Introduction
Oral tongue squamous cell carcinoma (OTSCC) is
difficult to detect because of its subtle early symptoms. The
incidence of OTSCC is caused by long-term mechanical and chemical
irritation of the oral cavity (1).
The early symptoms of OTSCC include foreign body sensation or
swallowing pain. With tumor growth, dysphagia, unclear enunciation
and deep ear pain may occur, resulting in heavy physical and mental
burdens (2,3). In recent years, with the application of
systemic chemotherapy and radiotherapy, the prognosis of patients
has significantly improved; however, the 5-year survival rate
remains low (4). It is therefore
crucial to identify novel key molecules that could provide
scientific targets for accurate diagnosis and precise treatment of
OTSCC, to ultimately improve the poor prognosis of patients.
Pyruvate kinase (PK) is the ultimate rate-limiting
enzyme of glycolysis, and the M2 splicing subtype of pyruvate
kinase (PKM2) is the key enzyme involved in the Warburg effect,
which participates in the transfer of phosphoenolpyruvate to
adenosine diphosphate (ATP) (5,6).
Accumulating evidence demonstrated that PKM2 is highly expressed in
many malignant tumors, such as liver, gastric and colon cancers,
and that its high expression is a necessary condition to ensure
Warburg effect (7,8). The Warburg effect allows tumor cells to
replenish their production capacity in the microenvironment, which
is characterized by hypoxia and mitochondrial oxidative
phosphorylation. The resulting high ATP/ADP ratio induces a rapid
proliferation of tumor cells and promotes malignant signal
transduction mechanisms (9).
Phosphorylation of PKM2 at tyrosine 105 (Y105) activates
Yes-associated protein (YAP), which then increases cancer stem-like
cell properties through Warburg effect to promote the malignant
proliferation of breast cancer cells (10). In addition, by interacting with
inositol-1,4,5-triphosphate receptor, methylated PKM2 can inhibit
the influx of calcium from endoplasmic reticulum to mitochondrion,
which leads to an increase in tumor cell proliferation, migration
and metastasis (11). However, the
effects of PKM2 on OTSCC have not been established.
The present study aimed to investigate the
expression of PKM2 in OTSCC tissues, and to determine its
prognostic value in patients with OTSCC. In addition, we analyzed
the effect of PKM2 on the proliferation and apoptosis of OTSCC
cells.
Materials and methods
Patients and samples
A total of 125 OTSCC tumor tissues and paired
adjacent tissues (>5 cm from the boundary of the tumor) were
collected from patients who underwent surgical resection between
June 2011 and August 2013 at the Suining Central Hospital. The
clinicopathological characteristics of patients were obtained from
the Suining Central Hospital. All clinical samples were stored in
liquid nitrogen (12). A follow-up
was implemented to evaluate the survival rate of patients 5 years
post-surgery. This study was approved by the Ethics Committee of
the Suining Central Hospital (approval no. 20110067, Suining,
China) and was conducted in accordance with the Declaration of
Helsinki.
Cell lines and transfection
The human oral squamous carcinoma cell lines SCC-9,
SCC-15, the human tongue squamous cell carcinoma cell line H357 and
the normal embryonic bovine tracheal epithelial cell line EBTr were
obtained from the American Type Culture Collection. All cells were
cultured in Dulbecco's modified Eagle's medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 100 units/ml penicillin and streptomycin
(Gibco; Thermo Fisher Scientific, Inc.) and placed at 37°C in a
humidified incubator containing 5% CO2.
A total of 1×105 SCC-9 and H357 cells
cultured in serum free DMEM were transfected with 100 nM of small
interfering (si)-PKM2 and si-negative control (NC; Guangzhou
RiboBio Co., Ltd.) according to the manufacturers' instructions
using Lipofectamine® 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C. The transfection efficiency was
confirmed using RT-qPCR 24 h after transfection and transfected
cells were used for following experiments.
RNA extraction and reverse
transcription quantitative (RT-q)PCR
Total RNA from OTSCC tissues, adjacent tissues and
cell lines was extracted using TRIzol™ reagent (Takara Bio, Inc.).
Total RNA was reverse transcribed into cDNA using a PrimeScript RT
Reagent kit (Takara Bio, Inc.) at 37°C for 10 min, 45°C for 30 min
and 75°C for 10 min. PrimeScript RT Reagent (Takara Bio, Inc.) was
used for reverse transcription and RT-qPCR was performed using SYBR
Premix Ex Taq II (Takara Bio, Inc.) and a LightCycler system (Roche
Diagnostics). The sequences of the primers were as follows: PKM2,
forward 5′-GGGTTCGGAGGTTTGATG-3′ and reverse
5′-ACGGCGGTGGCTTCTGT-3′; and GAPDH, forward
5′-ATGTTGCAACGGGAAGGA-3′ reverse 5′-AGGAAAAGCATCACCCGGAG-3′. The
following thermocycling conditions were used for the qPCR: Initial
denaturation at 90°C for 2 min; followed by 40 cycles at 95°C for
15 sec, 63°C for 30 sec and 75°C for 30 sec. The relative
expression levels were normalized to endogenous control and were
expressed as 2−ΔΔCq (13).
Immunohistochemistry (IHC) and
immunohistochemical staining
Tissue microarrays (TMA) were prepared from the 125
pairs of OTSCC and adjacent tissues. Briefly, tissues were fixed in
10% formalin for 12 h at room temperature and embedded in paraffin
blocks for 8 h at room temperature. TMA cores (1.5-mm diameter)
were constructed from formalin-fixed paraffin-embedded OTSCC and
adjacent tissues sections (4-µm thick). Tissue sections were
rehydrated in xylene and alcohol (100, 95 and 80%) and incubated
with 3% H2O2 for 30 min at 37°C. All sections
were incubated for 15 min with 5% goat serum (OriGene Technologies,
Inc.) to block nonspecific binding, followed by incubation with a
rabbit monoclonal PKM2 antibody (cat. no. ab38237; 1:100; Abcam) at
4°C overnight. Sections were subsequently incubated with
anti-rabbit secondary IgG antibody (1:100; cat. no. SAP-9100;
OriGene Technologies, Inc.) at 37°C for 30 min. After washing with
PBS, visualization was achieved using diaminobenzidine (Boster
Biological Technology).
PKM2 immunostaining was scored and examined by two
independent assessors. To determine the final staining scores, all
tissues were manually scored using the percentage of positively
stained cells and the intensity of the staining. The scoring
parameters included the staining intensity and the percentage of
positive cells. The staining intensity ranged from 0–3 as follows:
0, negative; 1, weak; 2, moderate; and 3, strong. The percentage of
positive cells ranged from 0–4 as follows: 0, negative or <5; 1,
6–25; 2, 26–50; 3, 51–75; and 4, 76–100%. The total score was equal
to the sum of staining intensity and percentage of positive cells.
Slides with a total score <4 were defined as low PKM2
expression, while slides with a score ≥4 were defined as high PKM2
expression (14).
Western blotting
Total protein was extracted using RIPA lysis buffer
(Beyotime Institute of Biotechnology). Total protein was quantified
using the Bradford protein assay (Bio-Rad Laboratories, Inc.) and
40 µg protein/lane was separated via 10% SDS-PAGE. The separated
proteins were subsequently transferred onto PVDF membranes that
were blocked with 5% skim milk powder at room temperature for 1 h.
The membranes were incubated with the following primary antibodies
at 4°C overnight: Anti-PKM2 (1:2,000; cat. no. ab85555; Abcam),
anti-large tumor suppressor kinase 1 (LATS1; 1:5,000; cat. no.
ab70562; Abcam), anti-yes-associated protein (YAP; 1:5,000; cat.
no. ab205270; Abcam), anti-phosphorylated (p)-YAP (1:1,000; cat.
no. ab205270; Abcam), anti-Bax (1:5,000; cat. no. ab32503; Abcam),
anti-Bcl-2 (1:1,000; cat. no. ab182858; Abcam), anti-Ki-67
(1:1,000; cat. no. ab92742; Abcam) and anti-GAPDH (1:5,000; cat.
no. ab9485; Abcam). Subsequently, the membranes were incubated with
secondary antibodies (1:5,000; cat. nos. ab6721 and ab6789; Abcam)
at room temperature for 1 h. Bands were visualized by enhanced
chemiluminescence (EMD Millipore). GAPDH was used as the loading
control. Protein expression was quantified using Quantity One
version 4.5 software (Bio-Rad Laboratories, Inc.).
Bioinformatic analysis
The gene expression profiles of 37 patients with
OTSCC, including 20 pairs of tumor and adjacent nontumor tissues,
were download from the Gene Expression Omnibus (GEO; GSE13601;
http://www.ncbi.nlm.nih.gov/geo/)
(15). This dataset contained the
highest number of paired tissue expression data of OTSCC in GEO
database. The data were normalized according to normalization
function between Arrays function contained in limma package in R
software (PMID: 25605792) (16), and
the expression of PKM2 was selected and visualized by GraphPad
Prism software. The clinicopathological parameters of patients
included in this dataset are presented in Table SI.
CCK-8 assay
Cells were seeded in a 96-well plate at the density
of 1×103 cells per 100 µl and cultured for 24 h at 37°C.
Cells were incubated with 10 µl of Cell Counting Kit-8 (CCK-8)
solution (Dojindo Molecular Technologies, Inc.) for 1 h. The
absorbance was read at 450 nm using a microplate reader.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
Cells were seeded in a 96-well plate
(1×105 cells) and cultured until they reached 30–50%
confluence. Cells were fixed with 4% polyformaldehyde at room
temperature for 30 min and the nuclei were permeabilized using 0.5%
Triton X-100 solution at room temperature for 30 min. Cells were
incubated with EdU (50 µM), 1× ApolloR reaction cocktail (100 µl)
and 1× Hoechst 33342 (100 µl) at room temperature for 30 min
according to the manufacturer's instructions. Cells were visualized
using fluorescent microscope (magnification, ×100). Cell
proliferation was analyzed according to the mean number of stained
cells by using ImageJ (version 1.41; National Institutes of
Health).
Flow cytometry analysis of cell
apoptosis and cell cycle
To evaluate cell apoptosis, SCC-9 or H357 cells
(1×105 cells) was seeded into 6 well plates. Once they
reached confluence, cells were collected and incubated with Annexin
V-FITC (5 µl) and propidium iodide (PI) solution (5 µl; Biogot
Technology Co., Ltd.) at room temperature for 15 min according to
the manufacturers' instructions. Cells were subsequently suspended
in 400 µl binding buffer. To evaluate the cell cycle, SCC-9 and
H357 cells were collected and fixed in 75% ethanol at −20°C
overnight. Then, the fixed cells were washed with PBS and incubated
with RNase A for 20 min at room temperature. These cells were
stained with PI and incubated in the dark for 30 min at 4°C. Cell
apoptosis and cell cycle progression were analyzed using flow
cytometry (BD Biosciences). The percentages of cells within each
phase of the cell cycle were analyzed with ModFit version 4.0
(Verity Software House, Inc.) and CellQuest version 5.1 (Thermo
Fisher Scientific, Inc.).
Statistical analysis
The data were analyzed using SPSS version 20.0
software (IBM Corp.) and GraphPad Prism version 6.0 software
(GraphPad Software Inc.). The overall survival (OS) of patients
with OTSCC was analyzed using Kaplan-Meier method and log-rank
test. Univariate and multivariate cox regression analyses were used
to analyze the prognostic significance of PKM2 expression. The
association between PKM2 expression and the clinicopathological
characteristics of patients with OTSCC patients was analyzed using
a χ2 test. Differences between two groups were analyzed
using Student's t-test. Comparisons between multiple groups were
performed using one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
PKM2 expression in OTSCC
The expression of PKM2 in OTSCC and normal adjacent
tissues was determined by RT-qPCR. The results demonstrated that
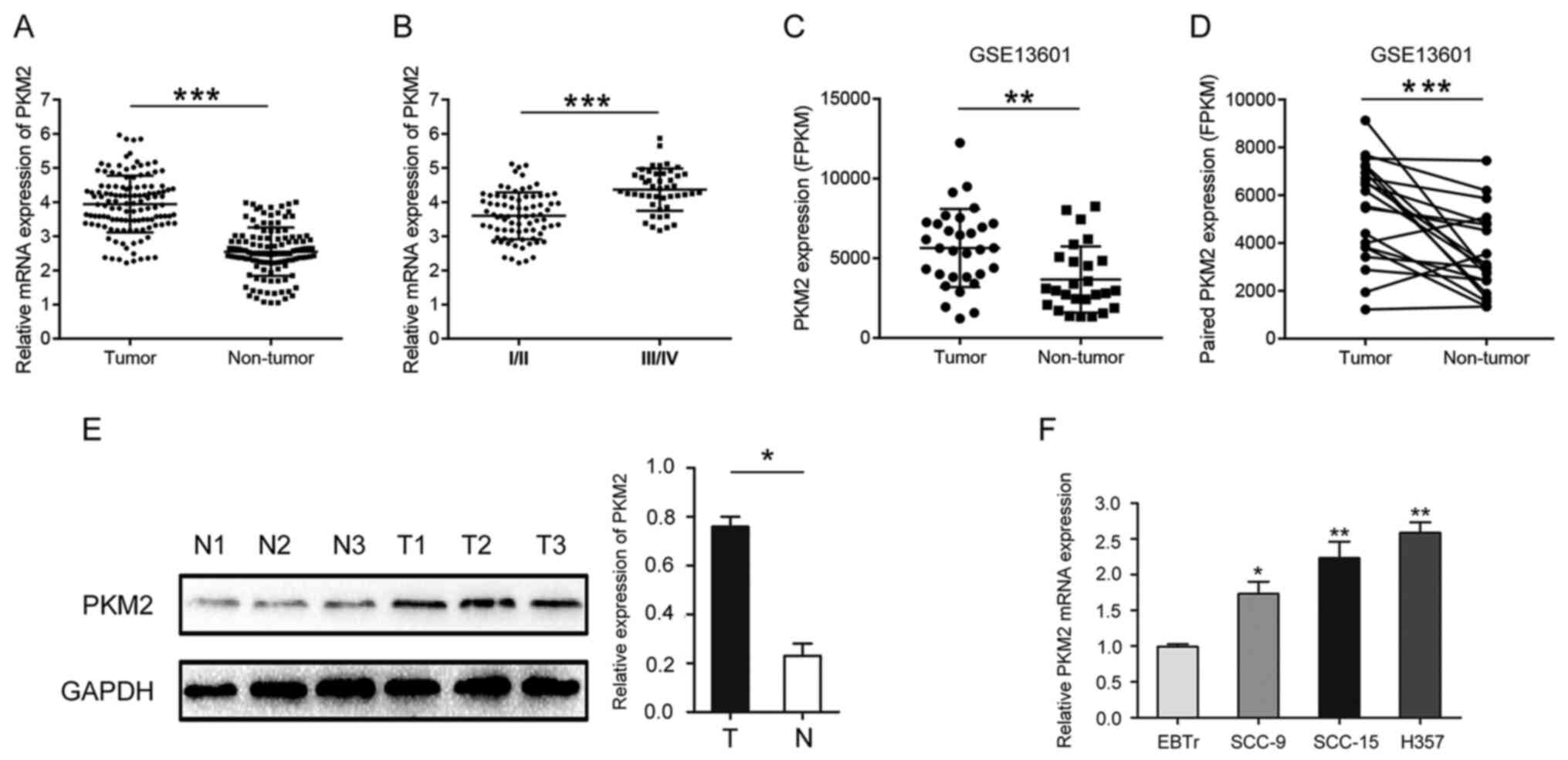
PKM2 expression was significantly higher in OTSCC tissues compared
with paired normal adjacent tissues (Fig. 1A). Furthermore, PKM2 expression was
significantly higher in stage III/IV OTSCC samples compared with
I/II OTSCC samples (Fig. 1B).
Similar results were obtained by analyzing microarray sequencing
results (GSE13601) downloaded from GEO public database using
Graphpad Prism software (Fig. 1C and
D). The results from western blotting confirmed that PKM2 was
highly expressed in OTSCC tissues compared with normal tissues
(Fig. 1E). The expression of PKM2
was also significantly increased in OTSCC cell lines compared with
normal cells (Fig. 1F).
Association between PKM2 expression
and the clinicopathological characteristics of patients with
OTSCC
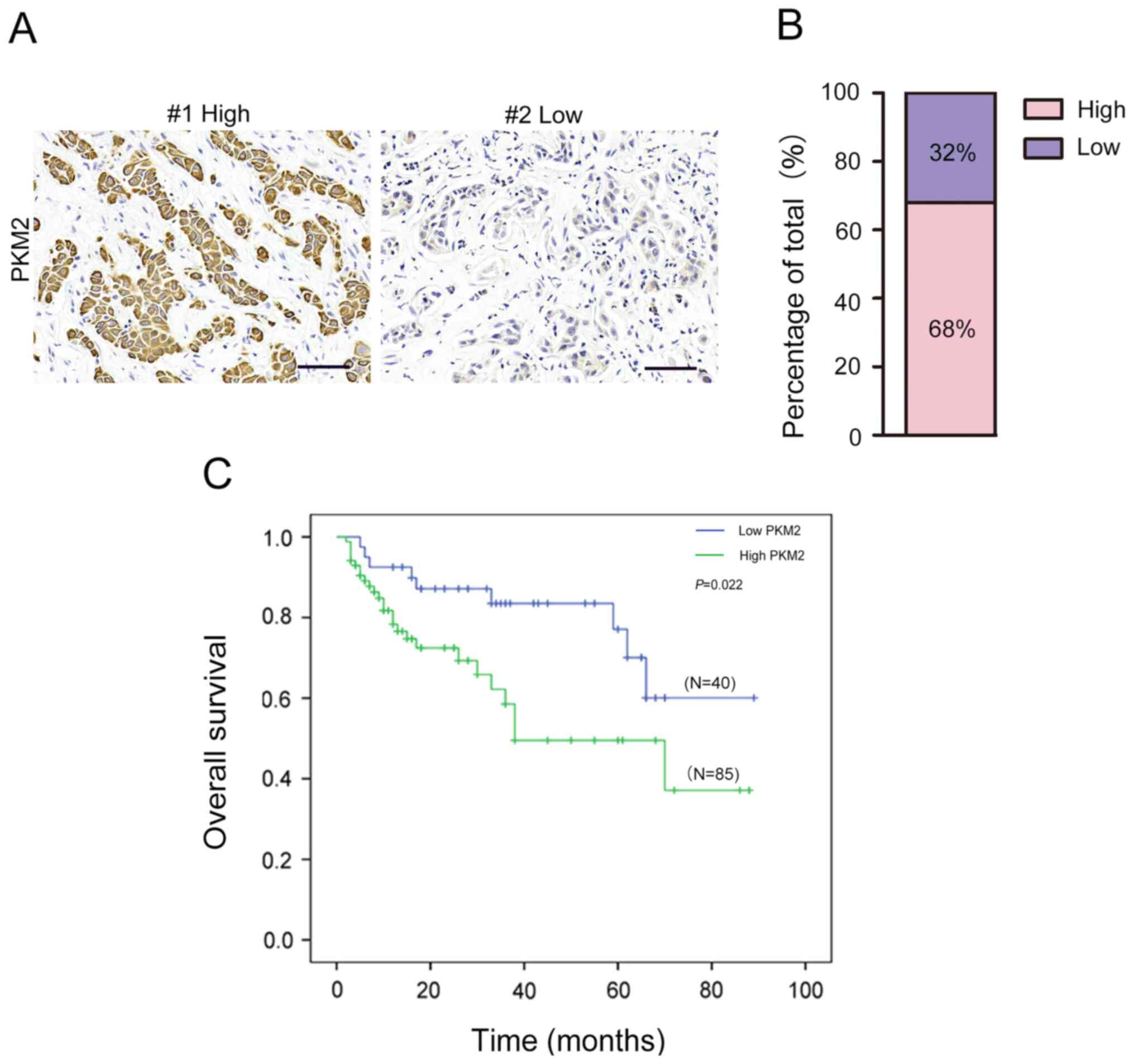
The results from IHC staining demonstrated that PKM2
expression was high in 85 out of the 125 OTSCC samples (68%) and
low in 40 out of 125 OTSCC samples (32%; Fig. 2A and B). Furthermore, high PKM2
expression was associated with TNM stage (Table I). However, there was no significant
association between PKM2 expression and age, sex, smoking, drinking
or tumor size (all P>0.05).
 | Table I.Association between PKM2 expression
and the clinicopathological characteristics of patients with oral
tongue squamous cell carcinoma. |
Table I.
Association between PKM2 expression
and the clinicopathological characteristics of patients with oral
tongue squamous cell carcinoma.
|
|
| PKM2
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | n | Low (n=40) | High (n=85) | P-value |
|---|
| Age, years |
|
|
| 0.963 |
|
<55 | 66 | 21 | 45 |
|
|
≥55 | 59 | 19 | 40 |
|
| Sex |
|
|
| 0.494 |
|
Male | 79 | 27 | 52 |
|
|
Female | 46 | 13 | 33 |
|
| Smoking |
|
|
| 0.443 |
| No | 38 | 14 | 24 |
|
|
Yes | 87 | 26 | 61 |
|
| Drinking |
|
|
| 0.881 |
| No | 45 | 15 | 30 |
|
|
Yes | 80 | 25 | 55 |
|
| TNM stage |
|
|
| 0.001 |
|
I/II | 61 | 28 | 32 |
|
|
III/IV | 65 | 12 | 53 |
|
| Tumor size |
|
|
| 0.103 |
|
T1-T2 | 68 | 26 | 42 |
|
|
T3-T4 | 57 | 14 | 43 |
|
Association between PKM2 expression
and OS in patients with OTSCC
The association between PKM2 expression and the
prognosis of patients with OTSCC was determined. The results from
Kaplan-Meier survival analysis revealed that patients with high
PKM2 expression had significantly shorter OS compared with those
with low PKM2 expression (Fig. 2C).
In addition, univariate analysis demonstrated that TNM stage
[P=0.009; confidence interval (CI):1.045–4.669, hazard ratio
(HR)=1.473] and PKM2 expression (P=0.007; CI:1.047–3.647; HR=1.227)
were significantly associated with OS in patients with OTSCC
(Table II). The results from
multivariate analysis demonstrated that TNM stage (P=0.011; CI,
0.978–2.604; HR=1.186) and PKM2 expression (P=0.013; CI,
0.768–2.964; HR=1.377) were independent prognostic factors for OS
in patients with OTSCC (Table
II).
 | Table II.Univariate and multivariate analysis
of the clinicopathological characteristics of patients with oral
tongue squamous cell carcinoma. |
Table II.
Univariate and multivariate analysis
of the clinicopathological characteristics of patients with oral
tongue squamous cell carcinoma.
|
|
| Univariate
analysis | Multivariate
analysis model |
|---|
|
|
|
|
|
|---|
| Variable | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex |
| 0.364
(0.408–1.273) | 0.534 |
|
|
|
Male | 70 |
|
|
|
|
|
Female | 46 |
|
|
|
|
| Age, years |
| 0.475
(0.339–1.567) | 0.663 |
|
|
|
<55 | 66 |
|
|
|
|
|
≥55 | 59 |
|
|
|
|
| Smoking |
| 1.203
(0.963–3.442) | 0.076 |
|
|
| No | 38 |
|
|
|
|
|
Yes | 87 |
|
|
|
|
| Drinking |
| 1.104
(0.861–2.776) | 0.537 |
|
|
| No | 45 |
|
|
|
|
|
Yes | 80 |
|
|
|
|
| TNM stage |
| 1.473
(1.045–4.669) | 0.009 | 1.186
(0.978–2.604) | 0.011 |
|
I/II | 61 |
|
|
|
|
|
III/IV | 65 |
|
|
|
|
| Tumor size |
| 0.864
(0.883–1.487) | 0.086 |
|
|
|
T1-T2 | 68 |
|
|
|
|
|
T3-T4 | 57 |
|
|
|
|
| PKM2
expression |
| 1.227
(1.047–3.647) | 0.007 | 1.377
(0.768–2.964) | 0.013 |
|
High | 85 |
|
|
|
|
|
Low | 40 |
|
|
|
|
PKM2 knockdown inhibits OTSCC cell
proliferation
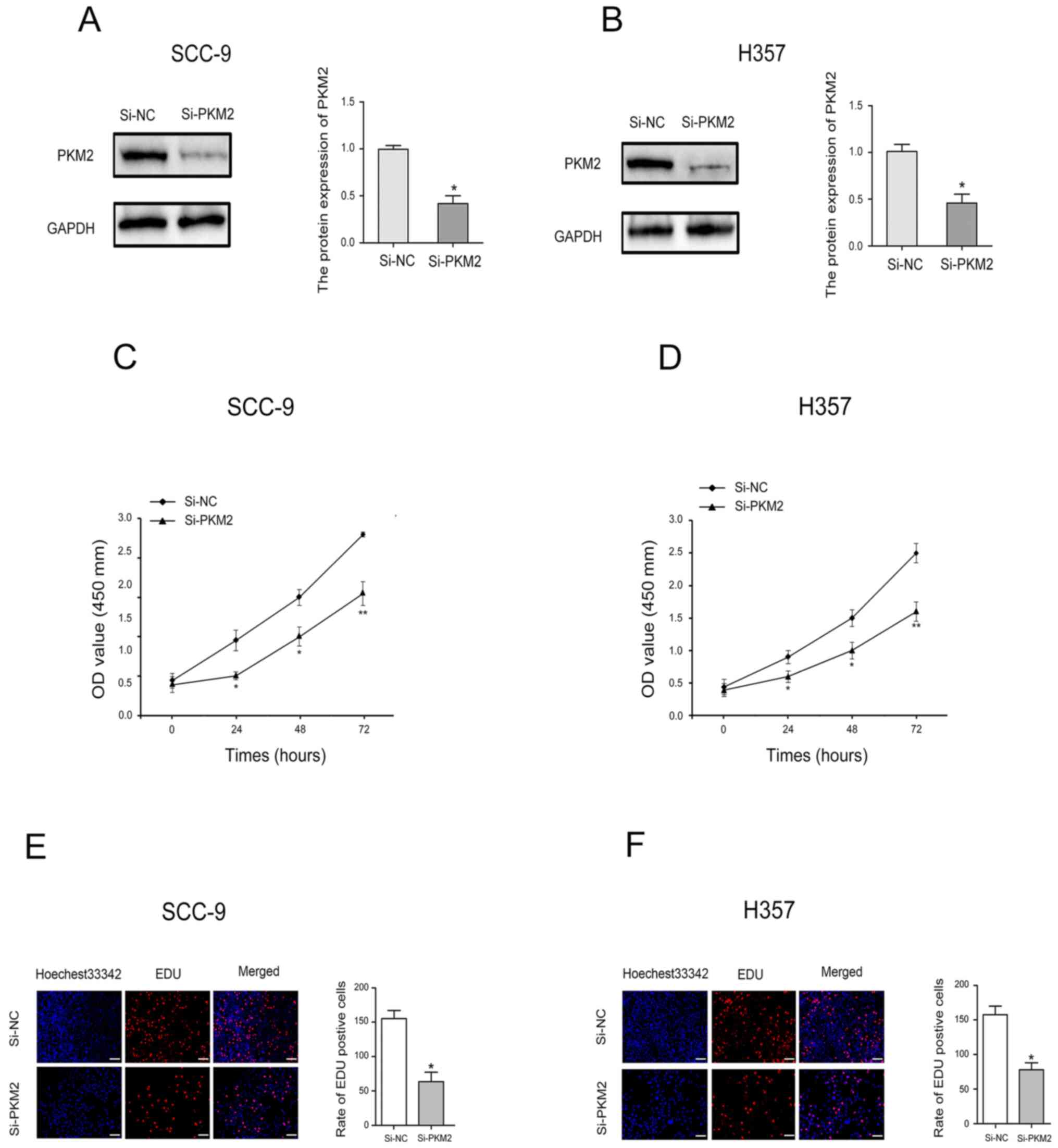
To determine the effect of PKM2 on OTSCC cell
proliferation, CCK-8 assay and EdU uptake experiments were
performed. Transfection with siRNA-PKM2 was used to significantly
decrease PKM2 expression in SCC-9 and H357 cells (Fig. 3A and B). The results from CCK-8 assay
revealed that PKM2 downregulation significantly inhibited SCC-9 and
H357 cell proliferation (Fig. 3C and
D). Furthermore, as presented in Fig. 3E and F, the number of SCC-9 and H357
cells incorporating EdU was significantly lower in PKM2 knockdown
group compared with the control group.
PKM2 knockdown inhibits cell cycle and
induces apoptosis of OTSCC cells
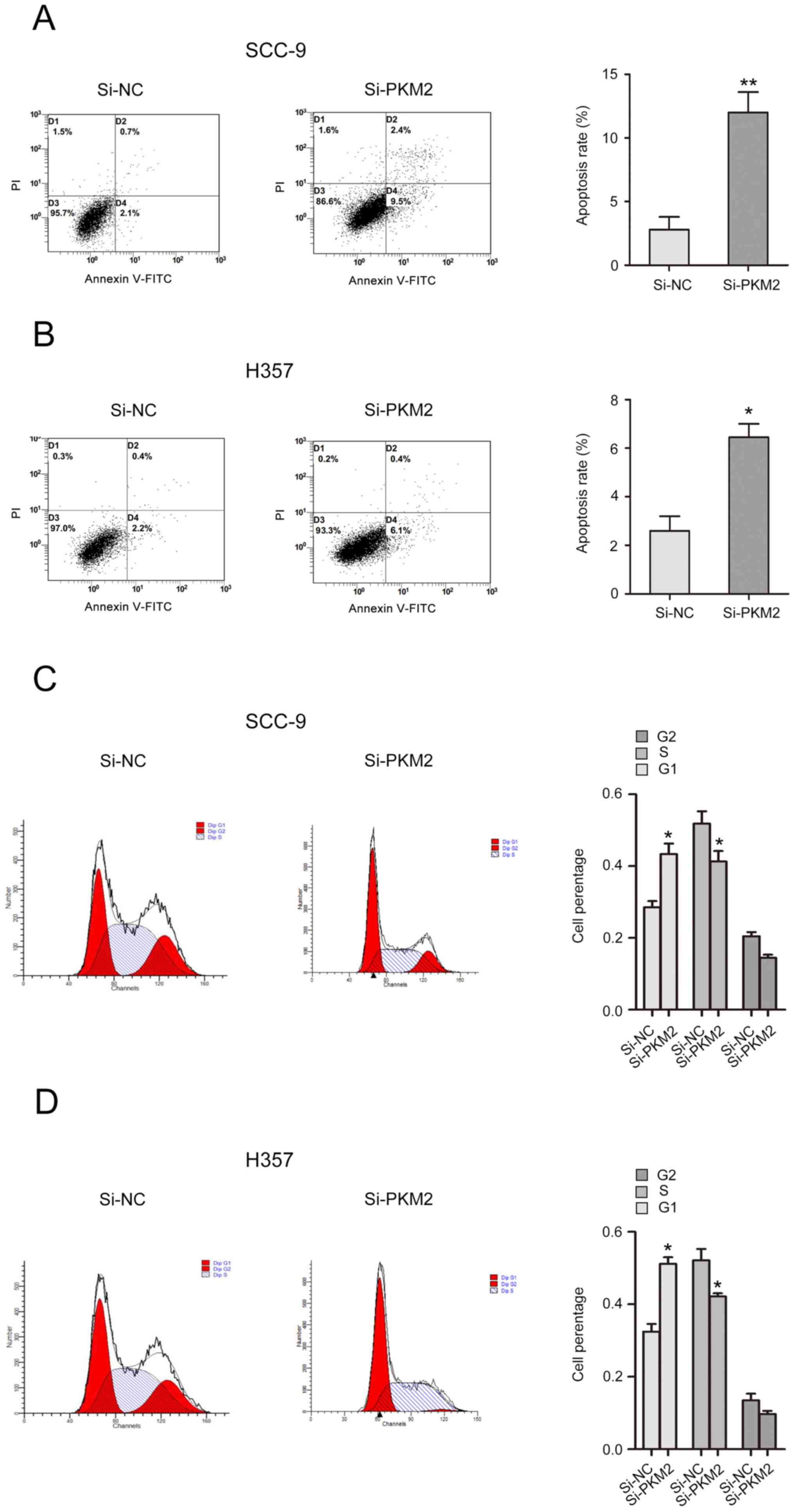
The effects of PKM2 on cell cycle and apoptosis of
OTSCC cells were analyzed. The results from flow cytometry
demonstrated that PKM2 silencing induced a significant increase in
SCC-9 and H357 cell apoptotic rate (Fig.
4A and B). In addition, the number of cells in G1 phase was
increased, while the number of cells in S phase was decreased in
si-PKM2 transfected cells compared with control cells (Fig. 4C and D).
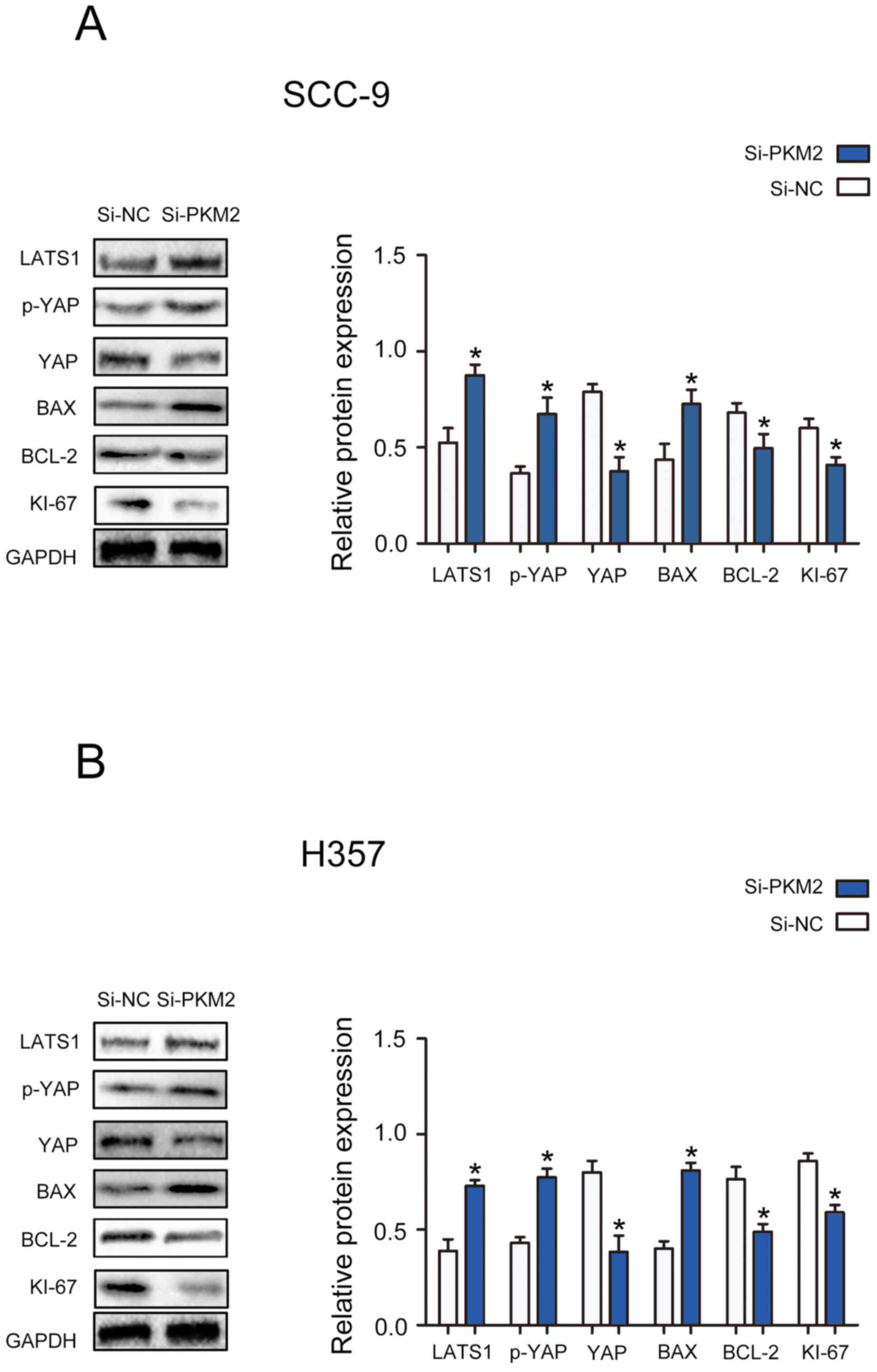
PKM2 knockdown modulates the
activation of the Hippo pathway in OTSCC cells
To determine the specific mechanism by which PKM2
might affect the proliferation and apoptosis of OTSCC cells, the
expression of proteins from the Hippo signaling pathway and of
proteins associated with cell proliferation and apoptosis was
determined. The results demonstrated that PKM2 silencing
significantly decreased the protein expression of YAP, Bcl-2 and
Ki-67, and significantly enhanced the protein expression of LATS1,
p-YAP and Bax in SCC-9 and H357 cells (Fig. 5A and B). Taken together, these
findings suggested that PKM2 may exert its function on OTSCC cell
proliferation and apoptosis in part through the Hippo pathway.
Discussion
Oral cancer is the general term for malignant tumors
that occur in the oral cavity. Each year, ~300,000 new cases are
diagnosed and this cancer ranks sixth among all malignant tumors
(17). OTSCC is a common form of
oral cancer that occurs in the gums, lips, tongue, soft and hard
palate or throat, with strong migration and invasion ability. OTSCC
seriously endanger human health (18). At present, a comprehensive treatment
approach is typically adopted for patients with OTSCC and includes
a combination of surgery, radiotherapy and chemotherapy. Although
surgery can remove tumors visible to the naked eye and can be
combined with radiotherapy and chemotherapy, active tongue
movement, abundant blood circulation and lymphatic reflux often
lead to poor prognosis and short-term survival (19). Due to the rapid growth of OTSCC, the
strong infiltration and the easy development of resistance to
radiotherapy and chemotherapy, the 5-year survival rate is only 50%
(20). Recently, studies have
reported that the occurrence and development of oral squamous cell
carcinoma is a complex process involving multiple genes, multiple
steps and multiple stages (21,22).
Given the increasing interest in molecular targeted therapies for
oral cancer, it is urgent to identify novel molecular targets that
would allow the accurate diagnosis and treatment of OTSCC, in order
to improve the survival time and quality of life of patients with
OTSCC.
The Warburg effect is the main way for generating
ATP in tumor cells via mitochondrial oxidative phosphorylation of
glucose produced by aerobic glycolysis. The Warburg effect is an
essential component of the metabolic rearrangement of tumor cells
(23). An important feature of tumor
cells is that under normal oxygen content, glucose intake and
lactic acid accumulation will also gradually increase (24). As glycolysis is the main source of
energy metabolism, a higher glycolytic capacity is obtained and
glucose is converted into lactic acid to produce ATP (25). The Warburg effect therefore allows
tumor cells to replenish their productivity under the
microenvironment, which is characterized by local hypoxia and
inhibition of mitochondrial oxidative phosphorylation. The Warburg
effect generates a high ATP/ADP ratio to allow the rapid
proliferation of tumor cells via macromolecular anabolism to
promote malignant signal transduction mechanisms maintained by
tumor cells (26). Previous studies
have confirmed that effective inhibition of the Warburg effect can
significantly inhibit the malignant growth of tumors and increase
the level of apoptosis of tumor cells (27,28). The
Warburg effect is essential for the energy metabolism rearrangement
of tumor cells, although its underlying mechanism requires further
investigation.
PK is the key enzyme of glycolysis and it catalyzes
the formation of pyruvate from phosphoenolpyruvate (29,30).
There are four major PK isoenzymes, named PKL, PKR, PKM1 and PKM2,
which are tissue specific (31). The
expression level of PKM2 has been associated with the clinical
stage and pathological grades of various types of tumor. PKM2
activates >100 proteins, including ERK1/2 (32). Sustained activation of PKM2 promotes
the proliferation and migration of tumor cells. Mitomycin 2, which
is a key regulator of mitochondrial fusion, interacts with PKM2,
which promotes mitochondrial fusion and production of phosphorus
oxide that weakens glycolysis (33).
In ovarian cancer, PKM2 overexpression leads to increased cell
proliferation and tumor growth, which might be related to the
effect of PKM2 on cell cycle progression, such as the decrease in
G1 phase and the significant increase in S phase (34). PKM2 is transported from the cytoplasm
to the mitochondria in case of increased oxidative stress (35). Once in the mitochondria, PKM2
interacts with Bcl-2 and phosphorylates Bcl-2 at T69 site. This
phosphorylation blocks the binding of Cul3-based E3 ligase to Bcl-2
and subsequent degradation of Bcl-2, and significantly inhibits the
malignant proliferation of tumor cells (35). Similarly, glycogen synthetase
kinase-3 β forms protein complexes with Hsp90 and PKM2, which
directly mediate the phosphorylation of T328 by Hsp90. T328
phosphorylation is essential to maintain the stability of PKM2,
promote the glycolysis and proliferation of HCC cells, and inhibit
apoptosis (36). miR-138 regulates
the expression of PKM2 to mediate superoxide dismutase 2 activity
and intracellular H2O2 levels to enhance the
metastatic potential of tongue squamous cell carcinoma (37). In head and neck squamous cell
carcinoma, the abnormal expression of PKM2 is associated with a
poor prognosis. In particular, nuclear PKM2 activates β-catenin
signaling and influences the proliferation and chemotherapeutic
sensitivity of head and neck squamous cell carcinoma cells
(38). In the biological process of
tongue squamous cell carcinoma growth, invasion and apoptosis, PKM2
expression is increased, reflecting the higher energy flow of tumor
cells and the dependence of their metabolism on aerobic glycolysis
and oxidative phosphorylation. The expression of PKM2 is also
related to the production of reactive oxygen species, glutamine and
lactic acid. It has been reported that PKM2 has multiple tumor
progression functions in oral squamous cell carcinoma (39). Taken together, these observations
suggest that PKM2 might be considered as an oncogene involved in
the promotion of tumor occurrence and development.
In the GEO database, the highest number of paired
OTSCC and normal adjacent tissues was seen in the GSE13601 dataset,
whereas only 4–12 paired mRNA expression profile was included in
other datasets (GSE3524, GSE2280 and GSE138206, GSE9844). In
addition, some OTSCC datasets only contained miRNA or long
non-coding-RNA expression profiles, such as GSE51829, GSE51700,
GSE98463, GSE137865, and some datasets only contained tumor mRNA
expression file. Subsequently, GSE13601 was the most appropriate
dataset to be used in the present study. The association between
PKM2 expression and the clinicopathological characteristics of
patients with OTSCC included in this dataset was therefore
analyzed. The results demonstrated that PKM2 was highly expressed
in patients with OTSCC. In addition, patients with OTSCC and high
PKM2 expression had worse OS, and high PKM2 expression was
associated with TNM stage. TNM stage and PKM2 expression were
therefore considered as independent prognostic factors for OS in
OTSCC. In addition, the results from in vitro experiments
demonstrated that PKM2 knockdown significantly inhibited the
proliferation and increased the apoptosis of OTSCC cells.
In summary, the findings from the present study
suggested that the high expression of PKM2 in OTSCC tissues may be
considered as an independent risk factor for the prognosis of
patients with OTSCC, and that PKM2 may therefore serve a crucial
role in promoting the proliferation of OTSCC cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the Key Research and
Development Projects of Sichuan Science and Technology Department
(grant no. 2018FZ0113), the China Stomatological Association
Western Medicine Stomatology Clinical Research Fund Project (grant
no. CSA-W2016-01), the Sichuan Science and Technology Support
Program (grant no. 2014SZ0038) and the Sichuan Provincial Primary
Health Service Development Research Fund (grant no.
SWFZ20-C-086).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
JL and SY conducted the experiments, analyzed the
data and wrote the manuscript. JL, LG and LZ conceived the study
and revised the manuscript critically for important intellectual
content. LG and SY made substantial contributions to patient and
tissue specimen collection and data interpretation. JL and SY
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Suining Central Hospital (approval no.
20110067). The patients signed written informed consent. All
specimens were handled and anonymized according to ethical and
legal standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar
|
|
2
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma-an update. Cancer J Clin.
65:401–421. 2015. View Article : Google Scholar
|
|
3
|
Gillison ML, Chaturvedi AK, Anderson WF
and Fakhry C: Epidemiology of human papillomavirus-positive head
and neck squamous cell carcinoma. J Clin Oncol. 33:32–37. 2015.
View Article : Google Scholar
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar
|
|
5
|
Tamada M, Suematsu M and Saya H: Pyruvate
kinase M2: Multiple faces for conferring benefits on cancer cells.
Clin Cancer Res. 18:5554–5561. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo W and Semenza GL: Emerging roles of
PKM2 in cell metabolism and cancer progression. Trends Endocrinol
Metab. 23:560–566. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dayton TL, Jacks T and Vander Heiden MG:
PKM2, cancer metabolism, and the road ahead. EMBO Rep.
17:1721–1730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Christofk HR, Vander Heiden MG, Harris MH,
Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL and
Cantley LC: The M2 splice isoform of pyruvate kinase is important
for cancer metabolism and tumour growth. Nature. 452:230–233. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burns JS and Manda G: Metabolic pathways
of the Warburg effect in health and disease: Perspectives of
choice, chain or chance. Int J Mol Sci. 18:27552017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou ZF, Li M, Zhang L, Zhao H, Şahin Ö,
Chen J, Zhao JJ, Songyang Z and Yu D: Oncogenic kinase-induced PKM2
tyrosine 105 phosphorylation converts nononcogenic PKM2 to a tumor
promoter and induces cancer stem-like cells. Cancer Res.
78:2248–2261. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu FB, Ma FF, Wang YY, Hao L, Zeng H, Jia
C, Wang Y, Liu P, Ong IM, Li B, et al: PKM2 methylation by CARM1
activates aerobic glycolysis to promote tumorigenesis. Nat Cell
Biol. 19:1358–1370. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li M, Bai YT, Han K, Li XD and Meng J:
Knockdown of ectodysplasin-A receptor-associated adaptor protein
exerts a tumor-suppressive effect in tongue squamous cell carcinoma
cells. Exp Ther Med. 19:3337–3347. 2020.PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu H, Zhang W, Wu Z, Liu Y, Shi Y, Gong J,
Shen W and Liu C: miR-29c-3p regulates DNMT3B and LATS1 methylation
to inhibit tumor progression in hepatocellular carcinoma. Cell
Death Dis. 10:482019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Estilo CL, O-charoenrat P, Talbot S, Socci
ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y,
Boyle JO, et al: Oral tongue cancer gene expression profiling:
Identification of novel potential prognosticators by
oligonucleotide microarray analysis. BMC Cancer. 9:112009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar
|
|
18
|
Solomon B, Young RJ and Rischin D: Head
and neck squamous cell carcinoma: Genomics and emerging biomarkers
for immunomodulatory cancer treatments. Semin Cancer Biol.
52:228–240. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Omura K: Current status of oral cancer
treatment strategies: Surgical treatments for oral squamous cell
carcinoma. Int J Clin Oncol. 19:423–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ali J, Sabiha B, Jan HU, Haider SA, Khan
AA and Ali SS: Genetic etiology of oral cancer. Oral Oncol.
70:23–28. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hussein AA, Forouzanfar T, Bloemena E, de
Visscher J, Brakenhoff RH, Leemans CR and Helder MN: A review of
the most promising biomarkers for early diagnosis and prognosis
prediction of tongue squamous cell carcinoma. Br J Cancer.
119:724–736. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim YJ and Kim JH: Increasing incidence
and improving survival of oral tongue squamous cell carcinoma. Sci
Rep. 10:78772020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu J: The Warburg metabolism fuels tumor
metastasis. Cancer Metastasis Rev. 38:157–164. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Icard P, Shulman S, Farhat D, Steyaert JM,
Alifano M and Lincet H: How the Warburg effect supports
aggressiveness and drug resistance of cancer cells? Drug Resist
Updat. 38:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wilde L, Roche M, Domingo-Vidal M, Tanson
K, Philp N, Curry J and Martinez-Outschoorn U: Metabolic coupling
and the reverse Warburg effect in cancer: Implications for novel
biomarker and anticancer agent development. Semin Oncol.
44:198–203. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liberti MV and Locasale JW: The Warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg Effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun L, Suo C, Li ST, Zhang H and Gao P:
Metabolic reprogramming for cancer cells and their
microenvironment: Beyond the Warburg Effect. Biochim Biophys Acta
Rev Cancer. 1870:51–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wiese EK and Hitosugi T: Tyrosine kinase
signaling in cancer metabolism: PKM2 paradox in the Warburg effect.
Front Cell Dev Bio. 6:792018. View Article : Google Scholar
|
|
30
|
van Niekerk G and Engelbrecht AM: Role of
PKM2 in directing the metabolic fate of glucose in cancer: A
potential therapeutic target. Cell Oncol (Dordr). 41:343–351. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Z, Deng X, Liu Y, Liu Y, Sun L and
Chen F: PKM2, function and expression and regulation. Cell Biosci.
9:522019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang W, Zheng Y and Xia Y:
ERK1/2-dependent phosphorylation and nuclear translocation of PKM2
promotes the Warburg effect. Nat Cell Biol. 14:1295–1304. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li T, Han J, Jia L, Hu X, Chen L and Wang
Y: PKM2 coordinates glycolysis with mitochondrial fusion and
oxidative phosphorylation. Protein Cell. 10:583–594. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miao Y, Lu M, Yan Q, Li SD and Feng YJ:
Inhibition of proliferation, migration, and invasion by knockdown
of pyruvate Kinase-M2 (PKM2) in ovarian cancer SKOV3 and OVCAR3
Cells. Oncol Res. 24:463–475. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang J, Cao R, Wang X, Zhang Y, Wang P,
Gao H, Li C, Yang F, Zeng R, Wei P, et al: Mitochondrial PKM2
regulates oxidative stress-induced apoptosis by stabilizing Bcl2.
Cell Res. 27:329–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu Q, Tu J, Dou C, Zhang J, Yang L, Liu X,
Lei K, Liu Z, Wang Y, Li L, et al: HSP90 promotes cell glycolysis,
proliferation and inhibits apoptosis by regulating PKM2 abundance
via Thr-328 phosphorylation in hepatocellular carcinoma. Mol
Cancer. 16:1782017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang W, He Q, Sun J, Liu Z, Zhao L, Lu Z,
Zhou X and Wang A: Pyruvate kinase M2 deregulation enhances the
metastatic potential of tongue squamous cell carcinoma. Oncotarget.
8:68252–68262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jing C, Qu X, Li Z, Wu C, Zhao M, Wang Y,
Sun S, Zhang S, Chen J, Qiao Y, et al: EGFRwt/vIII-PKM2-β-catenin
cascade affects proliferation and chemo-sensitivity in head and
neck squamous cell carcinoma. Am J Cancer Res. 7:2491–2502.
2017.PubMed/NCBI
|
|
39
|
Kurihara-Shimomura M, Sasahira T,
Nakashima C, Kuniyasu H, Shimomura H and Kirita T: The Multifarious
functions of pyruvate kinase M2 in oral cancer cells. Int J Mol
Sci. 19:29072018. View Article : Google Scholar : PubMed/NCBI
|