoter
region is crucial for the attachment of proteins, and polymorphisms
of the promoter region may affect the binding strength of
transcription complex, which may change PD-L1 expression (23).
The CNV of different genes is another genetic factor
affecting protein expression. Some studies have indicated that the
number of PD-L1 copies detected by fluorescence
in-situ hybridization (FISH) or quantitative (q)PCR is
associated with PD-L1 protein expression detected using IHC, which
is the only method approved in daily practice to assess PD-L1
expression in qualifying patients for immunotherapy (24–29).
qPCR is a simpler method compared with FISH for assessing
PD-L1 copies number. However, correlation between PD-L1
protein expression and PD-L1 copies number has not always
been statistically significant using these two methods (26,27).
Currently, IHC for assessing PD-L1 expression on TCs
is the gold standard in qualification of patients with NSCLC to
first-line immunotherapy (11,14,24).
However, anti-PD-1 or anti-PD-L1 blockade may be ineffective in
some patients with NSCLC with high percentage of TCs with PD-L1
expression (10,11,25). In
addition, immunotherapy may provide great benefits in patients
without expression of this protein (10,11,25).
Moreover, to the best of our knowledge, there are no clinical
trials comparing the efficacy of the first-line of immunotherapy
used in patients with a high percentage of PD-L1-positive TCs to
the second-line of immunotherapy used after first-line
chemotherapy, regardless of the status of PD-L1 expression on TCs.
It may be that both methods of treatments have similar effect on
overall survival (OS) of patients with NSCLC (10,11,14,24,25).
This may suggest that PD-L1 expression on TCs is a useful marker in
qualification of patients to first-line immunotherapy, but may not
be predictive of OS in patients treated with first- or second-line
immunotherapy. Hence, the present study investigated the molecular
background of PD-L1 expression on TCs in a large group of Caucasian
patients with NSCLC and aimed to identify new, potential predictive
factors for immunotherapy. In addition, the present study assessed
the effect of selected predictive factors and the type of treatment
[best supportive care (BSC), 1st, and 2nd line of immunotherapy] on
OS in patients with NSCLC.
Materials and methods
Studied group
A total of 673 locally advanced or advanced patients
with NSCLC without epidermal growth factor receptor (EGFR)
gene mutations and anaplastic lymphoma kinase (ALK) gene
rearrangement were enrolled. The present study was a multicenter
study, and materials were collected between March 2017 and February
2019 at the following centers: Independent Public Clinical Hospital
No. 4 (Lublin, Poland), Saint John of Dukla Oncology Centre of the
Lublin Region (Lublin, Poland), Specialist Hospital for Lung
Diseases (Zakopane, Poland), Poland Lord's Transfiguration Clinical
Hospital (Poznań, Poland), Mazovian Center for the Treatment of
Lung Diseases and Tuberculosis (Otwock, Poland), Provincial
Hospital Center of the Jeleniogórska Valley (Jelenia Góra, Poland),
Independent Public Provincial Hospital Pope John Paul II (Zamość,
Poland), Independent Public Group of Tuberculosis and Pulmonary
Diseases (Olsztyn, Poland) and Regional Hospital for Lung Diseases
(Szczecin-Zdunowo, Poland). The mean age of the patients was 67±7.7
years (age range, 36–88 years). There were 428 (64%) male and 245
(36%) female patients. The patients provided written consent prior
to enrollment in the study. The study was approved by the Ethics
Committee of the Medical University of Lublin (Lublin, Poland)
(approval no. KE-0254/95/2018). Adenocarcinoma (AC) was diagnosed
in 370 patients (55%), squamous cell carcinoma (SqCC) in 266
patients (40%), large cell carcinoma (LCC) in 9 patients (1%) and
NSCLC not otherwise specified (NOS) in 28 patients (4%). Genetic
and IHC assays were performed in formalin-fixed paraffin-embedded
(FFPE) tissue specimens (476; 71% cases) or in cell blocks (197;
29% cases) obtained via routine hospital procedures. FFPE material
was prepared from forceps biopsy carried out during bronchoscopy or
from surgical materials, while cell blocks were prepared mostly
from materials obtained in endobronchial ultrasound transbronchial
needle aspiration (EBUS-TBNA) or endoscopy ultrasound fine-needle
aspiration (EUS-FNA) procedures. 499 samples were primary tumor,
while 174 were lymph node metastases or distant metastases. Data on
methods of treatment and overall survival were available for 662
patients, who were included in the study. In the studied group, 131
(20%) patients received immunotherapy as one of the treatment lines
(as first-line therapy or after chemotherapy) and 489 (73%)
patients only chemotherapy. A total of 42 (6%) patients did not
receive any systemic treatment. Of the immunotherapy group, a total
of 57 (44%) patients received pembrolizumab as first-line therapy
and 74 (56%) patients received atezolizumab or nivolumab in the
second-line of treatment. A total of 44 of the aforementioned
patients had the second-line of chemotherapy after pembrolizumab
failure and 46 patients had the third-line of chemotherapy after
failure of atezolizumab or nivolumab. OS time was calculated from
diagnosis to death or last observation. Median follow-up was 24
months, with follow-up performed on the basis of patients' visits
to the hospital, analysis of data from the medical system and
telephone information.
IHC
Analysis of PD-L1 protein expression was performed
on FFPE or cell block materials cut into 3-µm-thick sections. These
were put on Thermo Scientific Superfrost Plus™ (Thermo Fisher
Scientific, Inc.) glass slides and preheated at 59°C on a hotplate
for at least 3 h prior to staining. The Ventana PD-L1 clone SP263
antibody (1.61 µg/ml; cat. no. 790-4905; Ventana Medical Systems,
Inc.; Roche Diagnostics) was used as a primary antibody for PD-L1
protein IHC staining. IHC was performed using automated Ventana
Benchmark GX equipment (Ventana Medical Systems, Inc.; Roche
Diagnostics) according to the manufacturer's instructions in a
closed, fixed programme. OptiView DAB IHC detection kit (Ventana
Medical Systems, Inc.; Roche Diagnostics) was used as a detection
system. Counterstaining using hematoxylin II (Ventana Medical
Systems, Inc.; Roche Diagnostics), was included in the staining
protocol. Rabbit monoclonal negative control immunoglobulin (10
µg/ml; cat. no. 790-4795; Ventana Medical Systems, Inc.; Roche
Diagnostics) was used as a negative control. After staining all
glass slides were washed and dehydrated in a series of 2 96%
ethanol and 2 ×ylene washing steps and then cover-slipped. Next,
the slides were assessed by 2 pathologists (Medical University of
Lublin, Lublin, and Specialist Hospital for Lung Diseases ‘Rebirth’
Klara Jelska, Zakopane; Poland) using an Olympus BX41 light
microscope. The assessment was based on whole stained slide
analysis, with at least 50 viable TCs available. The percentage of
PD-L1 positive TCs was determined according to the presence or
absence of staining of TCs, under consecutive magnifications (from
×40 to ×400) depending on the intensity of the staining. Comparison
of corresponding haematoxylin and eosin (H&E) and PD-L1 IHC
slides was made to make sure the material had enough TCs to get a
proper diagnosis.
DNA extraction
Genomic DNA (gDNA) was isolated from FFPE and cell
blocks using QIAamp DNA FFPE Tissue kit (Qiagen GmbH) according to
the manufacturer's instructions. The quality and quantity of DNA
was assessed using BioPhotometer UV/Vis Spectrophotometer
(Eppendorf). The quality (ratio 260/280) of the isolates was
1.7–2.0 and the concentration was in the range 20–192 ng/µl.
qPCR
A total of 2 polymorphisms, rs822335 (C_7590674_10;
cat. no. 4351379; Applied Biosystems; Thermo Fisher Scientific,
Inc.) and rs822336 (C_1348559_10; cat. no. 4351379; Applied
Biosystems; Thermo Fisher Scientific, Inc.) in the PD-L1
promoter region were studied. In the present study, 2 SNPs of PD-L1
promoter region were selected from the PubMed dbSNP database
(https://www.ncbi.nlm.nih.gov/snp/).
PCR mixture contained: 5.5 µl of Genotyping MasterMix (Applied
Biosystems; Thermo Fisher Scientific, Inc.), 4 µl of aforementioned
gDNA (5 ng/µl) and 0.5 µl of TaqMan SNP Assay (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The following conditions were used
for denaturation and enzyme activation: 95°C for 10 min, and 40
cycles of 95°C for 15 sec and 62°C for 60 sec.
CNV of PD-L1 were studied by qPCR using
TaqMan™ Copy Number Assay (Hs03704252_cn; cat. no. 4400291; Applied
Biosystems; Thermo Fisher Scientific, Inc.) and RNazeP was used as
a housekeeping gene (TaqMan™ Copy Number Reference Assay; cat. no.
4403326; Applied Biosystems; Thermo Fisher Scientific, Inc.). CNV
was scored using the 2−ΔΔCq method (28). The thermocycling conditions were as
follows: 10 min initial denaturation at 95°C followed by 40 cycles
of 95°C for 15 sec and 62°C for 90 sec. qPCR was performed on
Illumina Eco Real-Time PCR equipment (Illumina, Inc.). Each qPCR
reaction was performed in duplicate. qPCR results were analyzed
using the EcoStudy-v4.0.4420 software (Illumina, Inc.). Three
copies of PD-L1 were adopted as an exponent of abnormal
amplification.
Statistical analysis
Statistical analysis was carried out using
Statistica 13.1 (TIBCO Software, Inc.) and MedCalc (MedCalc
Software bvba) softwares. The data were presented as the mean ± SE.
Each qPCR reaction was performed in duplicate. The unpaired
Student's t-test was used to assess the differences in means of
PD-L1 copies number and percentage of tumor cells with PD-L1
expression in groups of patients with different SNPs and clinical
factors. Associations between genotypes of PD-L1 and
clinical factors, as well as PD-L1 CNV and protein expression were
examined using the Fisher's exact test. Correlation between
percentage of tumor cells with PD-L1 expression and PD-L1 CNV was
assessed using the Spearman correlation test. The Kaplan-Meier
log-rank method was used to analyze the OS time for patients with
different predictive factors: type of treatment, line of
immunotherapy, percentage of PD-L1-positive tumor cells, CNV and
SNPs of PD-L1. P<0.05 was considered to indicate a
statistically significant difference.
Results
PD-L1 protein expression analysis
In the entire patient group, mean percentage of
tumor cells with PD-L1 expression was 25±30%. The patients were
divided according to PD-L1 expression into 5 groups: i) Patients
without PD-L1 expression (0% of TCs with PD-L1 expression, group
1); ii) patients with any PD-L1 expression (≥1% of PD-L1 positive
tumor cells, group 2); iii) patients with ≥5% of PD-L1 positive TCs
(group 3); iv) patients with ≥10% of PD-L1 positive TCs (group 4);
and v) patients with ≥50% of PD-L1 positive TCs (group 5). Group 1
consisted of 162 (24%) patients; group 2, 511 patients (76%); group
3, 440 patients (65%); group 4, 355 (53%) and group 5, 169 (25%)
patients (Table I). Table I presents the characteristics of
patients from groups with different percentage of tumor cells with
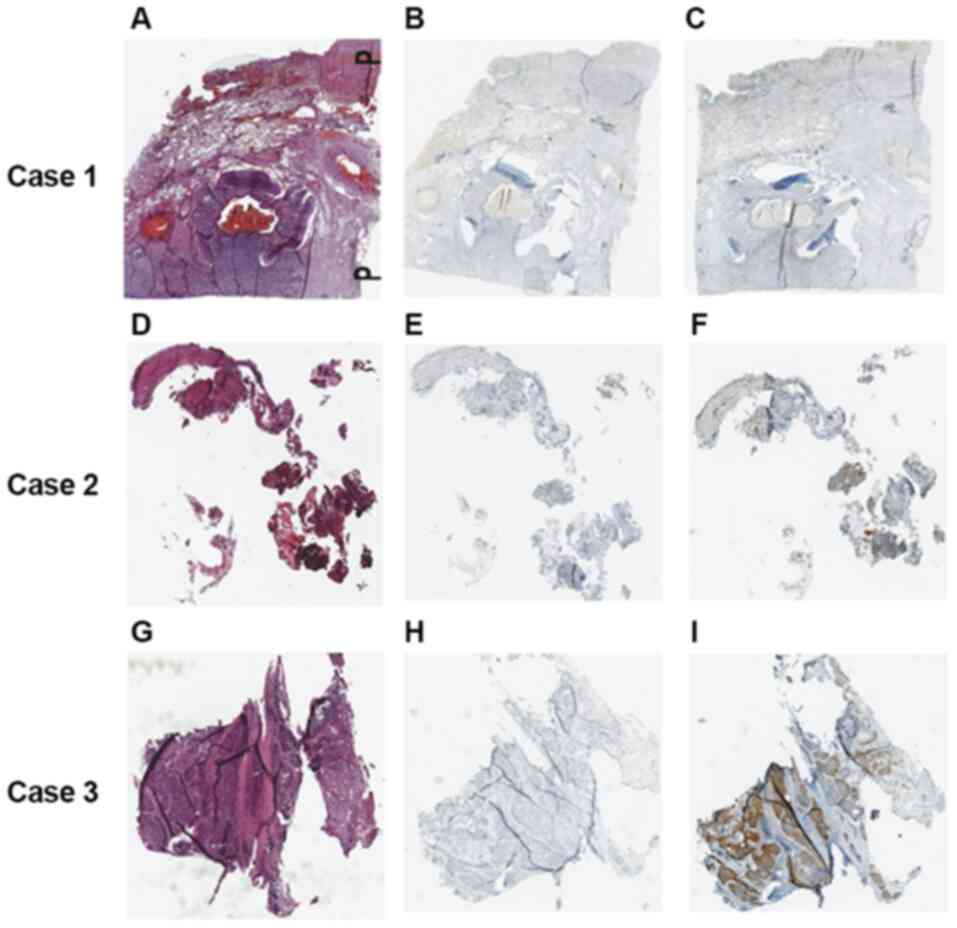
PD-L1 expression. Fig. 1 shows IHC
staining results of single patients from the groups described
above.
 | Table I.Clinicopathological features,
PD-L1 copies number and genotypes of PD-L1 promoter
region in patients with NSCLC with different expression of PD-L1
protein on TCs. |
Table I.
Clinicopathological features,
PD-L1 copies number and genotypes of PD-L1 promoter
region in patients with NSCLC with different expression of PD-L1
protein on TCs.
|
|
Percentage
of TCs with PD-L1 expression, n (%) |
|---|
|
|
|
|---|
| Feature (n, %) | 0% | ≥1% | χ2 | P-value | <5% | ≥5% | χ2 | P-value | <10% | ≥10% | χ2 | P-value | <50% | ≥50% | χ2 | P-value |
|---|
| Total | 162 (24) | 511 (76) | – | – | 233 (35) | 440 (65) | – | – | 318 (47) | 355 (53) | – | – | 504 (75) | 169 (25) | – | – |
| Age, years |
| <67
(330, 49) | 86 (26) | 244 (74) | 1.4 | 0.2 | 116 (35) | 214 (65) | 0.07 | 0.8 | 152 (46) | 178 (54) | 0.4 | 0.5 | 243 (74) | 87 (26) | 0.54 | 0.5 |
| ≥67
(343, 51) | 76 (22) | 267 (78) |
|
| 117 (34) | 226 (66) |
|
| 166 (48) | 177 (52) |
|
| 261 (76) | 82 (24) |
|
|
| Sex |
| Male
(428, 64) | 104 (24) | 324 (76) | 0.03 | 0.9 | 149 (35) | 279 (65) | 0.02 | 0.9 | 207 (48) | 221 (52) | 0.6 | 0.4 | 313 (74) | 116 (26) | 2.6 | 0.1 |
| Female
(245, 36) | 58 (24) | 187 (76) |
|
| 84 (34) | 161 (66) |
|
| 111 (45) | 134 (55) |
|
| 192 (79) | 53 (21) |
|
|
| Histopathological
diagnosis |
|
Adenocarcinoma, NSCLC NOS and
large-cell carcinoma (407, 60) | 121 (30) | 286 (70) | 18.04 | 0.00002 | 148 (36) | 259 (64) | 1.4 | 0.2 | 202 (49) | 205 (51) | 2.3 | 0.1 | 300 (74) | 107 (26) | 0.8 | 0.4 |
|
Squamous-cell carcinoma (266,
40) | 41 (15) | 225 (85) |
|
| 85 (32) | 181 (68) |
|
| 116 (44) | 150 (56) |
|
| 204 (77) | 62 (23) |
|
|
| Material |
| FFPE
(476, 71) | 107 (23) | 369 (77) | 2.3 | 0.1 | 155 (33) | 321 (67) | 3.04 | 0.08 | 213 (45) | 263 (55) | 3.04 | 0.08 | 349 (74) | 127 (26) | 2.4 | 0.122 |
| Cell
blocks (197, 29) | 55 (27) | 142 (73) |
|
| 78 (40) | 119 (60) |
|
| 105 (53) | 92 (47) |
|
| 155 (79) | 42 (21) |
|
|
| Tissue origin |
| Primary tumor (499,
74) | 107 (21) | 392 (79) | 7.3 | 0.007 | 163 (32) | 336 (68) | 3.3 | 0.07 | 226 (45) | 273 (55) | 3.0 | 0.08 | 364 (73) | 135 (27) | 4.5 | 0.03 |
| Lymph node or
distant metastases (174, 26) | 55 (32) | 119 (68) |
|
| 70 (40) | 104 (60) |
|
| 92 (53) | 82 (47) |
|
| 140 (81) | 34 (19) |
|
|
| rs822335 |
| CC
genotype (324, 48) | 63 (20) | 261 (80) | 7.3 | 0.007 | 92 (28) | 232 (72) | 11.0 | 0.0009 | 145 (45) | 179 (55) | 1.8 | 0.2 | 232 (72) | 92 (28) | 3.6 | 0.06 |
| CT+TT
genotype (349, 52) | 99 (28) | 250 (72) |
|
| 142 (40) | 208 (60) |
|
| 173 (50) | 176 (50) |
|
| 272 (78) | 77 (22) |
|
|
| rs822336 |
| CC
genotype (157, 23) | 37 (24) | 120 (76) | 0.03 | 0.9 | 52 (33) | 105 (67) | 0 | 1 | 84 (54) | 73 (46) | 0.3 | 0.6 | 117 (75) | 40 (25) | 0.1 | 0.7 |
| CG+GG
genotype (516, 77) | 125 (24) | 391 (76) |
|
| 171 (33) | 345 (67) | 0 | 1 | 234 (45) | 282 (55) |
|
| 377 (73) | 139 (27) | 0.131 |
|
| Number of
PD-L1 copies |
| <3
(571, 85) | 144 (25) | 427 (75) | 2.7 | 0.1 | 202 (35) | 369 (65) | 0.9 | 0.3 | 273 (48) | 298 (52) | 0.3 | 0.6 | 405 (71) | 147 (29) | 1.6 | 0.2 |
| ≥3
(102, 15) | 18 (18) | 84 (82) |
|
| 31 (30) | 71 (70) |
|
| 46 (45) | 56 (55) |
|
| 81 (79) | 21 (21) |
|
|
A significantly higher percentage of patients with
any expression of PD-L1 on TCs was observed in the SqCC group
compared with the AC group (χ2=18.039; P=0.0002;
Table I). In addition, it was also
observed that the presence of PD-L1 expression on ≥10% of TCs was
significantly more frequent in FFPE materials compared to
cellblocks (χ2=4.088; P=0.043; Table I). Tissue from primary tumors (most
commonly obtained by forceps biopsy or surgical resection) had
significantly higher expression of PD-L1 on ≥1% and ≥50% of TCs
compared with materials obtained from lymph node metastases (most
commonly obtained by EBUS-TBNA, EUS-FNA or other fine-needle
aspiration procedures) (χ2=7.296; P=0.007 and
χ2=4.506; P=0.034, respectively; Table I). In conclusion, the aforementioned
results demonstrated that histological materials are more relevant
for the analysis of PD-L1 expression compared with materials from
EBUS-TBNA or EUS-FNA.
CNV of PD-L1
The present study found that in 571 (85%) cases,
PD-L1 copies number was <3 copies and in 102 (15%) cases
it was ≥3. Tables I and II were generated to characterize groups of
patients with different PD-L1 copies number. It was
demonstrated that PD-L1 amplification was more common in
patients with SqCC compared with patients with other types of NSCLC
(χ2=5.51; P=0.02; Table
II). In addition, significantly higher number of PD-L1
copies in were demonstrated in SqCC compared with AC, NSCLC NOS, or
LCC (P<0.01; Fig. 2E).
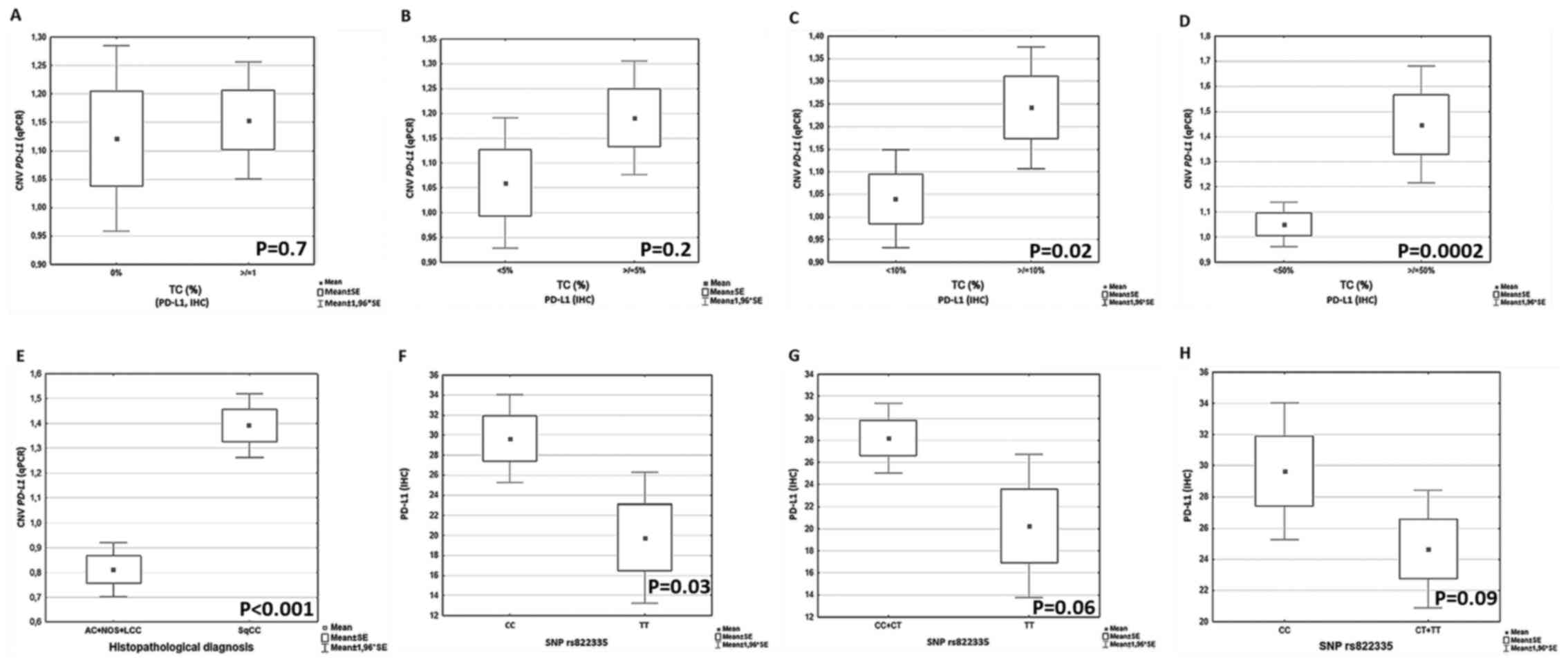 | Figure 2.Analysis of the relationship between
PD-L1 copies number and percentage of TCs expressing PD-L1
protein. Relationship between CNV and percentage of TCs with PD-L1
expression: (A) 0 vs. ≥1%, (B) <5 vs. ≥5%, (C) <10 vs. ≥10%
and (D) <50 vs. ≥50% TCs with PD-L1 expression. (E) Relationship
between CNV and histopathological diagnosis. Relationship between
TCs with PD-L1 expression and allelic variants of rs822335: (F) CC
vs. TT, (G) CC+CT vs. TT and (H) CC vs. CT+TT. PD-L1, programmed
death-ligand 1; TCs, tumor cells; CNV, copy number variant; SNP,
single nucleotide polymorphism; IHC, immunohistochemistry; q,
quantitative; AC, adenocarcinoma; NOS, not otherwise specified;
LCC, large cell carcinoma; SqCC, squamous cell carcinoma. |
 | Table II.Clinicopathological and demographic
features of patients according to PD-L1 copy number
variant. |
Table II.
Clinicopathological and demographic
features of patients according to PD-L1 copy number
variant.
| Feature (n, %) | <3 copies, n
(%) | ≥3 copies, n
(%) | χ2 | P-value |
|---|
| Total | 571 (85) | 102 (15) | – | – |
| Sex |
| Male
(428, 64) | 360 (81) | 68 (19) | 0.49 | 0.48 |
| Female
(245, 36) | 211 (86) | 34 (14) |
|
|
| Histopathological
diagnosis |
|
Adenocarcinoma, NSCLC NOS and
large-cell carcinoma (407, 60) | 356 (87) | 51 (13) | 5.51 | 0.02 |
|
Squamous cell carcinoma (266,
40) | 215 (81) | 51 (19) |
|
|
| Tissue origin |
| Primary
tumor (499, 74) | 424 (85) | 75 (15) | 0.29 | 0.59 |
| Lymph
node or distant metastases (174, 26) | 147 (84) | 27 (16) |
|
|
| Material |
| FFPE
(476, 71) | 405 (85) | 71 (15) | 0.07 | 0.79 |
| Cell
blocks (197, 29) | 166 (84) | 31 (16) |
|
|
A significant positive correlation was found between
PD-L1 copies number and percentage of TCs with PD-L1 protein
expression (R=0.2; P=0.01; data not shown). No significant
differences in number of PD-L1 copies between groups of
patients with 0% and any percentage of tumor cells with PD-L1
expression were found (P=0.7; Fig.
2A), as well as between groups of patients with <5% and ≥5%
of tumor cells with PD-L1 expression (P=0.2; Fig. 2B). However, a significantly higher
number of PD-L1 copies were observed in groups of patients
with ≥10% and ≥50% of PD-L1 positive cancer cells compared with
groups of patients with <10% and <50% of tumor cells with
PD-L1 expression (P=0.02 and P=0.0002; Fig. 2C and D, respectively). Differences in
the analyzed parameters are also shown in Table I.
SNPs of PDL-1 gene promoter
region
Tables I, III and IV
represent the genotype distribution in the two analyzed SNPs in the
studied group. The percentage of TCs with PD-L1 protein expression
was significantly lower in group of patients with TT genotype in
rs822335 compared with the CC genotype (P=0.03; Fig. 2F). A trend of lower percentage of TCs
with PD-L1 expression in group of patients with CT and TT genotypes
were observed compared with patients with CC genotype in rs822335
(P=0.09; Fig. 2H). Similarly, a
slightly lower percentage of PD-L1 TCs was observed in carriers of
TT genotype compared with patients with CC and CT genotypes
rs822335 (P=0.06; Fig. 2G).
 | Table III.Clinicopathological and demographic
features of patients according to rs822335 genotype. |
Table III.
Clinicopathological and demographic
features of patients according to rs822335 genotype.
| Feature (n, %) | CC, n (%) | CT, n (%) | TT, n (%) | χ2 | P-value |
|---|
| Total | 324 (48) | 255 (38) | 94 (14) | – | – |
| Sex |
| Male
(428, 64) | 213 (50) | 153 (36) | 62 (14) | 2.29 | 0.32 |
| Female
(245, 36) | 111 (45) | 102 (42) | 32 (13) |
|
|
| Histopathological
diagnosis |
|
Adenocarcinoma, NSCLC NOS and
large-cell carcinoma (407, 60) | 184 (45) | 165 (41) | 58 (14) | 4.34 | 0.11 |
|
Squamous-cell carcinoma (266,
40) | 140 (53) | 90 (34) | 36 (14) |
|
|
| Tissue origin |
| Primary
tumor (499, 74) | 249 (50) | 184 (37) | 66 (13) | 2.52 | 0.28 |
| Lymph
node or distant metastases (174, 26) | 75 (43) | 71 (41) | 28 (16) |
|
|
| Material |
| FFPE
(476, 71) | 243 (51) | 171 (36) | 62 (13) | 5.48 | 0.06 |
| Cell
blocks (197, 29) | 81 (41) | 84 (43) | 32 (16) |
|
|
 | Table IV.Clinicopathological and demographic
features of patients according to rs822336 genotype. |
Table IV.
Clinicopathological and demographic
features of patients according to rs822336 genotype.
| Feature (n, %) | CC, n (%) | CG, n (%) | GG, n (%) | χ2 | P-value |
|---|
| Total | 157 (24) | 258 (38) | 258 (38) | – | – |
| Sex |
| Male
(428, 64) | 89 (20) | 178 (42) | 161 (38) | 6.64 | 0.04 |
| Female
(245, 36) | 68 (28) | 80 (33) | 97 (39) |
|
|
| Histopathological
diagnosis |
|
Adenocarcinoma, NSCLC NOS and
large-cell carcinoma (407, 60) | 101 (25) | 157 (38) | 149 (37) | 1.79 | 0.41 |
|
Squamous-cell carcinoma (266,
40) | 56 (21) | 101 (38) | 109 (41) |
|
|
| Tissue origin |
| Primary
tumor (499, 74) | 118 (23) | 193 (39) | 188 (38) | 0.36 | 0.83 |
| Lymph
node or distant metastases (174, 26) | 39 (22) | 65 (37) | 70 (41) |
|
|
| Material |
| FFPE
(476, 71) | 110 (23) | 186 (39) | 180 (38) | 0.38 | 0.83 |
| Cell
blocks (197, 29) | 47 (24) | 72 (36) | 78 (40) |
|
|
OS analysis
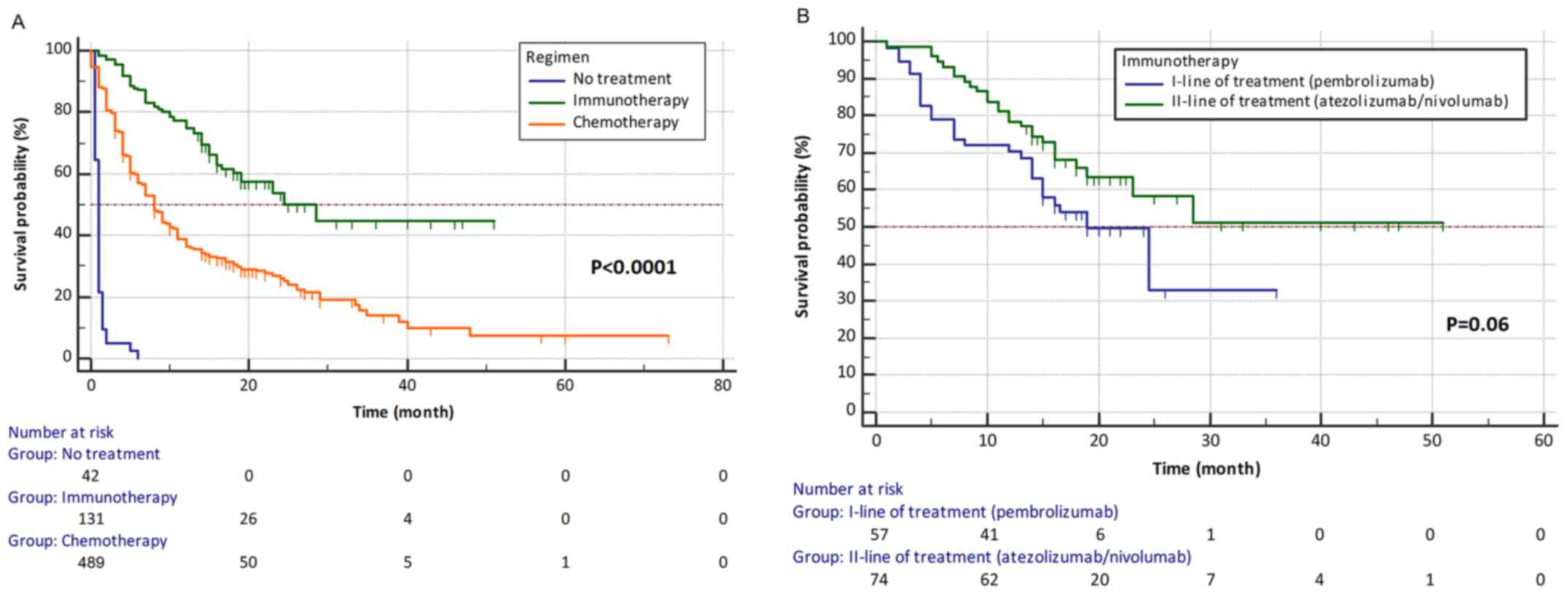
Median OS for the whole studied group was 9 months.
Significantly higher risk of death was observed in patients treated
with chemotherapy (median OS, 8 months) compared with those treated
with any line immunotherapy (median OS, 28 months) [P<0.0001;
hazard ratio (HR)=2.4768; 95% confidence interval (CI),
2.0120–3.0490; Fig. 3A]. In
addition, a slightly greater risk of death in patients receiving
first-line immunotherapy (median OS, 19 months) compared with those
treated with second-line immunotherapy (median OS, 28 months) was
observed (P=0.06; HR=1.6834; 95% CI, 0.9697–2.9222; Fig. 3B).
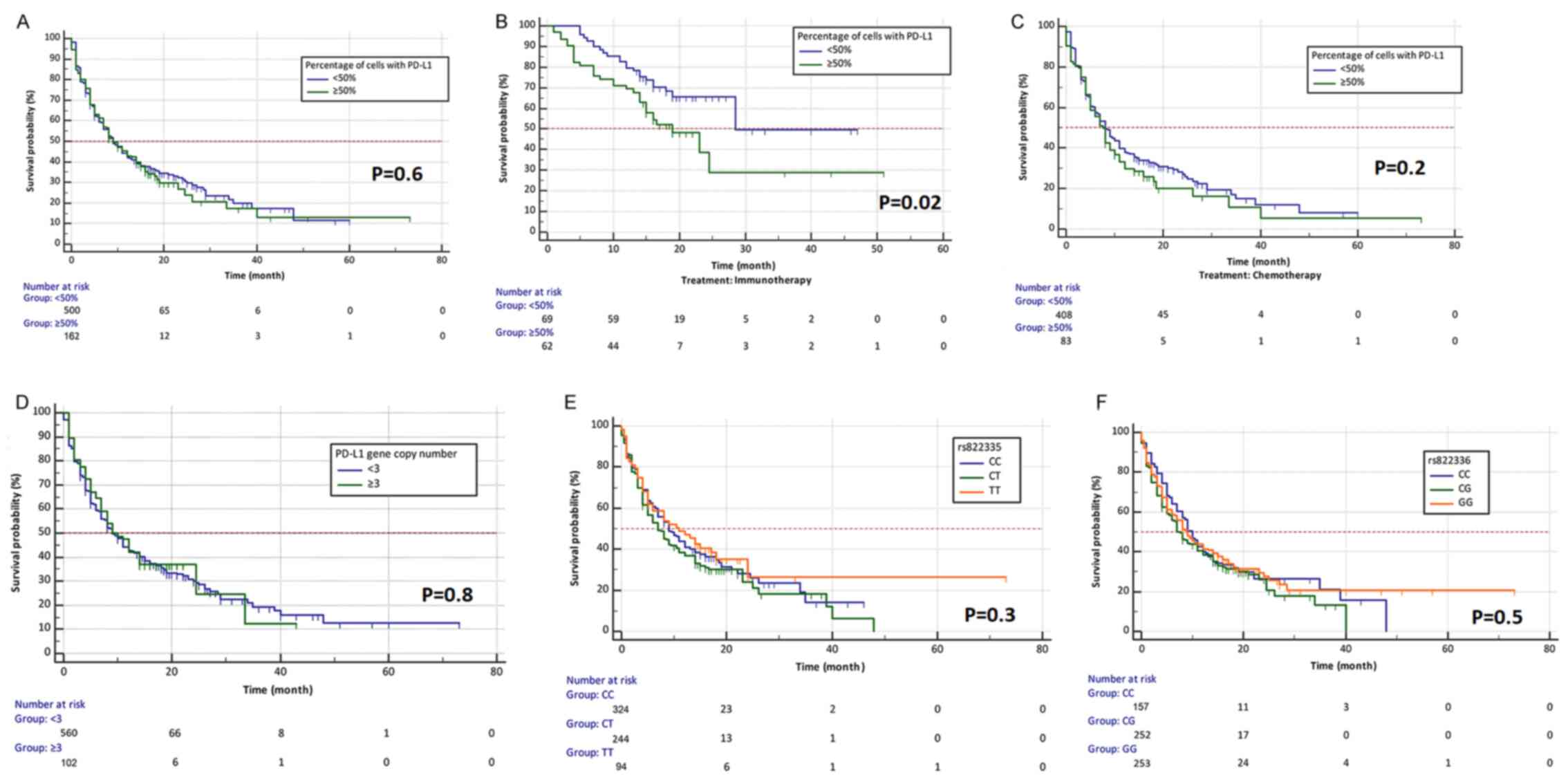
There were no statistically significant differences
in median OS between patients who had below and above 50% of PD-L1
positive TCs (median OS, 9 months; P=0.6; Fig. 4A). However, in the group of patients
treated with immunotherapy, patients with ≥50% PD-L1 positive tumor
cells had greater risk of death compared with patients with <50%
tumor cells with PD-L1 expression (P=0.02; HR=1.9299, 95% CI,
1.1221–3.3192; Fig. 4B). This
relationship was probably due to the surprisingly greater
effectiveness of second-line immunotherapy compared with first-line
immunotherapy (only patients with ≥50% of tumor cells with
expression of PD-L1 could receive pembrolizumab in the first-line
therapy). There were no differences in median OS depending on
percentage of TCs with PD-L1 expression in group of patients
treated with chemotherapy (P=0.2; Fig.
4C). No significant differences in risk of death in groups of
patients with PD-L1 copies number above and below 3 were
observed (P=0.8; Fig. 4D). No
relationship was found between CNV of PD-L1 and risk of
death in both groups of patients receiving chemotherapy or
immunotherapy. It was demonstrated that patients with the TT
genotype in rs822335 polymorphism had only slightly higher median
OS compared to patients with CC and CT genotypes (P=0.3; Fig. 4E). This was particularly evident in
patients receiving immunotherapy (P=0.1). Patients with different
genotypes in rs822336 polymorphism treated with chemotherapy or
immunotherapy had a similar risk of death (P=0.5; Fig. 4F).
Discussion
Differences in DNA of cancer cells, such as SNPs and
CNV, affect PD-L1 expression on tumor cells (15,16,27,29,30). In
the present study, 2 SNPs of PD-L1 promoter region were
selected from the PubMed dbSNP database (31). The results of the present study
indicated that the rs822335 polymorphism may be a predictor for
PD-L1 protein expression on TCs in Caucasian patients with NSCLC.
In the present study, the presence of T allele in rs822335
polymorphism of PD-L1 promoter was related to a lower
percentage of TCs with PD-L1 protein expression, which confirmed
our earlier on a smaller group of patients (29). In this previous study in 47 patients
with NSCLC, CC genotype in rs822335 predisposed to significantly
higher percentage of TCs with PD-L1 expression tested with 22C3
antibody (29). In the present
study, similar results were obtained with the use of the clone
SP263 antibody. In addition, the present study to the best of our
knowledge is one of the largest conducted in the world on this
topic (673 patients). Most of the existing studies about SNPs in
PD-L1 have been conducted in Asian populations (15–17). In
addition, a significant part of studied polymorphisms is located in
the 3′UTR of PD-L1 (16).
Large studies of the impact of PD-L1
polymorphisms on PD-L1 protein expression and on prognosis in
patients with NSCLC have been conducted by Ma et al
(16) and Lee et al (17). Ma et al (16) analyzed SNPs in genes coding the
immune-check point proteins [Cytotoxic T-Lymphocyte Associated
Protein 4 (CTLA-4), PD-1 and PD-L1] in 528 patients with NSCLC and
in 600 healthy people. Using PCR-RFLP (restriction fragments length
polymorphism) method, the aforementioned study demonstrated that
the distribution of polymorphic variants of CTLA-4 and
PD-1 genes in the group of patients with NSCLC and healthy
volunteers was similar (16).
However, the incidence of AC genotype and the presence of C allele
in rs2890658 polymorphism of PD-L1 was significantly higher
in patients with NSCLC compared with healthy subjects (16). In addition, the C allele was more
common in smokers compared with non-smokers (16). Further analysis indicated that this
allele increased the risk of lymph node metastases (16). Studies suggested that the CC genotype
in rs2890658 polymorphism may have a significant impact on the
development and progression of NSCLC (16,20,22).
In 2017, Lee et al (17) presented an analysis of SNPs of
PD-L1 in 354 patients with early stage NSCLC. The occurrence
of different alleles in rs822336, rs822337 and rs4143815 (the first
2 SNPs are located in the promoter and the third in the 3′UTR of
PD-L1) affected OS of patients NSCLC (11). In addition, they demonstrated that
low mRNA expression of PD-L1 was a negative prognostic
factor for patients with NSCLC (17). The luciferase test in the
aforementioned study demonstrated that the presence of the G allele
in rs4143815 provided lower transcription activity of PD-L1
compared with C nucleotide (17). In
addition, the presence of C allele in rs822336 and A allele in
rs822337 decreased promoter activity compared with the presence of
G allele in rs822336 and T allele in rs822337 polymorphism of
PD-L1 (17). However, no
significant correlation was observed between the relative mRNA
expression (measured by qPCR) for PD-L1 and the rs4143815,
rs822336 and rs822337 polymorphisms (17). The results presented by Lee et
al (17) are consistent with the
results of the present study, which demonstrated that the rs822336
polymorphism had no effect on PD-L1 expression on TCs.
In 2016, Lee et al (15) published a study on the relationship
between 12 SNPs of PD-1, PD-L1 and CTLA-4, and
response to first-line chemotherapy (paclitaxel and cisplatin), and
OS in 379 patients with advanced NSCLC. The rs2297136 polymorphism
in the 3′UTR of PD-L1 was significantly associated with
response to chemotherapy and median OS. In addition, rs4143815 in
the 3′UTR of the same gene was significantly associated with
response to chemotherapy (15). The
authors concluded that these SNPs may affect the expression of
PD-L1 as a result of modification of miR-324-5p, miR-570-3p
binding site in 3′UTR of mRNA (9).
Du et al (18)
also studied single nucleotide polymorphisms in the miR binding
sites located in the 3′UTR of PD-L1. They demonstrated that
rs2297136 and rs4742098 polymorphisms were associated with risk of
NSCLC, lymph node metastases, tumor invasion, and presence of
distant metastases (18). In
addition, they proved that both SNPs could influence the
development of lung cancer through the changes in action of
miR-296-5p, miR-138 and mRNA expression of PD-L1 (18).
Yeo et al (19) investigated the prognostic value of
rs4143815 (3′UTR), rs822336 (promoter), rs822337 (promoter)
polymorphisms of PD-L1 in 147 patients with NSCLC. Examined
group included 84 AC patients and 63 SqCC patients treated with
chemotherapy (19). The
aforementioned study used the pyrosequencing method for
PD-L1 genotyping. In AC cases, the GG genotype of rs4143815
polymorphism was associated with shorter OS and progression free
survival (PFS) compared with patients with other genotypes of this
SNP. CC genotype in rs4143815 significantly increased PD-L1
expression on TCs detected by the 22C3 antibody (three clones of
antibodies were used: SP142, 22C3 and SP263). No relationship
between the genotype of rs822336 and rs822337 and the expression of
PD-L1 was observed (19). The
results of the present study, regarding the rs822336 polymorphism
presented are consistent with the findings of the aforementioned
study.
To the best of our knowledge, the present study
demonstrated for the first time that patients with TT genotype of
rs822335 polymorphism had slightly higher median OS compared with
patients with CC and CT genotypes. This result is notable and
interesting, as the findings of the present study also demonstrated
that the presence of the TT genotype was associated with lower
percentage of PD-L1 positive TCs compared with the CC variant in
rs822335. The explanation for this phenomenon may be lower
effectiveness of the first-line immunotherapy used in patients with
PD-L1 expression on ≥50% of TC compared with the second-line
immunotherapy used in patients with lower percentage of PD-L1
positive TCs. Differences in effectiveness of first and second-line
immunotherapy could explain why patients with PD-L1 expression in
≥50% of TC had slightly lower median OS compared with patients with
<50% of PD-L1 positive TCs in the present study. However, the
findings of the present study require further investigation.
The higher effectiveness of immunotherapy over
chemotherapy is beyond doubt (7,24). This
fact is confirmed by the results of the present study and the
results of numerous clinical trials (11,13,14,24,25).
However, in clinical trials assessing the effectiveness of
first-line immunotherapy compared with first-line chemotherapy, a
number of patients after termination of chemotherapy received
second-line immunotherapy (crossover). KEYNOTE-024 (7) and KEYNOTE-042 (24) studies compared the effectiveness of
pembrolizumab versus platinum-based chemotherapy in patients with
≥50% or ≥1% of PD-L1 positive tumor cells, respectively. In
KEYNOTE-024, median OS was 30 months for patients treated with
pembrolizumab and 14 months in patients who received chemotherapy
(7). For a subgroup of patients with
≥50% of PD-L1 positive TCs enrolled in KEYNOTE-042 study, there was
also a significant benefit for pembrolizumab compared with
chemotherapy (median OS, 20 vs. 12 months) (24). Percentage of patients receiving a
subsequent PD-1 inhibitor after progression in chemotherapy arm was
65% in KEYNOTE-024 study and 20% in KEYNOTE-042 study (7,24).
The median OS in patients receiving the second-line
of immunotherapy is usually 10–12 months, which is calculated from
the start of the second-line of immunotherapy (7,24,25). The
time from diagnosis to death should be calculated for estimation of
OS of patients treated with chemotherapy followed by immunotherapy.
Second-line immunotherapy usually begins 10–12 months after
chemotherapy failure. Hence, OS calculated from the diagnosis in
patients treated with chemotherapy with subsequent immunotherapy
could reach 20–24 months. This would mean that first and
second-line immunotherapy have comparable efficacy (7,24,25). Of
course, it is unacceptable to use first-line chemotherapy instead
of immunotherapy in patients NSCLC with PD-L1 expression on ≥50% of
tumor cells. The best systemic treatment should be used at the
beginning of treatment. Numerous patients initially treated with
chemotherapy after progression are not able to receive further
treatment lines due to poor performance status. This phenomenon may
also explain the longer median OS in patients treated with
second-line immunotherapy compared with patients receiving
first-line immunotherapy. These two groups of patients may have
completely different clinical characteristics. Patients who receive
two lines of treatment are still in good performance status and the
course of the disease is usually slow. Progression (including
hyperprogression) and deterioration of performance status in
patients treated with pembrolizumab in the first-line of treatment
could prevent the use of second-line chemotherapy. Hence, patients
treated with the first-line pembrolizumab are patients who received
only one treatment line (7,24,25).
The present study demonstrated that PD-L1
copies number was higher in the group of patients with a higher
compared with a lower percentage of TCs with PD-L1 expression
detected by the IHC method. In the present study, there was
significant positive correlation between PD-L1 copies number
and percentage of TCs with PD-L1 expression. In addition,
PD-L1 copies number was higher in SqCC compared with ACC,
NOS and LCC. This was consistent with the fact that PD-L1
expression detected by the IHC method was significantly more
frequently found on TCs in patients with SqCC compared with
patients with other types of NSCLC. However, the present study
demonstrated that CNV of PD-L1 was not a good marker for the
estimation of OS in patients treated with immunotherapy,
chemotherapy or BSC. Yoshimura et al (26) stated that the assessment of
PD-L1 copies number alterations provided more reproducible
results than protein expression examination especially regarding
intratumoral heterogeneity. The aforementioned study was performed
in material from EBUS-TBNA of primary tumors, resected primary
tumors and resected metastases. They used fluorescent in
situ hybridization to score the CNV of PD-L1 and a
positive correlation between PD-L1 copies number and PD-L1
protein expression in corresponding materials was found (26). CNV of PD-L1 was less
heterogeneous compared with protein expression in different parts
of the tumor (26). Similarly, Inoue
et al (27) examined
differences in PD-L1 copies number between primary tumors
and synchronous regional lymph nodes metastases of patients with
NSCLC. The aforementioned study revealed that the analysis of CNV
of PD-L1 was highly consistent and reproducible compared
with PD-L1 expression assessment. Inoue et al (27) also studied PD-L1 CNV by FISH
method in correlation with PD-L1 expression on tumor cells detected
by the IHC method and demonstrated that there is a positive
correlation between PD-L1 CNV and protein expression on TCs.
Both, high PD-L1 copies number and high protein expression
were predictors of poor survival in untreated patients (27). However, they postulated that
increasing PD-L1 copies number may be a feasible,
alternative biomarker for prediction of response to anti-PD-1 or
anti-PD-L1 antibodies (27).
The aforementioned studies used the FISH method for
examination of PD-L1 status in TCs (26,27). The
alternative to FISH method is qPCR method used for CNV of
PD-L1 analysis by Ikeda et al (30) and the present study. Ikeda et
al (30) examined 94 surgically
resected lung cancer samples and found 5 samples (5%) with
PD-L1 amplification. Patients with PD-L1
amplification had poorer outcomes compared with patients with
normal number of PD-L1 (30).
The aforementioned study adopted 3 PD-L1 copies as the
cut-off threshold for recognition of PD-L1 amplification
(30). In the present study,
amplification of PD-L1 (≥3 copies) was observed in 15% of
patients with NSCLC. These discrepancies in percentage of patients
with PD-L1 amplification could result from the large size of
patients' group analyzed in the present study. George et al
(32) studied CNV of different genes
in patients with small cell lung cancer using whole-genome
sequencing and observed amplification of PD-L1, which may
indicate the sensitivity of small cell lung cancer patients to
immune-check point blockade. The aforementioned study included a
particularly important methodological approach, next generation
sequencing and analyzed DNA and mRNA simultaneously (32), which is definitely a good approach
enabling a deeper understanding of molecular background of tumor
escape mechanisms from immune surveillance in the context of the
implementation of immunotherapy in patients with lung cancer.
The present study demonstrated that the expression
of PD-L1 on tumor cells depends on extremely complex genetic
mechanisms. The findings of the present study demonstrated that the
rs822335 polymorphism and CNV of PD-L1 influenced PD-L1
expression on TCs, but was not related to the OS of locally
advanced or advanced patients with NSCLC. Of course, patients
benefit from the use of immunotherapy instead of chemotherapy. The
use of immunotherapy as the best first line of treatment depends on
the presence of PD-L1 expression on ≥50% of tumor cells (7,10,11).
Unfortunately, not all patients treated with pembrolizumab in the
first-line of treatment were able to receive a second-line of
therapy in the present study. Therefore, a longer OS calculated
from the time of diagnosis was recorded in patients treated with
the second-line of immunotherapy in whom the percentage of TCs with
PD-L1 expression was usually <50%. However, the present study
presents some limitations. For example, analysis of PD-L1 mRNA
expression and its epigenetic regulators should be further analyzed
to provide additional information. Therefore, further studies
should be performed to investigate the molecular background of
PD-L1 expression and to identify additional predictive factors for
immunotherapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
AG, PK, IC and TKuc conceived and designed the
study. TKuc, KK, TKub, KR, DS, KS, RR, AB, BJ, JP, SF, JBu, ASi,
AK, PS, JBl, JM, TG, RK and JS acquired the data. AG, TKuc, BJ, JP,
IP and KK performed the experiments. AG, TKuc, KK, KR, PK and JBu
were responsible for confirming the data authenticity. AG, BJ, ASz,
JP, KL and MG analyzed and interpreted the data. AG, PK and KL
drafted the manuscript. PK, JBl, RR, TG and JM revised the
manuscript for important intellectual content. All authors have
read and approved the manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Medical University of Lublin (Lublin, Poland; approval no.
KE-0254/95/2018). All patients provided written informed consent
prior to being enrolled in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
European Respiratory Society, . European
Lung White Book. www.erswhitebook.org/files/public/Chapters/19_lung_cancer.pdfJanuary
22–2021
|
|
2
|
Lung Cancer Europe, . Report on Lung
Cancer: Challenges in lung cancer in Europe. https://www.lungcancereurope.eu/wp-content/uploads/2017/10/LuCE-Report-final.pdfJanuary
22–2021
|
|
3
|
Navada S, Lai P, Schwartz AG and
Kalemkerian GP: Temporal trends in small cell lung cancer: Analysis
of the national Surveillance Epidemiology and End-Results (SEER)
database. J Clin Oncol. 24 (Suppl 18):70822006. View Article : Google Scholar
|
|
4
|
Bagley SJ, Bauml JM and Langer CJ:
PD-1/PD-L1 immune checkpoint blockade in non-small cell lung
cancer. Clin Adv Hematol Oncol. 13:676–683. 2015.PubMed/NCBI
|
|
5
|
Iwai Y, Hamanishi J, Chamoto K and Honjo
T: Cancer immunotherapies targeting the PD-1 signaling pathway. J
Biomed Sci. 24:262017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brahmer JR, Govindan R, Anders RA, Antonia
SJ, Sagorsky S, Davies MJ, Dubinett SM, Ferris A, Gandhi L, Garon
EB, et al: The Society for Immunotherapy of Cancer consensus
statement on immunotherapy for the treatment of non-small cell lung
cancer (NSCLC). J Immunother Cancer. 6:752018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al KEYNOTE-024 Investigators, : KEYNOTE-024 Investigators.
Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell
lung cancer. N Engl J Med. 375:1823–1833. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
García A, Recondo G, Greco M, de la Vega
M, Perazzo F, Recondo G, Avagnina A and Denninghoff V: Correlation
between PD-L1 expression (clones 28-8 and SP263) and histopathology
in lung adenocarcinoma. Heliyon. 6:e041172020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hirsch FR, McElhinny A, Stanforth D,
Ranger-Moore J, Jansson M, Kulangara K, Richardson W, Towne P,
Hanks D, Vennapusa B, et al: PD-L1 immunohistochemistry assays for
lung cancer: Results from phase 1 of the Blueprint PD-L1 IHC Assay
Comparison Project. J Thorac Oncol. 12:208–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wojas-Krawczyk K, Kalinka E, Grenda A,
Krawczyk P and Milanowski J: Beyond PD-L1-markers for lung cancer
immunotherapy. Int J Mol Sci. 20:E19152019. View Article : Google Scholar
|
|
11
|
Vokes EE, Ready N, Felip E, Horn L, Burgio
MA, Antonia SJ, Arén Frontera O, Gettinger S, Holgado E, Spigel D,
et al: Nivolumab versus docetaxel in previously treated advanced
non-small-cell lung cancer (CheckMate 017 and CheckMate 057):
3-year update and outcomes in patients with liver metastases. Ann
Oncol. 29:959–965. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Herbst RS, Baas P, Perez-Gracia JL, Felip
E, Kim DW, Han JY, Molina JR, Kim JH, Dubos Arvis C, Ahn MJ, et al:
Use of archival versus newly collected tumor samples for assessing
PD-L1 expression and overall survival: An updated analysis of
KEYNOTE-010 trial. Ann Oncol. 30:281–289. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bordoni R, Ciardiello F, von Pawel J,
Cortinovis D, Karagiannis T, Ballinger M, Sandler A, Yu W, He P,
Matheny C, et al: Patient-reported outcomes in OAK: A phase III
study of atezolizumab versus docetaxel in advanced non-small-cell
lung cancer. Clin Lung Cancer. 19:441–449.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Antonia SJ, Villegas A, Daniel D, Vicente
D, Murakami S, Hui R, Kurata T, Chiappori A, Lee KH, de Wit M, et
al PACIFIC Investigators, : overall survival with durvalumab after
chemoradiotherapy in stage III NSCLC. N Engl J Med. 379:2342–2350.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee SY, Jung DK, Choi JE, Jin CC, Hong MJ,
Do SK, Kang HG, Lee WK, Seok Y, Lee EB, et al: PD-L1 polymorphism
can predict clinical outcomes of non-small cell lung cancer
patients treated with first-line paclitaxel-cisplatin chemotherapy.
Sci Rep. 6:259522016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma Y, Liu X, Zhu J, Li W, Guo L, Han X,
Song B, Cheng S and Jie L: Polymorphisms of co-inhibitory molecules
(CTLA-4/PD-1/PD-L1) and the risk of non-small cell lung cancer in a
Chinese population. Int J Clin Exp Med. 8:16585–16591.
2015.PubMed/NCBI
|
|
17
|
Lee SY, Jung DK, Choi JE, Jin CC, Hong MJ,
Do SK, Kang HG, Lee WK, Seok Y, Lee EB, et al: Functional
polymorphisms in PD-L1 gene are associated with the prognosis of
patients with early stage non-small cell lung cancer. Gene.
599:28–35. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du W, Zhu J, Chen Y, Zeng Y, Shen D, Zhang
N, Ning W, Liu Z and Huang JA: Variant SNPs at the microRNA
complementary site in the B7 H1 3 untranslated region increase the
risk of non small cell lung cancer. Mol Med Rep. 16:2682–2690.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yeo MK, Choi SY, Seong IO, Suh KS, Kim JM
and Kim KH: Association of PD-L1 expression and PD-L1 gene
polymorphism with poor prognosis in lung adenocarcinoma and
squamous cell carcinoma. Hum Pathol. 68:103–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen YB, Mu CY, Chen C and Huang JA:
Association between single nucleotide polymorphism of PD-L1 gene
and non-small cell lung cancer susceptibility in a Chinese
population. Asia Pac J Clin Oncol. 10:e1–6. 2014. View Article : Google Scholar
|
|
21
|
Cheng S, Zheng J, Zhu J, Xie C, Zhang X,
Han X, Song B, Ma Y and Liu J: PD-L1 gene polymorphism and high
level of plasma soluble PD-L1 protein may be associated with
non-small cell lung cancer. Int J Biol Markers. 30:e364–e368. 2015.
View Article : Google Scholar
|
|
22
|
Nomizo T, Ozasa H, Tsuji T, Funazo T,
Yasuda Y, Yoshida H, Yagi Y, Sakamori Y, Nagai H, Hirai T, et al:
Clinical impact of single nucleotide polymorphism in PD-L1 on
response to nivolumab for advanced non-small-cell lung cancer
patients. Sci Rep. 7:451242017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vohra M, Sharma AR, Prabhu B N and Rai PS:
SNPs in sites for DNA methylation, transcription factor binding,
and miRNA targets leading to allele-specific gene expression and
Contributing to complex disease risk: A systematic review. Public
Health Genomics. 23:155–170. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho
BC, Turna HZ, Castro G Jr, Srimuninnimit V, Laktionov KK,
Bondarenko I, et al KEYNOTE-042 Investigators, : Pembrolizumab
versus chemotherapy for previously untreated, PD-L1-expressing,
locally advanced or metastatic non-small-cell lung cancer
(KEYNOTE-042): A randomised, open-label, controlled, phase 3 trial.
Lancet. 393:1819–1830. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pacheco JM, Gao D and Camidge DR: Extended
follow-up on KEYNOTE-024 suggests significant survival benefit for
pembrolizumab in patients with PD-L1 ≥50%, but unanswered questions
remain. Ann Transl Med. 7 (Suppl 3):S1272019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshimura K, Inoue Y, Karayama M, Tsuchiya
K, Mori K, Suzuki Y, Iwashita Y, Kahyo T, Kawase A, Tanahashi M, et
al: Heterogeneity analysis of PD-L1 expression and copy number
status in EBUS-TBNA biopsy specimens of non-small cell lung cancer:
Comparative assessment of primary and metastatic sites. Lung
Cancer. 134:202–209. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Inoue Y, Yoshimura K, Mori K, Kurabe N,
Kahyo T, Mori H, Kawase A, Tanahashi M, Ogawa H, Inui N, et al:
Clinical significance of PD-L1 and PD-L2 copy number gains in
non-small-cell lung cancer. Oncotarget. 7:32113–32128. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krawczyk P, Grenda A, Wojas-Krawczyk K,
Nicoś M, Kucharczyk T, Jarosz B, Reszka K, Pankowski J, Krukowska
K, Bożyk A, et al: PD-L1 gene copy number and promoter
polymorphisms regulate PD-L1 expression in tumor cells of non-small
cell lung cancer patients. Cancer Genet. 237:10–18. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ikeda S, Okamoto T, Okano S, Umemoto Y,
Tagawa T, Morodomi Y, Kohno M, Shimamatsu S, Kitahara H, Suzuki Y,
et al: PD-L1 is upregulated by simultaneous amplification of the
PD-L1 and JAK2 genes in non-small cell lung cancer. J Thorac Oncol.
11:62–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
NCBI Resource Coordinators, . Database
resources of the National Center for Biotechnology Information.
Nucleic Acids Res. 42:D7–D17. 2014. View Article : Google Scholar
|
|
32
|
George J, Saito M, Tsuta K, Iwakawa R,
Shiraishi K, Scheel AH, Uchida S, Watanabe SI, Nishikawa R, Noguchi
M, et al: Genomic amplification of CD274 (PD-L1) in small-cell lung
cancer. Clin Cancer Res. 23:1220–1226. 2017. View Article : Google Scholar : PubMed/NCBI
|