Introduction
It has been reported that gastric cancer is the
fifth most frequent malignancy worldwide and almost one million new
cases are estimated to occur each year, resulting in ~723,000
deaths per year globally (1,2). In >50% of cases, gastric cancer has
no noticeable symptoms, which may lead to advanced carcinoma with
multiple metastases upon diagnosis (3). Chemotherapy is still considered as the
primary therapy for advanced gastric cancer; however, its
disadvantages, such as low response rate and short duration of
clinical benefit, have limited its application (4). It is important to identify and develop
more specific targeted therapies to improve the prognosis of
gastric cancer. To achieve this, it is essential to screen and
identify the critical molecular pathways and signaling transduction
networks involved in the pathogenesis of gastric cancer using
in-depth studies.
In 2017, Padmanabhan et al (5) reported that epigenetic dysregulation
plays an essential role in the development of gastric cancer.
Post-translational histone modifications are involved in the
malignant progression of gastric cancer by regulating the
expression of oncogene and tumor suppressor genes. The occurrence
of gastric cancer is a result of the combination of environmental,
polygenic and epigenetic abnormalities. The epigenetic mechanisms
involved include DNA methylation (6), non-coding RNA (7) and histone translational modifications
(8). Among these regulations,
histone modifications, such as acetylation (9), methylation (10) and ubiquitination (11), are involved in the carcinogenesis of
gastric mucosa through the regulation of oncogene expression
(12) and protein-protein
interactions (13). For example,
while a group of genes including phosphatidylserine decarboxylase
proenzyme, SWI/SNF complex subunit SMARCC1 and vacuolar protein
sorting-associated protein 37A are aberrantly methylated, thus
aberrantly expressed, in gastric cancer (12), antisense-transcribed lncRNA HOXA11-AS
is upregulated and serves as a scaffold to form complex of
chromatin modification factors polycomb repressive complex 2,
lysine-specific histone demethylase 1A and DNA
(cytosine-5)-methyltransferase 1, thus regulating downstream gene
expression, including prostasin and Krueppel-like factor 2
(13).
Post-translational histone modifications are
involved in the malignant progression of gastric cancer by
regulating the expression of oncogene and tumor suppressor genes.
For instance, hypermethylation of histone 3 at lysine 9,
hypomethylation of histone 3 at lysine 4 and hypoacetylation of
histone 3 at lysine 9 are reported to be associated with the
silencing of tumor suppressor genes P16 and mutL homolog 1 (MLH1)
in gastric cancer cells (14).
Moreover, the aberrant expression of an oncogene, Fez family zinc
finger protein 1, is highly regulated by DNA methylation and
histone acetylation in gastric cancers (15). Histone methylation modification is an
essential regulatory mechanism in chromatin structure alteration
and gene transcription (16). As a
member of the histone demethylase family of proteins containing a
JmjC domain, lysine (K)-specific demethylase 6B (KDM6B) reverses
the dimethylation (H3K27me2) and trimethylation (H3K27me3) of
lysine at the 27th position in histone H3 and then activates the
expression of target genes, such as proinflammatory factors
including the p19 peptide of the chimeric cytokine IL-23,
granulocyte colony-stimulating factor and triggering receptor
expressed on myeloid cells 1 (17).
The overexpression of KMD6B is found in numerous types of tumor,
such as prostate cancer, diffuse large B cell lymphoma and renal
clear cell carcinoma, and it is associated with tumor progression
and poor prognosis (18–20).
The present study analyzed the expression profile of
KMD6B in gastric cancer and further explored the functions and the
potential underlying mechanisms of KDM6B in gastric cancer
development. Furthermore, the alterations in cell proliferation,
cycle distribution and the expression of cell cycle related
proteins after inhibiting KMD6B with its specific inhibitor GSK J4
were investigated. The present study reported novel evidence to
support the association between KMD6B-overexpression and gastric
cancer and evaluated KMD6B as a possible risk factor for gastric
cancer.
Materials and methods
Patients and ethics approval
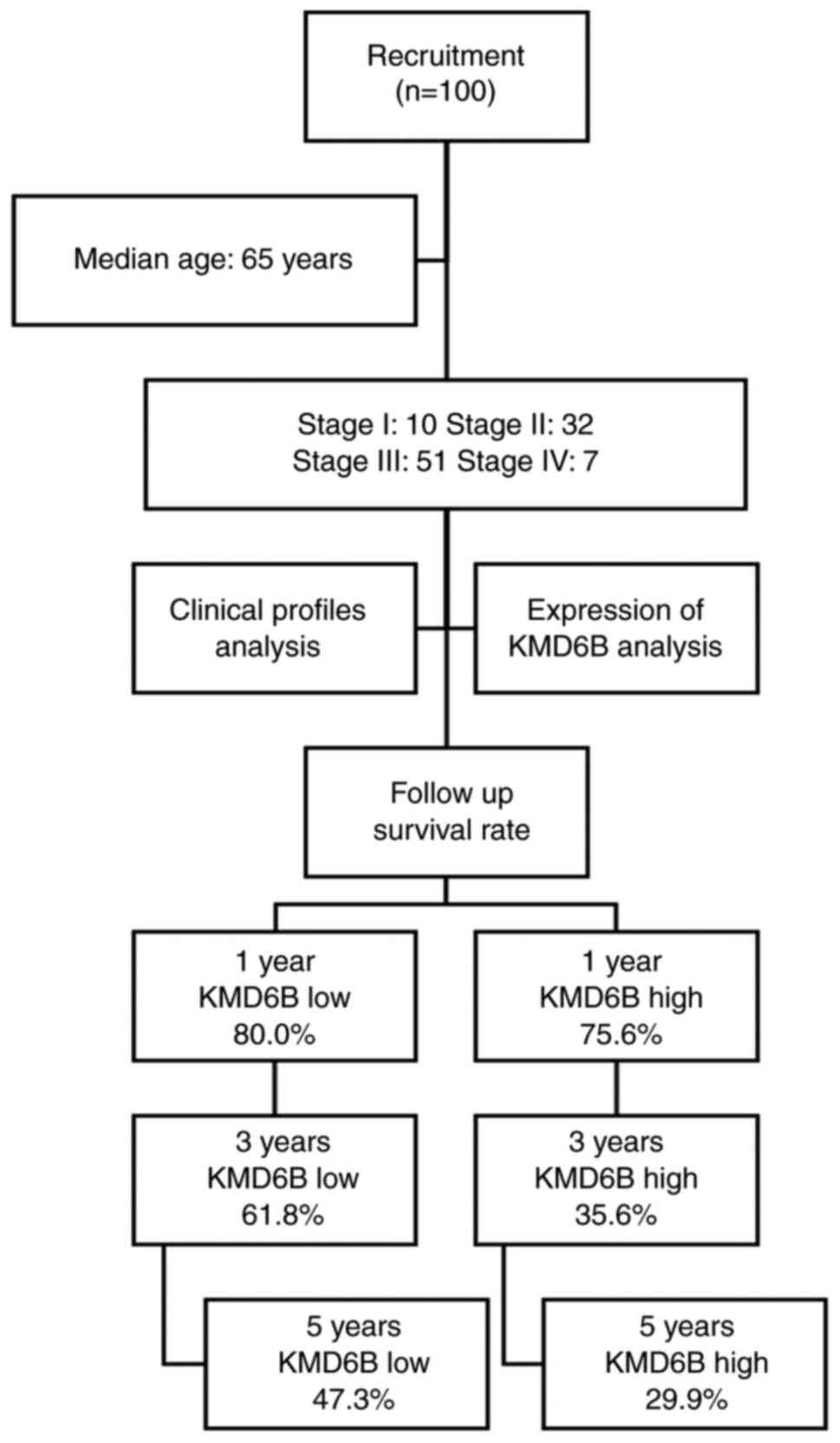
In total, 100 adult patients with cancer were
admitted to Cixi People's Hospital between March 2008 and December
2011. Patients with gastric cancer who underwent surgical resection
or gastroscopic biopsy were included in the present study. The age
range was from 32 to 81 years old, with the median age of 65 years.
Each case was diagnosed according to the World Health Organization
classification of digestive system tumors (2010 edition) (21) by two pathologists who were blinded to
patients' identification and followed up every six months in clinic
until December 2018. The detailed flow chart of the present study
is presented in Fig. 1. All patients
were newly diagnosed, without radiotherapy and chemotherapy or
other tumor histories. No other inclusion or exclusion criteria
were applied. The selected adjacent non-cancerous tissues were at
≥2-cm away from the edge of the cancer tissue. Tissues were fixed
in 10% neutral formalin, processed through the standard dehydration
and paraffin embedding protocol. Briefly, tissues were fixed in 10%
neutral formalin for 24 h at 4°C, followed by dehydrating in 70%
ethanol, two changes, 1 h each; 80% ethanol, one change, 1 h; 95%
ethanol, one change, 1 h; 100% ethanol, three changes, 1.5 h each;
and xylene, three changes, 1.5 h each. Then the tissues were
embedded in paraffin. Written informed consent for the use of
medical records of the patients was obtained at the time of
surgery. The study was approved by The Ethics Committee of Cixi
Hospital (Cixi, China; approval nos. 2008-005 and 2017-LS-25).
Cells and reagents
The human gastric cancer cell line HGC-27 was
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences and cultured in RPMI-1640 medium with
10% fetal bovine serum (both Thermo Fisher Scientific, Inc.). KMD6B
inhibitor GSK J4 was purchased from Selleckchem (purity ≥98%);
rabbit anti-human KMD6B, cyclin B1, Cdc2, p21 and GAPDH polyclonal
antibodies and horseradish peroxidase enzyme (HRP)-labeled goat
anti-rabbit IgG antibodies were purchased from Cell Signaling
Technology, Inc.
Immunohistochemistry (IHC) and
hematoxylin-eosin (HE) staining
IHC and HE staining were performed using the
Histostain™ kit (Thermo Fisher Scientific, Inc.) following
manufacturer's protocol. Briefly, paraffin embedded samples were
sectioned into 5-µm sections. Tissue slides were deparaffinized and
rehydrated by immersing the slides through the following solutions:
Xylene (three washes 5 min each) 100, 95, 70 and 50% ethanol (each
washed twice for 10 min each) and deionized water, two washes for 5
min each. The slides were then fixed in 10% paraformaldehyde for 10
min at room temperature and washed with PBS. Fixed samples were
treated with 3% H2O2 solution at room
temperature for 10 min followed by washing with PBS three times.
Antigen retrieval was performed by soaking the slides in boiling
0.01 M citrate buffer, pH 6.0 for 10 min, cooling to room
temperature and washing with PBS three times. Tissue sections were
incubated with serum blocking solution provided in the kit for 10
min, followed by incubation with rabbit primary antibody against
KMD6B (cat. no. EAB-2167; 1:50; Abcam) at room temperature for 1 h.
After washing with PBS, sections were incubated with biotinylated
broad-spectrum secondary antibody (Histostain®-Plus 3rd
Gen IHC Detection Kit, Invitrogen, #85-903) at room temperature for
10 min and washed with PBS as the manufacture suggested. After
incubation, sections were then incubated with streptavidin-enzyme
conjugate for 10 min at room temperature, washed with PBS and
incubated with substrate-chromogen mixture at room temperature for
5 min, washed with PBS again and counterstained with hematoxylin
for 1 min at room temperature following by thorough rinsing with
tap water. Sections were finally mounted and dried until
observation. Images were captured using a pathology microscopy
imaging system (Olympus Corporation). Qualitative staining refers
yellowish to brownish yellow staining as a positive marker in
sections and sections were divided into four categories depending
on staining intensity: 0, clear, 1, weak, 2, moderate and 3,
strong. Colored areas were 0, if positive cells percentage ≤1%; 1,
>1% to ≤25%; 2, >25% to ≤50% and 3, >50%. If the sum of
the intensity score and the positive percentage score was >3, it
was considered as high expression of KMD6B.
In vitro proliferation analysis
The viability of HGC-27 cells was determined by
staining the cells with trypan blue following the manufacturer's
instructions (Thermo Fisher Scientific, Inc.). The cells were
treated with either vehicle control (0 µM) or GSK J4 at 2 or 4 µM
for 24, 48 and 72 h. After treatment, cells were trypsinized and
resuspended in culture medium and then counted under the
microscope. For the colony formation assay, HGC27 cells were seeded
in six-well plate with 1×104 cells per well. GSK J4 was
added into the culture medium at the concentrations of 0, 2 and 4
µM. Cell culture medium with appropriate concentration of GSK J4
were refreshed every other day during the treatment. After 7 days
incubation at 37°C, the cells were fixed with 4% paraformaldehyde
solution for 10 min at room temperature and stained by 0.1%
crystallization purple for 15 min at 37°C. The formation of
colonies was analyzed (five fields randomly selected for counting
clones, which is defined as a colony ≥10 cells). For cell cycle
analysis, cells were trypsinized after treatment and were fixed
with 100% ethanol at −20°C for 10 min, followed by washing with TBS
at room temperature and rehydrating in PBS for 10 min. Cells were
then stained with propidium iodide (PI) at 1 µg/ml (BioLegend,
Inc.). Flow cytometry (BD FACSLyric™ Research System; BD
Biosciences, Inc.) was used to run the samples and the data were
analyzed using the ModFitLT software (ModFit5.0™;
VeritySoftwareHouse,). In the meantime, the PrestoBlue®
Cell Viability Reagent (Thermo Fisher Scientific, Inc.) was
employed for the cell viability and proliferation detection.
Briefly, 10 µl PrestoBlue reagent was added to 90 µl culture media
at 37°C in a cell culture incubator, protected from direct light
for 30 min. Next, 100 µl media collected from the culture wells was
used for absorbance quantification at 570 nm, using 600 nm as a
reference wavelength, using a plate reader.
Western blotting
Cell culture and drug treatment were performed as
aforementioned. Total cell lysate was extracted with RIPA buffer
(Thermo Fisher Scientific, Inc.), and the protein concentration was
determined using the BCA method. Then samples were analyzed by
using 12% SDS-PAGE with 20 µg loaded per lane. Then the proteins
were transferred to PVDF membranes at 300 mA constant current for
120 min. The membrane was blocked with 3% BSA for 1 h at room
temperature, then incubated with anti-cyclin B1 (cat. no. 12231;
Cell Signaling Technology Inc.), Cdc2 (cat. no. 28439; Cell
Signaling Technology, Inc.) and p21 (Cell Signaling Technology
Inc.). All the primary antibodies used were diluted at 1:1,000. for
2 h at room temperature, washed with TBS-T (0.1% Tween-20 in TBS)
for 10 min three times. Then the membranes were incubated with
HRP-labeled goat anti-rabbit IgG secondary antibody at room
temperature for 1 h and washed with TBST. The Pierce™ ECL Western
Blotting Substrate reagent (Thermo Fisher Scientific, Inc.) was
used to visualize following the manufacturer's instructions. GAPDH
was used as the internal control.
Statistical analysis
GraphPad Prism 8 (GraphPad Software, Inc.) was used
for data analysis. All data sets were tested for the normal
distribution. The χ2 test was used to compare the data
over the expressions or profiles (such as tissue origin, expression
level, age or sex). Fisher's exact tests were also used where
appropriate. Cox univariate analysis was performed to analyze
prognostic factors in patients as a whole. Cox multivariate
analysis was performed to analyze prognostic factors in male vs.
female patients. Clinical survival data was analyzed using
Kaplan-Meier analysis with the log-rank test performed. In
vitro experiment times was represented in each figure legends.
Data sets were analyzed with one-way ANOVA followed by Tukey's or
Dunnett's post hoc tests as appropriate. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of KMD6B in gastric cancer
tissues and the matching para-cancerous tissues
In total, 100 adult patients were included in the
present study, including 10 stage I, 32 stage II, 51 stage III and
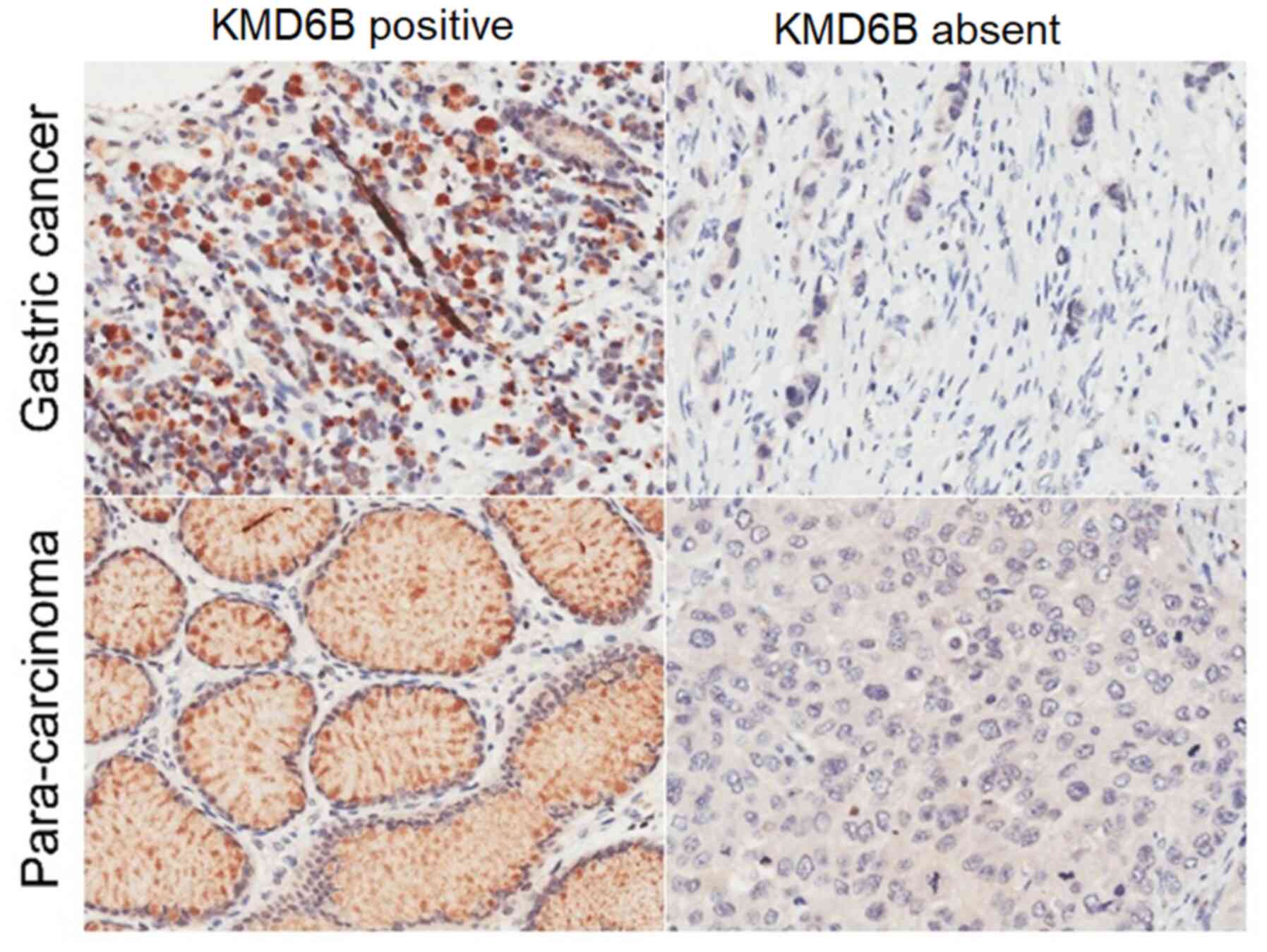
seven stage IV. To investigate whether the gastric cancer had
increased KDM6B expression compared with para-cancerous tissue, IHC
and HE staining and analysis were performed. There were 45 cases of
KMD6B high expression among 100 cases of gastric cancer tissues,
which accounted for 45.0% of the tested samples, seeing a
significant difference compared with 30 cases among adjacent
para-cancerous tissues, with the positive rate of 30.0% (P=0.028;
Table I) (Fig. 2).
 | Table I.Expression of KMD6B in gastric cancer
tissues and the matched para-cancerous tissues. |
Table I.
Expression of KMD6B in gastric cancer
tissues and the matched para-cancerous tissues.
|
|
| KMD6B expression, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Tissue origin | Value, n | High | Low | P-value |
|---|
| Cancer tissue | 100 | 45 (45) | 55 (55) | 0.028a |
| Para-cancerous
tissue | 100 | 30 (30) | 70 (70) |
|
Relationship between KMD6B expression
in gastric cancer and clinical profiles
Next, whether the overexpression of KMB6B had any
correlation with the clinical characteristics was investigated.
Demographic and clinical characteristics were analyzed, as shown in
Table II. KMD6B expression was not
associated with patient age, tumor size, location, degree of
differentiation, nerve and vascular invasion and T stage, with the
corresponding P-values, 0.917, 0.393, 0.611, 0.270, 0.685 and
0.191, respectively. The expression of KMD6B was associated with
sex, N, M and clinical stages, with P-values of 0.029, 0.020, 0.021
and 0.021, respectively.
 | Table II.Association between KMD6B expression
in gastric cancer and clinical profiles. |
Table II.
Association between KMD6B expression
in gastric cancer and clinical profiles.
|
|
| KMD6B expression,
n |
|
|---|
|
|
|
|
|
|---|
| Clinical
variable | Value, n | High | Low | P-value |
|---|
| Age, years |
|
|
| 0.917 |
|
<65 | 45 | 20 | 25 |
|
|
≥65 | 53 | 23 | 30 |
|
| Sex |
|
|
| 0.029a |
|
Male | 64 | 34 | 30 |
|
|
Female | 36 | 11 | 25 |
|
| Tumor size, cm |
|
|
|
|
|
<5 | 51 | 25 | 26 | 0.393 |
| ≥5 | 47 | 19 | 28 |
|
| Tumor location |
|
|
| 0.611 |
|
Cardia/fundus | 13 | 5 | 8 |
|
|
Body/antrum | 87 | 40 | 47 |
|
| Pathological
grade |
|
|
| 0.270 |
| I | 37 | 14 | 23 |
|
|
II/III | 63 | 31 | 32 |
|
| Nerve/vessel
invasion |
|
|
| 0.685 |
| No | 86 | 38 | 48 |
|
|
Yes | 14 | 7 | 7 |
|
| T stage |
|
|
| 0.191 |
|
1/2 | 19 | 6 | 13 |
|
|
3/4 | 81 | 39 | 42 |
|
| N stage |
| 0 | 27 | 7 | 20 | 0.020a |
|
1-3 | 73 | 38 | 35 |
|
| M stage |
|
|
| 0.021a |
| 0 | 92 | 38 | 54 |
|
| 1 | 8 | 7 | 1 |
|
| Clinical stage |
|
|
| 0.021a |
| I | 10 | 1 | 9 |
|
|
II/III/IV | 90 | 44 | 46 |
|
KMD6B expression may serve as gastric
cancer prognostic factor
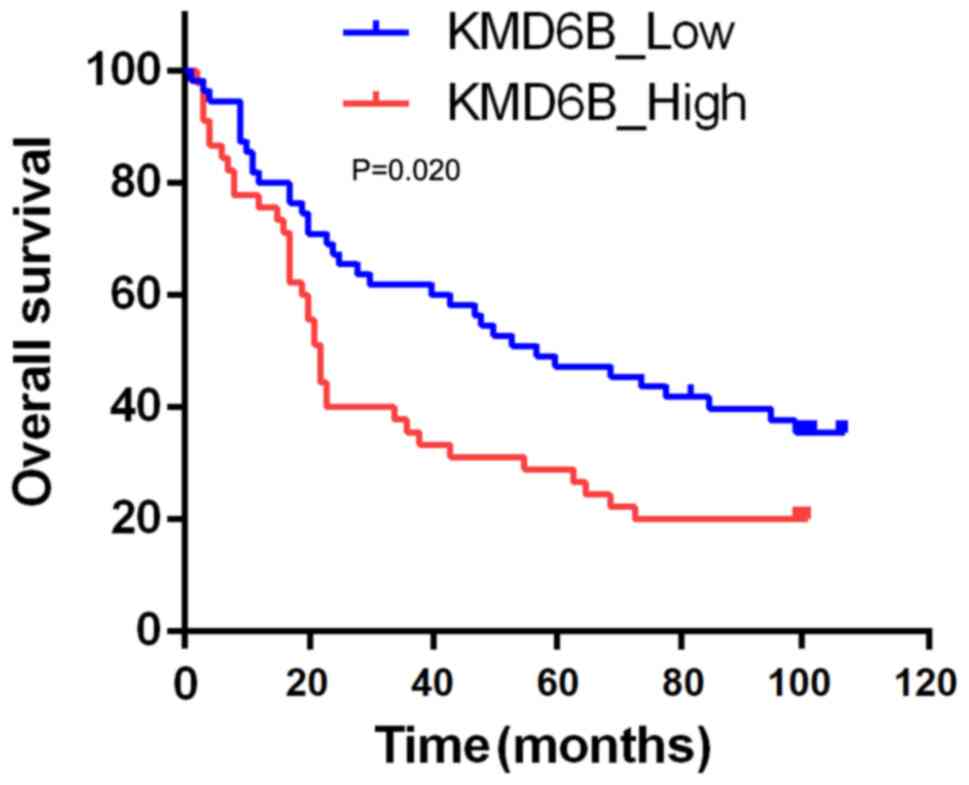
To investigate whether KMD6B has the potential to
predict patient prognosis, expression levels of KMD6B and patient
mortality rate were analyzed. In 45 cases that were considered as
KMD6B high expression, the 1-, 3- and 5-year cumulative survival
rates were 75.6, 35.6 and 29.9%, respectively. Whereas in the 55
cases with KMD6B low expression, the 1-, 3- and 5year cumulative
survival rates were 80.0, 61.8 and 47.3%, respectively (Fig. 1). The overall survival rate of
patients with KMD6B low expression was significantly higher
compared with that of the high expression group (P=0.020) (Fig. 3). These data indicated that the
expression of KMD6B might have the potential to serve as a
prognostic biomarker for gastric cancer.
A total of 11 other variables were also analyzed,
including age, sex, tumor size, tumor location, differentiation
degree, nerve/vascular invasion, T, N, M and clinical stage and
KMD6B expression level. As shown in Table III, Cox univariate analysis
suggested that nine out 11 variables could serve as prognostic
factors, including age (P=0.023), tumor size (P=0.013), degree of
differentiation (P=0.017), neurovascular invasion (P<0.001), T
stage (P=0.016), N stage (P=0.021), M stage (P<0.001), clinical
stage (P=0.031) and KMD6B expression level (P=0.023).
 | Table III.Cox univariate analysis of prognostic
factors of gastric cancer. |
Table III.
Cox univariate analysis of prognostic
factors of gastric cancer.
| Variables | HR | LCI | UCI | P-value |
|---|
| Age, years, <65
vs. ≥65 | 0.580 | 0.363 | 0.927 | 0.023a |
| Sex, female vs.
male | 0.614 | 0.378 | 0.997 | 0.048a |
| Tumor size, cm,
<5 vs. ≥ 5 | 0.551 | 0.344 | 0.883 | 0.013a |
| Tumor location,
cardia/fundus vs. body/antrum | 1.465 | 0.727 | 2.956 | 0.286 |
| Pathological grade,
I vs. II/III | 0.530 | 0.315 | 0.892 | 0.017a |
| Nerve/vessel
invasion, no vs. yes) | 0.323 | 0.177 | 0.587 |
<0.001b |
| T stage, 1/2 vs.
3/4 | 0.422 | 0.209 | 0.852 | 0.016a |
| N stage, 0 vs.
1/2/3 | 0.510 | 0.288 | 0.903 | 0.021a |
| M stage, 0 vs.
1 | 0.166 | 0.076 | 0.362 |
<0.001b |
| Clinical stage, I
vs. II/III/IV | 0.280 | 0.088 | 0.892 | 0.031a |
| KMD6B expression,
low vs. high | 0.580 | 0.363 | 0.927 | 0.023a |
Multivariate analysis reported eight significant
independent predictors, including age, sex, tumor size, tumor
location, the degree of differentiation, nerve/vascular invasion,
clinical stage and KMD6B expression. The results demonstrated that
tumor size (P=0.008), neurovascular invasion (P<0.001) and KMD6B
expression (P=0.007) were independent prognostic predictors of
gastric cancer (Table IV).
 | Table IV.Multivariate Cox analysis of
prognostic factors of gastric cancer, male vs. female. |
Table IV.
Multivariate Cox analysis of
prognostic factors of gastric cancer, male vs. female.
|
| Males | Females |
|
|---|
|
|
|
|
|
|---|
| Variable | HR | LCI | UCI | HR | LCI | UCI | P-value |
|---|
| Age, years, <65
vs. ≥65 | 0.631 | 0.385 | 1.249 | 0.711 | 0.429 | 0.965 | 0.118 |
| Tumor size, cm,
< 5 vs. ≥ 5 | 0.514 | 0.298 | 0.979 | 0.446 | 0.260 | 0.673 | 0.008a |
| Tumor location,
cardia/fundus vs. body/antrum | 1.842 | 0.895 | 4.577 | 2.078 | 0.967 | 3.671 | 0.076 |
| Pathological grade,
I vs. II/III | 0.818 | 0.521 | 1.475 | 0.762 | 0.383 | 1.291 | 0.410 |
| Nerve/vessel
invasion, no vs. yes | 0.401 | 0.187 | 0.722 | 0.253 | 0.165 | 0.496 |
<0.001a |
| Clinical stage, I
vs. II/III/IV | 0.629 | 0.210 | 1.802 | 0.515 | 0.130 | 2.056 | 0.368 |
| KMD6B expression,
low vs. high | 0.501 | 0.288 | 0.741 | 0.419 | 0.234 | 0.879 | 0.007a |
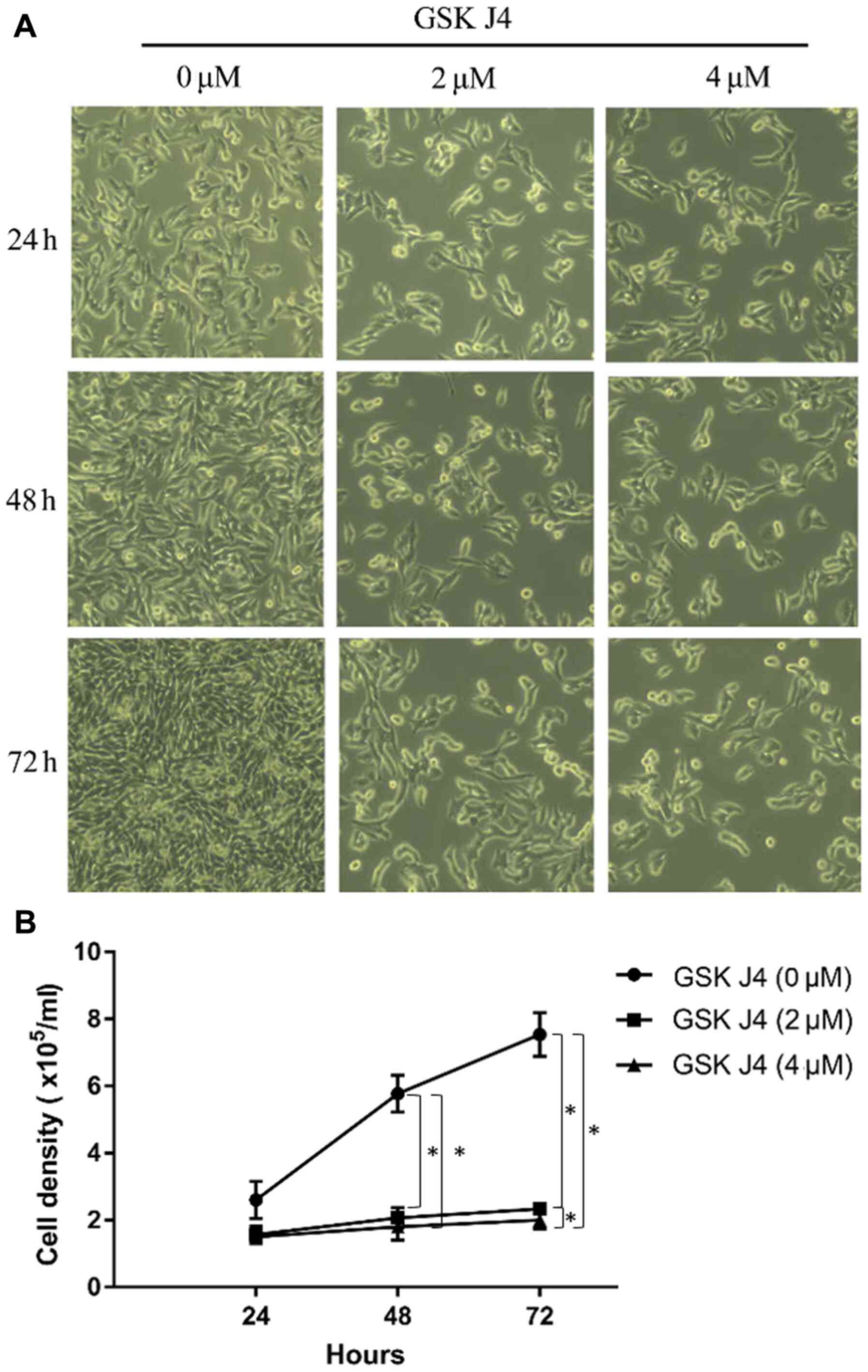
GSK J4 inhibits the proliferation of
gastric cancer HGC27 cells
The aforementioned data indicated that expression
pattern of KMD6B could serve as an independent prognostic factor in
gastric cancer. Due to its significantly upregulated expression in
gastric cancer tissues, it was speculated if overexpression also
contributes to the malignant development of gastric mucosa. Cell
proliferation was assessed by specific inhibition by GSK J4. The
gastric cancer cell line HGC27 was used to perform a proliferation
assay with 3 days of GSK J4 treatment. The density of HGC27 cells
in GSK J4 group was significantly less compared with that in the
control group (P<0.05; Fig. 4).
After 24-h treatment, cells showed different proliferation rates.
In vehicle control group, cell number reached
2.5±0.4×105, which was almost doubled that of the
starting cell numbers (1.8±0.1×105), whereas in GSK
J4-treated cells, both drug concentrations used inhibited cell
proliferation. The difference became more evident with increasing
doses. As shown in Fig. 4B, by the
end of treatment, the number of cells treated with GSK J4 4 µM was
~4-fold less compared with the vehicle treated group (7.6±0.4 vs.
1.9±0.2; P<0.05). GSK J4 2 µM treatment also showed the
significant inhibitory function compared with the control group
(2.2±0.1 vs. 1.9±0.2; P<0.05).
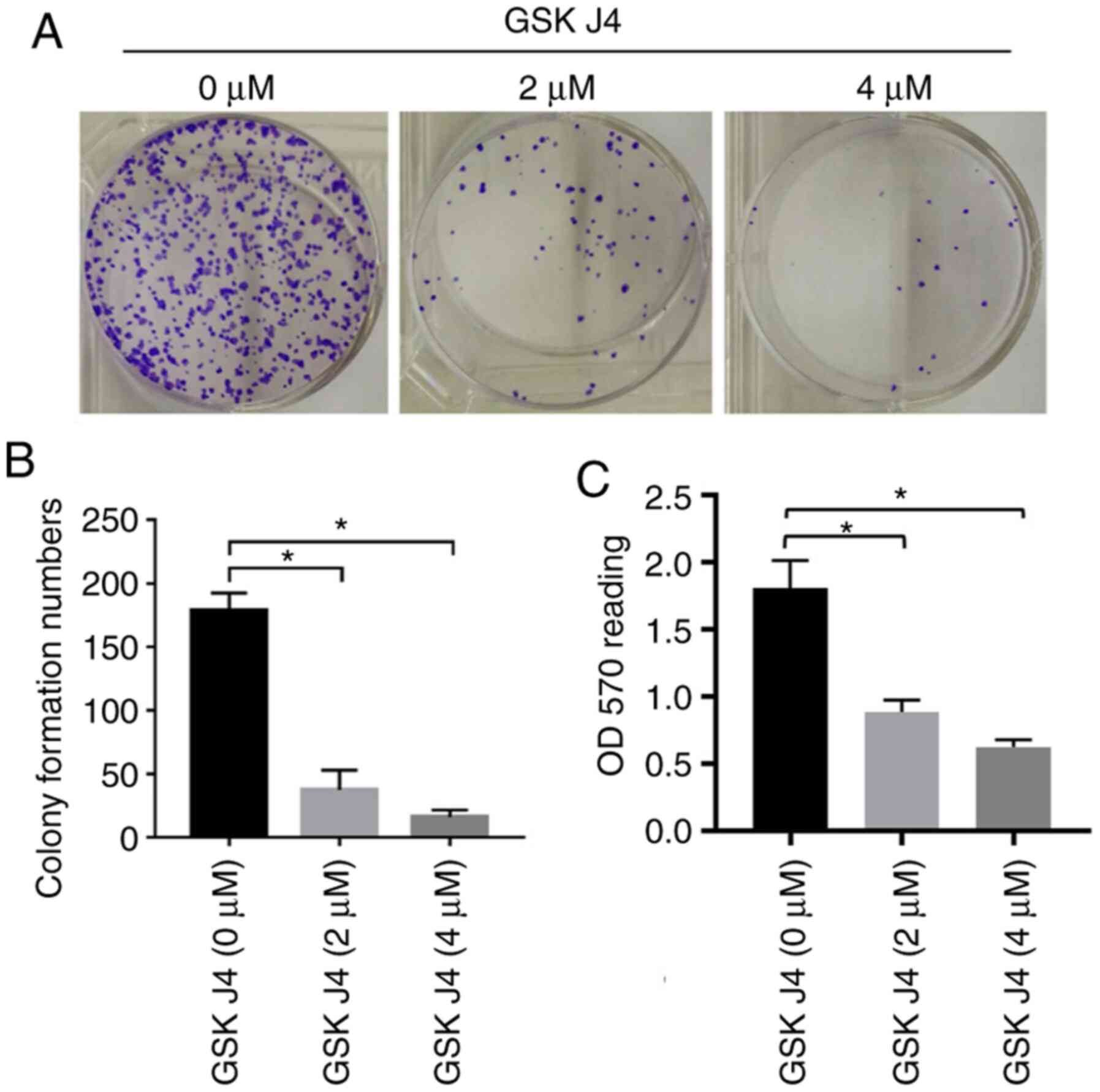
GSK J4 inhibits the colony formation
of gastric cancer cells
A colony formation assay was also used to test the
inhibitory effect of GSK J4 on gastric cancer cells. Following
treatment with GSK J4 for 7 days, the colony number was 37.3±15.5
and 16.0±5.6 in GSK J4 (2 µM) and GSK J4 (4 µM) groups,
respectively. However, there were 179.0±13.5 colonies in the
control group, suggesting that GSK J4 can inhibit the proliferation
of gastric cancer cells. Compared with the traditional the
formation of colonies, the resazurin-based PrestoBlue cell
viability assay represented the similar results, with significant
inhibition of cell proliferation between the GSK J4 2 and 4 µM
groups compared with the vehicle-treated group (0.887±0.088 vs.
1.810±0.206, P=0.0049, and 0.0.623±0.055 vs. 1.810±0.206, P=0.0013,
respectively; Fig. 5).
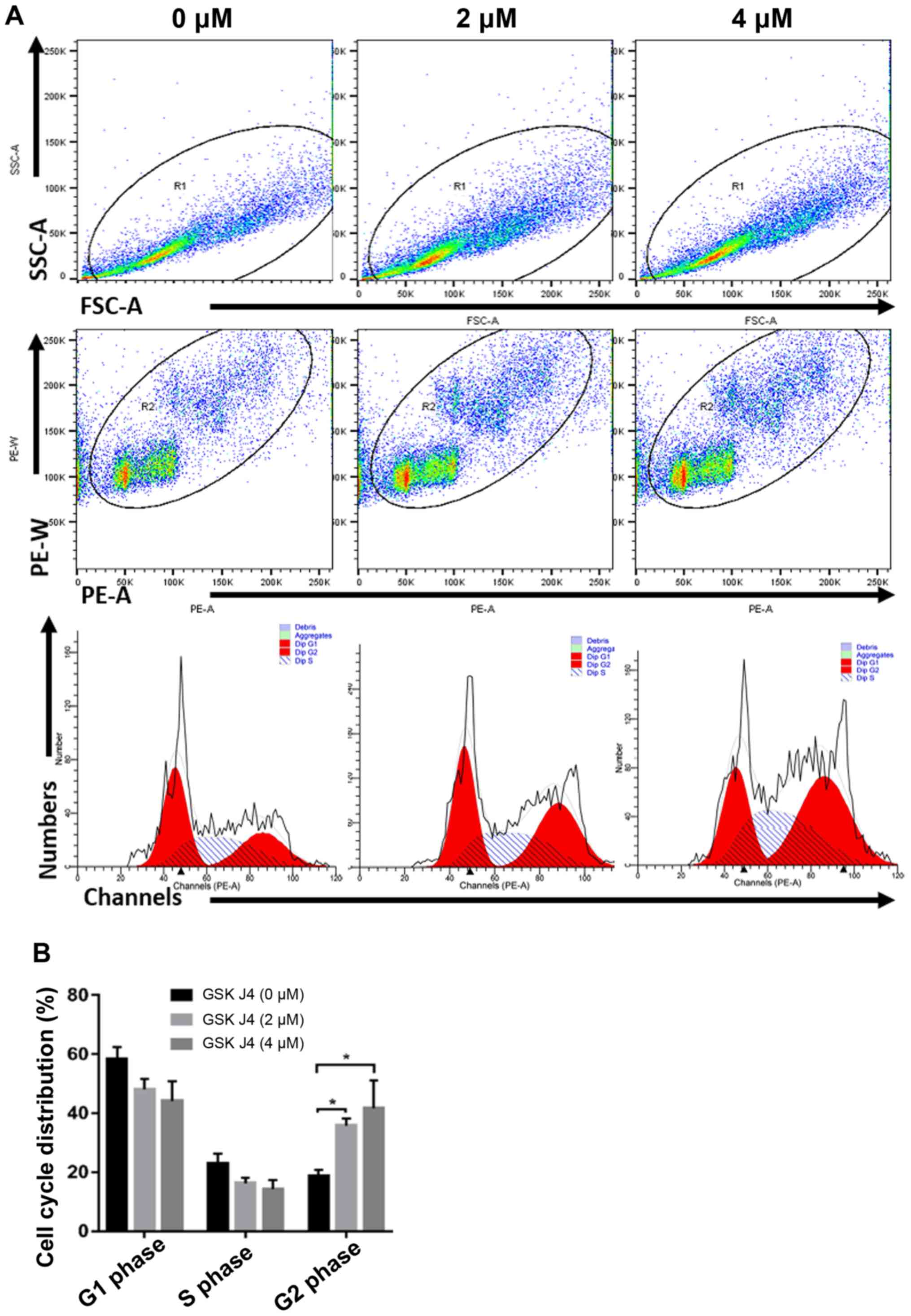
Effects of GSK J4 on cell cycle
distribution and related protein expression of gastric cancer
cells
Next, it was investigated how GSK J4 inhibited
gastric cancer cell proliferation. Flow cytometry was used to
analyze the cell cycle progression of cells treated with GSK J4.
Following treatment with GSK J4 2 and 4 µM for 24 h, the percentage
of HGC27 cells at G2 phase were 35.76±2.40 and
41.62±9.47% respectively, which was significantly increased
compared with the control group (18.80±2.05%) (both P<0.05). In
the G1 or S phase, the percentage of HGC27 cells after
GSK J4 treatment showed the trends of inhibition, compared with the
control group but no significant difference been observed (Fig. 6). This suggested that blocking the
demethylase activity of KMD6B can arrest the cell cycle at
G2/M phase.
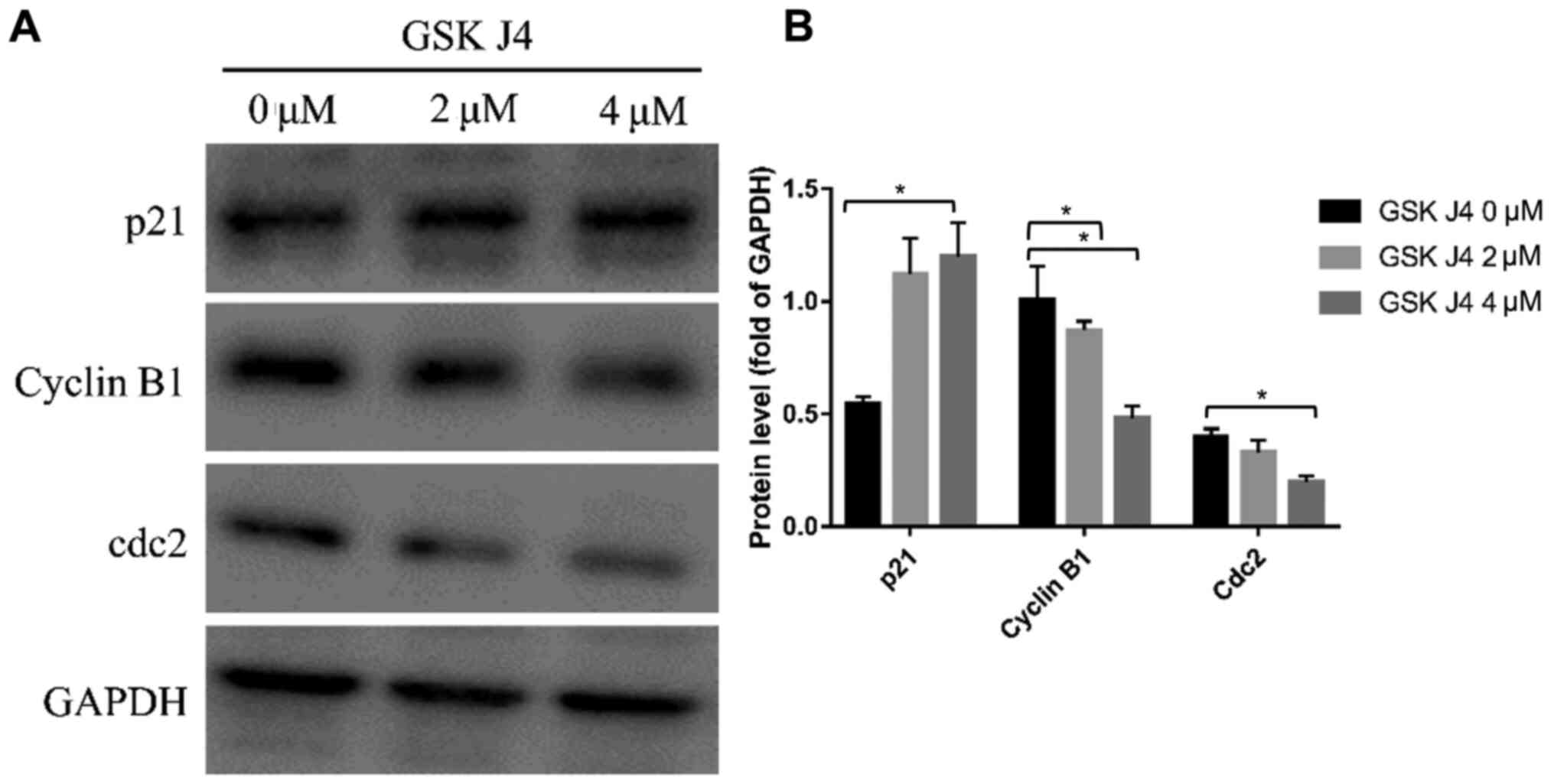
Cell cycle is restrictedly regulated by a set of
cell cycle regulating proteins. The expression level of those
proteins is closely related to the cell cycle status (22). Western blotting was performed to
analyze the expression level of key proteins involved in cell
cycle. As shown in Fig. 7A and B,
after 24-h treatment of GSK J4, the expression of cyclin B1 and
Cdc2 in HGC27 cells was significantly downregulated (both
P<0.05) while p21 was significantly upregulated compared with
the control group (P<0.05).
Discussion
In China, the morbidity and mortality of gastric
cancer are the third highest among all malignant tumors (2), of which >70% of patients are
diagnosed at the advanced stage (3).
Chemotherapy is the primary therapy for advanced gastric cancer
even though the response rate is low and the duration of
progression-free survival is short (4). Research has shown that different
causes, including environmental causes and genetic abnormalities,
contribute to the malignant transformation of gastric mucosa
leading to gastric cancer progression (6–10). Among
these genetic modifications, malfunction in epigenetic regulation
of oncogenes or tumor suppressor genes is an area of interest,
especially histone modifications including acetylation, methylation
and ubiquitination. Epigenetic changes are involved in cancer
progression by dysregulation of gene expression and/or
protein-protein interaction (9–13).
Intensive study of the pathogenesis of gastric cancer and screening
key molecules involved in epigenetic regulation in gastric cancer
will contribute to the development of targeted drugs and may
improve the prognosis of patients with gastric cancer.
KMD6B is a member of the histone demethylase family
of proteins containing the JmjC domain and requires Fe2+
and α-ketoglutarate as co-factors (23). KMD6B alters the expression of target
genes, such as those genes that are bivalently marked by H3K4me3
(tri-methylation at the 4th lysine residue of the histone H3) and
H3K27me3 (tri-methylation at the 27th lysine residue of the histone
H3) and associated with promoter-proximal, paused RNA polymerase II
(24), to induce cell carcinogenesis
by affecting the process of modifying factors binding and chromatin
remodeling (1). KMD6B also directly
regulates gene transcription by modifying H3K27 methylation in the
promoter region of the Ink4a/Arf locus, which encodes two distinct
proteins that intimately link the pRB and p53 tumor suppressor
pathways: p16INK4a and p14/p19ARF (25). KMD6B is overexpressed in different
types of tumors and is associated with tumor progression and poor
prognosis (26). KMD6B is
upregulated in prostate cancer and expressed at the highest level
in metastatic foci, and high KMD6B expression suggests a poor
prognosis (19). KMD6B is also
overexpressed in both primary and Epstein-Barr infection-related
Hodgkin's lymphoma (27). Patients
with renal clear cell carcinoma with KMD6B high expression have a
poor prognosis, and the expression of KMD6B is positively
correlated with the tumor size, lymph node metastasis and
pathological stage (18). The
present results indicated that KMD6B was highly expressed in 45% of
gastric cancer tissues. The protein level of KMD6B was
significantly associated with patient sex, N, M and clinical
stages. Survival analysis showed that KMD6B-overexpression was an
independent prognostic factor for gastric cancer. However, the
sample distribution among clinical stage was unbalanced (I to
II/III/IV, 1:9) in the study, which might be the reason why
clinical stage cannot be used as a prognostic marker in gastric
cancer in the present study. More patients with stage 1 are needed
in order to analyze the relationship between clinical phases and
gastric cancer prognosis. Similarly, the sex disproportion (male to
female, 16:9) might affect the relationship between KMD6B
expression and sex as well. Therefore, these factors require
further study with a larger sample size. Such are expected to
further demonstrate the impact of KMD6B on gastric cancer.
Targeted inhibition of the expression or demethylase
activity of KMD6B can induce cell cycle arrest, apoptosis and
differentiation, demonstrating the potent antitumor activity of
KMD6B (22,28–30). GSK
J4 inhibits KMD6B activity and decreases the self-renewal of breast
cancer stem cells by downregulating the expression of the key
transcription factors OCT4, NANOG and SOX2 (31,32). Ha
et al (33) reported that
KMD6B can promote G1/S phase arrest in THP-1 cells.
KMD6B mediates the malignant progression of diffuse large B cell
lymphoma by sustaining the activation of the NF-κB pathway via
interacting with the transcription factor IRF4 and also can promote
apoptosis resistance and cell proliferation (22,34,35). The
present study demonstrated that GSK J4 significantly inhibited the
proliferation of HGC27 gastric cancer cells as the percent of cells
in the G2 phase was increased in GSK J4-treated groups,
the expression of cyclin B1 and Cdc2 was significantly decreased
and p21 was upregulated.
The aforementioned experimental results suggested
that the inhibition of endogenous KMD6B demethylase activity can
induce G2/M arrest and inhibit cell proliferation,
suggesting KMD6B has potential as a therapeutic target for gastric
cancer. Previous studies have found that KMD6B can promote the
invasion and metastasis of hepatocellular carcinoma via mediating
the epithelial-mesenchymal transition (EMT) through upregulating
the expression of Slug (36,37). The present study has limitations, for
example the database was extracted from single center and more
cases are needed to validate the results. Moreover, clinical
outcomes associated with cancer-wide gene expression and web-based
platforms offering survival prediction data and cancer registry
risk estimates, such as SurvExpress (38), PROGgeneV2 (39), UALCAN (www.ualcan.path.uab.edu) and Oncomine (www.oncomine.org), should be used in future. Using
advanced genomic analysis tools could further improve our
understanding of the impact of aberrant KMD6B on the clinical
outcomes of gastric cancer (38–45).
Since metastasis is a key malignant characteristic of cancer, it
remains of considerable interest to further study the role of KMD6B
in mediating the EMT phenotype. In addition, the potential of KDM6B
targeted inhibition in preventing the invasion and metastasis of
gastric cancer should be investigated.
Acknowledgements
Not applicable.
Funding
The study was funded by The Fund Project Science and
Technology Plan Project of Cixi (grant no. CN2016023), The Medical,
Health Science and Technology Program of Zhejiang Province (grant
no. 2017KYB608) and The Traditional Chinese Medicine Research Fund
of Zhejiang Province (grant no. 2018ZA119).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW and YW conceived the present study. SW, YW, HZ,
MC and LZ designed and performed the experiments. YW, HZ, MC and LZ
analyzed and interpreted the data. SW and YW contributed to writing
the manuscript. SW and YW confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The study complied with the standards of the
Declaration of Helsinki and was approved by the Ethic Committee of
Cixi Hospital (Wenzhou, China; approval nos. 2008-005 and
2017-LS-25). Written informed consent was provided by all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen C, Wu M, Zhang W, Lu W, Zhang M,
Zhang Z, Zhang X and Yuan Z: MicroRNA-939 restricts Hepatitis B
virus by targeting Jmjd3-mediated and C/EBPα-coordinated chromatin
remodeling. Sci Rep. 6:359742016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang J, Gan L, Wu Z, Yan S, Liu X and Guo
W: The influence of marital status on the stage at diagnosis,
treatment, and survival of adult patients with gastric cancer: A
population-based study. Oncotarget. 8:22385–22405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yoo C, Ryu MH, Na YS, Ryoo BY, Lee CW and
Kang YK: Vorinostat in combination with capecitabine plus cisplatin
as a first-line chemotherapy for patients with metastatic or
unresectable gastric cancer: Phase II study and biomarker analysis.
Br J Cancer. 114:1185–1190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Padmanabhan N, Ushijima T and Tan P: How
to stomach an epigenetic insult: The gastric cancer epigenome. Nat
Rev Gastroenterol Hepatol. 14:467–478. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Liang J and Hou P: Hypermethylation
in gastric cancer. Clin Chim Acta. 448:124–132. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang M and Du X: Noncoding RNAs in
gastric cancer: Research progress and prospects. World J
Gastroenterol. 22:6610–6618. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang WY, Gu JL and Zhen TM: Recent
advances of histone modification in gastric cancer. J Cancer Res
Ther. 10 (Suppl):240–245. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun DF, Zhang YJ, Tian XQ, Chen YX and
Fang JY: Inhibition of mTOR signalling potentiates the effects of
trichostatin A in human gastric cancer cell lines by promoting
histone acetylation. Cell Biol Int. 38:50–63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Akiyama Y, Koda Y, Byeon SJ, Shimada S,
Nishikawaji T, Sakamoto A, Chen Y, Kojima K, Kawano T, Eishi Y, et
al: Reduced expression of SET7/9, a histone mono-methyltransferase,
is associated with gastric cancer progression. Oncotarget.
7:3966–3983. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang ZJ, Yang JL, Wang YP, Lou JY, Chen J,
Liu C and Guo LD: Decreased histone H2B monoubiquitination in
malignant gastric carcinoma. World J Gastroenterol. 19:8099–8107.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu X, Liu J, Xu X, Zhang C and Dai D:
Genome-wide analysis of histone modifications by ChIP-chip to
identify silenced genes in gastric cancer. Oncol Rep. 33:2567–2574.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z, et al: lncRNA HOXA11-AS Promotes
Proliferation and Invasion of Gastric Cancer by Scaffolding the
Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meng CF, Zhu XJ, Peng G and Dai DQ:
Re-expression of methylation-induced tumor suppressor gene
silencing is associated with the state of histone modification in
gastric cancer cell lines. World J Gastroenterol. 13:6166–6171.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song IS, Ha GH, Kim JM, Jeong SY, Lee HC,
Kim YS, Kim YJ, Kwon TK and Kim NS: Human ZNF312b oncogene is
regulated by Sp1 binding to its promoter region through DNA
demethylation and histone acetylation in gastric cancer. Int J
Cancer. 129:2124–2133. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Audia JE and Campbell RM: Histone
Modifications and Cancer. Cold Spring Harb Perspect Biol.
8:a0195212016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Perrigue PM, Najbauer J and Barciszewski
J: Histone demethylase JMJD3 at the intersection of cellular
senescence and cancer. Biochim Biophys Acta. 1865:237–244.
2016.PubMed/NCBI
|
|
18
|
Li Q, Hou L, Ding G, Li Y, Wang J, Qian B,
Sun J and Wang Q: KDM6B induces epithelial-mesenchymal transition
and enhances clear cell renal cell carcinoma metastasis through the
activation of SLUG. Int J Clin Exp Pathol. 8:6334–6344.
2015.PubMed/NCBI
|
|
19
|
Xiang Y, Zhu Z, Han G, Lin H, Xu L and
Chen CD: JMJD3 is a histone H3K27 demethylase. Cell Res.
17:850–857. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Shen L, Stupack DG, Bai N, Xun J,
Ren G, Han J, Li L, Luo Y, Xiang R, et al: JMJD3 promotes survival
of diffuse large B-cell lymphoma subtypes via distinct mechanisms.
Oncotarget. 7:29387–29399. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bosman FT, Carneiro F, Hruban RH and
Theise ND: WHO Classification of Tumours of the Digestive System.
3. 4th edition. International Agency for Research on Cancer; Lyon:
2010
|
|
22
|
Otto T and Sicinski P: Cell cycle proteins
as promising targets in cancer therapy. Nat Rev Cancer. 17:93–115.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shmakova A, Batie M, Druker J and Rocha S:
Chromatin and oxygen sensing in the context of JmjC histone
demethylases. Biochem J. 462:385–395. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W,
Lv R, Li X, Villen J, Gygi SP, et al: The histone H3 Lys 27
demethylase JMJD3 regulates gene expression by impacting
transcriptional elongation. Genes Dev. 26:1364–1375. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q,
Chen Y, Wang HY and Wang RF: Jmjd3 inhibits reprogramming by
upregulating expression of INK4a/Arf and targeting PHF20 for
ubiquitination. Cell. 152:1037–1050. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Poreba E, Broniarczyk JK and
Gozdzicka-Jozefiak A: Epigenetic mechanisms in virus-induced
tumorigenesis. Clin Epigenetics. 2:233–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Anderton JA, Bose S, Vockerodt M,
Vrzalikova K, Wei W, Kuo M, Helin K, Christensen J, Rowe M, Murray
PG, et al: The H3K27me3 demethylase, KDM6B, is induced by
Epstein-Barr virus and over-expressed in Hodgkin's lymphoma.
Oncogene. 30:2037–2043. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen J: The Cell-Cycle Arrest and
Apoptotic Functions of p53 in Tumor Initiation and Progression.
Cold Spring Harb Perspect Med. 6:a0261042016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Enyindah-Asonye G, Li Y, Xin W, Singer NG,
Gupta N, Fung J and Lin F: CD6 Receptor Regulates Intestinal
Ischemia/Reperfusion-induced Injury by Modulating Natural
IgM-producing B1a Cell Self-renewal. J Biol Chem. 292:661–671.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y, Singer NG, Whitbred J, Bowen MA, Fox
DA and Lin F: CD6 as a potential target for treating multiple
sclerosis. Proc Natl Acad Sci USA. 114:2687–2692. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan N, Xu L, Wu X, Zhang L, Fei X, Cao Y
and Zhang F: GSKJ4, an H3K27me3 demethylase inhibitor, effectively
suppresses the breast cancer stem cells. Exp Cell Res. 359:405–414.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sedrak H, El-Garem N, Naguib M, El-Zawahry
H, Esmat M and Rashed L: Vascular endothelial growth factor before
and after locoregional treatment and its relation to treatment
response in hepatocelluar carcinoma patients. Asian Pacific Journal
of Tropical Biomedicine. 5:1005–1009. 2015. View Article : Google Scholar
|
|
33
|
Ha SD, Cho W and Kim SO: HDAC8 Prevents
Anthrax Lethal Toxin-induced Cell Cycle Arrest through Silencing
PTEN in Human Monocytic THP-1 Cells. Toxins (Basel). 9:E1622017.
View Article : Google Scholar
|
|
34
|
Kennedy R and Klein U: Aberrant Activation
of NF-κB Signalling in Aggressive Lymphoid Malignancies. Cells.
7:E1892018. View Article : Google Scholar
|
|
35
|
Staudt LM: Oncogenic activation of
NF-kappaB. Cold Spring Harb Perspect Biol. 2:a0001092010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang B, Qi G, Tang F, Yuan S, Wang Z,
Liang X, Li B, Yu S, Liu J, Huang Q, et al: Aberrant JMJD3
Expression Upregulates Slug to Promote Migration, Invasion, and
Stem Cell-Like Behaviors in Hepatocellular Carcinoma. Cancer Res.
76:6520–6532. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yin X, Yang S, Zhang M and Yue Y: The role
and prospect of JMJD3 in stem cells and cancer. Biomed
Pharmacother. 118:1093842019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Aguirre-Gamboa R, Gomez-Rueda H,
Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R,
Rodriguez-Barrientos A, Tamez-Peña JG and Treviño V: SurvExpress:
An online biomarker validation tool and database for cancer gene
expression data using survival analysis. PLoS One. 8:e742502013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goswami CP and Nakshatri H: PROGgeneV2:
Enhancements on the existing database. BMC Cancer. 14:9702014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goldman M, Craft B, Swatloski T, Cline M,
Morozova O, Diekhans M, Haussler D and Zhu J: The UCSC Cancer
Genomics Browser: Update 2015. Nucleic Acids Res. 43D:D812–D817.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res.
45W:W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A Portal for Facilitating Tumor Subgroup Gene
Expression and Survival Analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46D:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|