Human sterile α motif and HD domain-containing
protein 1 (SAMHD1), also known as dendritic cell derived IFN-γ
induced protein, was first identified as interferon (IFN)-γ induced
protein in human dendritic cells (1). SAMHD1 has an N-terminal nuclear
localization signal (1–150), which contains the transferable
sequence (11KRPR14) (2), an N-terminal SAM domain, which
interacts with proteins, and an HD domain, which is responsible for
the hydrolysis of deoxyribonucleoside triphosphates (dNTPs)
(3). SAMHD1 is widely expressed in
most human tissues and cell lines (1). Rice et al (4) reported 14 mutations in exons of SAMHD1
gene in patients with Aicardi-Goutières syndrome (AGS), resulting
in alterations of the amino acid sequence (4). Mass spectrometry results have
demonstrated that SAMHD1 interacts with Vpx, which induces the
degradation of SAMHD1 via the ubiquitin-proteasome pathway
(5). Previous studies have reported
that SAMHD1 can block the replication of human immunodeficiency
virus type 1 (HIV-1) by hydrolyzing dNTPs, which depletes the pool
of dNTPs available for reverse transcription essential to DNA
replication of HIV-1 (6), hepatitis
B virus (HBV) (7,8) and herpes simplex virus 1 (9).
The maintenance of an appropriate intracellular
dNTPs pool is essential for cell cycle progression, and an
imbalance in the pool of dNTPs can enhance mutagenesis and DNA
replication, resulting in dysregulation of the cell cycle (10). SAMHD1 is the first dNTP
triphosphohydrolase (dNTPase) discovered in mammalian cells and is
crucial in maintaining the homeostasis of intracellular dNTPs
(6). dGTP-Mg2+-dGTP
binding at the allosteric site promotes the tetramerization of
SAMHD1 protein, leading to the formation of catalytically active
enzymes (11). A previous study
demonstrated that SAMHD1 has an essential role in regulating the
cell cycle in quiescent and cycling cells via regulation of the
intracellular concentration of dNTPs (12). In addition, genetic mutations of
SAMHD1 have been discovered in several types of cancer, including
chronic lymphocytic leukemia (CLL) (13), lung cancer (14) and colorectal cancer (15). Methylation at the promoter of SAMHD1
has been demonstrated to decrease its mRNA and protein expression
(14), and the functions of SAMHD1
are affected by acetylation (16)
and phosphorylation (17). In
particular, SAMHD1 is involved in DNA damage repair (DDR) (18), long interspersed element type 1
(LINE-1) suppression (19,20) and chemosensitivity to anticancer
drugs (21,22). In addition, genetic mutations of
SAMHD1 contribute to amino acid changes, which in part impair its
functions (11,15).
The present review aimed to summarize the underlying
mechanisms of SAMHD1 in cancer development and chemosensitivity to
anticancer drugs, and discussed numerous controversial
hypotheses.
The transcriptional regulation of SAMHD1 is the
result of methylation of its promoter. This has been observed in
several types of tumor tissue and cell line. Human SAMHD1 is
expressed at different levels in cell lines and tissues (1). de Silva et al (23) reported that the protein expression of
SAMHD1 is correlated with its mRNA expression level in THP-1 and
CD4+ T cells. In peripheral blood mononuclear cells from
patients with cutaneous T-cell lymphoma (CTCL), both the mRNA and
protein expression levels are significantly lower compared with
those in healthy donors (24), which
was also observed in lung cancer tissues compared with adjacent
normal tissues (14). The different
expression levels of SAMHD1 mRNA and protein in various types of
cancer are summarized in Table I.
The level of methylation has also been demonstrated to be inversely
associated with the expression of SAMHD1 in 466 skin cutaneous
melanoma samples from The Cancer Genome Atlas (TCGA) (25). Taken together, these findings
indicate that epigenetic regulation plays an important role in the
protein expression of SAMHD1. Results from methylation-specific PCR
demonstrated that the SAMHD1 promoter is methylated in lung
adenocarcinoma tissues compared with adjacent normal tissues
(14). Furthermore, in patients with
CTCL, methylated SAMHD1 has also been observed whereas it was
absent in healthy donors (24). The
DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-AzadC) has been
reported to increase the mRNA and protein expression levels of
SAMHD1 in the lung cancer cell line A549 (14), and Jurkat and Sup-T1 cells (acute
T-cell leukemia) (24). Conversely,
the promoter of SAMHD1 has been demonstrated to be in an
unmethylated state in THP-1 cells (23) and SAMHD1 protein expression does not
increase following treatment with 5-AzadC. Furthermore, decitabine,
which is a DNA methyltransferase inhibitor, has been demonstrated
to be a substrate of SAMHD1, and its anticancer effect in acute
myelocytic leukemia (AML) was found to be dependent of SAMHD1
protein expression (26). For other
tumors characterized by methylation of the SAMHD1 promoter, further
investigation is required to determine whether SAMHD1 could have a
two-sided effect on chemotherapy with decitabine.
Acetylation has been reported to be involved in the
post-transcriptional regulation of SAMHD1 expression. Trichostatin
A (TSA), a histone deacetylase inhibitor, can increase the mRNA and
protein expression of SAMHD1 in Jurkat and Sup-T1 cells (23). Lee et al (16) reported that SAMHD1 is acetylated at
the K405 site via arrest defective protein 1 (ARD1), which leads to
enhanced dNTPase activity (16).
Furthermore, SAMHD1 and ARD1 protein expression has been found to
be upregulated in hepatoma tissues compared with non-tumor tissues,
which indicates that SAMHD1 acetylated by ARD1 may participate in
tumorigenesis (16). However, the
K580 residue of SAMHD1, which is another acetylated site, could not
be acetylated by ARD1 in vitro (16). In particular, 5-AzadC and TSA were
found to notably increase the mRNA expression of SAMHD1; however,
only a slight increase in SAMHD1 protein expression was observed in
Jurkat and Sup-T1 cells (23).
Subsequently, besides methylation and acetylation, another
regulatory mechanism may exist in SAMHD1 transcription or
translation.
LINE-1 is the only active autonomous retrotransposon
in the human genome and has non-long terminal repeats, which can
encode its mRNA and two open reading frame (ORF) proteins, ORF1p
and ORF2p (46,47). In vitro, SAMHD1 has no effect
on the expression of LINE-1 mRNA and ORF1p protein but suppresses
expression of ORF2p (19). In
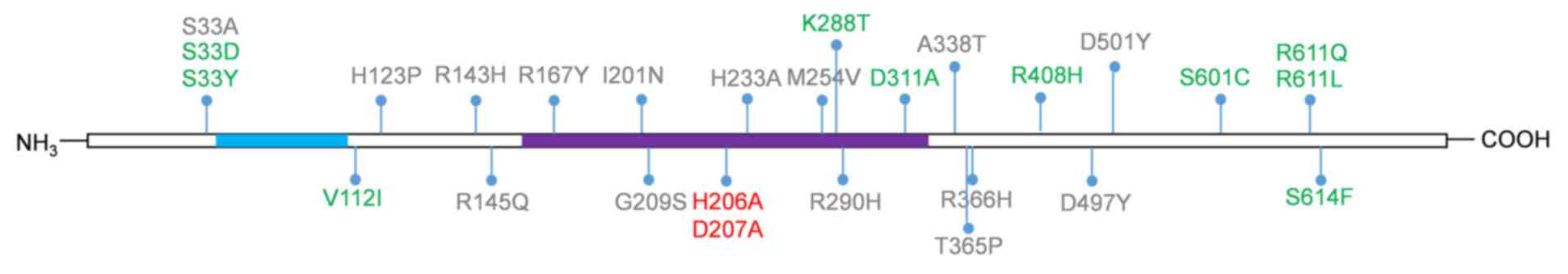
addition, a previous study has reported more mutants of SAMHD1
protein in various types of cancer (Fig.
2) (20). Furthermore, SAMHD1
single-nucleotide polymorphisms (SNPs) at seven codon positions
negatively regulate the retrotransposition of LINE-1 compared with
its wild-type equivalent (48).
Furthermore, SAMHD1 mutation deficiency of amino acids 162–335
attenuates the activity suppressing LINE-1; however, the mutant
lacking SAM domain enhances its ability against LINE-1 (19). The enzymatically active HD domain of
SAMHD1, SAM domain, is therefore crucial for the inhibition of
LINE-1 retrotransposon. The dNTPase-inactive SAMHD1 mutants
H206A/D207A and K288T can also inhibit the reverse transcription of
LINE-1 in colon cancer (20,48). Removing the nuclear localization
sequence of SAMHD1 or its K11A mutants could subsequently induce
its redistribution in the cytoplasm, leading to ORF2p deletion and
LINE-1 suppression, but with no effect on dNTPase activity
(49,50). Therefore, SAMHD1-mediated LINE-1
inhibition is independent of its dNTPase activity. In addition, the
activity of SAMHD1 against LINE-1 was found to be negatively
regulated by phosphorylation at Thr592 (51). Furthermore, exogenous expression of
wild-type SAMHD1, instead of H233A mutant, induces abnormal
distribution of ORF1p and promotes the formation of large
cytoplasmic foci, which suppresses LINE-1 retrotransposon dependent
of the expression of stress granule markers (Ras GTPase-activating
protein-binding protein 1 and Tia1 cytotoxic granule-associated RNA
binding protein) (52). Interaction
between SAMHD1 and ORF1p has not yet been discovered, and the
underlying mechanism of SAMHD1-induced redistribution of ORF1p into
stress granules requires further investigation.
RNA ribosomal 2-mediated dNTP biosynthesis and
SAMHD1-mediated dNTP hydrolysis are two different strategies used
to maintain dNTP homeostasis in mammalian cells (6,53) and
are essential for DDR and cell survival. In addition, DNA damage
also increases dNTPs pools through upregulating the activity of
ribonucleotide reductase (53).
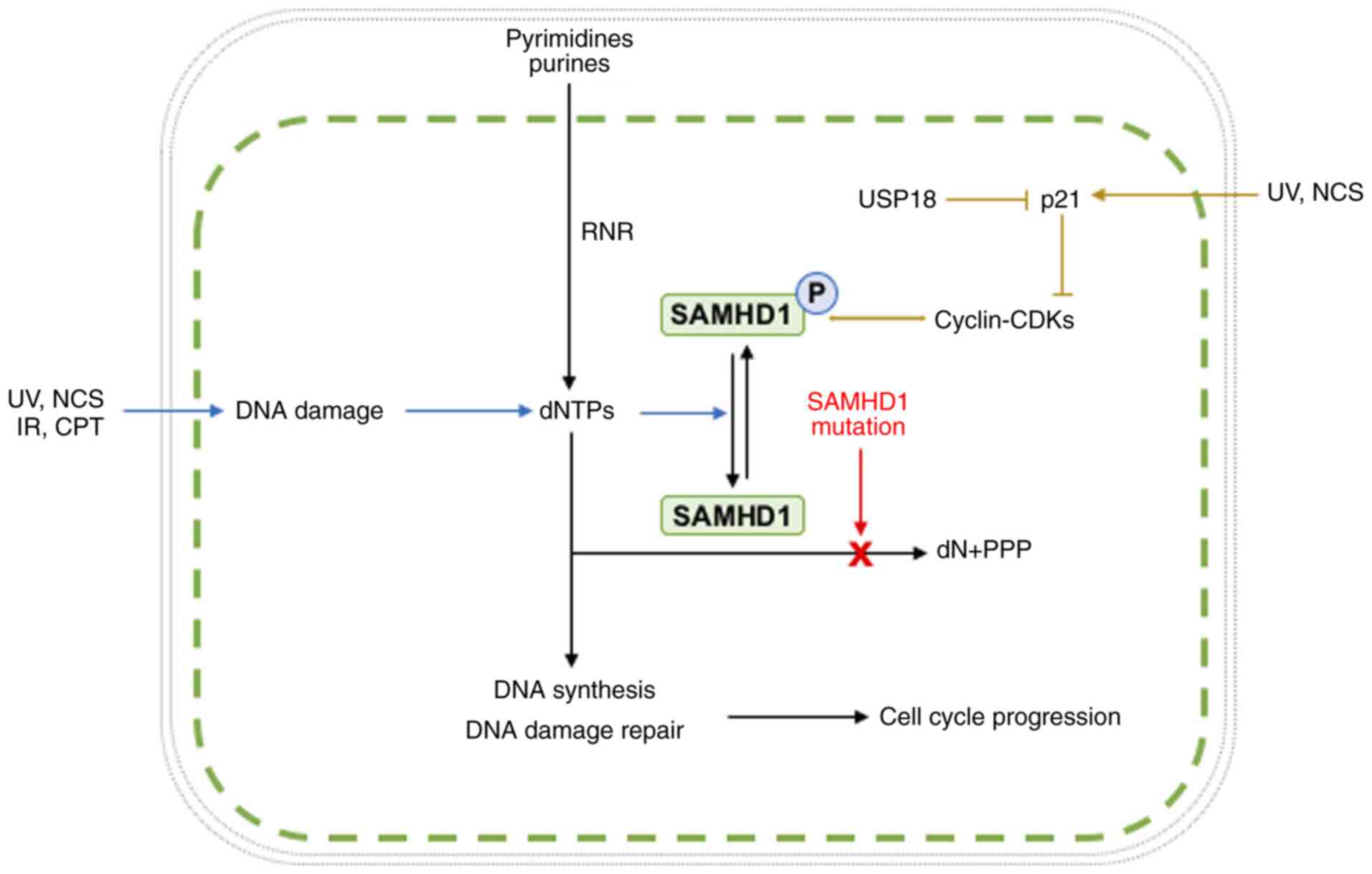
Etoposide, an anticancer drug inducing DNA double-strand breaks,
can also promote the dephosphorylation of SAMHD1 and the transition
from G1 to G0 phase in the cell cycle, which is accompanied by a
decrease in the protein expression of CDK1 (54). The cyclin A2/CDK1 complex interacts
with and phosphorylates SAMHD1 at the Thr592 site (31). Subsequently, etoposide-induced
dephosphorylation of SAMHD1 may result from etoposide-mediated
inhibition of CDK1 when cells return from the G1 phase into the G0
phase. DNA synthesis is impeded by SAMHD1 mutants lacking the
C-terminal end (618–626 residue), which is accompanied by the
phosphorylation of checkpoint kinase 1 (CHK1) at Ser345 and
activation of DNA damage checkpoints (33). A previous study reported that
treatment with neocarzinostatin increases p21 expression, which
inhibits CDK1/2, leading therefore to decreased phosphorylation of
SAMHD1 (54). Dephosphorylation of
SAMHD1 may thus be the result of increased p21 in DNA damage
induced by ultraviolet light or neocrazinostatin (Fig. 1). Another study reported that SAMHD1
knockdown can increase U2OS (osteosarcoma), MCF7 (breast cancer)
and HCT-116 (colon cancer) cell sensitivity to etoposide,
camptothecin and ionizing radiation, which can be rescued by
restoring the exogenous expression of SAMHD1 (55). Following treatment with camptothecin,
immunofluorescence shows that SAMHD1 is colocalized with gH2A
histone family member X and RAD51 in DNA damage sites, which are
markers of DNA damage and homologous recombination, respectively
(55). Furthermore, SAMHD1 is
colocalized with p53-binding protein 1 foci, which is an
indispensable regulatory protein involved in DDR, following
camptothecin-induced DNA double-strand breaks (13). SAMHD1 has been observed at
replication forks and interacts with double-strand break repair
protein MRE11 exonuclease, which is followed by activation of the
CHK1-ataxia telangiectasia and Rad3-related protein checkpoint and
restart of the replication forks, eventually leading to
acceleration of the normal fork progression, which could be
restored by T592E (phosphorylation) and K312A (dNTPase-inactive)
mutant of SAMHD1, but not by the T592A (non-phosphorylation) mutant
(18). In addition, SAMHD1 can
interact with and recruit CtBP-interacting protein to DNA damage
sites, followed by promotion of DNA end resection, which could not
be rescued by the gastric cancer-related K484T mutant of SAMHD1-
(55). Further experiment shows that
K484T mutant has no dNTPase activity and can not hydrolyze dNTPs
(55). Therefore, SAMHD1 can promote
DDR in cancer cells in response to DNA double-strand break-inducing
agents through a phosphorylation-dependent mechanism at the Thr592
site rather than a dNTPase-dependent mechanism.
At present, antiviral agents are nucleotide analogs
that incorporate into DNA and impede DNA synthesis in the virus
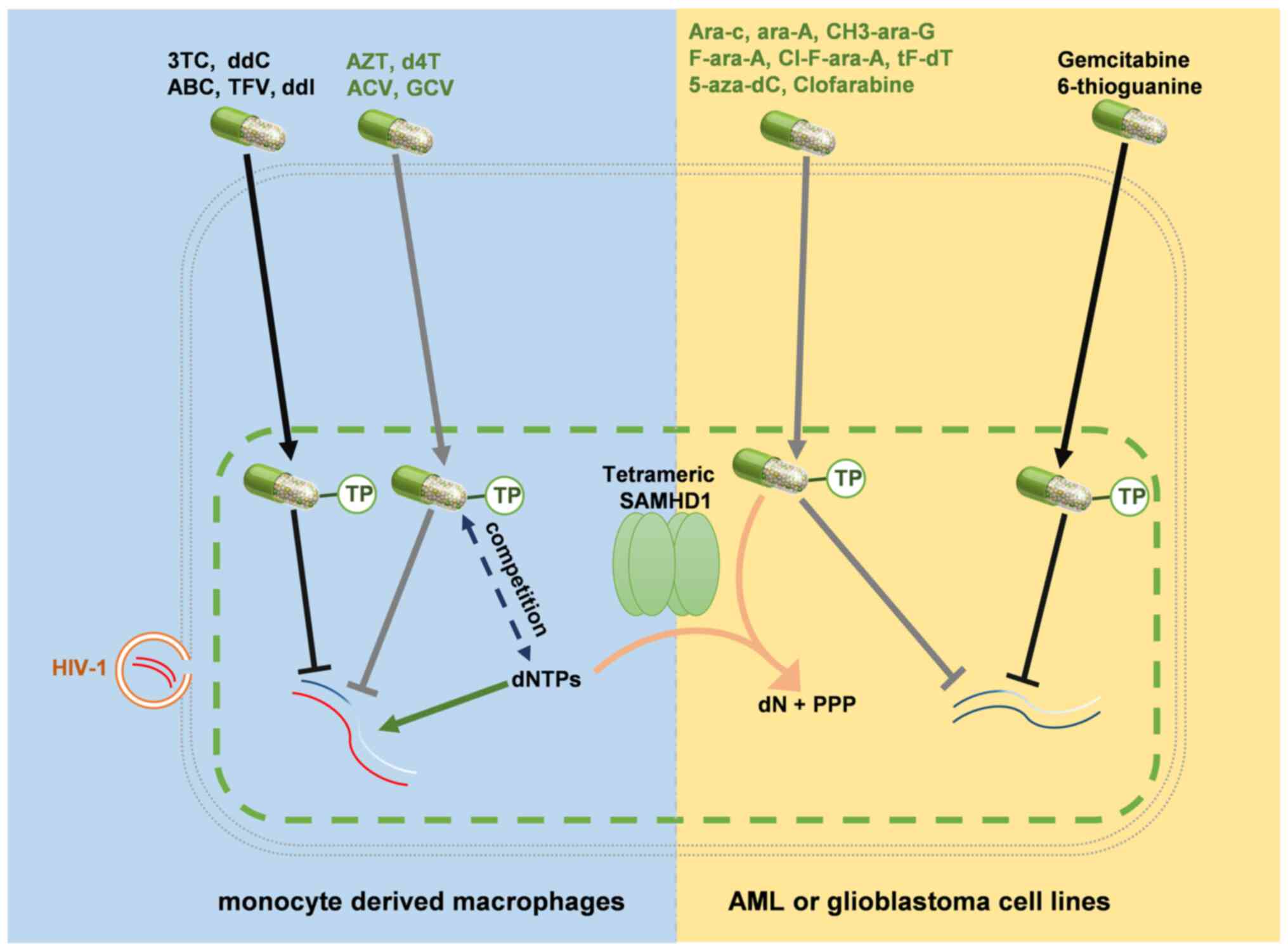
replication cycle. Ballana et al (56) demonstrated for the first time that
degradation of SAMHD1 via Vpx can decrease the sensitivity of
thymidine analogs against HIV-1, including zidovudine and stavudine
(Fig. 3). Based on the understanding
of dNTPase activity of SAMHD1, dNTP-based metabolite from
anticancer drugs became the focus in numerous studies on anticancer
drug chemosensitivity. Mutations in SAMHD1 have been found in 9.6%
of patients with relapsed or refractory CLL (57) and were demonstrated to interact with
proteins and implicated in DNA double-strand breaks (13). The antimetabolite cytosine
arabinoside (ara-C) blocks DNA polymerase and gets incorporated
into elongated DNA in the S phase of the cell cycle, leading to
reduced DNA replication and eventually cell death (58). Herold et al (21) revealed that intracellular ara-CTP, a
dCTP analog, is a substrate of SAMHD1 in THP-1 and HuT-78 cells.
Results from high-performance liquid-chromatography demonstrated
that ara-CTP is hydrolyzed to ara-C and inorganic triphosphate by
SAMHD1 in the presence of allosteric activators (59). Furthermore, it was also confirmed
that clofarabine is a substrate of SAMHD1 (60). In contrast to the research by Ballana
et al (56), Vpx-mediated
SAMHD1 degradation enhances the sensitivity of ara-CTP in AML and
cutaneous T-cell lymphoma cells (21). Another study by Herold et al
(22) reported that SAMHD1 could
also hydrolyze triphosphates of other nucleotide analogs, such as
vidarabine, nelarabine, fludarabin, decitabine and trifluridine
(22), whereas gemcitabine and
6-thioguanine are neither substrates nor activators of SAMHD1
(21,61). In a mouse model of AML, xenograft
tumors with SAMHD1 knockdown presented an improved response to
ara-C chemotherapy compared with those expressing SAMHD1 (21). The chemosensitivity of ara-C was also
found to be negatively regulated by SAMHD1 in patient-derived AML
blasts (21). In 143 patients with
AML who received ara-C-based chemotherapy, patients with low SAMHD1
expression levels had higher event-free survival rates than those
with high SAMHD1 expression levels (37 vs. 19%, respectively), as
well as higher 5-year overall survival rates (54 vs. 9%,
respectively) (62). Furthermore,
AML cells resistant to decitabine exhibit increased SAMHD1 protein
expression, which could be a result of SAMHD1 promoter
demethylation (26). In addition,
SAMHD1 may also be considered as a useful biomarker for predicting
the response to decitabine. Apart from its role in hematological
malignancies, it is also unclear whether SAMHD1 has a crucial
predictive function for the efficacy of nucleotide analogs in solid
tumors. Wang et al (63)
first reported that U251 (glioblastoma) cells with acquired
resistance to olaparib, talazoparib and simmiparib, exhibited
upregulated expression levels of SAMHD1 mRNA and protein than that
in parental cell lines. Furthermore, U251 cells with resistance to
olaparib displayed similar cross-resistance to 5-fluorouracil, and
the IC50 was increased by >3-fold (63). However, it remains unclear whether
upregulation of SAMHD1 expression is responsible for the
chemoresistance of other solid malignancies to 5-fluorouracil. The
structure and biochemical framework established by Knecht et
al (61) is useful to determine
novel anticancer drugs that could be hydrolyzed by SAMHD1.
Ribonucleotide reductase subunit R2 inhibitors (gemcitabine,
hydroxyurea and triapine) occupy the AS2 site and decrease the
ara-CTPase of SAMHD1, leading to enhanced chemosensitivity of AML
cells to ara-C (64). Hollenbaugh
et al (65) demonstrated that
Y374 in the catalytic pocket of SAMHD1 could be occupied by the
(2′S)-2′-methyl group of SMDU-TP, resulting in decreased dGTP
hydrolysis. Further investigation is therefore needed to determine
whether a combination of SAMHD1 inhibitors and nucleotide analogs
could improve chemosensitivity in the treatment of cancer.
Not applicable.
This work was supported by the National Natural
Science Foundation of China (grant no. 81572350).
Not applicable.
ZC and JH contributed to manuscript draft and figure
design. ZC, SY and AX provided critical revision of the manuscript.
Data sharing is not applicable. All authors read and approved the
final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Li N, Zhang W and Cao X: Identification of
human homologue of mouse IFN-gamma induced protein from human
dendritic cells. Immunol Lett. 74:221–224. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brandariz-Nuñez A, Valle-Casuso JC, White
TE, Laguette N, Benkirane M, Brojatsch J and Diaz-Griffero F: Role
of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac.
Retrovirology. 9:492012. View Article : Google Scholar
|
|
3
|
Wei W, Guo H, Han X, Liu X, Zhou X, Zhang
W and Yu XF: A novel DCAF1-binding motif required for Vpx-mediated
degradation of nuclear SAMHD1 and Vpr-induced G2 arrest. Cell
Microbiol. 14:1745–1756. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rice GI, Bond J, Asipu A, Brunette RL,
Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, et
al: Mutations involved in Aicardi-Goutieres syndrome implicate
SAMHD1 as regulator of the innate immune response. Nat Genet.
41:829–832. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Laguette N, Sobhian B, Casartelli N,
Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S,
Schwartz O and Benkirane M: SAMHD1 is the dendritic- and
myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx.
Nature. 474:654–657. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goldstone DC, Ennis-Adeniran V, Hedden JJ,
Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF,
Yap MW, et al: HIV-1 restriction factor SAMHD1 is a deoxynucleoside
triphosphate triphosphohydrolase. Nature. 480:379–382. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen Z, Zhu M, Pan X, Zhu Y, Yan H, Jiang
T, Shen Y, Dong X, Zheng N, Lu J, et al: Inhibition of Hepatitis B
virus replication by SAMHD1. Biochem Biophys Res Commun.
450:1462–1468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jeong GU, Park IH, Ahn K and Ahn BY:
Inhibition of hepatitis B virus replication by a dNTPase-dependent
function of the host restriction factor SAMHD1. Virology.
495:71–78. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim ET, White TE, Brandariz-Núñez A,
Diaz-Griffero F and Weitzman MD: SAMHD1 restricts herpes simplex
virus 1 in macrophages by limiting DNA replication. J Virol.
87:12949–12956. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Buj R and Aird KM: Deoxyribonucleotide
Triphosphate Metabolism in Cancer and Metabolic Disease. Front
Endocrinol (Lausanne). 9:1772018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji X, Wu Y, Yan J, Mehrens J, Yang H,
DeLucia M, Hao C, Gronenborn AM, Skowronski J, Ahn J and Xiong Y:
Mechanism of allosteric activation of SAMHD1 by dGTP. Nat Struct
Mol Biol. 20:1304–1309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Franzolin E, Pontarin G, Rampazzo C,
Miazzi C, Ferraro P, Palumbo E, Reichard P and Bianchi V: The
deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of
DNA precursor pools in mammalian cells. Proc Natl Acad Sci USA.
110:14272–14277. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clifford R, Louis T, Robbe P, Ackroyd S,
Burns A, Timbs AT, Wright Colopy G, Dreau H, Sigaux F, et al:
SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and
is involved in response to DNA damage. Blood. 123:1021–1031. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang JL, Lu FZ, Shen XY, Wu Y and Zhao LT:
SAMHD1 is down regulated in lung cancer by methylation and inhibits
tumor cell proliferation. Biochem Biophys Res Commun. 455:229–233.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rentoft M, Lindell K, Tran P, Chabes AL,
Buckland RJ, Watt DL, Marjavaara L, Nilsson AK, Melin B, Trygg J,
et al: Heterozygous colon cancer-associated mutations of SAMHD1
have functional significance. Proc Natl Acad Sci USA.
113:4723–4728. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee EJ, Seo JH, Park JH, Vo TTL, An S, Bae
SJ, Le H, Lee HS, Wee HJ, Lee D, et al: SAMHD1 acetylation enhances
its deoxynucleotide triphosphohydrolase activity and promotes
cancer cell proliferation. Oncotarget. 8:68517–68529. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tramentozzi E, Ferraro P, Hossain M,
Stillman B, Bianchi V and Pontarin G: The dNTP triphosphohydrolase
activity of SAMHD1 persists during S-phase when the enzyme is
phosphorylated at T592. Cell Cycle. 17:1102–1114. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Coquel F, Silva MJ, Técher H, Zadorozhny
K, Sharma S, Nieminuszczy J, Mettling C, Dardillac E, Barthe A,
Schmitz AL, et al: SAMHD1 acts at stalled replication forks to
prevent interferon induction. Nature. 557:57–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao K, Du J, Han X, Goodier JL, Li P,
Zhou X, Wei W, Evans SL, Li L, Zhang W, et al: Modulation of LINE-1
and Alu/SVA retrotransposition by Aicardi-Goutieres
syndrome-related SAMHD1. Cell Rep. 4:1108–1115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao W, Li G, Bian X, Rui Y, Zhai C, Liu P,
Su J, Wang H, Zhu C, Du Y, et al: Defective modulation of LINE-1
retrotransposition by cancer-associated SAMHD1 mutants. Biochem
Biophys Res Commun. 519:213–219. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Herold N, Rudd SG, Ljungblad L, Sanjiv K,
Myrberg IH, Paulin CB, Heshmati Y, Hagenkort A, Kutzner J, Page BD,
et al: Targeting SAMHD1 with the Vpx protein to improve cytarabine
therapy for hematological malignancies. Nat Med. 23:256–263. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herold N, Rudd SG, Sanjiv K, Kutzner J,
Bladh J, Paulin CBJ, Helleday T, Henter JI and Schaller T: SAMHD1
protects cancer cells from various nucleoside-based
antimetabolites. Cell Cycle. 16:1029–1038. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
de Silva S, Hoy H, Hake TS, Wong HK, Porcu
P and Wu L: Promoter methylation regulates SAMHD1 gene expression
in human CD4+ T cells. J Biol Chem. 288:9284–9292. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
de Silva S, Wang F, Hake TS, Porcu P, Wong
HK and Wu L: Downregulation of SAMHD1 expression correlates with
promoter DNA methylation in Sezary syndrome patients. J Invest
Dermatol. 134:562–565. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen W, Cheng P, Jiang J, Ren Y, Wu D and
Xue D: Epigenomic and genomic analysis of transcriptome modulation
in skin cutaneous melanoma. Aging (Albany NY). 12:12703–12725.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oellerich T, Schneider C, Thomas D, Knecht
KM, Buzovetsky O, Kaderali L, Schliemann C, Bohnenberger H,
Angenendt L, Hartmann W, et al: Selective inactivation of
hypomethylating agents by SAMHD1 provides a rationale for
therapeutic stratification in AML. Nat Commun. 10:34752019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
White TE, Brandariz-Nuñez A, Valle-Casuso
JC, Amie S, Nguyen LA, Kim B, Tuzova M and Diaz-Griffero F: The
retroviral restriction ability of SAMHD1, but not its
deoxynucleotide triphosphohydrolase activity, is regulated by
phosphorylation. Cell Host Microbe. 13:441–451. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Welbourn S, Dutta SM, Semmes OJ and
Strebel K: Restriction of virus infection but not catalytic dNTPase
activity is regulated by phosphorylation of SAMHD1. J Virol.
87:11516–11524. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hochegger H, Takeda S and Hunt T:
Cyclin-dependent kinases and cell-cycle transitions: Does one fit
all? Nat Rev Mol Cell Biol. 9:910–916. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takeda DY, Wohlschlegel JA and Dutta A: A
bipartite substrate recognition motif for cyclin-dependent kinases.
J Biol Chem. 276:1993–1997. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cribier A, Descours B, Valadão AL,
Laguette N and Benkirane M: Phosphorylation of SAMHD1 by cyclin
A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep.
3:1036–1043. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kretschmer S, Wolf C, König N, Staroske W,
Guck J, Häusler M, Luksch H, Nguyen LA, Kim B, Alexopoulou D, et
al: SAMHD1 prevents autoimmunity by maintaining genome stability.
Ann Rheum Dis. 74:e172015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan J, Hao C, DeLucia M, Swanson S,
Florens L, Washburn MP, Ahn J and Skowronski J:
CyclinA2-cyclin-dependent kinase regulates SAMHD1 protein
phosphohydrolase domain. J Biol Chem. 290:13279–13292. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu J, Qiao M, Chen Y, Tang H, Zhang W,
Tang D, Pi S, Dai J, Tang N, Huang A and Hu Y: Cyclin E2-CDK2
mediates SAMHD1 phosphorylation to abrogate its restriction of HBV
replication in hepatoma cells. FEBS Lett. 592:1893–1904. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pauls E, Badia R, Torres-Torronteras J,
Ruiz A, Permanyer M, Riveira-Muñoz E, Clotet B, Marti R, Ballana E
and Esté JA: Palbociclib, a selective inhibitor of cyclin-dependent
kinase4/6, blocks HIV-1 reverse transcription through the control
of sterile alpha motif and HD domain-containing protein-1 (SAMHD1)
activity. AIDS. 28:2213–2222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Badia R, Angulo G, Riveira-Muñoz E,
Pujantell M, Puig T, Ramirez C, Torres-Torronteras J, Martí R,
Pauls E, Clotet B, et al: Inhibition of herpes simplex virus type 1
by the CDK6 inhibitor PD-0332991 (palbociclib) through the control
of SAMHD1. J Antimicrob Chemother. 71:387–394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Allouch A, David A, Amie SM, Lahouassa H,
Chartier L, Margottin-Goguet F, Barré-Sinoussi F, Kim B,
Sáez-Cirión A and Pancino G: p21-mediated RNR2 repression restricts
HIV-1 replication in macrophages by inhibiting dNTP biosynthesis
pathway. Proc Natl Acad Sci USA. 110:E3997–E4006. 2013. View Article : Google Scholar
|
|
38
|
Pauls E, Ruiz A, Riveira-Muñoz E,
Permanyer M, Badia R, Clotet B, Keppler OT, Ballana E and Este JA:
p21 regulates the HIV-1 restriction factor SAMHD1. Proc Natl Acad
Sci USA. 111:E1322–E1324. 2014. View Article : Google Scholar
|
|
39
|
Osei Kuffour E, Schott K, Jaguva Vasudevan
AA, Holler J, Schulz WA, Lang PA, Lang KS, Kim B, Häussinger D,
König R and Münk C: USP18 (UBP43) abrogates p21-mediated inhibition
of HIV-1. J Virol. 92:e00592–18. 2018. View Article : Google Scholar
|
|
40
|
Schott K, Fuchs NV, Derua R, Mahboubi B,
Schnellbächer E, Seifried J, Tondera C, Schmitz H, Shepard C,
Brandariz-Nuñez A, et al: Dephosphorylation of the HIV-1
restriction factor SAMHD1 is mediated by PP2A-B55alpha holoenzymes
during mitotic exit. Nat Commun. 9:22272018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kueck T, Cassella E, Holler J, Kim B and
Bieniasz PD: The aryl hydrocarbon receptor and interferon gamma
generate antiviral states via transcriptional repression. Elife.
7:e388672018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bonifati S, Daly MB, St Gelais C, Kim SH,
Hollenbaugh JA, Shepard C, Kennedy EM, Kim DH, Schinazi RF, Kim B
and Wu L: SAMHD1 controls cell cycle status, apoptosis and HIV-1
infection in monocytic THP-1 cells. Virology. 495:92–100. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kodigepalli KM, Li M, Liu SL and Wu L:
Exogenous expression of SAMHD1 inhibits proliferation and induces
apoptosis in cutaneous T-cell lymphoma-derived HuT78 cells. Cell
Cycle. 16:179–188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Welbourn S, Miyagi E, White TE,
Diaz-Griffero F and Strebel K: Identification and characterization
of naturally occurring splice variants of SAMHD1. Retrovirology.
9:862012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kodigepalli KM, Bonifati S, Tirumuru N and
Wu L: SAMHD1 modulates in vitro proliferation of acute myeloid
leukemia-derived THP-1 cells through the PI3K-Akt-p27 axis. Cell
Cycle. 17:1124–1137. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Feng Q, Moran JV, Kazazian HH Jr and Boeke
JD: Human L1 retrotransposon encodes a conserved endonuclease
required for retrotransposition. Cell. 87:905–916. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Burns KH: Transposable elements in cancer.
Nat Rev Cancer. 17:415–424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
White TE, Brandariz-Nuñez A, Valle-Casuso
JC, Knowlton C, Kim B, Sawyer SL and Diaz-Griffero F: Effects of
human SAMHD1 polymorphisms on HIV-1 susceptibility. Virology.
460-461:34–44. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
White TE, Brandariz-Nuñez A, Valle-Casuso
JC, Amie S, Nguyen L, Kim B, Brojatsch J and Diaz-Griffero F:
Contribution of SAM and HD domains to retroviral restriction
mediated by human SAMHD1. Virology. 436:81–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Du J, Peng Y, Wang S, Hou J, Wang Y, Sun T
and Zhao K: Nucleocytoplasmic shuttling of SAMHD1 is important for
LINE-1 suppression. Biochem Biophys Res Commun. 510:551–557. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Herrmann A, Wittmann S, Thomas D, Shepard
CN, Kim B, Ferreirós N and Gramberg T: The SAMHD1-mediated block of
LINE-1 retroelements is regulated by phosphorylation. Mobile DNA.
9:112018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hu S, Li J, Xu F, Mei S, Le Duff Y, Yin L,
Pang X, Cen S, Jin Q, Liang C and Guo F: SAMHD1 inhibits LINE-1
retrotransposition by promoting stress granule formation. PLoS
Genetics. 11:e10053672015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ha°kansson P, Hofer A and Thelander L:
Regulation of mammalian ribonucleotide reduction and dNTP pools
after DNA damage and in resting cells. J Biol Chem. 281:7834–7841.
2006. View Article : Google Scholar
|
|
54
|
Jauregui P and Landau NR: DNA damage
induces a SAMHD1-mediated block to the infection of macrophages by
HIV-1. Sci Rep. 8:41532018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Daddacha W, Koyen AE, Bastien AJ, Head PE,
Dhere VR, Nabeta GN, Connolly EC, Werner E, Madden MZ, Daly MB, et
al: SAMHD1 promotes DNA end resection to facilitate DNA repair by
homologous recombination. Cell Rep. 20:1921–1935. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ballana E, Badia R, Terradas G,
Torres-Torronteras J, Ruiz A, Pauls E, Riveira-Muñoz E, Clotet B,
Martí R and Esté JA: SAMHD1 specifically affects the antiviral
potency of thymidine analog HIV reverse transcriptase inhibitors.
Antimicrob Agents Chemother. 58:4804–4813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Guièze R, Robbe P, Clifford R, de Guibert
S, Pereira B, Timbs A, Dilhuydy MS, Cabes M, Ysebaert L, Burns A,
et al: Presence of multiple recurrent mutations confers poor trial
outcome of relapsed/refractory CLL. Blood. 126:2110–2117. 2015.
View Article : Google Scholar
|
|
58
|
Grant S: Ara-C: Cellular and molecular
pharmacology. Adv Cancer Res. 72:197–233. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Schneider C, Oellerich T, Baldauf HM,
Schwarz SM, Thomas D, Flick R, Bohnenberger H, Kaderali L, Stegmann
L, Cremer A, et al: SAMHD1 is a biomarker for cytarabine response
and a therapeutic target in acute myeloid leukemia. Nat Med.
23:250–255. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Arnold LH, Kunzelmann S, Webb MR and
Taylor IA: A continuous enzyme-coupled assay for
triphosphohydrolase activity of HIV-1 restriction factor SAMHD1.
Antimicrob Agents Chemother. 59:186–192. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Knecht KM, Buzovetsky O, Schneider C,
Thomas D, Srikanth V, Kaderali L, Tofoleanu F, Reiss K, Ferreirós
N, Geisslinger G, et al: The structural basis for cancer drug
interactions with the catalytic and allosteric sites of SAMHD1.
Proc Natl Acad Sci USA. 115:E10022–E10031. 2018. View Article : Google Scholar
|
|
62
|
Rassidakis GZ, Herold N, Myrberg IH,
Tsesmetzis N, Rudd SG, Henter JI, Schaller T, Ng SB, Chng WJ, Yan
B, et al: Low-level expression of SAMHD1 in acute myeloid leukemia
(AML) blasts correlates with improved outcome upon consolidation
chemotherapy with high-dose cytarabine-based regimens. Blood Cancer
J. 8:982018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang YT, Yuan B, Chen HD, Xu L, Tian YN,
Zhang A, He JX and Miao ZH: Acquired resistance of phosphatase and
tensin homolog-deficient cells to poly(ADP-ribose) polymerase
inhibitor and Ara-C mediated by 53BP1 loss and SAMHD1
overexpression. Cancer Sci. 109:821–831. 2017. View Article : Google Scholar
|
|
64
|
Rudd SG, Tsesmetzis N, Sanjiv K, Paulin
CB, Sandhow L, Kutzner J, Hed Myrberg I, Bunten SS, Axelsson H,
Zhang SM, et al: Ribonucleotide reductase inhibitors suppress
SAMHD1 ara-CTPase activity enhancing cytarabine efficacy. EMBO Mol
Med. 12:e104192020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hollenbaugh JA, Shelton J, Tao S,
Amiralaei S, Liu P, Lu X, Goetze RW, Zhou L, Nettles JH, Schinazi
RF and Kim B: Substrates and Inhibitors of SAMHD1. PLoS One.
12:e01690522017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jones S, Zhang X, Parsons DW, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et
al: Core signaling pathways in human pancreatic cancers revealed by
global genomic analyses. Science. 321:1801–1806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kohnken R, Kodigepalli KM and Wu L:
Regulation of deoxynucleotide metabolism in cancer: Novel
mechanisms and therapeutic implications. Mol Cancer. 14:1762015.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
James CD, Prabhakar AT, Otoa R, Evans MR,
Wang X, Bristol ML, Zhang K, Li R and Morgan IM: SAMHD1 regulates
human papillomavirus 16-induced cell proliferation and viral
replication during differentiation of keratinocytes. mSphere.
4:e00448–19. 2019. View Article : Google Scholar
|
|
69
|
Stanland LJ and Luftig MA: The role of
EBV-induced hypermethylation in gastric cancer tumorigenesis.
Viruses. 12:12222020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cruz-Gregorio A, Aranda-Rivera AK and
Pedraza-Chaverri J: Human papillomavirus-related cancers and
mitochondria. Virus Res. 286:1980162020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cabello-Lobato MJ, Wang S and Schmidt CK:
SAMHD1 sheds moonlight on DNA double-strand break repair. Trends
Genet. 33:895–897. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Seamon KJ and Stivers JT: A
high-throughput enzyme-coupled assay for SAMHD1 dNTPase. J Biomol
Screen. 20:801–809. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mauney CH, Perrino FW and Hollis T:
Identification of inhibitors of the dNTP triphosphohydrolase SAMHD1
using a novel and direct high-throughput assay. Biochemistry.
57:6624–6636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jiang H, Li C, Liu Z and Shengjing
Hospital; Hu: Expression and relationship of SAMHD1 with other
apoptotic and autophagic genes in acute myeloid leukemia patients.
Acta Haematol. 143:51–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kodigepalli KM, Li M, Bonifati S, Panfil
AR, Green PL, Liu SL and Wu L: SAMHD1 inhibits epithelial cell
transformation in vitro and affects leukemia development in
xenograft mice. Cell Cycle. 17:2564–2576. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Herold N, Rudd SG, Sanjiv K, Kutzner J,
Myrberg IH, Paulin CBJ, Olsen TK, Helleday T, Henter JI and
Schaller T: With me or against me: Tumor suppressor and drug
resistance activities of SAMHD1. Exp Hematol. 52:32–39. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Rothenburger T, McLaughlin KM, Herold T,
Schneider C, Oellerich T, Rothweiler F, Feber A, Fenton TR, Wass
MN, Keppler OT, et al: SAMHD1 is a key regulator of the
lineage-specific response of acute lymphoblastic leukaemias to
nelarabine. Commun Biol. 3:3242020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yang CA, Huang HY, Chang YS, Lin CL, Lai
IL and Chang JG: DNA-sensing and nuclease gene expressions as
markers for colorectal cancer progression. Oncology. 92:115–124.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Shang Z, Qian L, Liu S, Niu X, Qiao Z, Sun
Y, Zhang Y, Fan LY, Guan X, Cao CX and Xiao H: Graphene
oxide-facilitated comprehensive analysis of cellular nucleic acid
binding proteins for lung cancer. ACS Appl Mater Interfaces.
10:17756–17770. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shi Y, Lv G, Chu Z, Piao L, Liu X, Wang T,
Jiang Y and Zhang P: Identification of natural splice variants of
SAMHD1 in virus-infected HCC. Oncol Rep. 31:687–692. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Merati M, Buethe DJ, Cooper KD, Honda KS,
Wang H and Gerstenblith MR: Aggressive CD8(+) epidermotropic
cutaneous T-cell lymphoma associated with homozygous mutation in
SAMHD1. JAAD Case Rep. 1:227–229. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Amin NA, Seymour E, Saiya-Cork K, Parkin
B, Shedden K and Malek SN: A quantitative analysis of subclonal and
clonal gene mutations before and after therapy in chronic
lymphocytic leukemia. Clin Cancer Res. 22:4525–4535. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhu KW, Chen P, Zhang DY, Yan H, Liu H,
Cen LN, Liu YL, Cao S, Zhou G, Zeng H, et al: Association of
genetic polymorphisms in genes involved in Ara-C and dNTP
metabolism pathway with chemosensitivity and prognosis of adult
acute myeloid leukemia (AML). J Transl Med. 16:902018. View Article : Google Scholar : PubMed/NCBI
|