Introduction
Prostate cancer (PCa) is the most common type of
cancer among men, worldwide (1.6 million cases in 2015) (1). Patients with localized PCa can achieve
long-term survival when treated with surgery, radiotherapy and
androgen-deprivation therapy. However, a large proportion of tumors
may progress to castration-resistant prostate cancer (CRPC), which
is an aggressive cancer type and typically results in metastasis
(2,3). Therefore, it is imperative to
investigate novel molecular targeted therapies for PCa that are
based on an in-depth knowledge of the signaling pathways underlying
prostate carcinogenesis.
Long non-coding (lnc)RNAs are a type of non-coding
RNA, that are >200 nucleotides in length. lncRNAs have been
associated with numerous cellular activities, including
proliferation, apoptosis, survival, differentiation and cell cycle
control (4). Both increased and
decreased expression levels of lncRNAs have been demonstrated to
play an important role in human tumorigenesis (4–6). For
example, in breast cancer, upregulation of ARNALI and lnc015192 or
downregulation of PDCD4-AS1 and ANCR has been implicated in tumor
progression (6). High-throughput RNA
sequencing has discovered a variety of novel lncRNAs in the past 10
years, which are also aberrantly regulated in PCa; however, only a
few have been functionally examined. For example, the expression
level of lnc01296 was associated with the preoperative blood level
of prostate-specific antigen (PSA), lymph node metastasis, and
tumor stage (7). Petrovics et
al (8) reported that the
overexpression of PCGEM1 in tumor cells promoted proliferation and
colony formation, and the increased expression level was associated
with a higher risk of developing PCa. In addition, Linc00963 was
associated with androgen-independent PCa, and Linc00963 knockdown
inhibited LNCaP and C4-2 cell viability, motility and invasiveness
(9).
lncRNA SNHG17 is located on 20q11.23 and is known as
an unfavorable prognostic factor in colorectal cancer, and the
overexpression of SNHG17 stimulated tumor cell proliferation by
epigenetically silencing P57 (10).
SNHG17 is also overexpressed by non-small cell lung cancer and
gastric cancer tissues, and promoted oncogenic phenotypes in the
two types of cancer (11,12). Microarray analysis found SNHG17 to be
one of four lncRNAs that was significantly upregulated in
metastatic and androgen-independent C4-2 tumor cells compared with
that in parental non-metastatic, androgen-dependent LNCaP cells
(9). This suggested that SNHG17
functions as an important regulator in human PCa. However, to the
best of our knowledge, the associations between SNHG17 and the
growth and aggressiveness of PCa have not been reported.
Wnt/β-catenin signaling is crucial for cell
polarity, tissue development and homeostasis. The binding of Wnt
ligands to the receptor Frizzled and the co-receptor low-density
lipoprotein receptor-related protein-5 or 6 activated Dishevelled,
which in turn leads to the inhibition of glycogen synthase
kinase-3β (GSK-3β). The inhibition of GSK-3β repressed β-catenin
phosphorylation to prevent its degradation by the
GSK-3β-adenomatous polyposis coli-axin multi-protein complex. As a
result, the stabilized β-catenin could translocate to the nucleus
and interact with T cell factor/lymphoid enhancer factor (TCF/LEF)
to initiate the transcription of target genes (e.g., c-myc
and cyclin D1), which are involved in the control of
cellular processes, such as growth, differentiation, and metabolism
(13,14). Hyperactive aberrant Wnt/β-catenin
signaling has been associated with different types of cancer
(14), including PCa (15).
In the present study, the SNHG17 mRNA expression
levels in PCa tumors and adjacent non-tumor tissue samples were
analyzed. The cellular and molecular mechanisms that underlie
SNHG17-mediated oncogenic properties in the C4-2 human PCa cell
line were also investigated. It was found that SNHG17 could be an
important regulator of tumor aggressiveness and that this oncogenic
effect was associated with β-catenin signaling activity.
Materials and methods
Acquisition of human tissues
The present study was approved by the Human Ethics
Committee of Qingdao Municipal Hospital (QMH;l Qingdao, China). The
patients were recruited from QMH and verbal informed consent was
provided by each patient. The diagnosis of PCa was performed using
both MRI radiography and prostate biopsy. Only patients displaying
multiple positive results in the biopsy were included in the study.
Patients that died from diseases other than PCa were excluded from
the study. Based on the criteria, a total of 58 patients (mean age,
68.5±4.2 years; range, 56.2-85.6 years) were selected for the
study. Paired PCa tumor samples and matched adjacent non-tumor
tissues (3-mm between tumor and non-tumor tissue) were obtained
from each patient undergoing radical prostatectomy between January
2013 and December 2018 at QMH. Clinical factors (age, sex, tumor
diameter and tumor grade) from these patients were used for
clinicopathological analysis. All specimens were immediately stored
at −80°C prior to RNA isolation. The integrity of the RNA was
confirmed using 1% agarose gel electrophoresis before the reverse
transcription-quantitative (RT-q)PCR assay. Based on the mean value
of the SNHG17 mRNA expression level (3.412±1.66) in the
tumor samples, the patients were divided into two groups: The high-
and the low-expression level groups.
Cell culture
The human prostate cancer cell lines LNCaP [clone
FGC; CRL-1740; American Type Culture Collection (ATCC)] and C4-2
(CRL-3314; ATCC), and the normal human prostate epithelial cell
line, HPrEC (PCS-440-010; ATCC) were cultured in DMEM (Invitrogen;
Thermo Fisher Scientific, Inc.) supplemented with 10% FCS
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified incubator with 5% CO2. For SNHG17 knockdown,
recombinant lentivirus-mediated SNHG17-short hairpin (sh)RNA
vectors (Lv-SNHG17-shRNA; Sangon Biotech, Co., Ltd.) were prepared.
These vectors were ready-to-use viral particles, which contained a
pool of three expression constructs, each encoding target-specific
shRNA. The three SNHG17 shRNA sequences are shown in Table SI. When the C4-2 cells reached 80%
confluency, they were transduced with Lv-SNHG17-shRNA viral
particles at a multiplicity of infection of 25 in the presence of
polybrene (8 µg/ml; Sigma-Aldrich; Merck KGaA). After 16 h at 37°C,
the virus-containing medium was removed and fresh complete medium
was added. The transduced cells were then split at a ratio of 1:5
and treated with puromycin (5 µg/ml; Thermo Fisher Scientific,
Inc.) for 2 weeks at 37°C in a humidified incubator with 5%
CO2. The cells transduced with negative control (NC)
shRNA lentivirus harboring a scrambled shRNA sequence were used as
the control. For the overexpression of SNHG17, cDNA encoding the
human SNHG17 gene was amplified using PCR and subcloned into the
pcDNA3.1 expression vector (Invitrogen; Thermo Fisher Scientific,
Inc.). When the C4-2 cells had reached 80% confluency they were
transfected with pcDNA-SNHG17 (2 µg/100 µl medium) using FuGENE
reagent (Roche Diagnostics) at 37°C for 24 h, according to the
manufacturer's protocol. The C4-2 cells transfected with pcDNA3.1
empty vector were used as the control. After 24 h, the cells were
used for the subsequent experimentation. The cell lines were
routinely tested for contamination.
Proliferation, viability, and
apoptosis assays
The proliferation of SNHG17-knockdown,
SNHG17-overexpressing, or control C4-2 tumor cells was assessed
using an MTT assay kit (Abcam) according to the manufacturer's
protocol. Dimethyl sulfoxide was used to dissolve the formazan
product. The absorbance at 570 nm was measured. Cells were cultured
and measured every 24 h for 5 days. For treatment,
SNHG17-overexpressing cells were cultured in DMEM-10% FCS and
treated with ICG001 (Selleck Chemicals) at a dosage of 10 µM.
Cell viability was determined using the
CellTiter-Glo assay kit (Promega Corporation). Briefly, the
SNHG17-knockdown or control C4-2 cells were seeded at
1×104, 5×104 and 1×105 cells/well
in one 96-well plate, then the assay reagent was added to each well
to induce cell lysis, and the luminescence signal was measured
using a microplate reader. The viability was evaluated based on the
output of relative luminescence units (RLU), which correlates with
viable cell numbers.
For the apoptosis assay, the SNHG17-knockdown,
SNHG17-overexpressing, or control C4-2 cells were cultured under
normal (DMEM; 10% FCS) or serum starvation conditions (DMEM; 0.5%
FCS) for 48 h. Caspase 3 activity was analyzed with a colorimetric
assay kit (Novus Biologicals) and the absorbance at 405 nm was
recorded using a microplate reader (Synergy HTX, BioTek Instruments
Inc.). Comparison of the absorbance allows the determination of the
fold change in caspase 3 activity. For treatment,
SNHG17-overexpressing cells were cultured in normal or
serum-starved medium and treated with ICG001 at 10 µM for 48 h.
Immunofluorescence (IF) and
immunocytochemistry (ICC)
The SNHG17-knockdown or control C4-2 tumor cells
were fixed in methanol for 10 min at room temperature (RT) and
blocked with 10% normal horse serum (Sigma-Aldrich; Merck KGaA) for
1 h at RT. After washing with PBS, the cells were incubated
overnight at 4°C with a rabbit antibody against human β-catenin
(1:100, cat. no. 8480, Cell Signaling Technology, Inc.). The cells
were then washed with PBS and further incubated with
FITC-conjugated secondary antibody (1:200, cat. no. ab97063, Abcam)
for 45 min at RT. The cells were then incubated in DAPI-containing
mounting medium for nuclear staining (Abcam) for 5 min at RT. Cells
were monitored using a fluorescence microscope (Carl Zeiss, ×200).
For ICC, the SNHG17-knockdown or control C4-2 cells were incubated
with a rabbit antibody against human TCF1 (1:100, cat. no. 2203,
Cell Signaling Technology, Inc.) at 4°C overnight. After washing
with PBS, cells were treated with biotinylated donkey anti-rabbit
secondary antibody (1:500, cat. no. ab208000, Abcam) for 1 h at RT.
Cells were counterstained with eosin solution (Sigma-Aldrich) for
30 sec at RT. Nuclear staining of TCF1 was monitored using a light
microscope (Olympus, ×200).
Invasion assay
The SNHG17-knockdown or control C4-2 tumor cell
invasion was evaluated using the Boyden chamber system in 24-well
plates. The filter inserts, with 8-µm pore size, were pre-coated
with Matrigel for 30 min at 37°C. The tumor cells were cultured in
DMEM with 10% FCS at 37°C. After 5 h, the cells were cultured in
fresh DMEM, with 1% FCS for 24 h. The cells were then harvested and
plated on the upper chamber of the Matrigel-coated 24-well
Transwell filters (2×105 cells/filter) and incubated in
DMEM with 1% FCS at 37°C. DMEM-20% FCS medium was added to the
lower chamber. After 24 h, unmigrated cells from the upper chamber
were removed and the inserts were stained with eosin for 30 sec at
RT. The invading cells were counted under a light microscope
(Olympus, ×200).
Docetaxel sensitivity assay
For the chemotherapeutic resistance assay, the
SNHG17-knockdown or control C4-2 cells were plated into three
96-well plates at 5×104 cells/well. After 6 h, the cells
were treated with docetaxel at dosages of 0, 1, 2.5, 5 or 10 nM for
1, 3 or 5 days. The cell viability was determined using a Cell
Counting Kit-8 kit (Sigma-Aldrich; Merck KGaA). The CCK-8 solution
provided from the kit was added to each well and incubated at 37°C
for 2 h. The absorbance at 450 nm was measured for each well.
TCF reporter assay
The SNHG17-knockdown and control C4-2 cells were
co-transfected with TOPflash (Sigma-Aldrich; Merck KGaA) and pRL
(Promega Corporation), or FOPflash (Sigma-Aldrich; Merck KGaA) and
pRL, using FuGENE reagent (Roche Diagnostics), according to the
manufacturer's protocols. TOPflash is a β-catenin-responsive
firefly luciferase reporter plasmid, which contains three copies of
the TCF binding site upstream of the thymidine kinase minimal
promoter and luciferase open reading frame. FOPflash is a non
β-catenin-responsive plasmid that contains mutated TCF binding site
(16). pRL is a constitutively
expressing Renilla luciferase plasmid used as an internal
control for transfection efficiency (17). The medium was removed and replaced
with fresh medium containing 5 mM LiCl (or NaCl as a control) 24 h
post-transfection. Cell lysates were extracted using reporter lysis
buffer (Promega Corporation) 24 h after treatment. Luciferase
activity was detected using dual-luciferase assay kits (Promega
Corporation). Both firefly and Renilla luminescence signals
were recorded. The firefly luciferase activities of TOPflash and
FOPflash were normalized to the Renilla luciferase activity
of pRL, respectively. The ratio of TOPflash activity to FOPflash
activity (TOPflash/FOPflash) was calculated for the measurement of
TCF reporter activity.
RT-qPCR and western blot analysis
Total RNA was extracted from the specimens cells
using TRIzol® (Thermo Fisher Scientific, Inc.). The RNA
was then reverse transcribed into cDNA using the SuperScript III
reverse transcriptase kit (Thermo Fisher Scientific, Inc.) at 50°C
for 50 min. qPCR was conducted using SYBR green master mix (Thermo
Fisher Scientific, Inc.). The cycling conditions were: 50°C for 2
min; 95°C for 10 min; then 95°C for 15 sec and 60°C for 1 min for
40 cycles. PCR products were analyzed with the ABI PRISM 7900HT
Sequence Detections System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The relative mRNA level of the target gene was
determined using ABI software (RQ Manager, version 1.2). The
threshold cycle of the target gene was normalized to that of the
endogenous GAPDH transcript (ΔΔCt). The fold change was determined
using the formula 2−ΔΔCq method (18). The designed primers for PCR are
listed in Table I.
 | Table I.Primers designed for quantitative
PCR. |
Table I.
Primers designed for quantitative
PCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| SNHG17 |
TGCTTGTAAGGCAGGGTCTC |
ACAGCCACTGAAAGCATGTG |
| CTNNB1 |
TCTTGCCCTTTGTCCCGCAAATCA |
TCCACAAATTGCTGCGTCCCA |
| TCF1 |
CGGGACAGAGGACCATTACA |
CCACCTGCCTCGGCCTGCCAAAGT |
| TCF4 |
CTGCCTTAGGGACGGACAAAG |
TGCCAAAGAAGTTGGTCCATTTT |
| LEF1 |
CTTTATCCAGGCTGGTCTGC |
TCGTTTTCCACCATGTTTCA |
| c-myc |
TTCGGGTAGTGGAAAACCAG |
AGTAGAAATACGGCTGCACC |
| Cyclin D1 |
TCTGGCATTTTGGAGAGGAAG |
CATCTACACCGACAACTCCATC |
| Axin2 |
CAAGGGCCAGGTCACCAA |
CCCCCAACCCATCTTCGT |
| GAPDH |
GAAGGTGAAGGTCGGAGTC |
GAAGATGGTGATGGGATTTC |
For the western blot analysis, the cell lysates were
extracted from SNHG17-knockdown or control C4-2 cells with RIPA
lysis buffer (Sigma-Aldrich; Merck KGaA) and 25 µg of each protein
sample was separated using 8% SDS-PAGE. After electrophoresis, the
proteins were transferred to a PVDF membrane (Bio-Rad Laboratories,
Inc.), then washed with TBS/0.1% Tween-20 (TBST) and blocked with
1% BSA (Sigma-Aldrich; Merck KGaA) for 30 min at RT. Subsequently,
the membrane was incubated with rabbit anti-human β-catenin
(1:1,000, cat. no. 9562, Cell Signaling Technology, Inc.) and
rabbit anti-human β-tubulin antibody (1:1,000, cat. no. 2146, Cell
Signaling Technology, Inc.) overnight at 4°C. The PVDF membrane was
then incubated with an HRP-conjugated secondary antibody (1:3,000,
cat. no. 7074, Cell Signaling Technology, Inc.) for 1 h at RT.
Protein expression was detected using an enhanced chemiluminescence
kit (Bio-Rad Laboratories, Inc.). Densitometry of the bands was
measured with ImageJ software (v1.51j8; National Institutes of
Health).
Xenograft mouse model
A total of 14 BALB/c female nude mice (8-10 weeks
old; weight, 21.18±1.23 g) were used in the present study. The C4-2
tumor cells were washed with PBS and mixed with PBS/Matrigel (1:1)
at a density of 104/µl. The animals were anesthetized
using an intraperitoneal injection of a mixture of ketamine (100
mg/kg) and xylazine (10 mg/kg). Next, 1×106 (100 µl)
SNHG17-knockdown or control C4-2 tumor cells (7 animals/group) were
subcutaneously injected into the flanks of the mice. Local tumor
growth was examined using a caliper every 5 days. The tumor volume
(mm3) was calculated using the ‘width2 ×
length/2’ formula. At 50 days after tumor cell injection, the mice
were sacrificed using CO2, with a fill rate of 30% of
the chamber volume/min, to the existing air inside the chamber to
minimize the animals' distress. The death of mice was confirmed by
examining the respiratory rate and cardiac arrest. All tumor
samples were harvested immediately following the sacrifice of the
animals at day 50. During the entire experiment, all the animals
were maintained in a standard mouse facility maintained at 22–24°C,
50–60% humidity, a 12-h light/dark cycle, and free access to water
and food. The animal study was reviewed and approved by the Animal
Care Committee of Qingdao University.
Immunohistochemistry (IHC)
Tumors were harvested and fixed in 4%
paraformaldehyde at 4°C for 2 days. The samples were embedded in
paraffin and 5-µm thick sections were made. For IHC analysis, tumor
sections were deparaffinized in xylene and were rehydrated via
graded alcohol (100%, 95%, 90%, 80% and 70%) and distilled water.
After washing with PBS, sections were incubated with proteinase K
(Dako) for 10 min at RT for antigen retrieval. Sections were washed
with PBS and blocked with horse serum (Sigma-Aldrich; Merck KGaA)
for 30 min at RT. Sections were then incubated with rabbit
anti-Ki67 (1:200, cat. no. ab16667, Abcam), rabbit anti-β-catenin,
(1:100, cat. no. 8480, Cell Signaling Technology, Inc.), or rabbit
anti-TCF1 (1:100, cat. no. 2203, Cell Signaling Technology, Inc.),
respectively, overnight at 4°C. Sections were washed with PBS and
incubated with biotinylated donkey anti-rabbit secondary antibody
(1:1,000, cat. no. ab208000, Abcam) for 1 h at RT. After washing
with PBS, sections were incubated with DAB substrate (Vector Lab)
for 5 min at RT, and were counterstained with Mayer's hematoxylin
solution (Sigma-Aldrich; Merck KGaA) for 10 sec at RT. Positive
stains were monitored under a light microscope (Olympus, ×100).
Statistical analysis
A two-tailed paired Student's t-test or one-way
ANOVA followed by Tukey's post hoc test was used for comparisons
between two groups or among multiple groups, respectively. A
χ2 test was used to analyze the clinicopathology
characteristics. A log-rank test was used to analyze the
Kaplan-Meier survival curve. Experiments were performed in
triplicate and repeated three times. Data are presented as the mean
± standard deviation. All statistical analyses were conducted using
the GraphPad Prism software (v5.0; GraphPad Software, Inc.).
Results
SNHG17 upregulation is associated with
poor outcomes in patients with PCa
To investigate the role of SNHG17 in prostate
carcinogenesis, the expression level of SNHG17 was determined in 58
pairs of human PCa tumor and matched adjacent non-tumor tissues
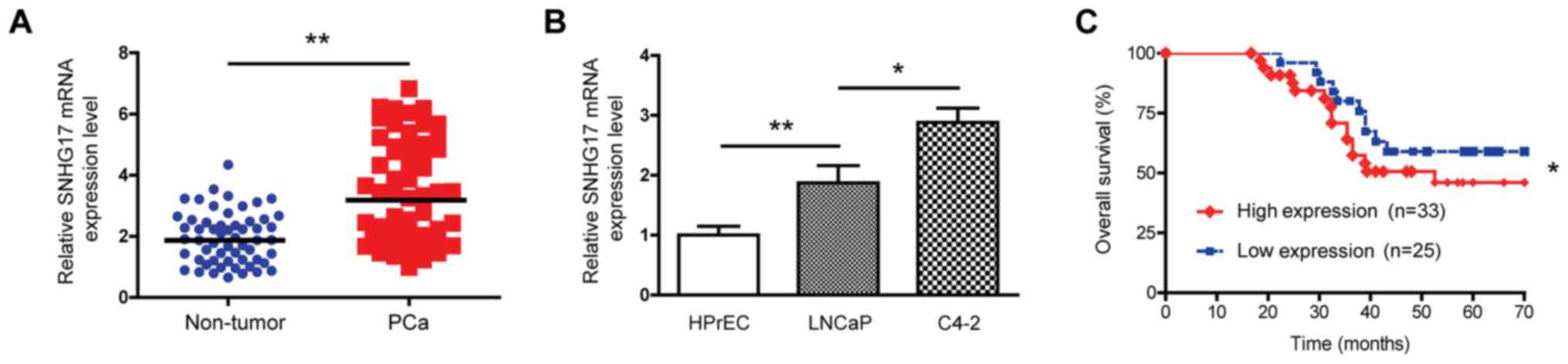
using RT-qPCR. As shown in Fig. 1A,
the expression level of SNHG17 mRNA was higher in the PCa
samples compared with that in the non-tumor tissues, suggesting
that this lncRNA was upregulated in PCa tumors. The mRNA expression
level of SNHG17 was also compared between tumor and epithelial
cells, and the results showed that the mRNA expression level of
SNHG17 was elevated in the LNCaP and C4-2 tumor cells
compared with that in the normal HPrEC cells (Fig. 1B). Notably, the aggressive and
metastatic C4-2 tumor cells exhibited a 1.54-fold increase in the
SNHG17 mRNA expression level compared with that in the
parental less aggressive, non-metastatic LNCaP cells, suggesting
that high SNHG17 expression level might be associated with tumor
aggressiveness. These findings indicated that SNHG17 was
upregulated in human PCa.
Next, the association between the SNHG17 mRNA
expression level and survival time in patients with PCa was also
analyzed. Based on the mean SNHG17 mRNA expression level
(3.412±1.66), the patients were divided into high-(n=33) and
low-expression level groups (n=25). As shown in Fig. 1C, the Kaplan-Meier survival curve
indicated that compared with that in those with low expression
levels, patients with high SNHG17 mRNA expression levels exhibited
poor overall survival time. The clinicopathological analysis showed
that most patients suffered from advanced PCa. For example, among
the 58 patients, 32 patients displayed a Gleason score of ≥7; 40
patients had T3+T4 stage PCa; 22 patients had lymph node
metastasis, and 19 patients developed distant metastasis. Notably,
high mRNA expression levels of SNHG17 were associated with
advanced histological grade, tumor stage and metastasis. However,
no association was found between the SNHG17 mRNA expression level
and age, tumor size or Gleason score (Table II). These findings suggested that
increased SNHG17 expression levels in tumors may be a predictor of
poor patient outcomes.
 | Table II.Analysis of the clinicopathological
characteristics. |
Table II.
Analysis of the clinicopathological
characteristics.
|
|
| Expression level of
SNHG17 |
|---|
|
|
|
|
|---|
| Characteristic | Number | Low (n=25) | High (n=33) | P-value |
|---|
| Mean age,
years |
|
|
|
|
|
<65 | 16 | 6 | 10 | 0.594 |
|
≥65 | 42 | 19 | 23 |
|
| Tumor diameter,
cm |
|
|
|
|
|
<2.0 | 27 | 13 | 14 | 0.469 |
|
≥2.0 | 31 | 12 | 19 |
|
| Gleason score |
|
|
|
|
|
<6 | 26 | 12 | 14 | 0.672 |
| ≥7 | 32 | 13 | 19 |
|
| Histological
grade |
|
|
|
|
|
II+III | 30 | 18 | 12 | 0.007 |
| IV | 28 | 7 | 21 |
|
| Tumor stage |
|
|
|
|
| T2 | 18 | 13 | 5 | 0.003 |
|
T3+T4 | 40 | 12 | 28 |
|
| Lymph node
metastasis |
|
|
|
|
|
Yes | 22 | 4 | 18 | 0.003 |
| No | 36 | 21 | 15 |
|
| Distant
metastasis |
|
|
|
|
|
Yes | 19 | 4 | 15 | 0.018 |
| No | 39 | 21 | 18 |
|
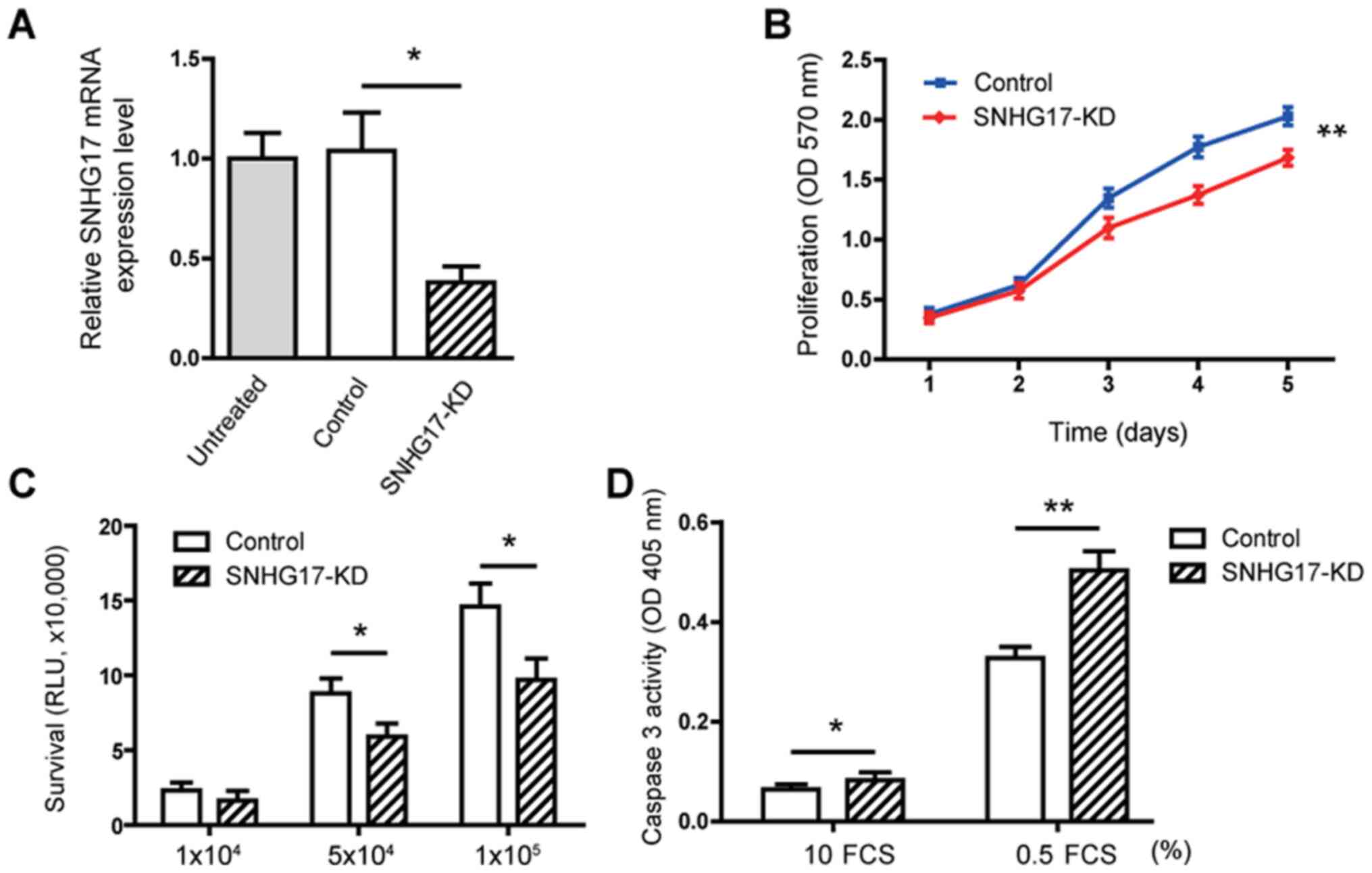
SNHG17 increases the proliferation,
viability but reduces apoptosis in the C4-2 tumor cells
To investigate the role of SNHG17 in modulating PCa
oncogenic properties, a knockdown approach was used in the C4-2
tumor cells to determine the function of SNHG17 at the cellular
level. RT-qPCR analysis showed that knockdown of SNHG17
resulted in a 68% reduction in SNHG17 mRNA expression level
(Fig. 2A). The MTT assays showed
that knockdown of SNHG17 significantly (P<0.01) decreased the
proliferation of the C4-2 cells (Fig.
2B). Tumor cell viability was also decreased in cells treated
with SNHG17 shRNA (Fig. 2C).
Subsequently, caspase 3 activity was measured and it was found that
SNHG17 knockdown only slightly increased caspase 3 activity in
cells cultured under normal conditions (10% FCS). However, SNHG17
knockdown significantly stimulated caspase 3 activity in the cells
cultured under serum starvation conditions (0.5% FCS) (Fig. 2D). These data suggested that SNHG17
increases the proliferation and viability, but reduces apoptosis in
PCa tumor cells.
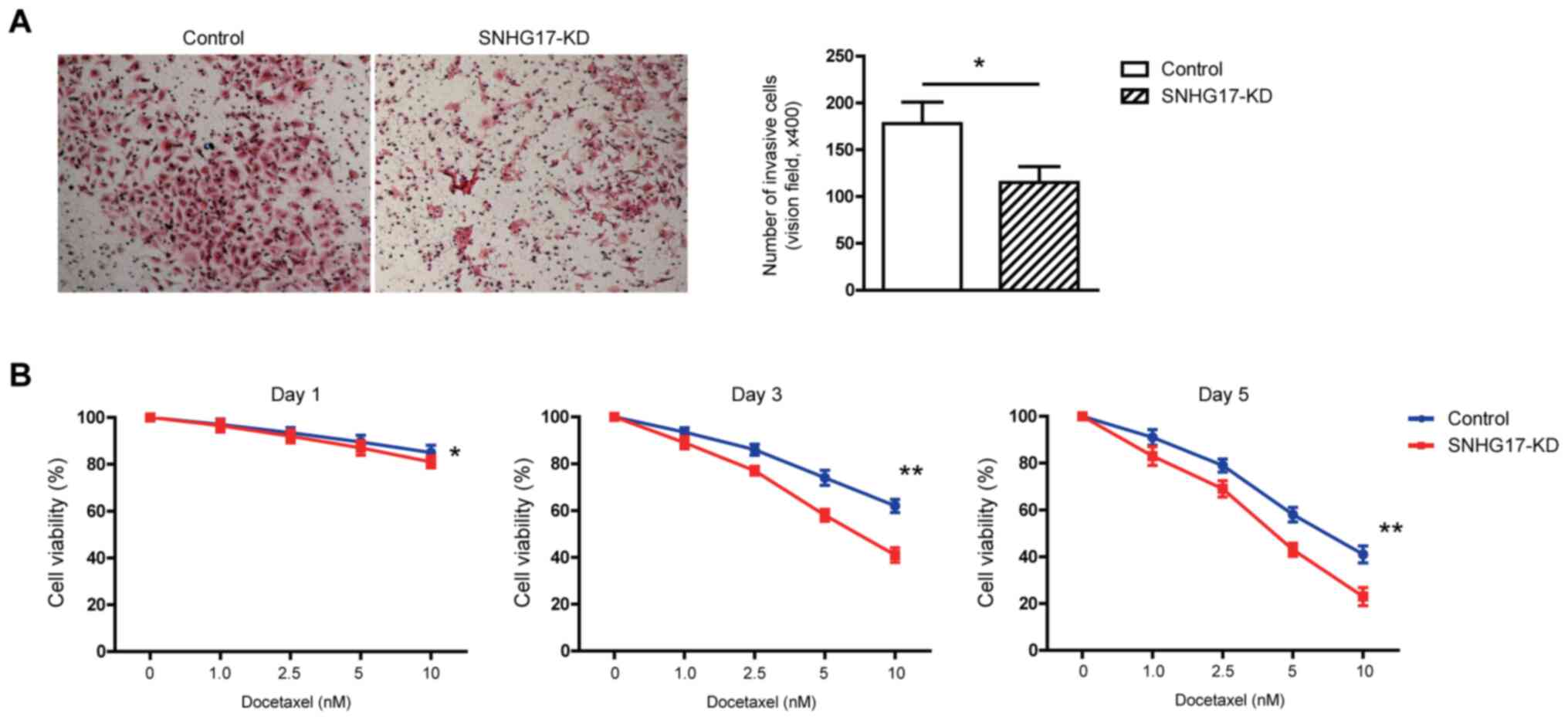
SNHG17 increases invasion and
chemotherapeutic resistance in the C4-2 tumor cells
Invasive ability is an essential marker of tumor
cell aggressiveness (19).
Therefore, a Transwell chamber was used to investigate how altering
the SNHG17 mRNA expression level in tumor cells affected their
invasive ability. It was found that SNHG17 knockdown significantly
decreased the number of invading C4-2 tumor cells compared with
that in the control group (Fig. 3A),
suggesting that SNHG17 increased tumor cell invasion.
Since 2004, docetaxel has been used as an important
chemotherapeutic agent for treating patients with PCa and
metastatic CRPC (20); however, most
patients with CRPC, who are treated with docetaxel, eventually
become refractory, due to the development of drug resistance
(21). Therefore it was subsequently
investigated whether SNHG17 was associated with chemotherapeutic
resistance in the PCa tumor cells, by treating androgen-independent
C4-2 cells with various concentrations of docetaxel over different
time points. It was found that docetaxel reduced the viability of
the tumor cells in a dose-dependent manner. Notably, this reduction
was further enhanced by the knockdown of SNHG17 (Fig. 3B). These observations suggested that
SNHG17 decreased the cellular sensitivity of tumor cells in
response to docetaxel treatment, leading to increased
chemotherapeutic resistance.
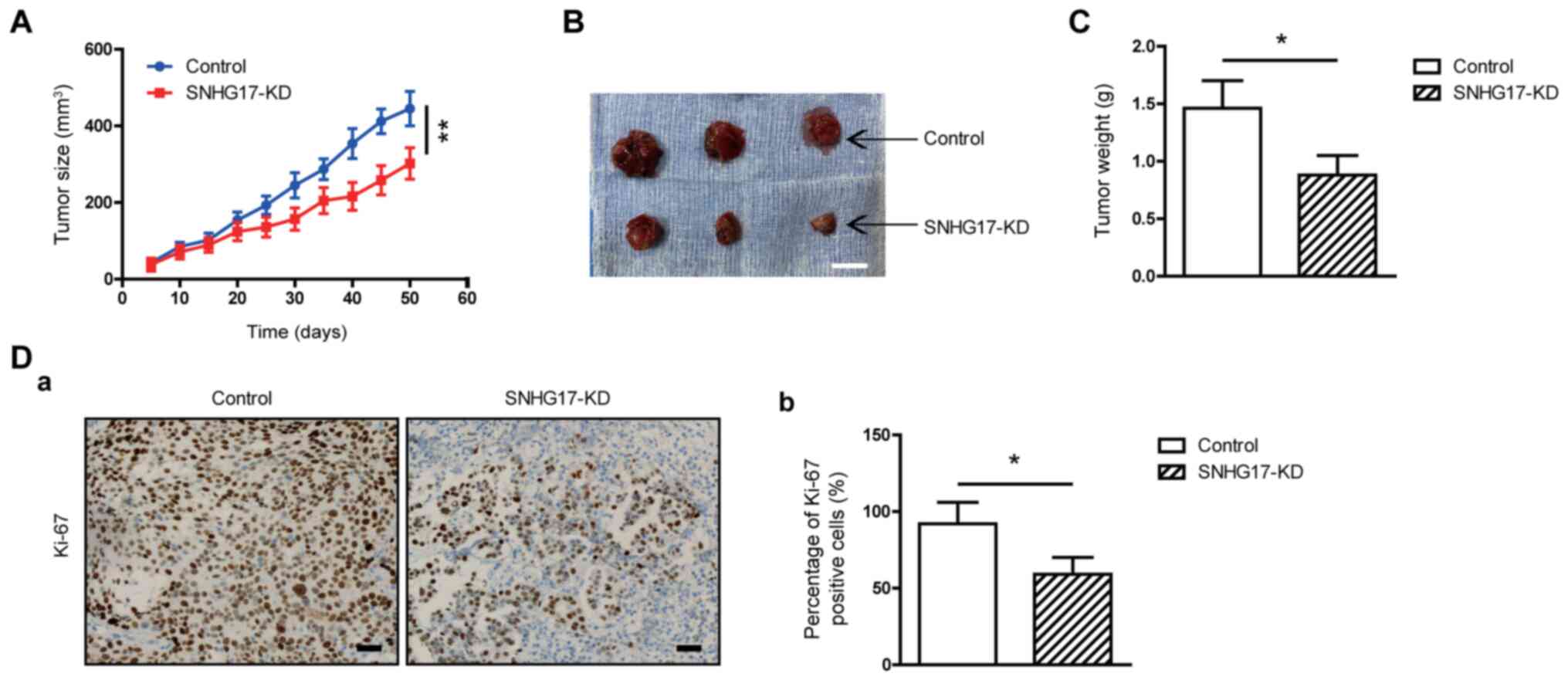
SNHG17 stimulates in vivo tumor
growth
As aforementioned it was found that SNHG17 increased
tumor cell viability and invasion in vitro. To determine
whether SNHG17 also increased tumor growth in vivo, a
subcutaneous xenograft mouse model was used and C4-2 tumor cells
were injected into the flanks of the BALB/c female nude mice. After
50 days, the tumor sizes in the mice injected with SNHG17-knockdown
cells were smaller compared with that in mice that were injected
with the control cells (Fig. 4A and
B). The mean weight of the tumor samples was also lower in mice
injected with the SNHG17-knockdown tumor cells compared with that
in animals injected with the control cells (Fig. 4C). Immunostaining for Ki-67 was also
performed to further confirm that SNHG17 knockdown inhibited tumor
cell proliferation in vivo. As shown in Fig. 4D, the number of Ki-67-positive cells
was reduced by ~36% in tumors, that had developed from
SNHG17-knockdown cells compared with that in the control group.
These results suggest that SNHG17 promotes in vivo tumor
growth.
SNHG17 facilitates tumor growth via
β-catenin activity
As hyperactivity of the Wnt/β-catenin pathway has
been associated with human PCa (15), it was determined whether the
SNHG17-induced tumor-promoting effect was associated with the
activation of this pathway. Notably, SNHG17 knockdown decreased the
expression level of β-catenin, at both the mRNA and protein levels
in the C4-2 tumor cells (Fig. 5A and
B). The IF analysis revealed that β-catenin fluorescence was
reduced in the tumor cells following knockdown of SNHG17 (Fig. 5C). The immunocytochemistry results
showed that there were fewer TCF1-positive cells in the
SNHG17-knockdown group compared with that in the control group
(Fig. 5D). Furthermore, the TCF
reporter assay showed that SNHG17 knockdown not only inhibited
baseline luciferase activity, but also LiCl-enhanced luciferase
activity (Fig. 5E). In addition, the
gene expression levels of several Wnt signal molecules, such as
TCF1, TCF4, LEF1, c-myc, cyclin D1 and axin2, were
decreased in SNHG17-knockdown C4-2 cells (Fig. 5F). These data indicated that SNHG17
upregulated β-catenin/TCF transcription.
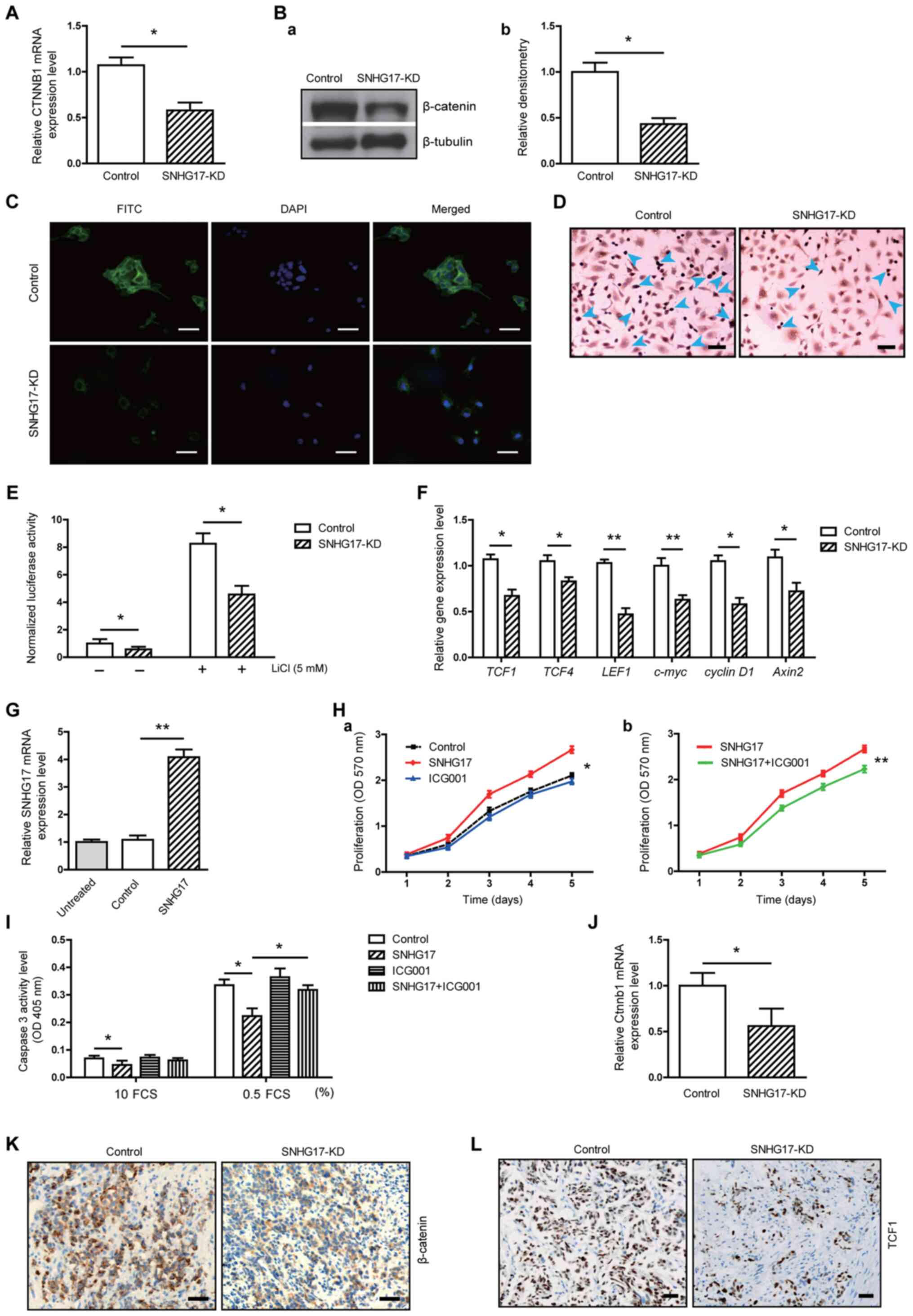 | Figure 5.SNHG17 facilitates C4-2 tumor growth
via β-catenin. (A) RT-qPCR and (B-a) western blot analysis revealed
that β-catenin mRNA and protein expression level was decreased in
the SNHG17-KD cells, respectively (n=3). (B-b) Densitometry
analysis of β-catenin (normalized to β-tubulin using ImageJ
software). (C) Immunofluorescence staining of β-catenin and (D)
immunocytochemical staining of TCF1 in the control or SNHG17-KD
tumor cells. (E) TCF reporter assay in the control or SNHG17-KD
cells, treated with/without LiCl (5 mM) for 24 h (n=3). RT-qPCR was
performed to determine the mRNA expression level of (F) different
genes in the control or SNHG17-KD tumor cells and (G) SNHG17 in
cells transfected with pcDNA-SNHG17 vector (n=3). MTT assay was
used to determine cell proliferation in the (H-a) control,
SNHG17-overexpressing and control cells treated with ICG001 (10 µM)
and (H-b) in SNHG17-overexpressing cells treated with/without
ICG001 (n=4). (I) Caspase 3 activity assay was measured in the
control or SNHG17-KD cells, treated with/without ICG001 (10 µM) for
48 h (n=3). (J) RT-qPCR was used to measure the mRNA expression
level of β-catenin in the tumor samples (n=3). Immunohistochemistry
staining of (K) β-catenin and (L) TCF1 in the tumor samples. For C:
Scale bar, 50 µm; For D, K and L: Scale bar, 100 µm. The data are
presented as the mean ± SD. *P<0.05, **P<0.01. RT-qPCR,
reverse transcription-quantitative PCR; KD, knockdown; OD, optical
density. |
To further confirm that the SNHG17-induced oncogenic
effect was mediated by its association with β-catenin, SNHG17 was
also overexpressed in the C4-2 cells and treated them with ICG001,
an inhibitor of the Wnt/β-catenin signaling pathway. As shown in
Fig. 5G, transfection with
pcDNA-SNHG17 resulted in a 3.78-fold increase of SNHG17 mRNA
expression level compared to that in the control cells. The MTT
assay showed that SNHG17 overexpression significantly increased
tumor cell proliferation, whereas ICG001 alone only slightly
reduced the proliferation rate (Fig.
5Ha). In contrast, ICG001 significantly attenuated
SNHG17-accelerated proliferation (Fig.
5Hb). The overexpression of SNHG17 also reduced the caspase 3
activity level of C4-2 cells, whereas this reduction was reversed
in the presence of ICG001 (Fig. 5I).
To verify the association between SNHG17 and the β-catenin pathway
during in vivo tumor growth, RT-qPCR analysis was performed
and a reduction of β-catenin mRNA expression level in the tumors
from SNHG17-knockdown cells was found compared with that in the
control group (Fig. 5J).
Immunohistochemistry analysis further revealed that the β-catenin
staining intensity was decreased in tumors derived from
SNHG17-knockdown cells compared with that in the control group
(Fig. 5K). The population of TCF1+
cells was also notably decreased in tumors derived from
SNHG17-knockdown cells (Fig. 5L).
Taken together, these results suggested that SNGH17 could
facilitate tumor growth via its association with β-catenin-mediated
TCF-dependent transcriptional activity.
Discussion
In the present study, the role of SNHG17 in human
PCa was characterized. SNHG17 mRNA expression level was found to be
increased in tumor tissues and its mRNA expression level was also
associated with poor patient outcomes. Functionally, SNHG17
increased tumor aggressiveness by promoting cell proliferation,
invasion and resistance to chemotherapy. Notably, this modulating
effect was mediated via β-catenin signaling activity. The results
from the present study suggested that SNHG17 could be a critical
regulator of human PCa aggressiveness.
The role of lncRNA in human different types of
cancer has received considerable attention in recent years. Several
lncRNAs, such as PCA3, PCAT1, CCAT2, GASS, and Linc01296, have been
found to play either an oncogenic or tumor-suppressive role in PCa
(22). SNHG17 is also a novel lncRNA
and has been associated with lung (11), breast (23) and gastric (12) cancers. Based on a microarray, SNHG17
was shown to be among the few lncRNAs, that were upregulated in
metastatic and androgen-independent C4-2 tumor cells compared with
that in the parental non-metastatic, androgen-dependent LNCaP cells
(9). However, it is unclear whether
SNHG17 is associated with PCa progression.
To investigate this issue, 58 pairs of PCa tumor and
matched adjacent non-tumor tissues were analyzed. The mRNA
expression level of SNHG17 was increased in the PCa samples
compared with that in the non-tumor tissues, suggesting that SNHG17
may play a role in PCa. Notably, patients with high tumor mRNA
expression levels of SNHG17 had a poorer overall survival rate
compared with patients with lower SNHG17 mRNA expression levels.
The clinicopathological analysis further showed that high SNHG17
mRNA expression level was associated with indicators of cancer
aggressiveness, including advanced histological grade, tumor stage
and metastasis. These results showed that increased SNHG17 mRNA
expression level could be a good predictor of poor outcomes in
patients with PCa. In the present study, the overall survival was
relatively lower compared with that in reported statistics
(24). This may be due to the fact
that all the patients selected in the present study had advanced
PCa. For example, among the 58 patients, 32 patients displayed a
Gleason score of ≥7; 40 patients had T3+T4 stage PCa; 22 patients
had lymph node metastasis, and 19 patients developed distant
metastasis (Table II). In addition,
all patients showed multiple positive results using prostate biopsy
prior to radical prostatectomy. It should also be noted that only
patients expressing high-level SNHG17 displayed a shortened overall
survival rate, whereas patients expressing low-level SNHG17
significantly had a prolonged survival rate.
In addition, one limitation of the present
clinicopathological study is that it did not include PSA doubling
time (PSADT) data; PSADT has been found to be a useful marker to
predict patient outcomes. Takeuchi et al (25) reported that patients with longer
preoperative PSADTs (>24 months) displayed favorable
pathological findings (80% of patients with T2 stage and 55% of
patients with a Gleason score of 2–6), and a higher PSA
non-recurrence rate compared with patients who had shorter
preoperative PSADTs (<24 months). In androgen-independent PCa,
patients with a mean PSADT of 12.7 months before deferred
antiandrogen therapy showed an improved response compared with that
in patients with a mean PSADT of 7.5 months (26). These studies suggest that PSADT is a
strong predictor of patient outcomes. Therefore, the inclusion of
PSADT data may provide a more creditable clinicopathological
evaluation regarding the association between SNGH17 and PCa.
Another limitation is that only 58 patients were included in the
present study. This sample size could meet the requirement for
meaningful statistical assessments in clinicopathological and
survival analyses; however, a larger number of samples would be
preferred for more reliable evaluation.
The SNHG17 mRNA expression level between tumor cell
lines and normal epithelial cells was also compared in the present
study. SNHG17 had higher mRNA expression levels in the LNCaP and
C4-2 tumor cells compared with that in the normal HPrEC cells.
Notably, SNHG17 mRNA expression level was significantly increased
in the metastatic C4-2 cell line compared with that in the
non-metastatic parental LNCaP tumor cell line. These in
vitro and in vivo results indicated that SNHG17 was
associated with the aggressiveness of human PCa. To clarify the
mechanism by which SNHG17 regulated the oncogenic properties of
PCa, a knockdown approach was used and the modulatory effect of
SNHG17 on tumor cell behavior was investigated. SNHG17 has been
shown to increase the proliferation of lung and breast cancer cells
(11,23). Consistent with these studies, it was
found that SNHG17 knockdown decreased cell proliferation and
viability, but increased caspase 3 activity in the C4-2 cells,
indicating that SNGH17 could exert a growth-promoting effect in the
human PCa tumor cells. In a xenograft mouse model, it was also
found that SNHG17 knockdown suppressed tumor growth in vivo.
Ki-67 staining revealed that the inhibition of tumorigenic activity
was associated with decreased cancer cell proliferation. Based on
these results, SNHG17 could promote PCa progression, at least
partly through its positive regulation of tumor cell growth.
Cancer cell invasion is a key factor that
contributes to tumor aggressiveness, including metastasis (18). SNHG17 has been reported to induce
tumor cell invasion in breast (23)
and gastric (12) cancers.
Consistent with the previous studies, it was found that SNHG17
knockdown reduced the invasive ability of the C4-2 tumor cells,
suggesting that this lncRNA could also promote the invasion of
prostate tumor cells. Resistance to chemotherapy is another key
factor leading to poor prognosis in patients with cancer (27). Docetaxel has been an important
chemotherapy agent to treat patients with PCa and metastatic CRPC
(20); however, most patients with
CRPC treated with docetaxel eventually develop drug resistance
(19). In the present study, it was
found that SNHG17 knockdown significantly reduced the viability of
the C4-2 tumor cells following docetaxel treatment, suggesting that
SNHG17 reduced the cellular sensitivity of PCa cells in response to
docetaxel. Based on the results from the present study, it is
possible that SNHG17 promoted the aggressiveness of prostate tumor
cells by enhancing both their invasive ability and their resistance
to chemotherapy.
Furthermore, it was found that SNHG17 facilitated
tumor growth and progression in association with β-catenin
signaling activity. Increasing evidence has indicated that lncRNAs
regulate various cellular processes through the
activation/inactivation of signaling pathways. For example, the
lncRNA PICART1 suppressed gastric tumor cell proliferation by
positively regulating the PI3K/AKT and MAPK/ERK pathways (28). Rani et al (29) reported that LncND induced
neuroblastoma cell proliferation and neuronal differentiation via
its crosstalk with the Notch signaling pathway. The interaction
between lncRNAs and the Wnt signaling pathway has also received
increasing attention (30). Several
lncRNAs, such as lncSOX4 and SLCO4A1-AS1, have been reported to
facilitate tumor cell motility and metastasis through a
β-catenin-dependent mechanism in osteosarcoma and colorectal cancer
(31,32). Silencing of HOXA11-AS negatively
regulated hepatocellular carcinoma stem cell growth and
self-renewal via the inactivation of the Wnt signaling pathway
(33). In the present study, it was
found that SNHG17 knockdown inhibited the accumulation of
β-catenin. Notably, the TCF reporter activity and the mRNA
expression level of several key β-catenin signaling molecules, such
as TCF1, TCF4, LEF1, axin2, c-myc, and cyclin D1, were
downregulated in the SNHG17-knockdown tumor cells. These findings
support our hypothesis that SNHG17 stabilized β-catenin and
subsequently activated the TCF-dependent transcription of Wnt
target genes. Notably, the SNHG17-induced tumor cell growth and
SNHG17-reduced cell apoptosis were attenuated following treatment
with the β-catenin inhibitor, ICG001. The animal study from the
present study also showed that β-catenin and TCF1 protein
expression level was inhibited in the tumor samples from mice
treated with SNHG17-knockdown cells. Taken together, these results
indicated that SNHG17 induced PCa tumor growth in association with
β-catenin/TCF activity. The present study has identified the
SNHG17/β-catenin/TCF axis in PCa tumor cells; however, the
possibility that other mechanisms may also be involved cannot be
excluded. For example, Kim et al (34) reported that pimozide repressed the
proliferation of PCa tumor cell lines (PC3 and DU145) via the
generation of reactive oxygen species (ROS). Takeuchi et al
(35) also reported that ROS was
associated with the proapoptotic activity of bladder cancer cells.
Therefore, whether SNHG17 exerts its tumor-promoting effect by
targeting ROS merits further investigation.
In conclusion, the results from the present study
demonstrated that lncRNA SNHG17 mRNA expression level was increased
in human PCa and, thus, represents a potential biomarker of poor
prognosis. Mechanistically, SNHG17 increased cell proliferation,
viability, invasion and resistance to chemotherapy. The
SNHG17-induced oncogenic effect was mediated via β-catenin
activity. The present study revealed that SNHG17 could be
critically implicated in PCa and may be considered as a potential
molecular target for PCa treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Qingdao
Science and Technology Foundation (grant nos. 19-6-1-44-nsh and
20-3-4-41-nsh).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Author's contributions
HJZ and HZ designed the study. HJZ, HJD and PW
performed experiments and collected the data. HJZ, HJD and HZ
performed the data analysis. HJZ and HZ drafted the initial
manuscript. HJZ, HJD, and HZ confirmed the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal procedures were reviewed and approved by
the Animal Care Committee of Qingdao University (approval no.
201803233A). The present study was also approved by the Human
Ethics Committee of Qingdao Municipal Hospital (approval no. 05243)
and verbal informed consent was provided by each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GSK-3β
|
glycogen synthase kinase-3β
|
|
HPrEC
|
human prostate epithelial cells
|
|
lncRNAs
|
long non-coding RNAs
|
|
LEF
|
lymphoid enhancer factor
|
|
PCa
|
prostate cancer
|
|
TCF
|
T cell factor
|
References
|
1
|
Global Burden of Disease Cancer
Collaboration, ; Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional, and national cancer incidence, mortality, years
of life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chandrasekar T, Yang JC, Gao AC and Evans
CP: Mechanisms of resistance in castration-resistant prostate
cancer (CRPC). Transl Andro Urol. 4:365–380. 2015.PubMed/NCBI
|
|
3
|
Mansinho A, Macedo D, Fernandes I and
Costa L: Castration-Resistant prostate cancer: Mechanisms, targets
and treatment. Adv Exp Med Biol. 1096:117–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aird J, Baird AM, Lim MCJ, McDermott R,
Finn SP and Gray SG: Carcinogenesis in prostate cancer: The role of
long non-coding RNAs. Noncoding RNA Res. 3:29–38. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Weiss M, Plass C and Gerhauser C: Role of
lncRNAs in prostate cancer development and progression. Biol Chem.
395:1275–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang T, Hu H, Yan G, Wu T, Liu S, Chen W,
Ning Y and Lu Z: Long non-coding RNA and breast cancer. Technol
Cancer Res Treat. 18:15330338198438892019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu J, Cheng G, Zhang C, Zheng Y, Xu H,
Yang H and Hua L: Long noncoding RNA LINC01296 is associated with
poor prognosis in prostate cancer and promotes cancer-cell
proliferation and metastasis. Onco Targets Ther. 10:1843–1852.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Petrovics G, Zhang W, Makarem M, Street
JP, Connelly R, Sun L, Sesterhenn IA, Srikantan V, Moul JW and
Srivastava S: Elevated expression of PCGEM1, a prostate-specific
gene with cell growth-promoting function, is associated with
high-risk prostate cancer patients. Oncogene. 23:605–611. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Han S, Jin G, Zhou X, Li M, Ying
X, Wang L, Wu H and Zhu Q: Linc00963: A novel, long non-coding RNA
involved in the transition of prostate cancer from
androgen-dependence to androgen-independence. Int J Oncol.
44:2041–2049. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma Z, Gu S, Song M, Yan C, Hui B, Ji H,
Wang J, Zhang J, Wang K and Zhao Q: Long non-coding RNA SNHG17 is
an unfavourable prognostic factor and promotes cell proliferation
by epigenetically silencing P57 in colorectal cancer. Mol Biosyst.
13:2350–2361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu T, Yan S, Jiang L, Yu S, Lei T, Yang D,
Lu B, Wei C, Zhang E and Wang Z: Gene amplification-driven long
noncoding RNA SNHG17 regulates cell proliferation and migration in
human non-small-cell lung cancer. Mol Ther Nucleic Acids.
17:405–413. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang G, Xu Y, Wang S, Gong Z, Zou C,
Zhang H, Ma G, Zhang W and Jiang P: LncRNA SNHG17 promotes gastric
cancer progression by epigenetically silencing of p15 and p57. J
Cell Physiol. 234:5163–5174. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Willert K and Nusse R: Wnt proteins. Cold
Spring Harb Perspect Biol. 4:a0078642012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murillo-Garzon V and Kypta R: WNT
signalling in prostate cancer. Nat Rev Urol. 14:683–696. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kumar A, Zloza A, Moon RT, Watts J,
Tenorio AR and Al-Harthi L: Active beta-catenin signaling is an
inhibitory pathway for human immunodeficiency virus replication in
peripheral blood mononuclear cells. J Virol. 82:2813–2820. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gedaly R, Galuppo R, Daily MF, Shah M,
Maynard E, Chen C, Zhang X, Esser KA, Cohen DA, Evers BM, et al:
Targeting the Wnt/beta-catenin signaling pathway in liver cancer
stem cells and hepatocellular carcinoma cell lines with FH535. PLoS
One. 9:e992722014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krakhmal NV, Zavyalova MV, Denisov EV,
Vtorushin SV and Perelmuter VM: Cancer invasion: Patterns and
mechanisms. Acta Naturae. 7:17–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Petrylak DP, Tangen CM, Hussain MH, Lara
PN Jr, Jones JA, Taplin ME, Burch PA, Berry D, Moinpour C, Kohli M,
et al: Docetaxel and estramustine compared with mitoxantrone and
prednisone for advanced refractory prostate cancer. N Engl J Med.
351:1513–1520. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lohiya V, Aragon-Ching JB and Sonpavde G:
Role of chemotherapy and mechanisms of resistance to chemotherapy
in metastatic Castration-Resistant prostate cancer. Clin Med
Insights Oncol. 10 (Suppl 1):S57–S66. 2016.PubMed/NCBI
|
|
22
|
Ramnarine VR, Kobelev M, Gibb EA, Nouri M,
Lin D, Wang Y, Buttyan R, Davicioni E, Zoubeidi A and Collins CC:
The evolution of long noncoding RNA acceptance in prostate cancer
initiation, progression, and its clinical utility in disease
management. Euro Urol. 76:546–559. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Du Y, Wei N, Hong J and Pan W: Long
non-coding RNASNHG17 promotes the progression of breast cancer by
sponging miR-124-3p. Cancer Cell Int. 20:402020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Eggener SE, Badani K, Barocas DA,
Barrisford GW, Cheng JS, Chin AI, Corcoran A, Epstein JI, George
AK, Gupta GN, et al: Gleason 6 prostate cancer: Translating biology
into population health. J Urol. 194:626–634. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takeuchi H, Ohori M and Tachibana M:
Clinical significance of the prostate-specific antigen doubling
time prior to and following radical prostatectomy to predict the
outcome of prostate cancer. Mol Clin Oncol. 6:249–254. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shulman MJ, Karam JA and Benaim EA:
Prostate-specific antigen doubling time predicts response to
deferred antiandrogen therapy in men with androgen-independent
prostate cancer. Urology. 63:732–736. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luqmani YA: Mechanisms of drug resistance
in cancer chemotherapy. Med Princ Pract. 14 (Suppl 1):S35–S48.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li JF, Li WH, Xue LL and Zhang Y: Long
non-coding RNA PICART1 inhibits cell proliferation by regulating
the PI3K/AKT and MAPK/ERK signaling pathways in gastric cancer. Eur
Rev Med Pharmacol Sci. 23:588–597. 2019.PubMed/NCBI
|
|
29
|
Rani N, Nowakowski TJ, Zhou H, Godshalk
SE, Lisi V, Kriegstein AR and Kosik KS: A primate lncRNA mediates
notch signaling during neuronal development by sequestering miRNA.
Neuron. 90:1174–1188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu XY, Hou PF, Li TT, Quan HY, Li ML, Lin
T, Liu JJ, Bai J and Zheng JN: The roles of Wnt/β-catenin signaling
pathway related lncRNAs in cancer. Int J Biol Sci. 14:2003–2011.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu J, Han Z, Sun Z, Wang Y, Zheng M and
Song C: LncRNA SLCO4A1-AS1 facilitates growth and metastasis of
colorectal cancer through β-catenin-dependent Wnt pathway. J Exp
Clin Cancer Res. 37:2222018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tian Z, Yang G, Jiang P, Zhang L, Wang J
and Sun S: Long non-coding RNA Sox4 promotes proliferation and
migration by activating Wnt/β-catenin signaling pathway in
osteosarcoma. Pharmazie. 72:537–542. 2017.PubMed/NCBI
|
|
33
|
Guo JC, Yang YJ, Zheng JF, Zhang JQ, Guo
M, Yang X, Jiang XL, Xiang L, Li Y, Ping H and Zhuo L: Silencing of
long noncoding RNA HOXA11-AS inhibits the Wnt signaling pathway via
the upregulation of HOXA11 and thereby inhibits the proliferation,
invasion, and self-renewal of hepatocellular carcinoma stem cells.
Exp Mol Med. 51:1–20. 2019. View Article : Google Scholar
|
|
34
|
Kim U, Kim CY, Lee JM, Ryu B, Kim J, Shin
C and Park JH: Pimozide inhibits the human prostate cancer cells
through the generation of reactive oxygen species. Front Pharmacol.
10:15172020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takeuchi H, Taoka R, Mmeje CO, Jinesh GG,
Safe S and Kamat AM: CDODA-Me decreases specificity protein
transcription factors and induces apoptosis in bladder cancer cells
through induction of reactive oxygen species. Urol Oncol.
34:337.e11–8. 2016. View Article : Google Scholar : PubMed/NCBI
|