Introduction
Osteosarcoma is the most common malignant bone tumor
occuring in adolescents or children under the age of 20 years,
accounting for ~5% of the total number of pediatric tumors
(1,2). Osteosarcoma can occur in all parts of
the bone tissue, although it is more common in the distal femur,
proximal tibia and proximal humerus and metaphysis (2). At present, the cause and underlying
mechanism of osteosarcoma remain unclear. Neoadjuvant chemotherapy,
surgery and radiation therapy are common treatments for patients
with osteosarcoma, which allow patients to reach a 5-year overall
survival rate of 65–70% (3,4). However, 25–30% of patients with
osteosarcoma present with metastases, including lung and bone
metastases, which significantly decrease their 5-year survival rate
(4). Studying the pathogenesis of
osteosarcoma would therefore help the development of potential
novel targets and new treatment strategies.
Long non-coding RNA (lncRNAs) are defined as
transcripts of >200 nucleotides in length that are not
translated into proteins because they do not have a development
reading frame. The Human Genome Project considers lncRNAs as
‘garbage sequences’ that have accumulated during human evolution.
However, during recent years, increasing evidence has demonstrated
that lncRNAs are dysregulated in numerous diseases, especially in
cancers, and that they serve crucial role in the regulation of
various pathophysiological processes, including cell proliferation,
apoptosis, necrosis, and autophagy (5–7). The
lncRNA PTPRG-AS1 has not only been found to be overexpressed in
tumor tissues, including breast cancer (8) and ovarian epithelial cancer (9), but has also been demonstrated to be
involved in the regulation of radiosensitivity, metastasis and
proliferation of tumor cells (10,11).
However, the expression of PTPRG-AS1 and its role in osteosarcoma
remain unclear. The present study aimed to determine PTPRG-AS1
expression in patients with osteosarcoma, and to investigate the
association between PTPRG-AS1 expression and the prognosis of
patients with osteosarcoma. In addition, the function of PTPRG-AS1
on the metastasis of osteosarcoma cells was also assessed.
Materials and methods
Patients and tissues
Between January 2014 and January 2019, 106 pairs of
osteosarcoma tumor tissue and adjacent healthy tissue were
collected from patients at the Peking University International
Hospital. The clinicopathological characteristics, including sex,
age, anatomical site, histological grade, histologic subtype and
clinical stage were collected for each patient with osteosarcoma.
The histological subtype and clinical stage of patients with
osteosarcoma were divided as before (12,13). The
age of the 106 patients ranged from 5 to 32 years (mean age, 11
years). All other clinicopathological characteristics are presented
in Table I. All tissues were
collected before chemotherapy, radiation or other treatment
procedures were conducted, and we followed-up on each patient for
up to 5 years after tissue collection. All patients or their
guardians provided written informed consent, and the study protocol
was approved by the Peking University International Hospital Ethics
Committee (Beijing, China; approval no. 2018-042 (BMR). Fresh
tissues were stored in liquid nitrogen until subsequent use to
detect PTPRG-AS1 expression.
 | Table I.PTPRG-AS1 expression and
clinicopathological characteristics of patients with
osteosarcoma. |
Table I.
PTPRG-AS1 expression and
clinicopathological characteristics of patients with
osteosarcoma.
|
|
| PTPRG expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Case number | Low (n=51) | High (n=55) | χ2 | P-value |
|---|
| Sex |
|
Female | 50 | 26 | 24 | 0.573 | 0.449 |
|
Male | 56 | 25 | 31 |
|
|
| Age, years |
|
<14 | 45 | 23 | 22 | 0.044 | 0.834 |
|
≥14 | 61 | 28 | 33 |
|
|
| Anatomical
site |
|
Femur | 50 | 20 | 30 | 3.611 | 0.461 |
|
Tibia | 21 | 13 | 8 |
|
|
|
Humerus | 15 | 7 | 8 |
|
|
|
Pelvis | 12 | 6 | 6 |
|
|
|
Other | 8 | 5 | 3 |
|
|
| Histological
grade |
| G1 | 23 | 11 | 12 | 0.368 | 0.832 |
| G2 | 38 | 15 | 13 |
|
|
| G3 | 45 | 25 | 20 |
|
|
| Histologic
subtype |
|
Osteoblastic | 54 | 20 | 25 | 4.066 | 0.397 |
|
Chondroblastic | 15 | 10 | 5 |
|
|
|
Fibroblastic | 13 | 9 | 4 |
|
|
|
Telangiectatic | 13 | 6 | 7 |
|
|
|
Other | 11 | 6 | 5 |
|
|
| TNM stage |
| I | 23 | 7 | 16 | 9.357 | 0.025 |
| II | 20 | 7 | 13 |
|
|
|
III | 34 | 17 | 17 |
|
|
| IV | 29 | 20 | 9 |
|
|
| Lymph node
metastasis |
|
Yes | 24 | 7 | 17 | 4.461 | 0.035 |
| No | 82 | 44 | 38 |
|
|
| Distant
metastasis |
|
Yes | 29 | 10 | 19 | 5.799 | 0.016 |
| No | 77 | 41 | 26 |
|
|
Reverse transcription quantitative
(RT-q)PCR
A Cell/tissue Total RNA Isolation kit (cat. no.
RC101-01; Vazyme Biotech Co., Ltd.) was used to extract total RNA
from tissues and cells. A 20 µl of RT-qPCR system was prepared as
described in the qPCR master mix kit instructions (cat. no. A6001;
Promega Corporation). The following thermocycling conditions were
used for qPCR: 95°C for 2 min, followed by 40 cycles of 95°C for 5
sec and 60°C for 30 sec, and a final extension at 72°C for 5 min.
The relative expression of the gene was calculated using the
2−ΔΔCq method (14), and
β-actin was used as a loading control. The sequences of the primers
were as follows: PTPRG-AS1-2, forward, 5′-CCCTTGAGTGGTCCTCTGTTC-3′
and reverse, 5′-GAGCCGGATTTGTCCCAACT-3′; and β-actin, forward,
5′-AGCCCATCCTTCGAGTACAAA-3′ and reverse,
5′-TCTTGGTGCGATAACTGGTGG-3′.
Cell culture and transfection
hFOB1.19, U2OS, SJSA1 and Saos-2 cell lines were
purchased from the American Type Culture Collection, cultured in
DMEM medium (cat. no. 11965092; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (cat. no. 16140071; Thermo Fisher
Scientific, Inc.), and placed at 37°C in a humidified incubator
containing 5% CO2. Cells (2.5×106) were
transfected with 50 nmol/l small interfering (si)RNA using
Lipofectamine 2000 reagent (cat. no. 11668019; Thermo Fisher
Scientific, Inc.) at 37°C for 6 h, according to the manufacturer's
instructions. The sequences of the siRNAs used were as follows:
si-negative control (NC), forward, 5′-AGGUAGGCCCCUAUCAGCCGGC-3′ and
reverse, 5′-AUGUGUCGAAUUCGCGUACG-3′; si-PTPRG-AS1-1, forward,
5′-UUCAAAUAUAUUUACUGAGCA-3′ and reverse,
5′-CUCAGUAAAUAUAUUUGAAUG-3′; and si-PTPRG-AS1-2, forward,
5′-AUUAUGAUGAAUGUUAACGGG-3′ and reverse,
5-CGUUAACAUUCAUCAUAAUUU-3′. After 72 h of transfections, cells were
collected for subsequent experiments.
Transwell assay
Transwell chambers (cat. no. 140629; Thermo Fisher
Scientific, Inc.) were used to evaluate the migratory and invasive
abilities of cancer cells following to the manufacturer's
instructions. Briefly, 0.5×106 cells/ml in DMEM medium
were added into the upper chamber of a 24-well Transwell chamber.
The upper chamber was pre-coated with Extra Matrigel (cat. no.
BD354248; Becton-Dickinson and Company) to evaluate the invasive
ability of cancer cells. Culture medium containing 20% FBS was
added into the lower Transwell chamber. Cells were placed at 37°C
in a humidified incubator containing 5% CO2 for 24 h.
Subsequently, medium was removed from lower chamber, cells were
washed twice with PBS and membrane was fixed with methanol for 5
min at room temperature and allowed to dry for 30 min. Once
membrane was dried, it was stained with crystal violet for 20 min
at room temperature and cells were imaged using a light microscope
(×200). ImageJ software [v1.8.0; National Institutes of Health
(NIH)] was used to quantify the gray value of spots.
Fluorescence in situ hybridization
(FISH)
FISH was performed as previously described (15). Briefly, after cells (50,000 cells in
200 µl DMEM medium added 10% FBS) or tissues were fixed with 4%
paraformaldehyde (cat. no. LGLS0005; Loogene) for 10 min at room
temperature, they were incubated with a fluorescent probe
(Genomeditech Co., Ltd.) that can bind to the human version of the
PTPRG-AS1 gene at 37°C for 16 h. For cells, the nuclei were
counterstained with 5 µg/ml DAPI for 5 min at room temperature. All
samples were visualized using confocal microscopy (LAS AF Lite 4.0,
Leica; magnification, ×800).
Western blotting
A Whole Cell Extraction Kit (cat. no. OP-0003;
Epigentek Group, Inc.) was used to extract total protein from the
osteosarcoma cells. Protein concentration was determined using a
BCA Protein Quantification Kit (cat. no. E112-01; Vazyme Biotech
Co., Ltd.). Subsequently, 45 µg proteins were separated by 10%
SDS-PAGE and were transferred onto PVDF membranes. Membranes were
blocked using 5% skimmed milk at room temperature for 1 h and
incubated with the primary antibodies (diluted with 5% skimmed
milk) against E-cadherin (cat. no. ab15148; 1:1,000; Abcam),
N-cadherin (cat. no. ab18203; 1:1,000; Abcam), and matrix
metalloproteinase-9 (MMP-9) antibody (cat. no. ab38898; 1:2,000;
Abcam) and β-actin (cat. no. sc-69879; 1:3,000; Santacruze)
overnight at 4°C Membranes were then incubated with goat anti-mouse
IgG H&L (HRP) (cat. no. ab6789; 1:2,000; Abcam) or goat
anti-rabbit IgG H&L (HRP) (cat. no. ab6721; 1:2,000; Abcam) at
room temperature for 2 h. Enhanced chemiluminescence reagent (cat.
no. WBKLS0100; Beijing Xinjingke Biotechnologies Co., Ltd.) was
used to detect the signal on the membrane. ImageJ software (v1.8.0;
NIH) was used to quantify the gray value of protein bands.
Statistical analysis
SPSS 20.0 software (IBM Corp.) was used to analyze
the data. Student t-test and χ2 test were used to
compare differences between two groups, and one-way ANOVA followed
by Tukey's post hoc test was used to compare differences between
multiple groups. The multivariate analysis model was used to
analyze the association between marker expression levels and
clinicopathological characteristics of patients. Log-rank
(Mantel-Cox) test was used to compare the survival of patients with
high and low levels of PTPRG-AS1 expression. P<0.05 was
considered to indicate a statistically significant difference.
Results
PTPRG-AS1 is highly expressed in
osteosarcoma cells and tissues
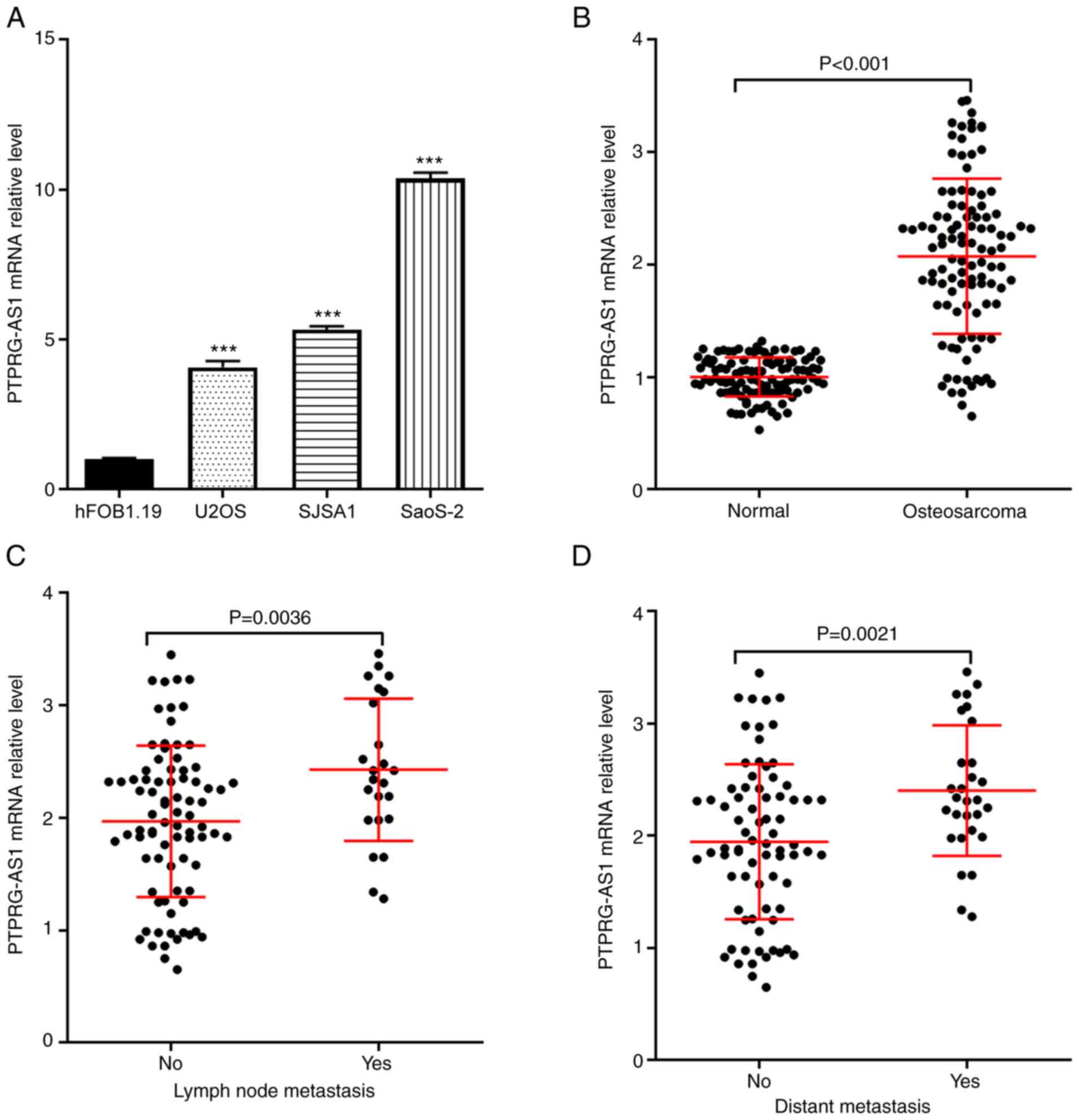
As presented in Fig.
1A, the expression of PTPRG-AS1 was detected in normal human
osteoblast (hFOB1.19) and human osteosarcoma cells (U2OS, SJSA1 and
Saos2). The results demonstrated that PTPRG-AS1 expression level in
human osteosarcoma cells was significantly higher compared with
normal human osteoblasts. Furthermore, 106 pairs of human
osteosarcoma and normal adjacent tissues were collected and used to
detect PTPRG-AS1 expression levels using RT-qPCR. The results
demonstrated that PTPRG-AS1 expression level in osteosarcoma
tissues was significantly higher compared with normal adjacent
tissues (Fig. 1B). In addition,
according to the presence of lymph node metastasis or distant
metastasis, the 106 patients with osteosarcoma were divided into
two groups (presence or absence). The results demonstrated that
PTPRG-AS1 expression in patients with lymph node metastasis (n=24)
was significantly higher than in patients without lymph node
metastasis (n=82; Fig. 1C). In
addition, compared with patients without distant metastasis (n=77),
PTPRG-AS1 expression in patients with distant metastasis (n=29) was
significantly higher (Fig. 1D).
Association between PTPRG-AS1
expression and the clinicopathological characteristics of patients
with osteosarcoma
According to the expression of PTPRG-AS1 in
osteosarcoma tissues, the 106 patients were divided into two
groups, the low PTPRG-AS1 expression group (n=51; PTPRG-AS1
expression < mean of PTPRG-AS1 expression in the 106 patients)
and the high PTPRG-AS1 expression group (n=55; PTPRG-AS1 expression
≥ mean of PTPRG-AS1 expression in the 106 patients). The
association between PTPRG-AS1 expression and the
clinicopathological characteristics of patients, including sex,
age, anatomical site and histological grade, was subsequently
evaluated. As presented in Table I,
the expression of PTPRG-AS1 was significantly associated with
Tumor-Node-Metastasis (TNM) stage (P=0.025), lymph node metastasis
(P=0.035) and distant metastasis (P=0.016) in patients with
osteosarcoma, but was not significantly associated with sex
(P=0.449), age (P=0.834), anatomical site (P=0.461), histological
grade (P=0.832) and histological subtype (P=0.397).
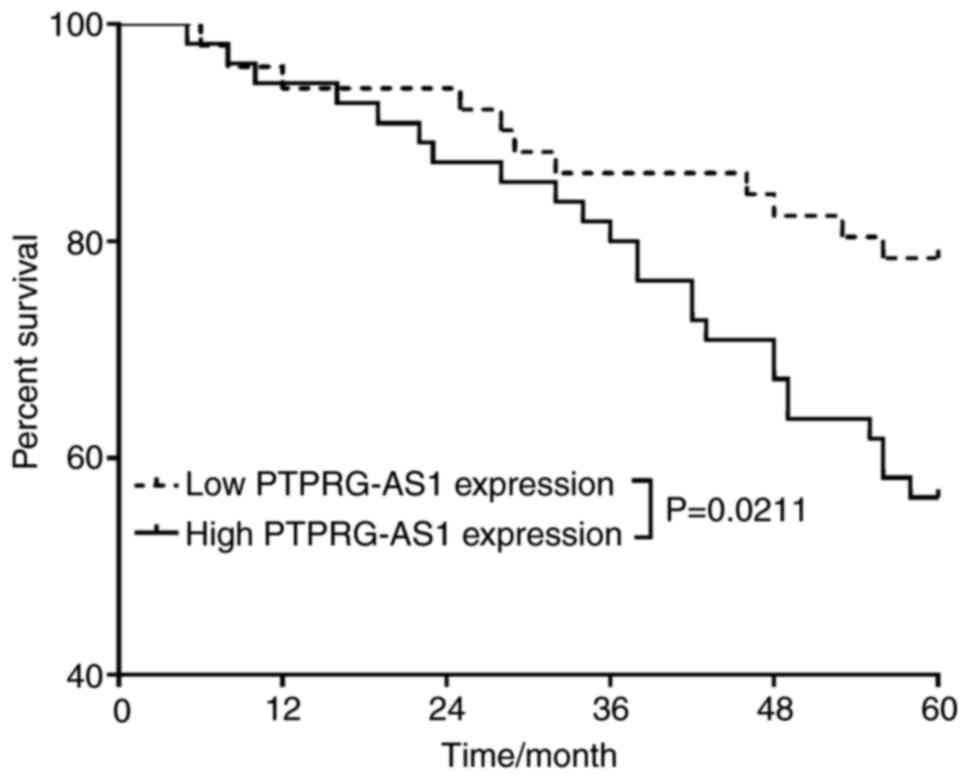
PTPRG-AS1 can predict the poor
prognosis of patient with osteosarcoma
The 106 patients were followed up at least once
every four months or when the patients came to the hospital for
review five years after surgery. The 5-year overall survival of
patients was recorded, and the factors influencing the survival of
the patients were analyzed using univariate and multivariate
analyses. As presented in Table II,
histological grade [odd ratio (OR)=1.659; 95% confidence interval
(CI, 1.844-2.064], TNM stage (OR=1.353; 95% CI, 1.232-2.564), lymph
node metastasis (OR=0.985; 95% CI, 0.421-1.654), distant metastasis
(OR=3.127; 95% CI, 1.846-4.325) and PTPRG-AS1 expression level
(OR=3.012; 95% CI, 1.564-4.219) were identified as independent risk
factors that could affect the 5-year survival of patients with
osteosarcoma. Further analysis of the effect of PTPRG-AS1
expression level on the 5-year overall survival rate of patients
with osteosarcoma demonstrated that 78.43% (40/51) of patients with
low PTPRG-AS1 expression were still alive 5 years after surgery,
whereas only 56.36% (31/55) of patients with high PTPRG-AS1
expression were still alive 5 years after surgery. The difference
between the 5-year survival of patients with osteosarcoma and the
difference in PTPRG-AS1 expression was significant (P=0.0211;
Fig. 2).
 | Table II.Analysis of the influencing factors
on the survival of patients with osteosarcoma. |
Table II.
Analysis of the influencing factors
on the survival of patients with osteosarcoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
| 95% CI | OR | P-value | 95% CI | OR | P-value |
|---|
| Sex | 1.567-4.215 | 2.423 | 0.382 | – | – | – |
| Age | 0.567-3.342 | 0.984 | 0.412 | – | – | – |
| Anatomical
site | 3.129-4.031 | 3.672 | 0.058 | – | – | – |
| Histological
grade | 0.876-2.324 | 1.561 | 0.042 | 1.844-2.064 | 1.659 | <0.001 |
| Histological
subtype | 1.524-3.659 | 2.227 | 0.067 | – | – | – |
| TNM stage | 0.912-3.021 | 1.025 | 0.042 | 1.232-2.564 | 1.353 | 0.049 |
| Lymph node
metastasis | 1.324-2.845 | 1.622 | 0.024 | 0.421-1.654 | 0.985 | 0.019 |
| Distant
metastasis | 1.652-5.126 | 3.241 | <0.001 | 1.846-4.325 | 3.127 | <0.001 |
| PTPRG-AS1
levels | 1.226-5.324 | 2.984 | 0.016 | 1.564-4.219 | 3.012 | 0.011 |
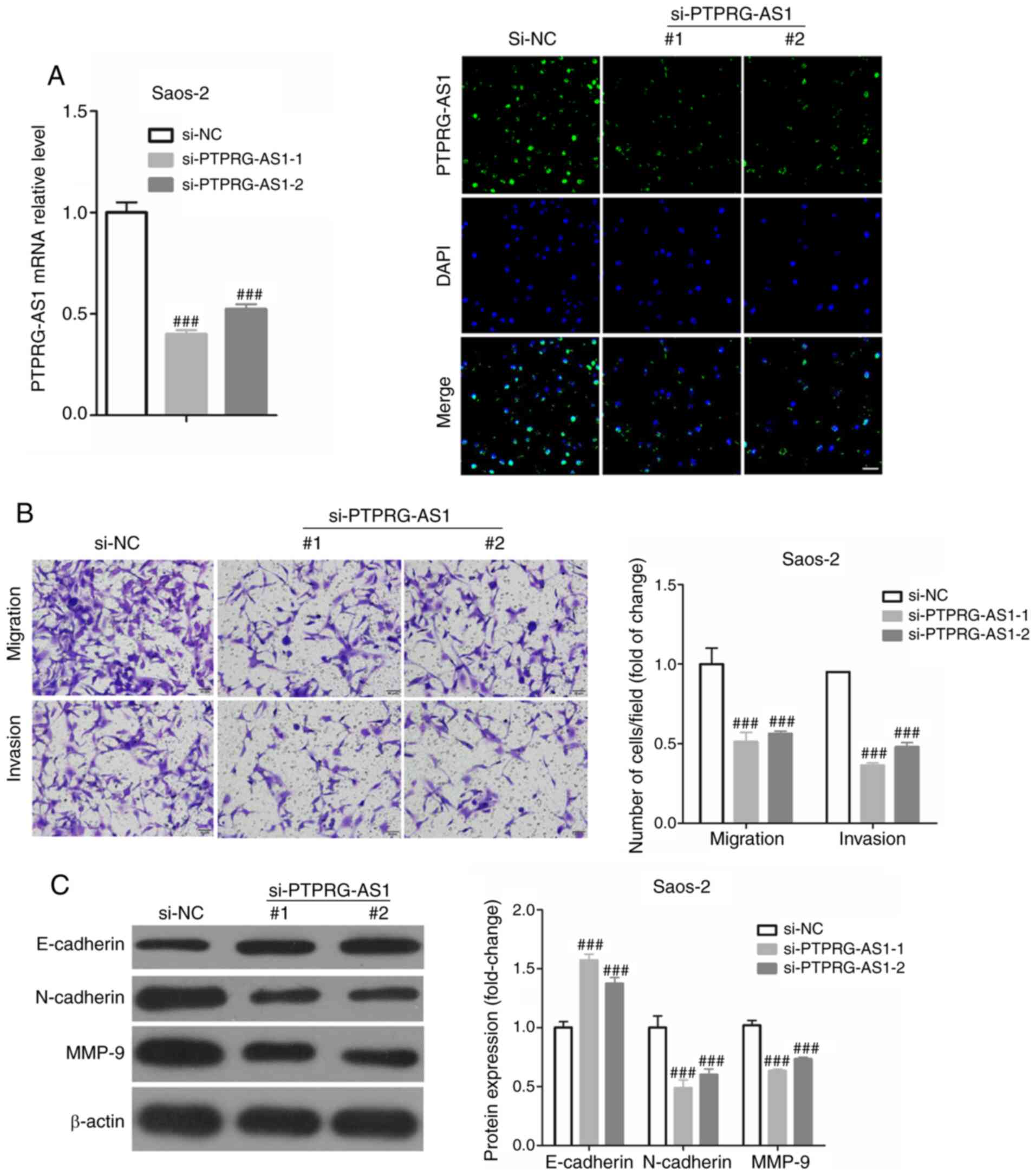
PTPRG-AS1 promotes the migratory and
invasive abilities of osteosarcoma cells
The metastasis of cancer cells includes migration
and invasion (16), and we used
different Transwell chamber to assess invasion and migration in
osteosarcoma cells. PPRRG-AS1 expression was knocked down in
osteosarcoma cells (Saos-2 and SJSA1) using si-PPRRG-AS1-1 and −2
and the efficiency of the transfection was verified by RT-qPCR and
FISH staining. As presented in Fig.
3A, the results from RT-qPCR demonstrated that si-PPRRG-AS1-1
and si-PPRRG-AS1-2 significantly decreased PPRRG-AS1 expression up
to 60 and 52%, respectively, in osteosarcoma cells. Therefore,
si-PPRRG-AS1-1 and si-PPRRG-AS1-2 were used to knockdown PPRRG-AS1
expression in subsequent experiments. To investigate whether
PTPRG-AS1 could regulate the migratory and invasive abilities of
osteosarcoma cells, Transwell assays were performed using Saos-2
cells transfected or not with si-PPRRG-AS1-1 and si-PPRRG-AS1-2.
The results demonstrated that PTPRG-AS knockdown could
significantly decrease the migratory and invasive abilities of
osteosarcoma cells (Fig. 3B).
Furthermore, PTPRG-AS knockdown could significantly increase the
expression of E-cadherin and decrease the expressions of N-cadherin
and MMP-9 in Saos-2 cells (Fig. 3C).
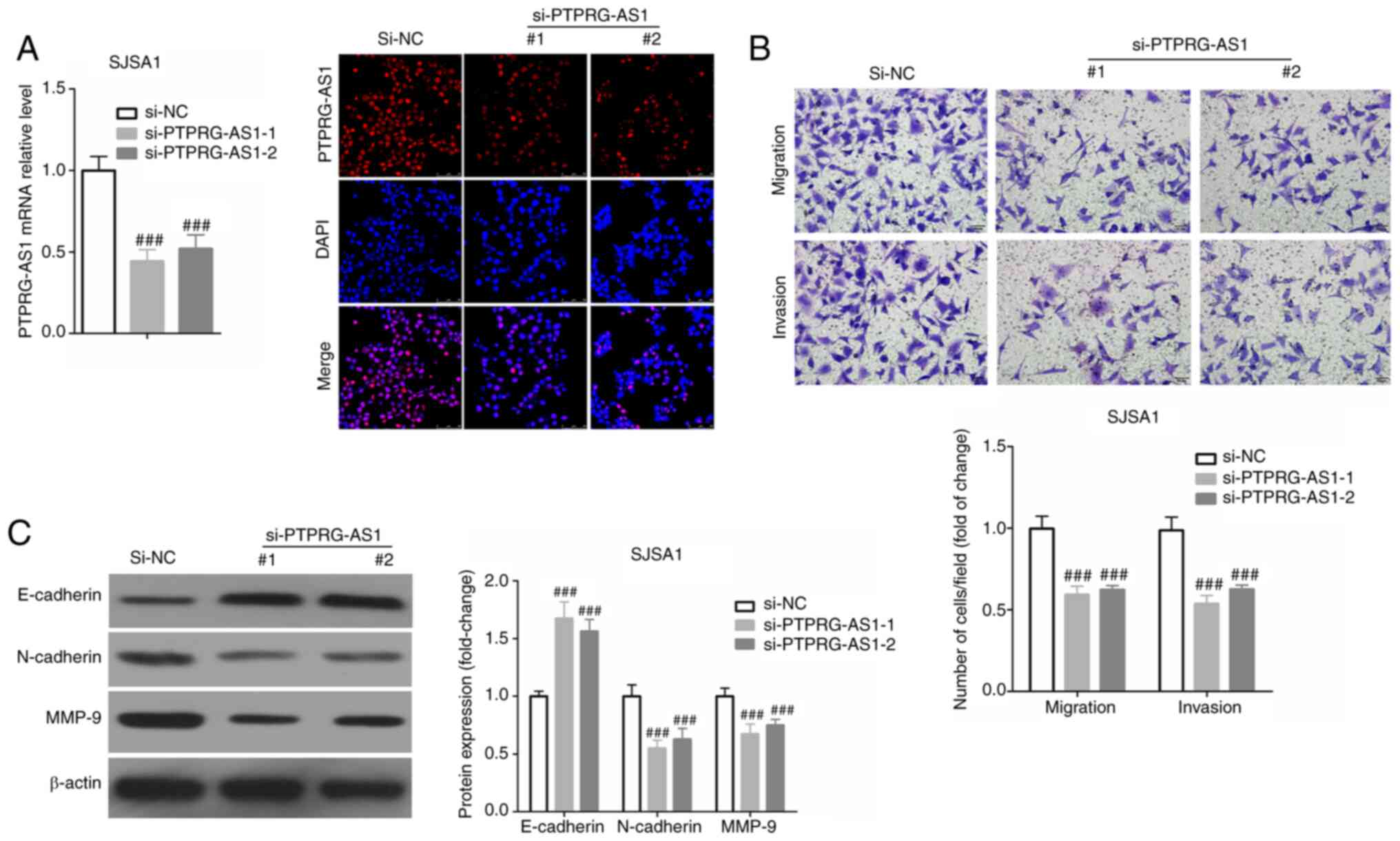
Similar results were obtained following SJSA1 cell transfection
with si-PPRRG-AS1-1 and si-PPRRG-AS1-2 (Fig. 4).
Discussion
Metastasis is one of the main characteristics of
cancer cells and cancer-associated mortality often results from
metastases rather than from primary tumor growth (17,18).
Osteosarcoma, the most common type of primary bone tumor in
children, is highly metastatic, and ~25–30% of patients with
osteosarcoma develop metastases (4).
Understanding the underlying mechanism of osteosarcoma metastasis
to tissues and organs is essential to identify novel therapeutic
targets (19). An increasing number
of studies have reported that lncRNAs serve crucial roles in the
regulation of osteosarcoma cell metastasis. For example, the lncRNA
HULC is highly expressed in osteosarcoma cells and tissues and can
promote cell metastasis in osteosarcoma (20). Furthermore, Zhao et al
(21) reported that the lncRNA
HNF1A-AS1 could promote osteosarcoma cell metastasis by activating
the Wnt/β-catenin signaling pathway. Zhou et al (22) demonstrated that the lncRNA SNHG12
promotes the tumorigenesis and metastasis of osteosarcoma through
the upregulation of Notch2 by sponging miR-195-5p. Similarly, GAS5
(23), ZFAS1 (24) and MALAT1 (25) have also been demonstrated to be
involved in the regulation of osteosarcoma cell metastasis.
The present study demonstrated that PTPRG-AS1
expression was elevated in osteosarcoma cells and tissues, and that
PTPRG-AS1 expression was higher in patients with lymph node
metastasis or distal metastasis. Importantly, PTPRG-AS1 expression
level in osteosarcoma tissues was not only significantly associated
with lymph node metastasis or distal metastasis, but was also
significantly associated with the 5-year survival of patients with
osteosarcoma. These results suggested that PTPRG-AS1 may act as an
oncogene in osteosarcoma and may therefore be associated with
osteosarcoma cell metastasis. PTPRG-AS1 is located on human
chromosome 3p14.2 and has been found to play an important role in
the regulation of tumor cell behavior. Yi et al (11) reported that PTPRG-AS1 expression is
significantly elevated in nasopharyngeal carcinoma tissues using
gene sequencing technology, and that PTPRG-AS1 can promote the
expression of protein regulator of cytokinesis 1 (PRC1) by binding
to miR-194-3p, while PRC1 could enhance the resistance of
nasopharyngeal carcinoma cells to radiotherapy and promote the
migration of nasopharyngeal carcinoma cells. These results
indicated that PTPRG-AS1 serves an indirect role as an oncogene
through PRC1. In addition, Xu et al (10) demonstrated that PTPRG-AS1 can
regulate the growth of glioma cells by sponging miR-185-5 in
vitro.
To evaluate the effect of PTPRG-AS1 on osteosarcoma
cell metastasis, the expression of PTPRG-AS1 in osteosarcoma cells
was knocked down in the present study, and the changes in the
migratory and invasive abilities of cells were determined. The
results demonstrated that PTPRG-AS1 silencing could significantly
decrease the invasive and migratory abilities of osteosarcoma cells
in vitro. PTPRG-AS1 knockdown also significantly increased
the expression of E-cadherin and decreased the expression of
N-cadherin and MMP-9 in osteosarcoma cells. Tumor metastasis is an
extremely complicated multi-step process, which includes tumor cell
detachment from the primary focal site, invasion of surrounding
tissue, entry in the circulatory system, escape from immune
surveillance, attachment to distant luminal beds, extravasation
into target organ tissue and formation of several secondary tumors
(26,27). Cell adhesion plays therefore a
crucial role in the invasion and metastasis of cancer cells.
Firstly, the separation of cancer cells from the primary cancer is
associated with the reduction of homogeneous adhesion of cancer
cells, and changes in the adhesion to the stroma is also an
important factor that favors tumor metastasis (26,27).
Secondly, the formation of homogeneous or heterogeneous tumor plugs
in the lumen also results from adhesion (26,27). In
addition, during tumor cell migration out of the lumen, adhesion to
the lumen endothelium and underlying basement membrane is also an
important step during the metastatic process (26,27).
E-cadherin, N-cadherin and MMP-9 are important
proteins involved in epithelial-mesenchymal transition (EMT), and
EMT is defined as the transformation of epithelial cells to
mesenchymal cells (28,29). EMT can alter cell adhesion and
enhance cell ability to metastasize and invade, by inducing stem
cell characteristics, decreasing apoptosis and aging, and promoting
immunosuppression, which not only play a key role in the
developmental process, but also participate in tissue healing,
organ fibrosis and cancer development (30–32).
E-cadherin and N-cadherin are both subtypes of cadherin, which is a
calcium-dependent transmembrane glycoprotein that mainly mediates
homogeneous adhesion between cells (33,34). A
hydrophobic gene in E-cadherin is located in the transmembrane
region and an amino terminus is located outside the cell membrane,
while the hydroxyl end is located in the cytoplasm and is connected
to actin (33). Overall, E-cadherin
plays an important role in maintaining cell morphology and
regulating cell adhesion (33).
However, N-cadherin acts as a promoter to initiate tumor invasion
in most malignant tumors, because N-cadherin can decrease cell
adhesion by changing cell morphology, and ultimately promotes EMT
(34,35). One of the hallmarks of EMT is the
loss of epithelial cell integrity, which is accompanied by a
decrease in adhesion connections between epithelial cells. The
proteolytic digestive function of MMPs is one of the driving
factors that cause a decrease in the adhesion connections between
epithelial cells (36,37).
In summary, the results from the present study
demonstrated that PTPRG-AS1 upregulation in osteosarcoma may
promote osteosarcoma cell metastasis, resulting ultimately in the
poor prognosis of patients with osteosarcoma. However, the
underlying mechanisms by which PTPRG-AS1 may promote osteosarcoma
cell metastasis remain unclear and require further
investigation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
BW was responsible for the conception and design of
the study. RG and PY performed the experiments and analyzed the
data. RG and BW confirmed the authenticity of all the raw data. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Peking
University International Hospital Ethics Committee (Beijing, China;
approval no. 2018-042 (BMR), and written informed consent was
provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Pediatric and Adolescent Osteosarcoma. Springer;
pp. 3–13. 2009, View Article : Google Scholar
|
|
2
|
Lindsey BA, Markel JE and Kleinerman ES:
Osteosarcoma Overview. Rheumatol Ther. 4:25–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harrison DJ, Geller DS, Gill JD, Lewis VO
and Gorlick R: Current and future therapeutic approaches for
osteosarcoma. Expert Rev Anticancer Ther. 18:39–50. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Anderson ME: Update on survival in
osteosarcoma. Orthop Clin North Am. 47:283–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang DQ, Fu P, Yao C, Zhu LS, Hou TY, Chen
JG, Lu Y, Liu D and Zhu LQ: Long non-coding RNAs, novel culprits,
or bodyguards in neurodegenerative diseases. Mol Ther Nucleic
Acids. 10:269–276. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Niu ZS, Niu XJ and Wang WH: Long
non-coding RNAs in hepatocellular carcinoma: Potential roles and
clinical implications. World J Gastroenterol. 23:5860–5874. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tehrani SS, Karimian A, Parsian H,
Majidinia M and Yousefi B: Multiple functions of long non-coding
RNAs in oxidative stress, DNA damage response and cancer
progression. J Cell Biochem. 119:223–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iranpour M, Soudyab M, Geranpayeh L,
Mirfakhraie R, Azargashb E, Movafagh A and Ghafouri-Fard S:
Expression analysis of four long noncoding RNAs in breast cancer.
Tumour Biol. 37:2933–2940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang X, Yan M, Chengjiang W, et al:
Analysis of LncRNAs and mRNAs expression profiles in ovarian
epithelial cancer cell lines by gene microarray. Chinese Journal of
Clinical Laboratory Science. 36:384–387,400. 2018.PubMed/NCBI
|
|
10
|
Xu C, Li Z, He T, Yuan B and Ding B: Long
noncoding RNA PTPRG-AS1 regulates growth of glioma cells by
sponging miR-185-5p. RSC Advances. 9:10870–10880. 2019. View Article : Google Scholar
|
|
11
|
Yi L, Ouyang L, Wang S, Li SS and Yang XM:
Long noncoding RNA PTPRG-AS1 acts as a microRNA-194-3p sponge to
regulate radiosensitivity and metastasis of nasopharyngeal
carcinoma cells via PRC1. J Cell Physiol. 234:19088–19102. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cates JM: Comparison of the AJCC, MSTS,
and modified spanier systems for clinical and pathologic staging of
osteosarcoma. Am J Surg Pathol. 41:405–413. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Laurini JA and Cooper K: Pathologic
staging of bone and soft tissue tumors: What is new in the eighth
edition of the American Joint Committee on Cancer Staging Manual?
AJSP. 23:149–156. 2018.
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP
and Gao YF: LncSox4 promotes the self-renewal of liver
tumour-initiating cells through Stat3-mediated Sox4 expression. Nat
Commun. 7:125982016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhuang J, Huang Y, Zheng W, Yang S, Zhu G,
Wang J, Lin X and Ye J: TMEM100 expression suppresses metastasis
and enhances sensitivity to chemotherapy in gastric cancer. Biol
Chem. 401:285–296. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mumprecht V and Detmar M:
Lymphangiogenesis and cancer metastasis. J Cell Mol Med.
13:1405–1416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol
Cancer. 14:482015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang J and Zhang W: New molecular insights
into osteosarcoma targeted therapy. Curr Opin Oncol. 25:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun XH, Yang LB, Geng XL, Wang R and Zhang
ZC: Increased expression of lncRNA HULC indicates a poor prognosis
and promotes cell metastasis in osteosarcoma. Int J Clin Exp
Pathol. 8:2994–3000. 2015.PubMed/NCBI
|
|
21
|
Zhao H, Hou W, Tao J, Zhao Y, Wan G, Ma C
and Xu H: Upregulation of lncRNA HNF1A-AS1 promotes cell
proliferation and metastasis in osteosarcoma through activation of
the Wnt/β-catenin signaling pathway. Am J Transl Res. 8:3503–3512.
2016.PubMed/NCBI
|
|
22
|
Zhou S, Yu L, Xiong M and Dai G: LncRNA
SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by
upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res
Commun. 495:1822–1832. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y and Kong D: LncRNA GAS5 represses
osteosarcoma cells growth and metastasis via sponging MiR-203a.
Cell Physiol Biochem. 45:844–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu G, Wang L, Han H, Li Y, Lu S, Li T and
Cheng C: LncRNA ZFAS1 promotes growth and metastasis by regulating
BMI1 and ZEB2 in osteosarcoma. Am J Cancer Res. 7:1450–1462.
2017.PubMed/NCBI
|
|
25
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Radisky DC: Epithelial-mesenchymal
transition. J Cell Sci. 118:4325–4326. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakajima S, Doi R, Toyoda E, Tsuji S, Wada
M, Koizumi M, Tulachan SS, Ito D, Kami K, Mori T, et al: N-cadherin
expression and epithelial-mesenchymal transition in pancreatic
carcinoma. Clin Cancer Res. 10:4125–4133. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nguyen PT, Kudo Y, Yoshida M, Kamata N,
Ogawa I and Takata T: N-cadherin expression is involved in
malignant behavior of head and neck cancer in relation to
epithelial-mesenchymal transition. Histol Histopathol. 26:147–156.
2011.PubMed/NCBI
|
|
36
|
Radisky ES and Radisky DC: Matrix
metalloproteinase-induced epithelial-mesenchymal transition in
breast cancer. J Mammary Gland Biol Neoplasia. 15:201–212. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Malemud CJ: Matrix metalloproteinases
(MMPs) in health and disease: An overview. Front Biosci.
11:1696–1701. 2006. View
Article : Google Scholar : PubMed/NCBI
|