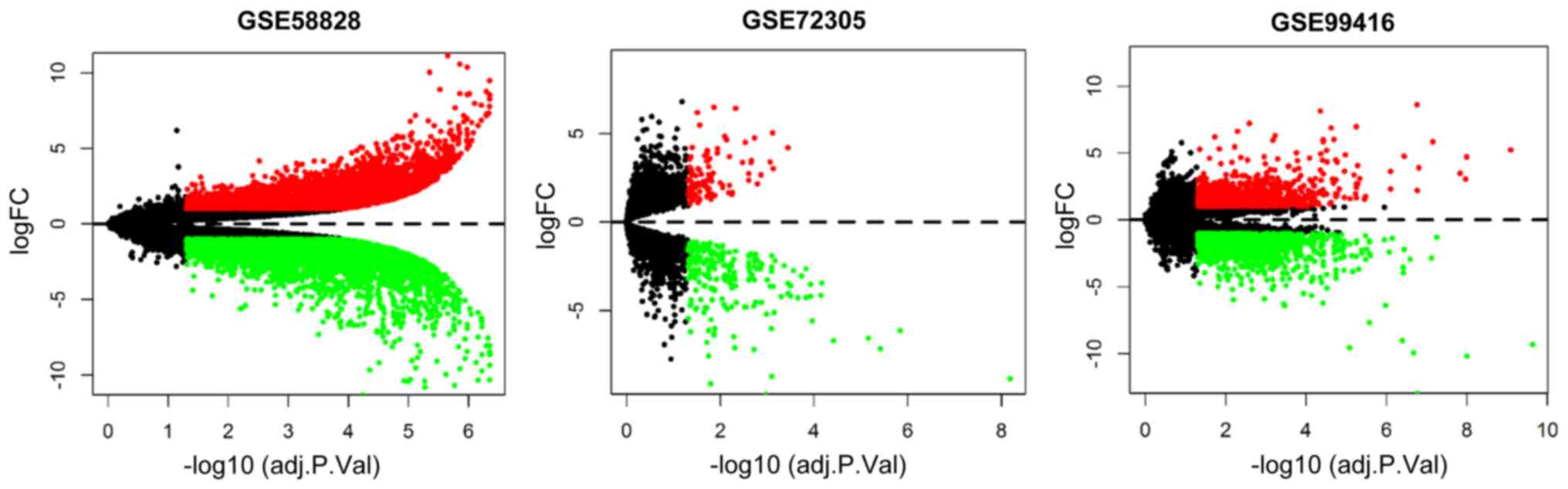
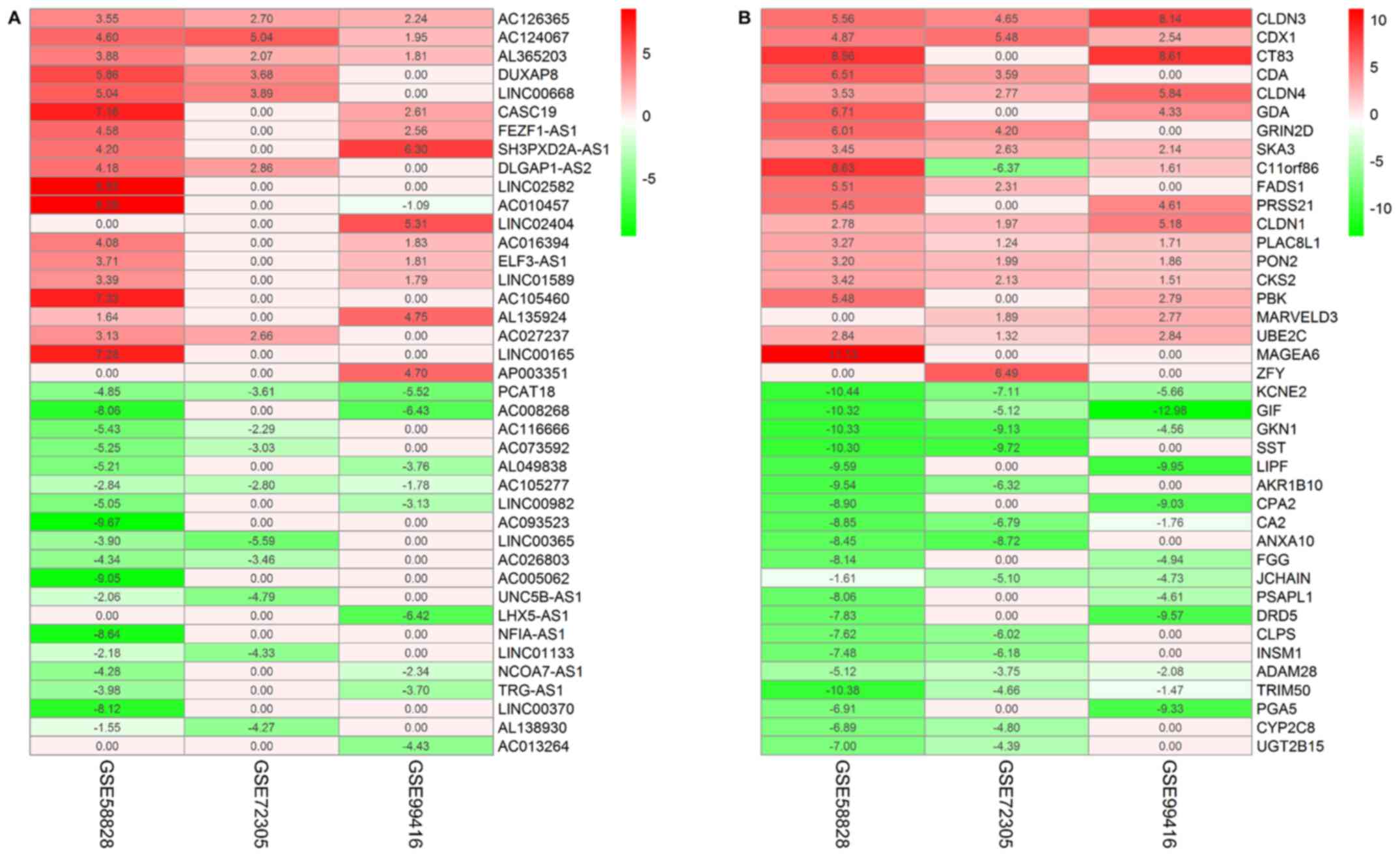
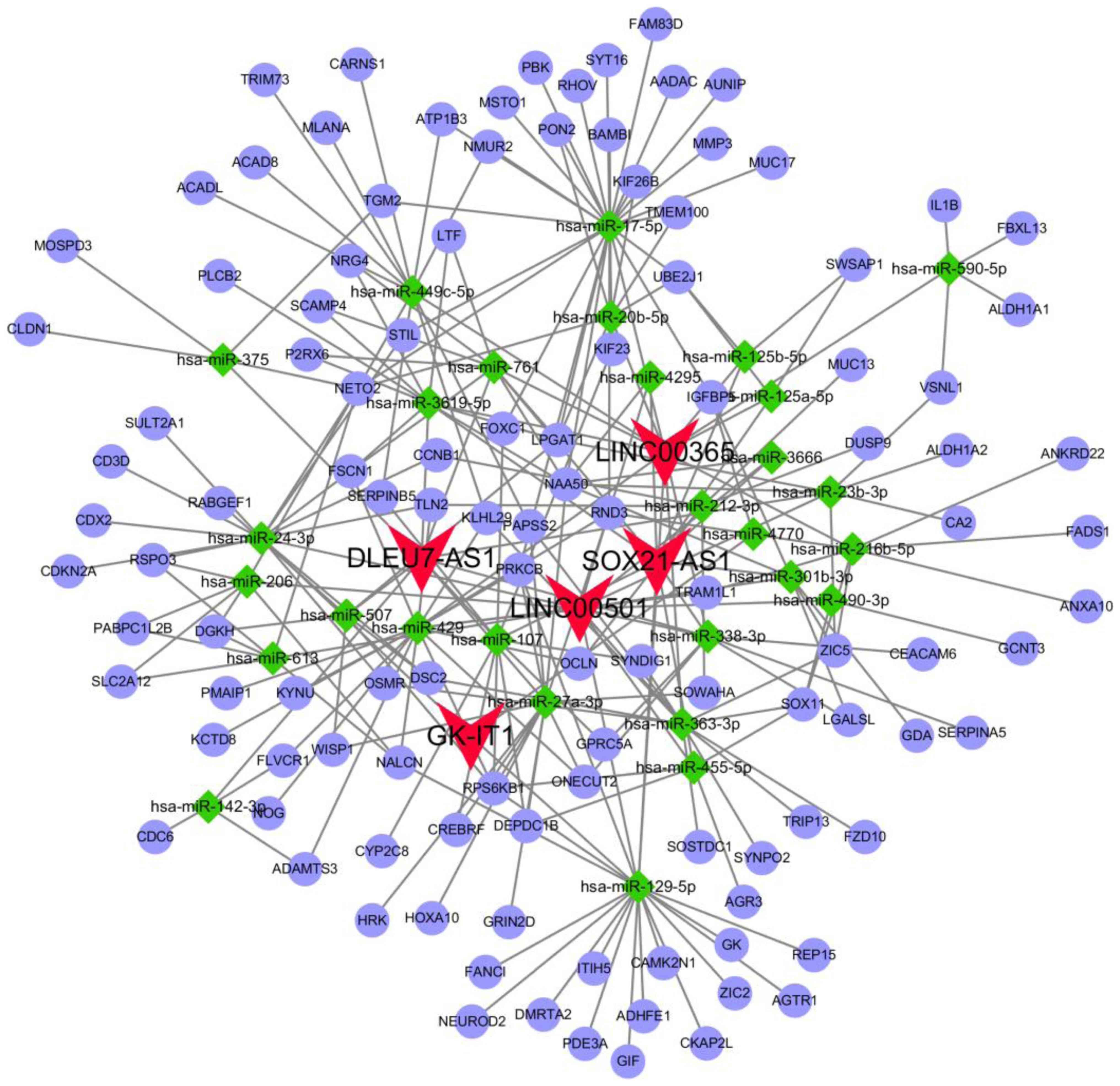
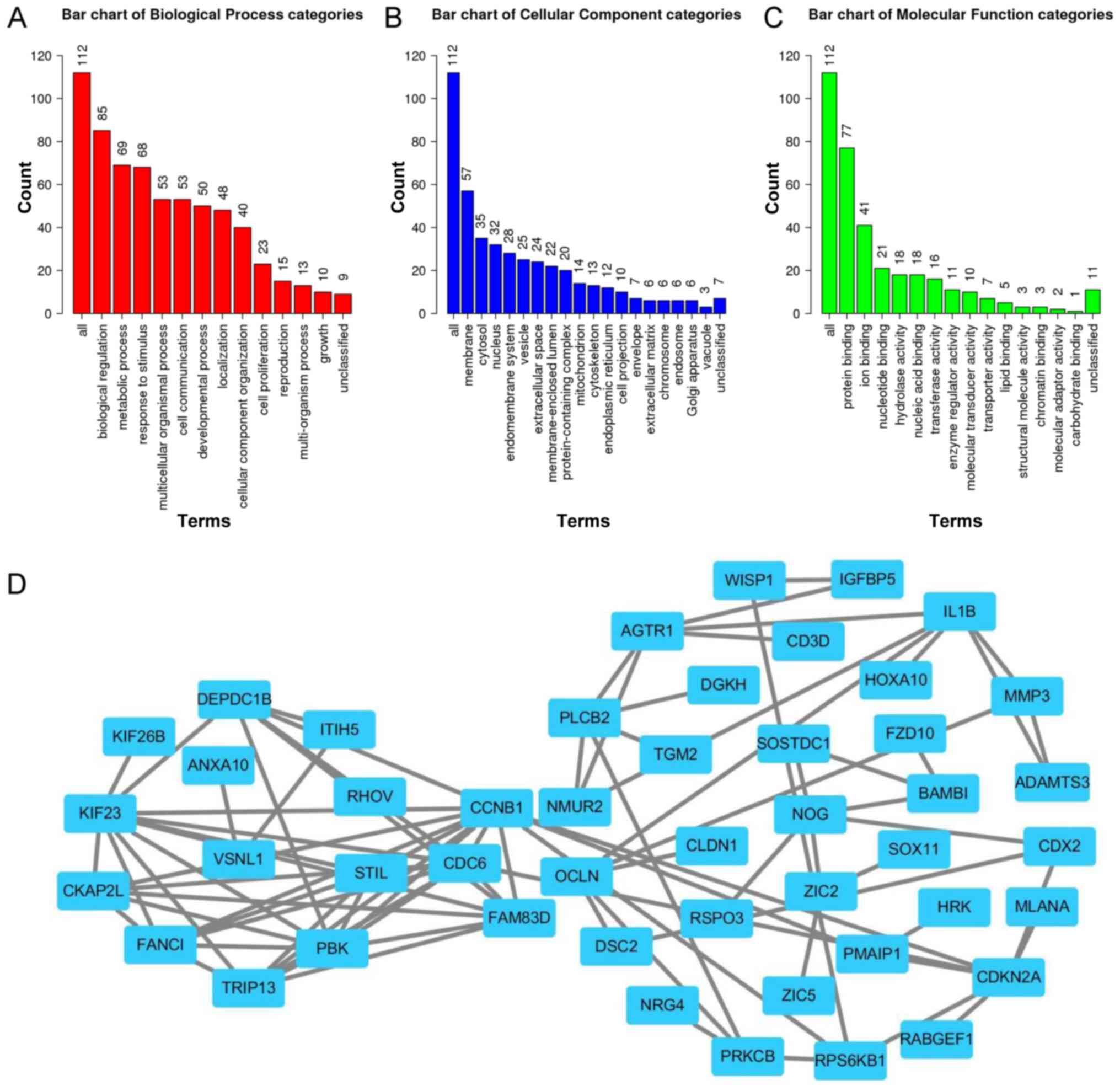
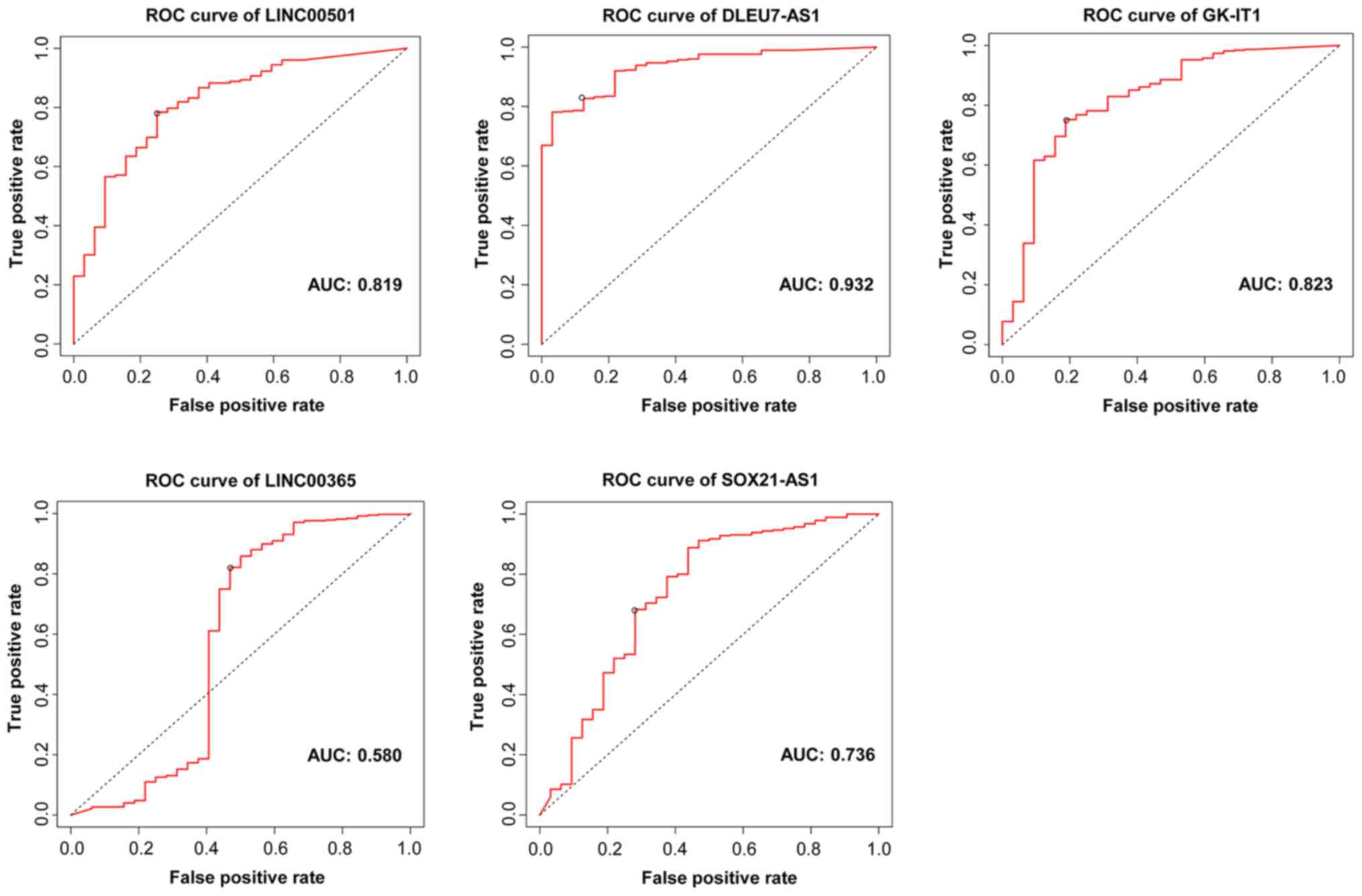
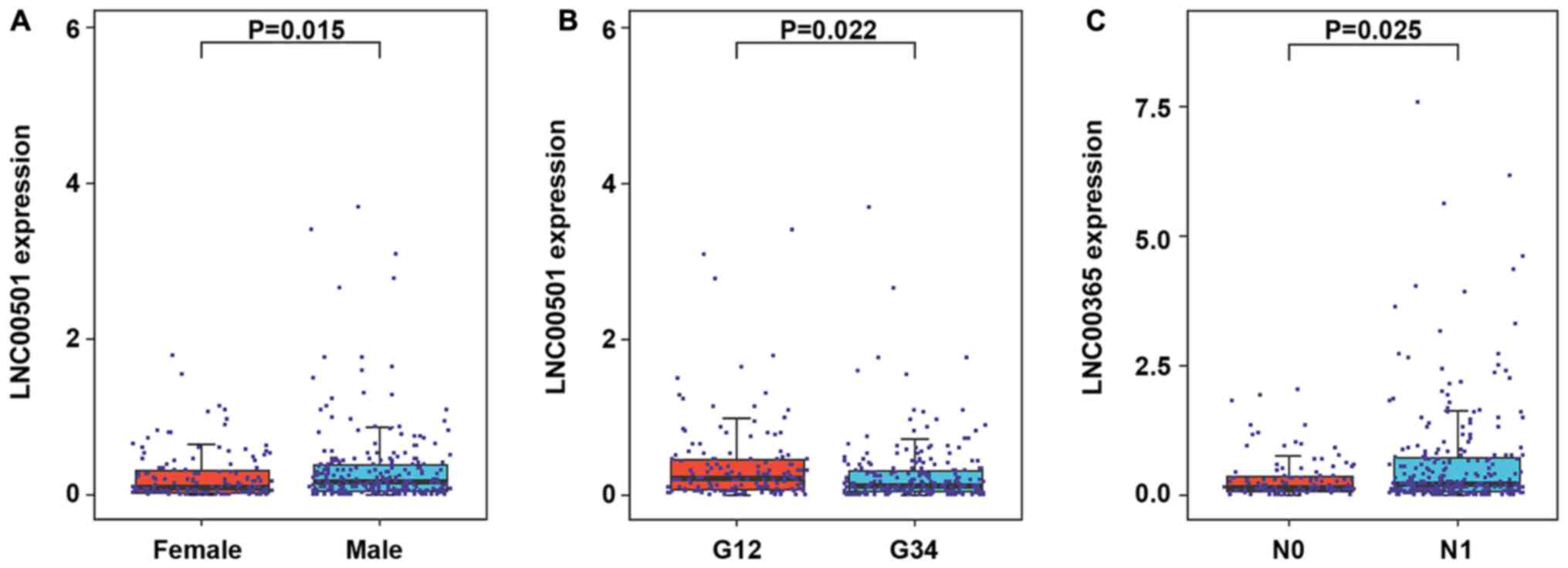
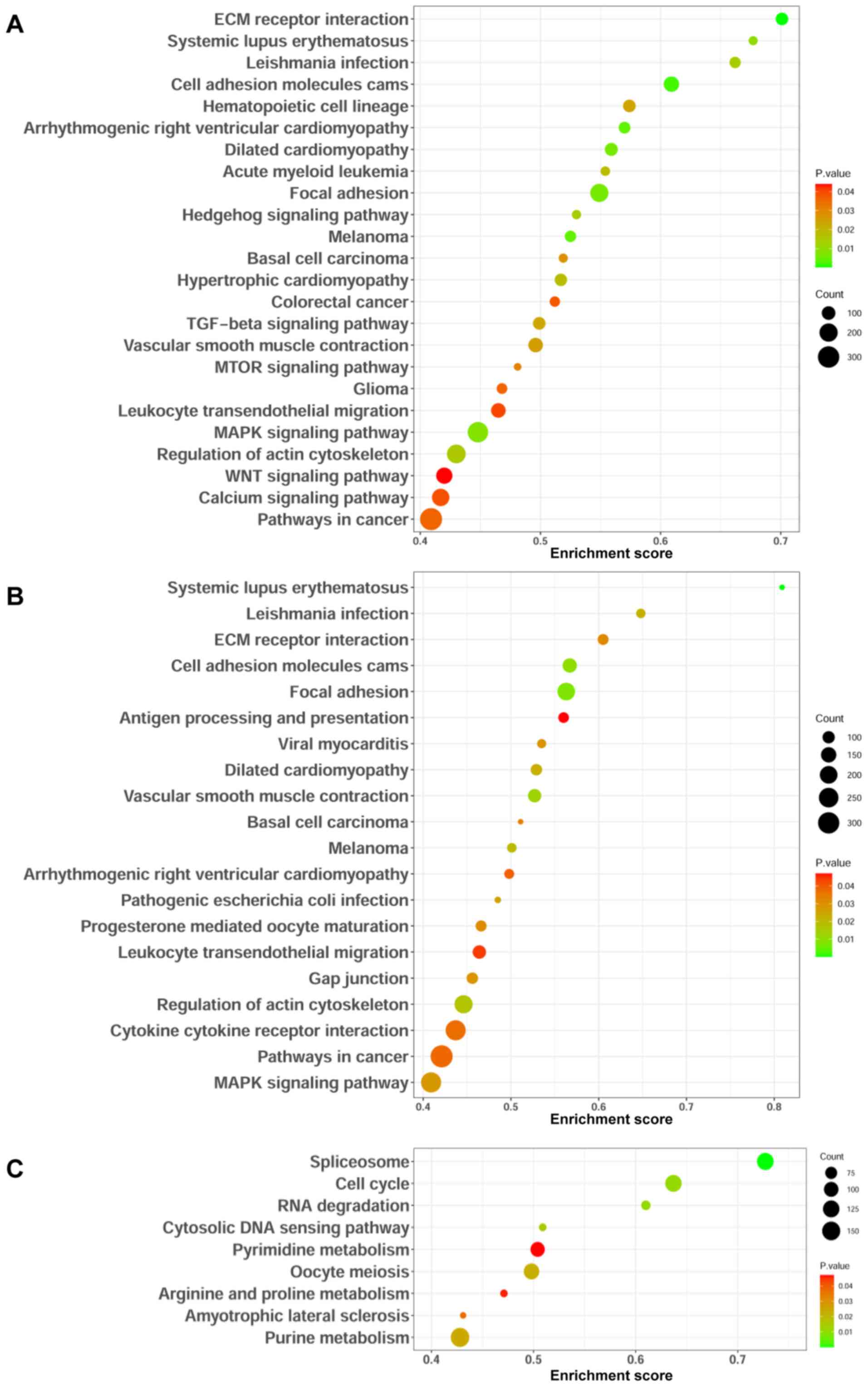
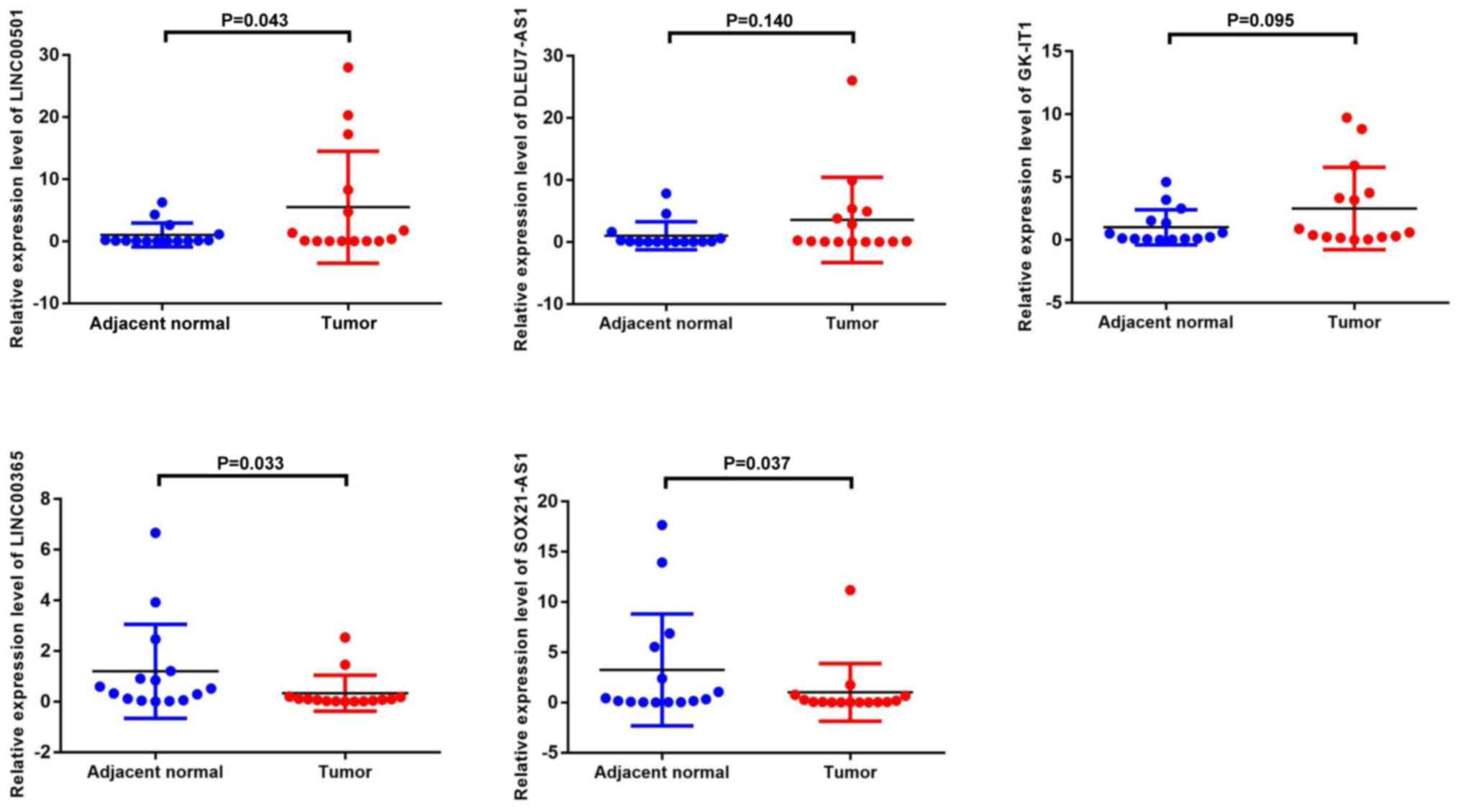
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forrest ME and Khalil AM: Review:
Regulation of the cancer epigenome by long non-coding RNAs. Cancer
Lett. 407:106–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY,
Liu Q, Zhao G and Zhang ZZ: The lncRNA UCA1 promotes proliferation,
migration, immune escape and inhibits apoptosis in gastric cancer
by sponging anti-tumor miRNAs. Mol Cancer. 18:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Deng W, Zhang Y, Cai J, Zhang J, Liu X,
Yin J, Bai Z, Yao H and Zhang Z: lncRNA-ANRIL promotes gastric
cancer progression by enhancing NF-kB signaling. Exp Biol Med
(Maywood). 244:953–959. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ba MC, Ba Z, Long H, Cui SZ, Gong YF, Yan
ZF, Lin KP, Wu YB and Tu YN: lncRNA AC093818.1 accelerates gastric
cancer metastasis by epigenetically promoting PDK1 expression. Cell
Death Dis. 11:642020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Russo F, Fiscon G, Conte F, Rizzo M, Paci
P and Pellegrini M: Interplay between long noncoding RNAs and
MicroRNAs in cancer. Methods Mol Biol. 1819:75–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu HT, Ma RR, Lv BB, Zhang H, Shi DB, Guo
XY, Zhang GH and Gao P: lncRNA-HNF1A-AS1 functions as a competing
endogenous RNA to activate PI3K/AKT signalling pathway by sponging
miR-30b-3p in gastric cancer. Br J Cancer. 122:1825–1836. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su YZ, Cui MF, Du J and Song B: lncRNA
DCST1-AS1 regulated cell proliferation, migration, invasion and
apoptosis in gastric cancer by targeting miR-605-3p. Eur Rev Med
Pharmacol Sci. 24:1158–1167. 2020.PubMed/NCBI
|
|
11
|
Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G,
Zhao Q, Wu D, Gong W, Du M, et al: lncRNA MT1JP functions as a
ceRNA in regulating FBXW7 through competitively binding to
miR-92a-3p in gastric cancer. Mol Cancer. 17:872018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luan M, Shi SS, Shi DB, Liu HT, Ma RR, Xu
XQ, Sun YJ and Gao P: TIPRL, a Novel tumor suppressor, suppresses
cell migration, and invasion through regulating AMPK/mTOR Signaling
pathway in gastric cancer. Front Oncol. 10:10622020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX,
Wu ZH, Chen XW, Gao P, Miao ZF and Wang ZN: Non-coding RNAs
participate in the regulatory network of CLDN4 via ceRNA mediated
miRNA evasion. Nat Commun. 8:2892017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41((Database Issue)):
D991–D995. 2013.
|
|
15
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA Pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e411. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Leek JT, Johnson WE, Parker HS, Jaffe AE
and Storey JD: The sva package for removing batch effects and other
unwanted variation in high-throughput experiments. Bioinformatics.
28:882–883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao Y, Wang J, Jaehnig EJ, Shi Z and
Zhang B: WebGestalt 2019: Gene set analysis toolkit with revamped
UIs and APIs. Nucleic Acids Res. 47:W199–W205. 2019. View Article : Google Scholar
|
|
23
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar
|
|
24
|
Martin IG, Dixon MF, Sue-Ling H, Axon AT
and Johnston D: Goseki histological grading of gastric cancer is an
important predictor of outcome. Gut. 35:758–763. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Washington K: 7th edition of the AJCC
cancer staging manual: Stomach. Ann Surg Oncol. 17:3077–3079. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lei S, He Z, Chen T, Guo X, Zeng Z, Shen Y
and Jiang J: Long noncoding RNA 00976 promotes pancreatic cancer
progression through OTUD7B by sponging miR-137 involving EGFR/MAPK
pathway. J Exp Clin Cancer Res. 38:4702019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H,
Kong J, Ding K, Shen HM, Wu H, et al: Long non-coding RNA linc00673
regulated non-small cell lung cancer proliferation, migration,
invasion and epithelial mesenchymal transition by sponging
miR-150-5p. Mol Cancer. 16:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
30
|
Yao N, Fu Y, Chen L, Liu Z, He J, Zhu Y,
Xia T and Wang S: Long non-coding RNA NONHSAT101069 promotes
epirubicin resistance, migration, and invasion of breast cancer
cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene.
38:7216–7233. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan H, Jin X, Liao C, Qiao L and Zhao W:
MicroRNA-301b-3p accelerates the growth of gastric cancer cells by
targeting zinc finger and BTB domain containing 4. Pathol Res
Pract. 215:1526672019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yan R, Li K, Yuan DW, Wang HN, Zhang Y,
Dang CX and Zhu K: Downregulation of microRNA-4295 enhances
cisplatin-induced gastric cancer cell apoptosis through the
EGFR/PI3K/Akt signaling pathway by targeting LRIG1. Int J Oncol.
53:2566–2578. 2018.PubMed/NCBI
|
|
33
|
Deng M, Qin Y, Chen X, Wang Q and Wang J:
MiR-206 inhibits proliferation, migration, and invasion of gastric
cancer cells by targeting the MUC1 gene. Onco Targets Ther.
12:849–859. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu Y, Tang L, Zhang Q, Zhang Z and Wei W:
MicroRNA-613 inhibits the progression of gastric cancer by
targeting CDK9. Artif Cells Nanomed Biotechnol. 46:980–984. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun T, Wang C, Xing J and Wu D: miR-429
modulates the expression of c-myc in human gastric carcinoma cells.
Eur J Cancer. 47:2552–2559. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu Y, Shao QS, Yao HB, Jin Y, Ma YY and
Jia LH: Overexpression of FOXC1 correlates with poor prognosis in
gastric cancer patients. Histopathology. 64:963–970. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu H, Zhang Y, Hao X, Kong F, Li X, Yu J
and Jia Y: GPRC5A overexpression predicted advanced biological
behaviors and poor prognosis in patients with gastric cancer.
Tumour Biol. 37:503–510. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Y, Chen H, Dong P, Xie G, Zhou Y, Ma
Y, Yuan X, Yang J, Han L, Chen L and Shen L: KIF23 activated
Wnt/β-catenin signaling pathway through direct interaction with
Amer1 in gastric cancer. Aging (Albany NY). 12:8372–8396. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen EB, Qin X, Peng K, Li Q, Tang C, Wei
YC, Yu S, Gan L and Liu TS: HnRNPR-CCNB1/CENPF axis contributes to
gastric cancer proliferation and metastasis. Aging (Albany NY).
11:7473–7491. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ding L, Yang L, He Y, Zhu B, Ren F, Fan X,
Wang Y, Li M, Li J, Kuang Y, et al: CREPT/RPRD1B associates with
Aurora B to regulate Cyclin B1 expression for accelerating the G2/M
transition in gastric cancer. Cell Death Dis. 9:11722018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang M, Ma X, Shi H, Hu L, Fan Z, Pang L,
Zhu F, Yang X, Xu W, Liu B, et al: FAM83D, a microtubule-associated
protein, promotes tumor growth and progression of human gastric
cancer. Oncotarget. 8:74479–74493. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao B, Zhang J, Chen X, Xu H and Huang B:
Mir-26b inhibits growth and resistance to paclitaxel chemotherapy
by silencing the CDC6 gene in gastric cancer. Arch Med Sci.
15:498–503. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cao Y, Xiong JB, Zhang GY, Liu Y, Jie ZG
and Li ZR: Long Noncoding RNA UCA1 Regulates PRL-3 expression by
sponging MicroRNA-495 to promote the progression of gastric cancer.
Mol Ther Nucleic Acids. 19:853–864. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cui Y, Pu R, Ye J, Huang H, Liao D, Yang
Y, Chen W, Yao Y and He Y: lncRNA FAM230B promotes gastric cancer
growth and metastasis by regulating the miR-27a-5p/TOP2A Axis. Dig
Dis Sci. Sep 10–2020.(Epub ahead of print). doi:
10.1007/s10620-020-06581-z. View Article : Google Scholar
|
|
45
|
Zhang M, Dong BB, Lu M, Zheng MJ, Chen H,
Ding JZ, Xu AM and Xu YH: miR-429 functions as a tumor suppressor
by targeting FSCN1 in gastric cancer cells. Onco Targets Ther.
9:1123–1133. 2016.PubMed/NCBI
|
|
46
|
Chen ZL, Qin L, Peng XB, Hu Y and Liu B:
INHBA gene silencing inhibits gastric cancer cell migration and
invasion by impeding activation of the TGF-β signaling pathway. J
Cell Physiol. 234:18065–18074. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xiang Z, Li J, Song S, Wang J, Cai W, Hu
W, Ji J, Zhu Z, Zang L, Yan R and Yu Y: A positive feedback between
IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric
cancer metastasis. J Exp Clin Cancer Res. 38:3142019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gao J, Zhao C, Liu Q, Hou X, Li S, Xing X,
Yang C and Luo Y: Cyclin G2 suppresses Wnt/β-catenin signaling and
inhibits gastric cancer cell growth and migration through Dapper1.
J Exp Clin Cancer Res. 37:3172018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang L, Kang W, Lu X, Ma S, Dong L and
Zou B: lncRNA CASC11 promoted gastric cancer cell proliferation,
migration and invasion in vitro by regulating cell cycle pathway.
Cell Cycle. 17:1886–1900. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hou Y, Yu Z, Tam NL, Huang S, Sun C, Wang
R, Zhang X, Wang Z, Ma Y, He X and Wu L: Exosome-related lncRNAs as
predictors of HCC patient survival: A prognostic model. Am J Transl
Res. 10:1648–1662. 2018.PubMed/NCBI
|
|
51
|
Lou X, Li J, Yu D, Wei YQ, Feng S and Sun
JJ: Comprehensive analysis of five long noncoding RNAs expression
as competing endogenous RNAs in regulating hepatoma carcinoma.
Cancer Med. 8:5735–5749. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ye J, Zhang J, Lv Y, Wei J, Shen X, Huang
J, Wu S and Luo X: Integrated analysis of a competing endogenous
RNA network reveals key long noncoding RNAs as potential prognostic
biomarkers for hepatocellular carcinoma. J Cell Biochem.
120:13810–13825. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhu Y, Bian Y, Zhang Q, Hu J, Li L, Yang
M, Qian H, Yu L, Liu B and Qian X: LINC00365 promotes colorectal
cancer cell progression through the Wnt/β-catenin signaling
pathway. J Cell Biochem. 121:1260–1272. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang L, Yan X, Yu S, Zhong X, Tian R, Xu
L, Bian X and Su J: LINC00365-SCGB2A1 axis inhibits the viability
of breast cancer through targeting NF-κB signaling. Oncol Lett.
19:753–762. 2020.PubMed/NCBI
|
|
55
|
Yan XY, Zhang JJ, Zhong XR, Yu SH, Xu L,
Tian R, Sun LK and Su J: The LINC00365/SCGB2A1 (Mammaglobin B) axis
down-regulates NF-κB signaling and is associated with the
progression of gastric cancer. Cancer Manag Res. 12:621–631. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang R, Li Y, Du P, Zhang X, Li X and
Cheng G: Hypomethylation of the lncRNA SOX21-AS1 has clinical
prognostic value in cervical cancer. Life Sci. 233:1167082019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang X, Zhao X, Li Y, Zhou Y and Zhang Z:
Long noncoding RNA SOX21-AS1 promotes cervical cancer progression
by competitively sponging miR-7/VDAC1. J Cell Physiol.
234:17494–17504. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wei C, Wang H, Xu F, Liu Z and Jiang R:
lncRNA SOX21-AS1 is associated with progression of hepatocellular
carcinoma and predicts prognosis through epigenetically silencing
p21. Biomed Pharmacother. 104:137–144. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lu X, Huang C, He X, Liu X, Ji J, Zhang E,
Wang W and Guo R: A Novel long non-coding RNA, SOX21-AS1, indicates
a poor prognosis and promotes lung adenocarcinoma proliferation.
Cell Physiol Biochem. 42:1857–1869. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wei AW and Li LF: Long non-coding RNA
SOX21-AS1 sponges miR-145 to promote the tumorigenesis of
colorectal cancer by targeting MYO6. Biomed Pharmacother.
96:953–959. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yang CM, Wang TH, Chen HC, Li SC, Lee MC,
Liou HH, Liu PF, Tseng YK, Shiue YL, Ger LP and Tsai KW: Aberrant
DNA hypermethylation-silenced SOX21-AS1 gene expression and its
clinical importance in oral cancer. Clin Epigenetics. 8:1292016.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang J, Hou T, Qi X, Wang J and Sun X:
SOX21-AS1 is associated with clinical stage and regulates cell
proliferation in nephroblastoma. Biosci Rep. 39:BSR201906022019.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu XB, Han C and Sun CZ: Long non-coding
RNA DLEU7-AS1 promotes the occurrence and development of colorectal
cancer via Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci.
22:110–117. 2018.PubMed/NCBI
|
|
64
|
Yu Y, Chen X and Cang S: Cancer-related
long noncoding RNAs show aberrant expression profiles and competing
endogenous RNA potential in esophageal adenocarcinoma. Oncol Lett.
18:4798–4808. 2019.PubMed/NCBI
|
|
65
|
Pan H, Guo C, Pan J, Guo D, Song S, Zhou Y
and Xu D: Construction of a competitive endogenous RNA network and
identification of Potential regulatory axis in gastric cancer.
Front Oncol. 9:9122019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Peng J, Zhu Y, Dong X, Mao X, Lou Y, Mu Y,
Xue D and Zhou H: Construction and analysis of lncRNA-associated
ceRNA network identified potential prognostic biomarker in gastric
cancer. Transl Cancer Res. 8:1116–1128. 2019. View Article : Google Scholar
|
|
67
|
Qi M, Yu B, Yu H and Li F: Integrated
analysis of a ceRNA network reveals potential prognostic lncRNAs in
gastric cancer. Cancer Med. 9:1798–1817. 2020. View Article : Google Scholar : PubMed/NCBI
|