Introduction
Head and neck squamous cell carcinoma (HNSC) is the
sixth most common malignancy in the world (1), which predominantly develops from
squamous cell epithelia according to Chaturvedi et al in
2013 (2). The main risk-factors of
HNSC are cigarette smoking, excessive alcohol use and the presence
of human papillomavirus (HPV). Although the overall survival (OS)
time and quality of life have been enhanced by improved standard
treatment and supportive care (3),
HNSC prognosis remains poor with a 5-year OS rate of ~50% worldwide
by 2011 (4).
In recent years, some biomarkers have been
identified and used in the diagnosis of HNSC (5). For example, matrix metalloproteinases
(MMPs), which promote tumor invasion and metastasis, have been
found to be significantly increased in the serum of patients with
head and neck cancer and are promising biomarkers for the diagnosis
of HNSC (6). In addition, DNA
methylation is a major epigenetic change that often precedes the
malignant proliferation of cells. The identification of DNA
methylation changes is of great significance for the early
detection of tumors (7). At present,
the methylation status of genes such as p16, cyclin-dependent
kinase and stratifin have been associated with HNSC (8,9).
However, the sensitivity and specificity of these biomarkers
reported in literatures are quite different.
Since immune-associated mechanisms have a critical
role in HNSC, immunotherapies represent a promising strategy for
treatment (10,11). Immune checkpoints can respond to
pathogens by regulating the balance of immune stimuli and
inhibitory signals, or as regulators of mutant/overexpressing T
cell immune responses (12,13). Previous research has demonstrated
that the interaction between programmed cell death protein-1 (PD-1)
and PD ligand-1 is a critical immune checkpoint. Inhibiting PD-1
has been found to exhibit high treatment efficacy for melanoma and
is now approved for the treatment of HNSC (14,15).
However, current anti-PD-1 immunotherapies do not generate good
responses from patients with advanced HNSC. According to reports,
the median OS time was 7.5 months, and some patients show
resistance (16,17). Additionally, several studies have
reported that patients with a greater number of tumor-infiltrating
lymphocytes display increased survival in HPV-positive and
-negative oropharyngeal disease (18–21).
Therefore, there is a need to elucidate the specific immune
phenotypes of tumor-immune relationships and identify novel
immunological targets for the treatment of HNSC.
As a protein-coding gene, transmembrane channel-like
8 (TMC8) is not yet fully understood. TMC6 and TMC8 null mutations
have been shown to result in severe susceptibility to cutaneous
(β-type) HPV infections, causing a rare syndrome termed
epidermodysplasia verrucciformis (EV) (22,23).
Moreover, an association between TMC8 variants and susceptibility
to skin and cervical cancer has been observed (23,24).
Notably, patients with HNSC are often accompanied by HPV infection,
but the expression level of TMC8 and its relationship with patient
prognosis and clinical stage has not yet been reported. Previous
studies have shown that T lymphocytes exhibit high levels of TMC8
gene expression, indicating that TMC8 plays a multifunctional role
in the mechanisms associated with tumor infiltration (25,26).
However, the impact of this gene on the OS time of patients with
HNSC and the underlying function of TMC8 in tumor-immune
interactions remains unclear.
Oral squamous cell cancer is a typical
representative of HNSC, accounting for the majority of HNSC
(1,4). As shown in the workflow of Fig. 1, the expression of TMC8 was examined
in oral cancer cell lines, clinical specimens and The Cancer Genome
Atlas (TCGA) database and its relationship with the OS time of
patients with HNSC was also evaluated. Moreover, its function and
role in the immune cell network, as well as its impact on prognosis
in multiple cancers (pan-cancers) were analyzed. The purpose of the
present study was to explore the role of TMC8 in the immune
microenvironment and to further clarify its impact on the incidence
and outcome of HNSC.
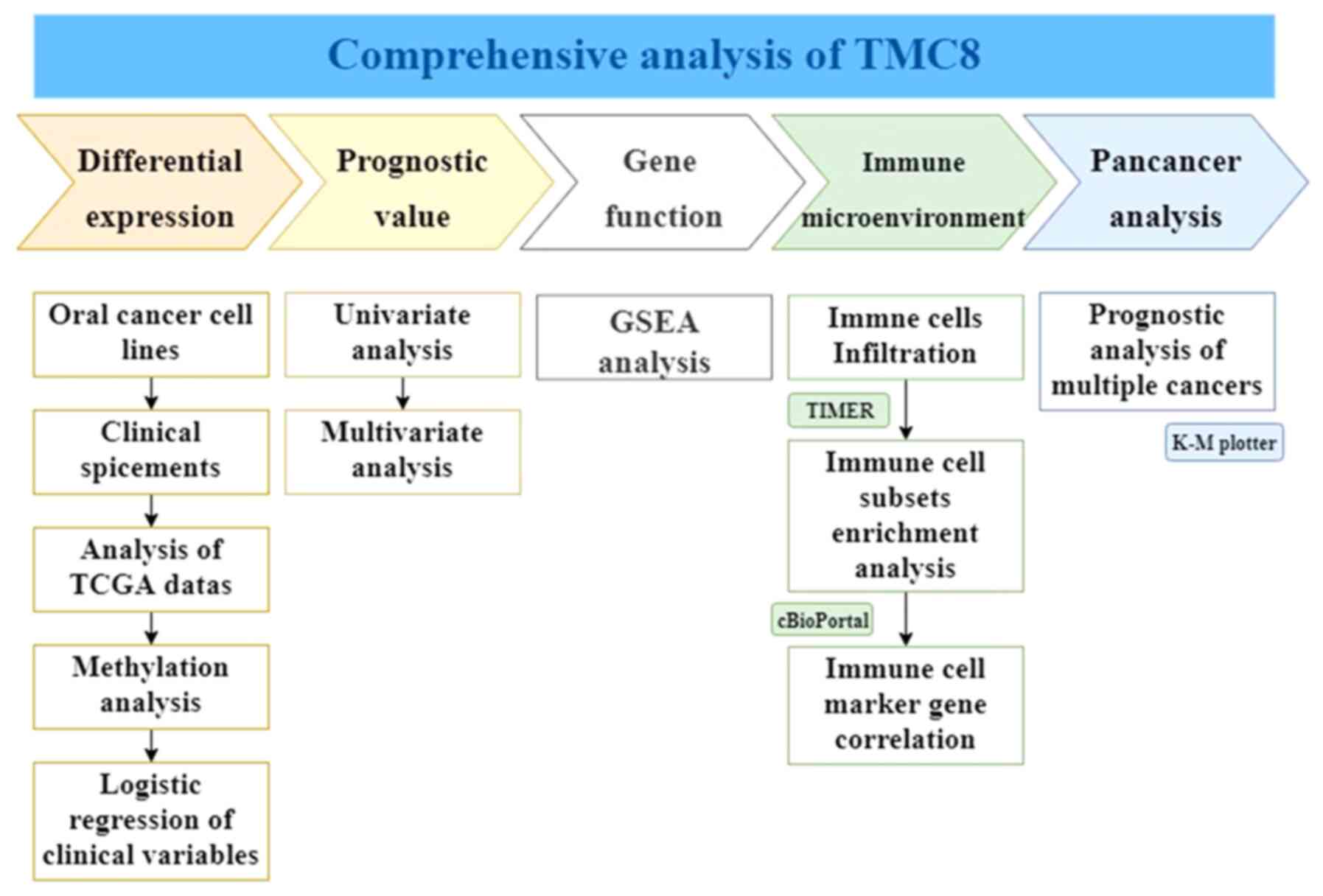 | Figure 1.Workflow of the study. A
comprehensive analysis of TMC8 was conducted. In the first line,
from left to right, the five parts of differential expression,
prognostic value of head and neck squamous cancer, gene function,
immune infiltration relationship and prognostic effect on
pan-cancers are analyzed and are marked with different colors.
Below each part, the specific analysis content is listed. TMC8,
transmembrane channel-like 8; GSEA, gene set enrichment analysis;
TCGA, The Cancer Genome Atlas; K-M, Kaplan-Meier; TIMER, Tumor
Immune Estimation Resource. |
Materials and methods
Acquisition of clinical specimens
Oral squamous cell carcinoma (OSCC) and adjacent
normal tissues were collected from patients with HNSC at the Peking
University Shenzhen Hospital (Shenzhen, China) between January 2017
and December 2019. HNSC was diagnosed and classified by two
independent pathologists based on the World Health Organization
classification system (27). The
specimens were all taken from the tongue cancer resection process,
and the distance between tumor tissue and adjacent normal tissue
was greater than 2 cm. Specimens from patients with a history of
preoperative chemotherapy were excluded. The study was approved by
The Ethics Committee of Peking University Health Science Center
(Guangdong, China). Informed consent was obtained from patients
before the study began.
Immunochemical staining
The above mentioned, obtained from the operating
room without pre-embedded OSCC samples were fixed in 10% neutral
buffered formalin for 24 h at room temperature, dehydrated in
gradient alcohol solution (50, 70, 80, 95, 95 and 100 alcohol, each
for 1 h) and paraffin-embedded. Paraffin-embedded tumor sections
with a thickness of 5-µm were deparaffinized with xylene I for 15
min, xylene II for 10 min, and rehydrated with 100% ethanol and
100, 95, 95 and 80% ethanol for 5 min each. Sections were then
washed twice for 5 min each with dH2O. Slides were heated in a
microwave in 1X citrate antigen retrieval Solution (cat. no.
MVS-0066; MBX Bioscience) until boiling, followed by incubation at
the boiling state for 20 min. After cooling, sections were washed
in PBS three times for 5 min each. Antigen retrieval was achieved
by blocking with 3% H2O2 for 30 min at room
temperature. The aforementioned washing steps were repeated and
then blocking was performed using goat serum (cat. no. ZLI-9022;
OriGene Technologies, Inc.) at room temperature for 20 min. Then
the sections were incubated with antibodies against TMC8 (cat. no.
ab69859; 1:75; Abcam) overnight at 4°C. The slides were then
incubated with a biotin-labeled secondary antibody UltraSensitive™
SP (Mouse/Rabbit) IHC kit (cat. no. KIT-9710; MBX Biotechnologies,
Inc.) to TMC8 for 30 min at room temperature. The aforementioned
washing steps were repeated and the slices were stained with DAB
(cat. no. DAB-0031; MBX Biotechnologies, Inc.) at room temperature
for ~1 min. All images shown are wide-field light microscopy images
acquired at sufficient resolution.
Cell culture
Human oral squamous cell carcinoma cell lines SCC9,
SCC15, SCC25 and CAL27 were obtained from the American Type Culture
Collection. Human oral keratinocyte (HOK) cells were obtained from
The Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. SCC15, SCC25, CAL27 and HOK cells were maintained in DMEM
supplemented with 10% FBS and 1% penicillin and streptomycin
sulfate (all Gibco; Thermo Fisher Scientific, Inc.). SCC9 cells
were cultured in 1:1 DMEM/F12 (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS, 1% penicillin and streptomycin sulfate,
1% sodium pyruvate (Gibco; Thermo Fisher Scientific, Inc.) and 400
ng/ml hydrocortisone (MedChemExpress). All cells were cultured at
37°C and 5% CO2.
RNA extraction and reverse
transcription quantitative PCR (RT-qPCR)
Total RNA from frozen tissues or cultured cells was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
A PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time)
(cat. no. RR047A; Takara Bio, Inc.) was used for
reverse-transcribing the RNA into cDNA, as per the manufacturer's
instructions. RT-qPCR was performed with TB Green®
Premix Ex Taq™ II (Tli RNaseH Plus) (cat. no. RR820A; Takara Bio,
Inc.) and was monitored using an ABI PRISM™ 7500 Sequence Detection
system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Thermocycling conditions were as follows: Initial denaturation at
95°C for 5 min, followed by followed by denaturation at 95°C for 5
sec; annealing at 58°C for 30 sec and elongation at 72°C for 20
sec, for 40 cycles. The following primers were used for qPCR:
β-Actin, forward: 5′-AAACTGGAACGGTGAAGGTG-3′ and reverse:
5′-AGTGGGGTGGCTTTTAGGAT-3′; TMC8, forward:
5′-GAACTACCCTCCCAACACG-3′ and reverse: 5′-TGCTCTTGTCTCTGCCAATG-3′.
Comparative quantification was performed with either the ΔCq or the
2−ΔΔCq method (28).
RT-qPCR was used to determine the expression of TMC8 in SCC9,
SCC15, SCC25 and CAL27 cell lines. Data from three independent
experiments were obtained, the mean value ± standard deviation was
calcualted and unpaired Student's t-test was used.
Acquisition of mRNA data
TMC8 gene expression and methylation data, as well
as the corresponding clinical information, were downloaded from
TCGA website (https://portal.gdc.cancer.gov) for HNSC and estimated
as log2 (x+1) transformed RNA sequencing by
expectation-maximization normalized counts (29). A total of 528 patients with HNSC were
sampled, containing 44 patients with adjacent non-tumorous tissue
as the control group. All data were processed using R studio
software version 3.5.3 (30). The
‘ESTIMATE’ R package was used to predict the presence of
infiltrating stromal/immune cells in tumor tissues using gene
expression data (31).
Gene Expression Profiling Interactive
Analysis (GEPIA) and survival analysis
The online database GEPIA (http://gepia.cancer-pku.cn/index.html) was used to
analyze the differential expression of the TMC8 and its prognostic
value.
Methylation and gene expression
analyses
DNA methylation data from TCGA contained β-values
for 485,577 CpG sites. The β-value was calculated as (M/M+U) and
ranged from zero to one, where M was the frequency of the
methylated allele and U was the frequency of the unmethylated
allele. Therefore, higher β-values indicated higher levels of
methylation. Levels of TMC8 methylation between HNSC and normal
tissues were compared. In addition, the association between TMC8
expression and its DNA methylation status was investigated.
Oncomine database analysis
The Oncomine database (https://www.oncomine.org/resource/login.html) was
screened for the expression of the TMC8 gene in several types of
cancer. A threshold P-value of 0.001 and fold-change ≥2 was used to
assess statistical significance.
Gene Set Enrichment Analysis
(GSEA)
To identify potential biological mechanism of TMC8,
GSEA was conducted to detect whether a previously defined set of
genes showed statistically significant differential expression.
Firstly, the TMC8 high expression group was selected based on the
median expression (cut-off value 0.453101463) of TMC8, and the
genes are sorted according to expression differences to form a gene
list. The annotated gene sets C2.CP (186 gene sets) and C5.BP
(5,910 gene sets) MSigDB datasets from the Broad Institute
(https://www.gsea-msigdb.org/gsea/msigdb/index.jsp)
were selected as the reference gene sets. These preset gene sets
represented different biological processes or signal pathways. Then
the GSEA algorithm could determine whether the members of this
reference gene set were randomly distributed in the TMC8 high
expression group gene list, or were mainly enriched at the front or
end sides of the list. The third step was to calculate the
enrichment score of the gene set and perform a permutation test of
significance to obtain the P-value and the false discovery rate
(FDR). FDR <25% and P<0.05 in the enrichment of MSigDB
Collection (c2.cp.kegg.v6.2.symbols) were considered to be
significantly enriched.
Tumor Immune Estimation Resource
(TIMER) database analysis
The TIMER database can be used as a comprehensive
resource for systematically analyzing immune infiltrates between
several types of cancer (https://cistrome.shinyapps.io/timer/). Multiple
deconvolution algorithms including TIMER (32), CIRBSORT (33) and xCell (34) are used to analyze the expression of
TMC8 and the degree of infiltration of immune cells, including i)
CD8+ T cells; ii) CD4+ T cells; iii) B cells;
iv) dendritic cells (DCs); v) neutrophils; and vi) macrophages and
their subgroups. The correlation between the infiltration levels of
above-mentioned immune cells and the expression of TMC8 was
obtained.
Co-expression analysis in
cBioPortal
For cancer genomics, cBioPortal (https://www.cbioportal.org) is an open-access,
open-source resource for the interactive exploration of
multidimensional cancer genomics data sets. The correlation between
the hub gene expression and gene markers of immune cells were
explored. The gene markers of the immune cells included markers of:
i) CD8+ T Cells; ii) T cells (general); iii) B cells;
iv) monocytes; v) tumor-associated macrophages (TAMs); vi) M1
macrophages; vii) M2 macrophages; viii) neutrophils; ix) natural
killer (NK) cells; x) DCs; xi) T helper (Th)-1 cells; xii) Th2
cells; xiii) follicular helper T (Tfh) cells; xiv) Th17 cells; xv)
regulatory T cells (T regs); and xvi) exhausted T cells (34). Spearman's rank correlation
coefficient method was used to identify the correlation
coefficient.
Prognostic analysis of
pan-cancers
Kaplan-Meier plotter (http://kmplot.com/analysis/) is capable of assessing
the effect of 54,000 genes on survival in 21 types of cancer
(35). The association between the
hub gene expression and survival in all 21 types of cancer were
analyzed using Kaplan-Meier plotter. All possible cut-off values
between the lower and upper quartiles were computed and the best
performing threshold was used as a cut-off value. The hazard ratio
(HR) with 95% confidence intervals (CI) and log-rank P-value were
also calculated.
Statistical analysis
Survival curves were created using GEPIA and
Kaplan-Meier plots. Three independent repeated unpaired and paired
t-tests were carried out in GraphPad Prism (GraphPad Software, Inc.
version 7.04) and R software (30)
(version 3.5.3), and the results were shown as mean ± standard
deviation. The Oncomine results were presented with P-values,
fold-changes and ranks. The Kaplan-Meier plots and GEPIA results
were displayed with the HR and P-values obtained using log-rank
tests. Correlation between TMC8 expression and the
clinicopathological characteristics were analyzed using logistic
regression. Potential prognostic factors were determined with a
univariate Cox analysis and the associations between TMC8
expression and survival in conjunction with other clinical features
were verified with a multivariate Cox analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of TMC8 in oral cancer
specimens and adjacent normal tissue
HNSC specimens from 25 patients were analyzed in the
present study, including 17 males and 8 females. The age of this
group ranged from 43 to 81 years with a median age of 67.
Immunohistochemical staining of OSCC tissue specimens and adjacent
normal tissues revealed that TMC8 was strongly stained in tumor
tissues, but the staining intensity was lower in adjacent tissues
(Fig. 2A-D).
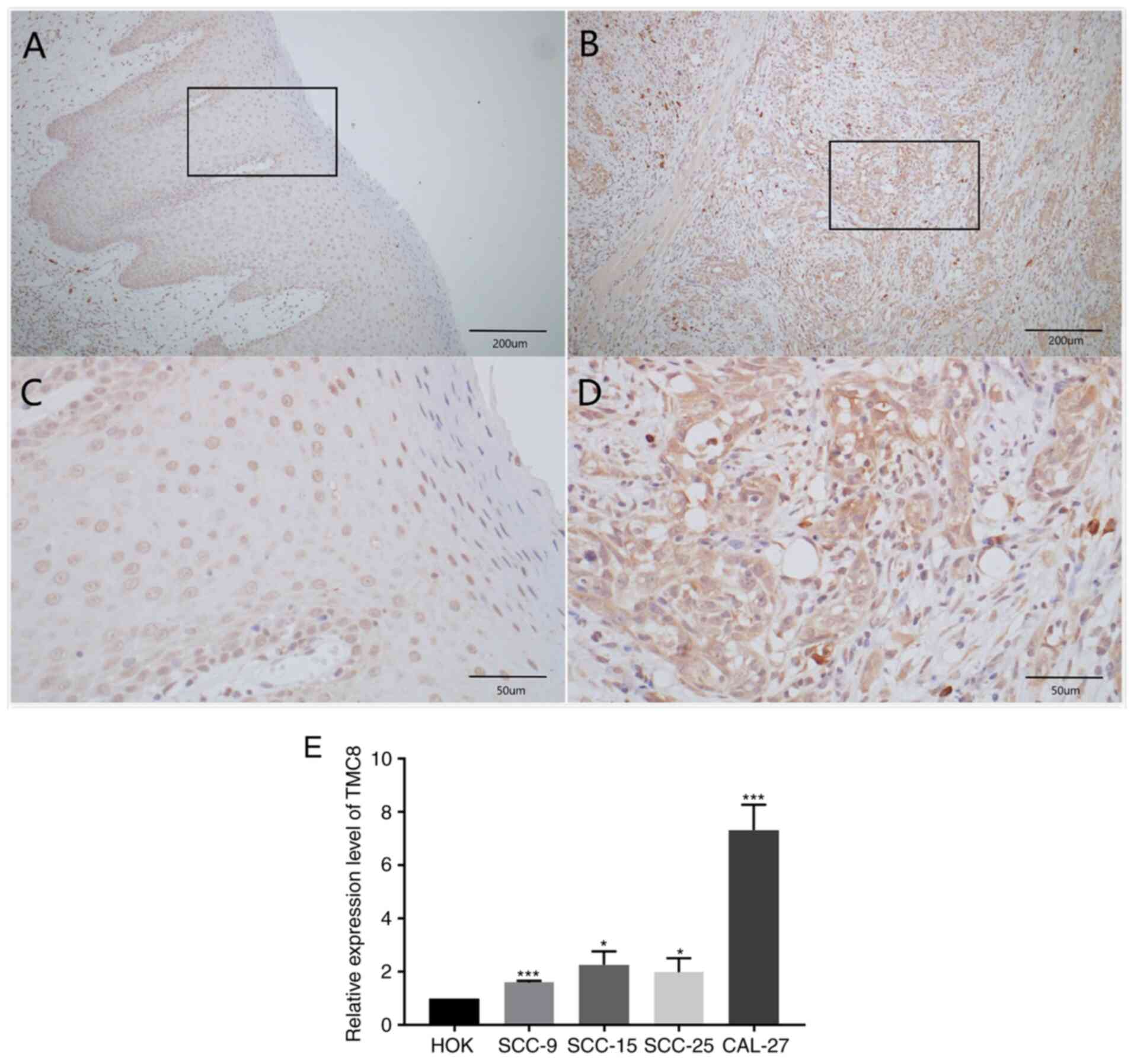 | Figure 2.TMC8 is upregulated in OSCC.
Representative immunohistochemical staining performed for detecting
the expression of TMC8 from tumor-tissue specimens and adjacent
normal tissues of patients with OSCC. (A and B) Normal mucosal
epithelial cells, lightly stained with TMC8 antibody.
(magnification, ×100 or 400, respectively). (C and D) Oral cancer
mucosal epithelial cells, lightly stained with TMC8 antibody.
(magnification, ×100 or 400, respectively). (E) Compared with HOK,
the expression levels of TMC8 in SCC9, SCC15, SCC25 and CAL27 cell
lines were significantly upregulated. *P<0.05 and ***P<0.001
vs. HOK. TMC8, transmembrane channel-like 8; OSCC, oral squamous
cell carcinoma; HOK, human oral keratinocyte. |
Expression of TMC8 is increased in
oral cancer cell lines
Fig. 2E shows that,
compared with HOK, the expression levels of TMC8 in SCC9, SCC15,
SCC25 and CAL27 cell lines were significantly upregulated.
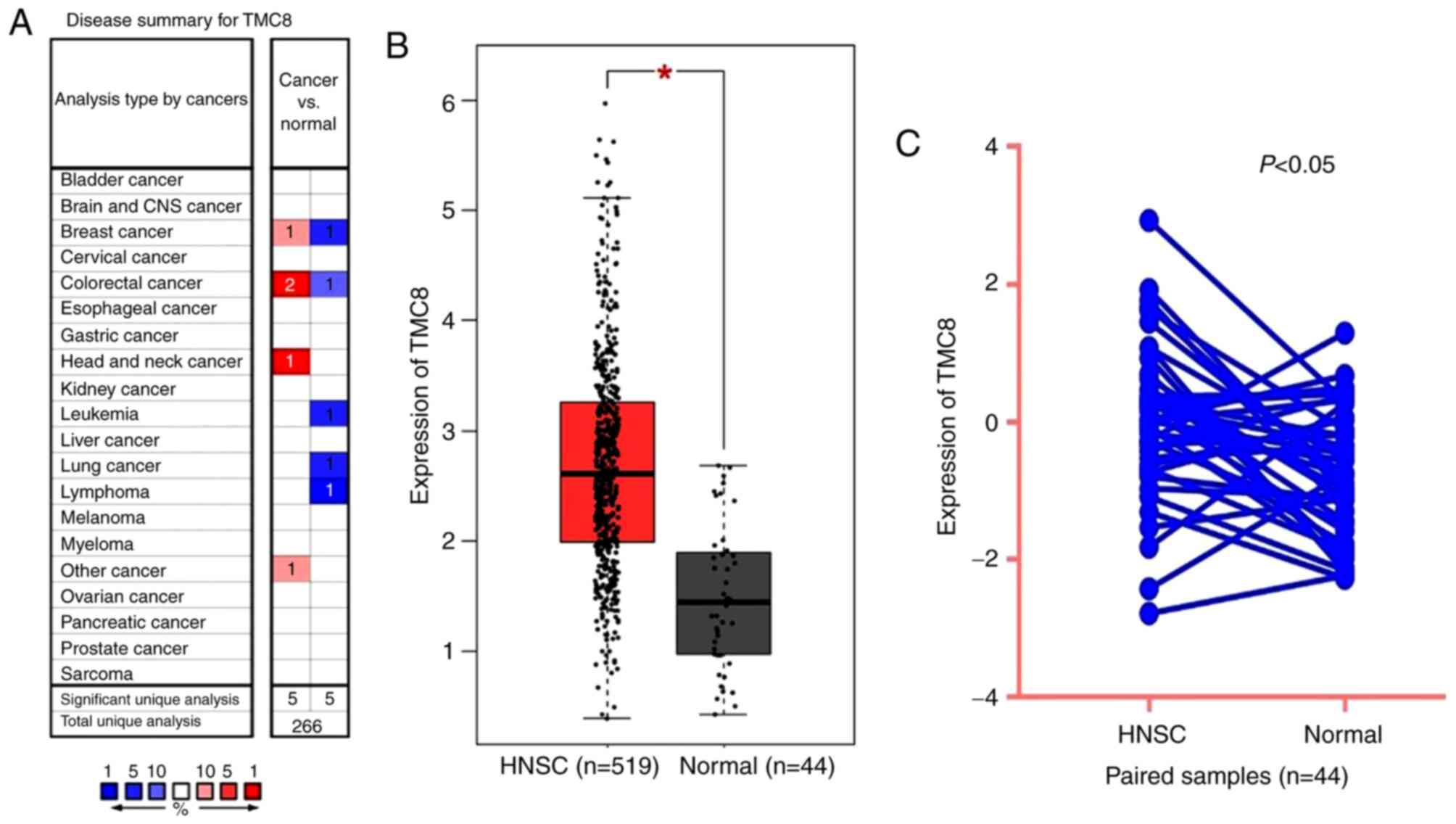
TMC8 is upregulated in HNSC of TCGA
data
As shown in Fig. 3A,
TMC8 expression was higher in colorectal, breast and head and neck
cancer compared with that of normal tissues. However, lower TMC8
expression was also observed in colorectal and breast cancer in
addition to lung, leukemia and lymphoma cancer in some data sets.
The differential level of TMC8 expression between tumor and normal
tissues in HNSC for TCGA data is shown in Fig. 3B and C. The results indicated that
TMC8 was overexpressed in HNSC samples (P<0.001) and paired
samples (P<0.05).
Methylation analysis
In order to explore the possible reasons for the
upregulation of TMC8 in HNSC, the relationship between its
methylation status and its expression was further analyzed.
Overall, 26 methylation sites out of a total of 31 were found to be
significantly hypomethylated. Among them, cg00447208, cg01246266,
cg03190661, cg08470991, cg19056418 and cg20943461, which were all
located in the promoter region, showed significantly decreased
levels of methylation in HNSC compared with normal samples.
Additionally, they were significantly negatively correlated with
the expression of TMC8 (all P<0.05). The relationship between
methylation sites and expression of TMC8 is shown in Table I.
 | Table I.Relationship between methylation
sites and expression of transmembrane channel-like 8. |
Table I.
Relationship between methylation
sites and expression of transmembrane channel-like 8.
| Methylation
site | Cor. | P-value |
|---|
|
cg00447208a | −0.24 |
5.79×10−08 |
|
cg01246266a | −0.16 |
2.63×10−04 |
|
cg03190661a | −0.26 |
3.10×10−09 |
|
cg08470991a | −0.19 |
1.74×10−05 |
|
cg19056418a | −0.28 |
1.39×10−10 |
|
cg20943461a | −0.31 |
1.73×10−12 |
| cg01125010 | 0.13 |
2.20×10−03 |
| cg01791634 | −0.18 |
4.01×10−05 |
| cg02909991 | −0.17 |
6.01×10−05 |
| cg02911077 | −0.20 |
4.09×10−06 |
| cg03596178 | 0.06 | 0.21 |
| cg03742808 | −0.24 |
5.20×10−08 |
| cg04947157 | −0.24 |
3.02×10−08 |
| cg05637296 | −0.29 |
2.67×10−11 |
| cg06248406 | −0.04 | 0.35 |
| cg06643271 | −0.17 |
1.16×10−04 |
| cg08852879 | 0.09 | 0.04 |
| cg09413013 | −0.10 | 0.03 |
| cg11493223 | −0.18 |
2.17×10−05 |
| cg12798338 | −0.24 |
5.76×10−08 |
| cg14210726 | 0.29 |
1.72×10−11 |
| cg16214492 | −0.23 |
8.22×10−08 |
| cg16301617 | −0.26 |
3.79×10−09 |
| cg16935597 | −0.16 |
1.55×10−04 |
| cg18901278 | −0.17 |
6.72×10−05 |
| cg21282054 | −0.26 |
1.11×10−09 |
| cg22563987 | 0.13 |
4.16×10−03 |
| cg22833809 | −0.16 |
2.60×10−04 |
| cg24109860 | −0.06 | 0.17 |
| cg24988684 | −0.24 |
3.65×10−08 |
| cg26003388 | −0.22 |
4.63×10−07 |
Association with TMC8 expression and
clinicopathologic variables
As shown in Table
II, the increased expression of TMC8 was significantly
correlated with the tumor origin (oropharynx vs. oral cavity;
P=0.003), histological grade (G3/G4 vs. G1/G2; P=0.04), HPV
infection status (positive vs. negative; P=0.001), immune-score
(high vs. low, P<0.001), as well as the stromal-score (high vs.
low; P<0.001). However, no significant differences between TMC8
expression and i) age; ii) alcohol use; iii) metastasis; iv) lymph
node infiltration; v) T classification; vi) clinical stage; vii)
lymphovascular invasion; viii) perineural invasion; or ix) nodal
extra-capsular spread were observed.
 | Table II.Correlation between TMC8 expression
and the clinicopathological characteristics of patients with head
and neck squamous cancer (logistic regression). |
Table II.
Correlation between TMC8 expression
and the clinicopathological characteristics of patients with head
and neck squamous cancer (logistic regression).
| Clinical
characteristics | Total, n | Odds ratio in TMC8
expression hazard ratio (CI) | P-value |
|---|
| Age,
continuous | 520 | 1.01
(0.99–1.02) | 0.23 |
| Drinking, yes vs.
no | 509 | 0.96
(0.65–1.43) | 0.84 |
| Smoke, yes vs.
no | 520 | 1.10
(0.78–1.56) | 0.60 |
| Tumor origin,
oropharynx vs. oral cavity | 520 | 1.74
(1.20–2.51) |
3.20×10−3a |
| Distance
metastasis, positive vs. negative | 496 | 0.94
(0.51–1.74) | 0.85 |
| Lymph nodes,
positive vs. negative | 498 | 0.82
(0.58–1.17) | 0.28 |
| T classification,
T1/T2 vs. T3/T4 | 504 | 0.71
(0.49–1.02) | 0.07 |
| Stage, I/II vs.
III/IV | 506 | 0.75
(0.28–1.98) | 0.57 |
| HPV, positive vs.
negative | 121 | 3.98
(1.78–9.39) |
1.10×10−3a |
| Lymphovascular
invasion, positive vs. negative | 348 | 1.39
(0.89–2.16) | 0.15 |
| Perineural
invasion, positive vs. negative | 362 | 0.75
(0.49–1.13) | 0.17 |
| Nodal extracapsular
spread, positive vs. negative | 356 | 0.77
(0.49–1.21) | 0.25 |
| Histological grade,
G3/G4 vs. G1/G2 | 498 | 1.51
(1.02–2.27) | 0.04 |
| Immune score, high
vs. low | 520 | 5.17
(3.57–7.55) |
6.81×10−18a |
| Stromal score, high
vs. low | 520 | 1.95
(1.38–2.77) |
1.72×10−4a |
Survival outcomes and multivariate
analysis
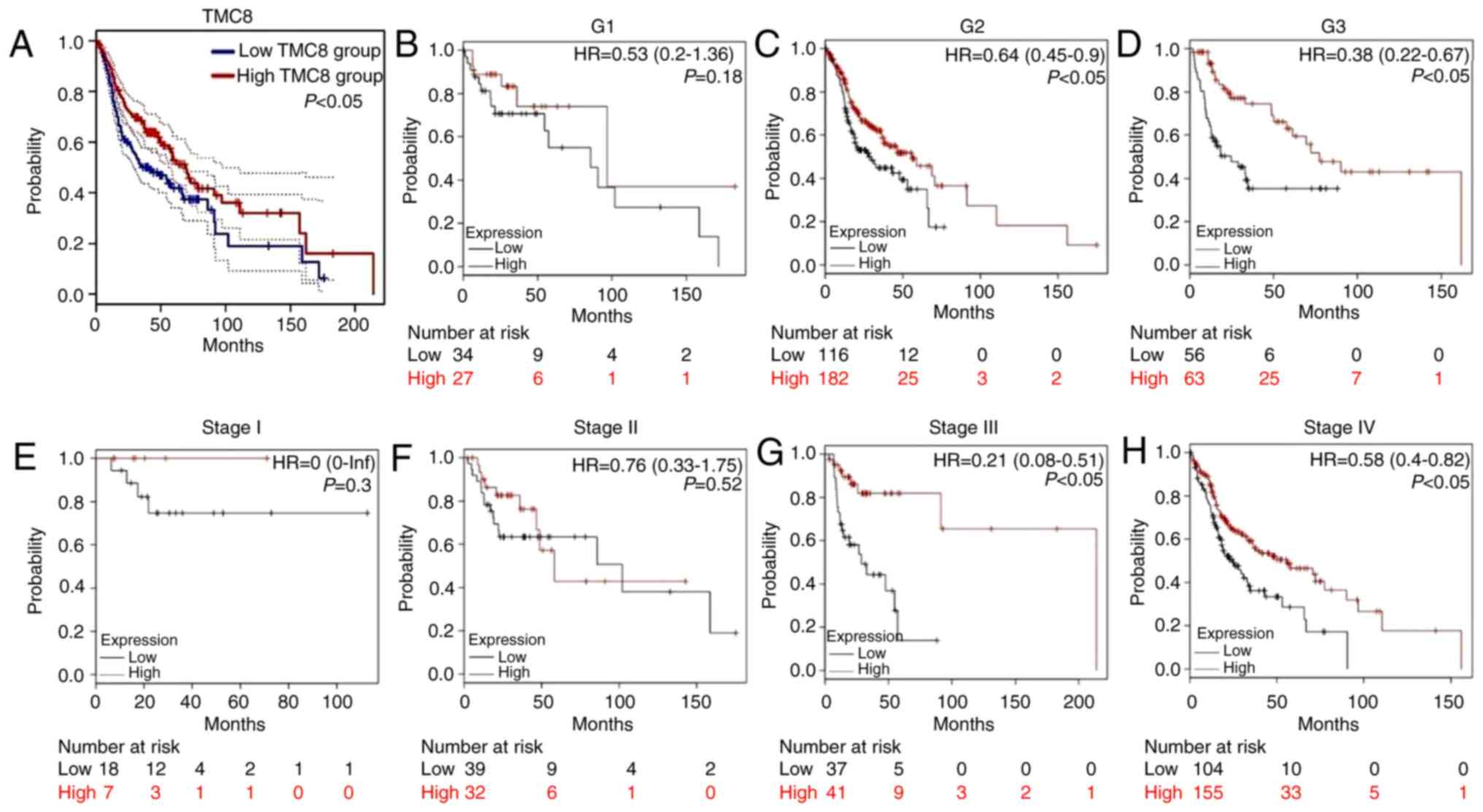
As shown in Fig. 4A,
the analysis of HNSC cases in TCGA showed that the 5-year OS time
of the TMC8-high group was significantly greater compared with that
of the low-expression group (P<0.05). Fig. 4B-H further shows the relationship
between TMC8 expression and clinical features. In the cases of
histopathological grade II–III and clinical stage III–IV,
overexpression of TMC8 was significantly correlated with improved
OS time. This result suggested that in advanced HNSC cases, high
expression of TMC8 may improve the prognosis.
Univariate analysis (Table III) revealed that
TMC8-overexpression was significantly associated with an improved
OS time (HR: 0.80; 95% CI: 0.70–0.90; P=0.003). Other
clinicopathological variables were associated with increased
survival including advanced age, positive marginal status, B cell
infiltration and immune score. In the multivariate analysis, TMC8
remained independently associated with OS, with an HR of 0.80 (CI:
0.68–0.95; P=0.01), In conjunction with the advanced age, clinical
stage and positive marginal status.
 | Table III.Univariate and multivariate analyses
of OS time using the Cox proportional hazard regression model
(n=415). |
Table III.
Univariate and multivariate analyses
of OS time using the Cox proportional hazard regression model
(n=415).
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameter | HR | CI, 95% | P-value | HR | CI, 95% | P-value |
|---|
| Age | 1.02 | 1.01–1.04 |
9.94×10−4a | 1.02 | 1.006–1.035 |
4.00×10−3a |
| Alcohol-use | 0.96 | 0.71–1.29 | 0.77 | 1.03 | 0.741–1.425 | 0.87 |
| Sex | 0.81 | 0.59–1.10 | 0.18 | 0.87 | 0.614–1.222 | 0.41 |
| Clinical stage | 1.32 | 0.93–1.88 | 0.12 | 1.45 | 1.005–2.078 |
0.47×10−2a |
| Margin status | 1.61 | 1.11–2.34 | 0.01a | 1.69 | 1.146–2.480 | 0.01a |
| Histological
grade | 1.08 | 0.78–1.49 | 0.63 | 1.24 | 0.881–1.752 | 0.22 |
| B cell | 0.09 | 0.01–0.64 | 0.02a | 0.16 | 0.011–2.298 | 0.18 |
| CD4+ T
cell | 0.28 | 0.05–1.61 | 0.16 | 0.03 | 0.008–7.332 | 0.42 |
| CD8+ T
cell | 0.55 | 0.20–1.49 | 0.24 | 0.77 | 0.130–4.509 | 0.77 |
| Neutrophil | 0.81 | 0.15–4.52 | 0.81 | 3.93 | 0.229–67.382 | 0.35 |
| Macrophage | 1.43 | 0.32–6.39 | 0.64 | 3.49 | 0.328–36.985 | 0.30 |
| Dendritic cell | 0.88 | 0.47–1.66 | 0.70 | 3.75 | 0.745–18.908 | 0.11 |
| Immune-score | 1.00 |
9.998×10−3−9.99×10−3 | 0.04a | 1.00 |
9.997×10−3−1.000×10−3 | 0.14 |
| Stromal-score | 1.00 |
9.998×10−3−1.000×10−3 | 0.13 | 1.00 |
9.999×10−3−1.000×10−3 | 0.15 |
| TMC8 | 0.80 | 0.70–0.90 |
3.00×10−4a | 0.80 | 0.68–0.95 |
0.01a |
GSEA identifies a TMC8-associated
signaling pathway
TMC8-associated pathway enrichment analysis results
showed that 28 gene sets were significantly enriched when the
adjusted P-value was <5%. As shown in Table IV, the following immune-associated
biological processes are enriched in response to the increased TMC8
phenotype: i) ‘Intestinal immune network for IgA production’; ii)
‘primary immunodeficiency’; iii) ‘leishmania infection’; iv)
‘cytokine-cytokine receptor interaction’; v) ‘natural killer
cell-mediated cytotoxicity’; vi) ‘hematopoietic cell lineage’; vii)
‘autoimmune thyroid disease’; viii) ‘T cell receptor signaling
pathway’; ix) ‘antigen processing and presentation’; and x)
‘cell-adhesion molecules’. These signaling pathways may be the
mechanisms involved in TMC8 function.
 | Table IV.Gene sets enriched analysis of
upregulated TMC8 in head and neck squamous cancer. |
Table IV.
Gene sets enriched analysis of
upregulated TMC8 in head and neck squamous cancer.
| Gene sets | NES | Adjusted
P-value |
|---|
| Intestinal immune
network for IgA production | 2.19 | 0.01 |
| Primary
immunodeficiency | 2.13 | 0.01 |
| Leishmania
infection | 2.08 | 0.01 |
| Cytokine-cytokine
receptor interaction | 2.01 | 0.01 |
| Natural killer cell
mediated cytotoxicity | 2.01 | 0.01 |
| Hematopoietic cell
lineage | 2.01 | 0.01 |
| Autoimmune thyroid
disease | 1.98 | 0.01 |
| T cell receptor
signaling pathway | 1.99 | 0.01 |
| Antigen processing
and presentation | 1.96 | 0.01 |
| Cell-adhesion
molecules | 1.97 | 0.01 |
| Type I diabetes
mellitus | 1.88 | 0.03 |
| Chemokine signaling
pathway | 1.89 | 0.03 |
| Systemic lupus
erythematosus | 1.86 | 0.03 |
| B cell receptor
signaling pathway | 1.86 | 0.03 |
| Asthma | 1.85 | 0.03 |
| Tryptophan
metabolism | 1.83 | 0.04 |
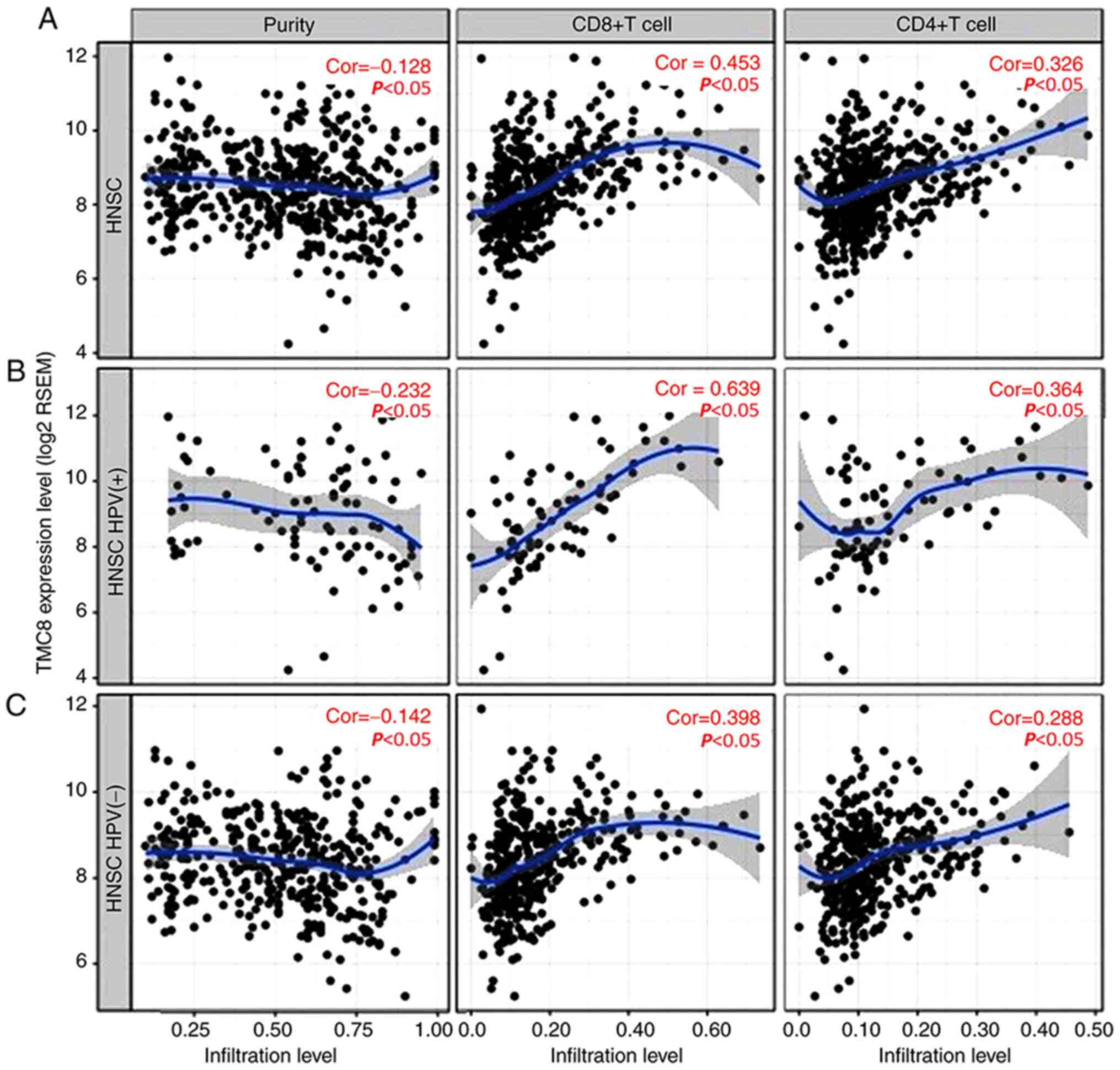
Relationship between TMC8 and immune
cells infiltration
There was a significant negative correlation between
TMC8 expression and tumor purity (Fig.
5A). In addition, the level of TMC8 expression was significant
correlated with high levels of CD4+ T cell and
CD8+ T cell infiltration in HNSC. Similarly, there were
weak to moderate positive correlations with the level of
infiltrating lymphocytes in HPV-positive HNSC samples, as well as
in HPV-negative HNSC samples (correlation coefficient 0.142 to
0.639, all P<0.05; Fig. B and C).
To avoid bias, the results of six other algorithms
were listed, each of which provided the correlation coefficient
between infiltration of different subsets of immune cells and
expression of TMC8. As shown in Table
V, different algorithms acquired similar results. TMC8 was
positively correlated with T cell subsets, such as Th1, Th17, T
regs and Tfh.
 | Table V.Relationship between transmembrane
channel-like 8 and immune cell subgroups by different
algorithm. |
Table V.
Relationship between transmembrane
channel-like 8 and immune cell subgroups by different
algorithm.
|
| Correlation
coefficients |
|---|
|
|
|
|---|
| Subsets of immune
cells | CIBERSORT | Xcell | TISIDB | EPIC | MCPCOUNTER | QUANTISEQ |
|---|
| CD4+ T
cell | – | – | – | – | – | – |
|
CD4+ T cell | – | – | – | 0.18 | – | −0.30a |
|
CD4+ naive | −0.17 | 0.37a |
| – | – | – |
|
CD4+ Th1 | – | 0.13 | 0.42a | – | – | – |
|
CD4+ Th2 | – | 0.11 | 0.05 | – | – | – |
|
CD4+ Th17 | – | – | 0.33a | – | – | – |
|
Tfh | 0.67a | – | 0.36a | – | – | – |
|
Treg | 0.44a | 0.16 | 0.26a | – | – | 0.52a |
|
CD4+ memory | – | 0.25a | – | – | – | – |
|
CD4+ memory
resting | 0.22a | – | – | – | – | – |
|
CD4+ central
memory | – | −0.10 | 0.04 | – | – | – |
|
CD4+ memory
activated | 0.26a | – | – | – | – | – |
|
CD4+ effector
memory | – | 0.00 | 0.17 | – | – | – |
| γδ | 0.10 | 0.18 | – | – | – | – |
| CD8+ T
cell | – | – | – | – | – | – |
| T cell
CD8+ | 0.60a | 0.41a | – | 0.13 | 0.58a | 0.54a |
| T cell
CD8+ naive | – | 0.02 | 0.50a | – | – | – |
| T cell
CD8+ central memory | – | 0.52a | −0.06 | – | – | – |
| T cell
CD8+ effector memory | – | 0.35a | – | – | – | – |
| B cell | – | – | – | – | – | – |
| B
cell | – | – | – | 0.41a | 0.48a | 0.53a |
| B cell
naive | 0.21 | 0.27a | – | – | – | – |
| B cell
plasma | 0.24a | 0.25a | – | – | – | – |
| B cell
memory | 0.22a | 0.39a | 0.12 | – | – | – |
| Macrophage | – | – | – | – | – | – |
|
Macrophage | – | 0.26a | 0.34a | 0.43a | 0.27a | – |
|
Macrophage M1 | 0.49a | 0.31a | – | – | – | 0.17 |
|
Macrophage M2 | 0.45a | 0.20 | – | – | – | 0.40a |
| Dendritic cell | – |
| – | – | – | – |
| Myeloid
dendritic cell | – | 0.33a | – | – | 0.51a | 0.08 |
| Myeloid
dendritic cell activated | −0.05 | 0.48a | 0.25a | – | – | – |
| Myeloid
dendritic cell resting | 0.19 | – | – | – | – | – |
| NK cell | – | – | – | – | – | – |
| NK
cell | – | −0.13 | 0.34a | 0.19 | 0.52a | 0.32a |
| NK cell
resting | −0.09 | – | – | – | – | – |
| NK cell
activated | 0.37a | – | – | – | – | – |
| Neutrophil | – | – | – | – | – | – |
|
Neutrophil | −0.09 | 0.22a | −0.05 | – | −0.03 | 0.06 |
| Monocyte | – | – | – | – | – | – |
|
Monocyte | 0.11 | −0.02 | 0.19 | – | 0.27a | 0.11 |
| Mast cell | – | – |
| – | – | – |
| Mast
cell | – | −0.03 | 0.27a | – | – | – |
| Mast
cell resting | −0.10 | – | – | – | – | – |
| Mast
cell activated | 0.25a | – | – | – | – | – |
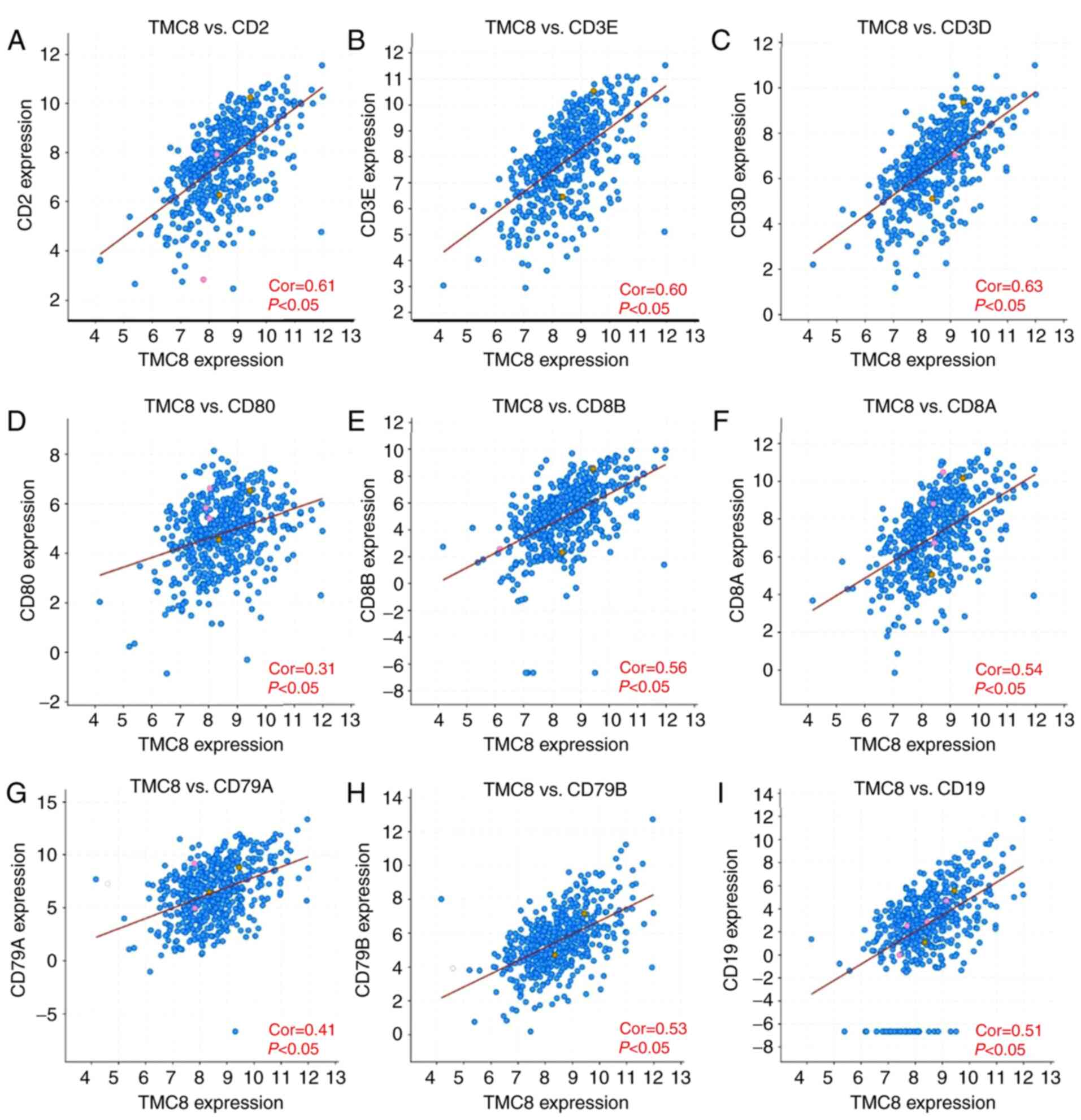
TMC8 expression and immune marker
correlation analysis
To further verify the relationship between TMC8 and
immune cells, the correlation between marker genes and TMC8
expression in different cells was calculated. As shown in Fig. 6A-C, TMC8 is strongly correlated with
T cell (general) marker genes, including CD3D, CD3E and CD2 (all
coefficients >0.6 and P<0.05). TMC8 was significantly
correlated with marker genes of CD8+ cells (all
P<0.05 and coefficients >0.3; Fig.
6D-F) and B cells (all coefficients >0.4 and P<0.05;
Fig. 6G-I). In addition, the level
of expression for a majority of markers on M1 (nitric oxide
synthase 2 and interferon regulatory factor 5), M2 macrophages
(CD163, V-set and immunoglobulin domain-containing protein 4 and
membrane-spanning 4-domains subfamily A member 4A), TAMs (C-C motif
chemokine 2, CD68 and interleukin 10), monocytes, DCs (human
leukocyte antigen [HLA]-DPB1, HLA-DRA and HLA-DPA1) and other
subclasses of T cells were significantly correlated with TMC8
expression, details shown in Table
SI.
Prognostic analysis of TMC8 on
pan-cancers
As shown in Fig. 7,
in five types of tumors: i) Bladder cancer; ii) cervical squamous
cell carcinoma; iii) pheochromocytoma and paraganglioma; iv)
sarcoma; and v) thymoma, high expression of TMC8 predicted a
significantly increased OS time and improved prognosis (all
P<0.05).
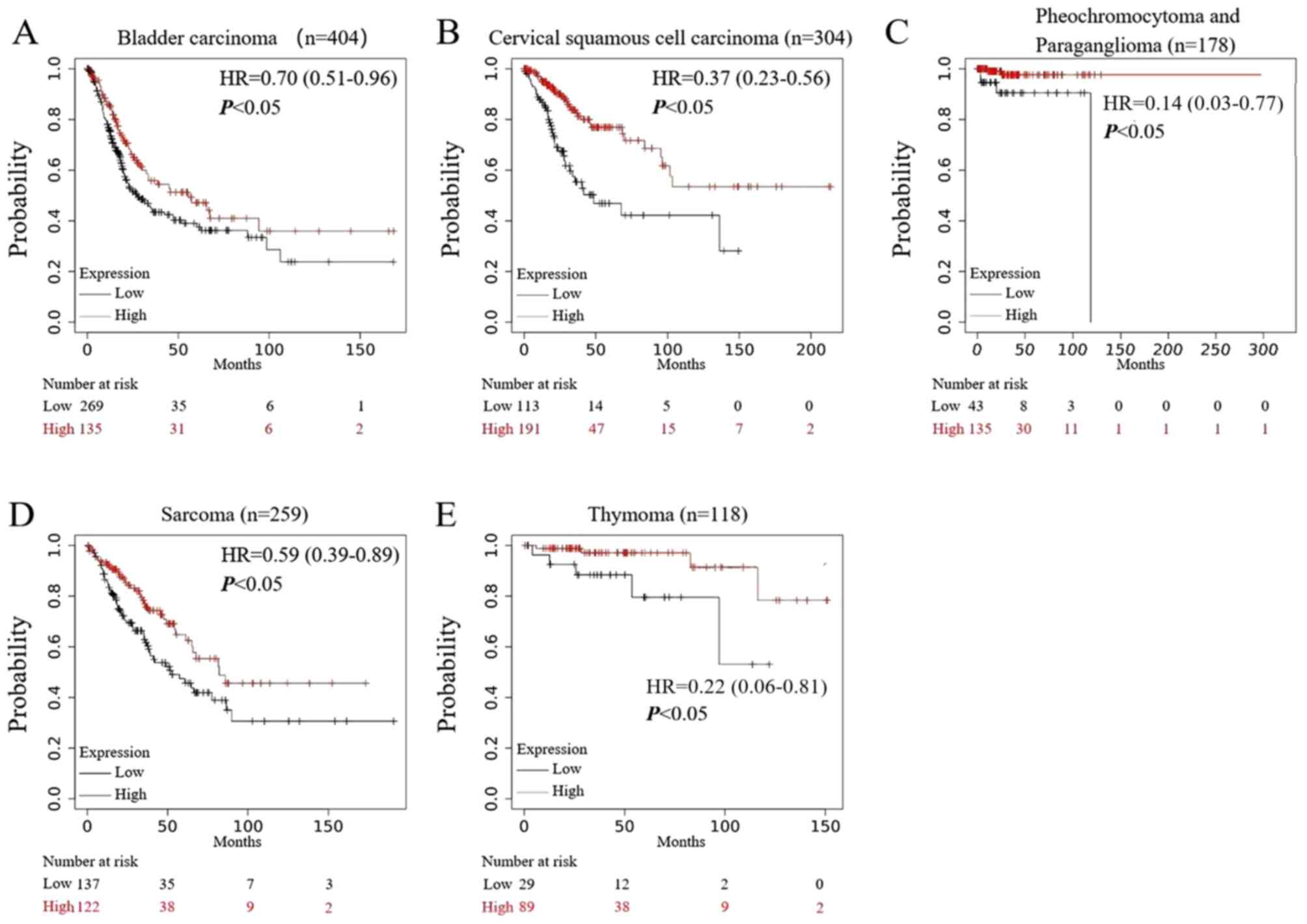 | Figure 7.Prognostic analysis of TMC8 in
multiple types of cancer. (A-E) In bladder carcinoma, cervical
squamous cell carcinoma, pheochromocytoma and paraganglioma,
sarcoma and thymoma, high expression of TMC8 was associated with
good prognosis (P=2.7×10−2, P=1.6×10−5,
P=8.2×10−3, P=0.01 and P=1.3×10−2,
respectively). TMC8, transmembrane channel-like 8; HR, hazard
ratio. |
Discussion
The present study analyzed the levels of TMC8
expression in HNSC and its prognostic association. The expression
of TMC8 was upregulated in patients with HNSC and the methylation
level in the promoter was significantly lower compared with that in
normal tissues, which may be the reason for the change in TMC8
expression. The increased expression level of TMC8 in HNSC was
further validated using tissue specimens. GSEA analysis was used to
clarify the function of TMC8 and the results showed that it was
involved in immune-associated pathways.
TMC8 expression was found to be significantly
positively correlated with the infiltration of CD4+ T
cells, CD8+ T cells, B cells, macrophages and DCs and
their respective subgroups. These results indicated that TMC8 may
enhance the infiltration of cytotoxic T cells and B lymphocytes by
affecting CD4+ T cells. Finally, the impact of TMC8 on
the prognosis of various malignancies was evaluated and the results
showed that TMC8 could significantly affect the OS of various types
of cancer, including HNSC. The findings of the present study showed
that TMC8, as one of the genes involved in HPV immune responses
(36) may play a very complex role
in the immune network and ultimately inhibit the development of
tumors.
The relationship between HPV and skin malignancies
has been found in patients with EV (37). These patients usually develop
invasive skin squamous cell carcinoma (38). In a previous study (38) germline mutations of two genes, EVER1
and EVER2, also known as TMC6 and TMC8, were found in patients with
EV. The inability of patients with EV to effectively clear HPV from
their own skin is mainly due to cell-mediated immunodeficiency
(39). Defects in the TMC8 gene have
been previously suggested to promote an environment favorable to
HPV replication in addition to the persistence of skin cancer-prone
lesions (23). A study investigating
a common polymorphism (rs7208422) in the TMC8 gene in patients with
skin cancer has found that the polymorphism is not only associated
with positive serological tests for skin HPV types, but also with
an increased risk of skin squamous cell carcinoma (40). In the present study, TMC8 was found
to be highly expressed in HNSC. At present, no study has been
conducted to detect the changes in the expression level of TMC8 in
HNSC, to the best knowledge of the authors. Two previous studies
have revealed that common single nucleotide polymorphisms in TMC8
increase the risk of HNSC (36,41). In
the current study, the TIMER algorithm showed a negative
correlation between TMC8 expression and tumor purity, suggesting
that TMC8 was expressed more frequently in tumor-infiltrating
lymphocytes and mesenchymal cells compared with in squamous cancer
cells. The mesenchymal cells around the tumor mainly include immune
cells, for example, macrophages, T cells and neutrophils (42). Previously published data indicate
that TMC8 is highly expressed in various types of hematopoietic
cells, including CD4+ and CD8+ T lymphocytes,
B cells and NK cells (25,26,43).
Therefore, the high expression of TMC8 in mesenchymal cells may
contribute to its upregulation in HNSC. To determine whether TMC8
is highly expressed in tumor cells, immunohistochemical staining
and detection of TMC8 expression in SCC cell lines using qPCR was
used in the present study. These results confirmed that
TMC8-upregulation was not only due to the enrichment of T cells in
tumor stroma, but also synchronously upregulated in tumor cells as
the SCC cell line tested by qPCR avoids the interstitial cells
contained in the tumor tissue.
The present study then explored the underlying
causes of the observed transcriptional changes and focused on the
frequency of methylation changes. The downregulation of gene
expression caused by hypermethylation of CpG islands in gene
promoter regions is a common gene silencing mechanism in epigenetic
regulation (44). A total of six
methylation sites of TMC8 located in the promoter region showed
significant hypomethylation in patients with HNSC, which was a
notable reason for TMC8-downregulation. The DNA modification of
different structural elements, such as the promoter, coding region
or distal enhancer region of the gene, as well as the combined
action of transcription factors and microRNA, constitute a complex
regulatory system for gene transcription (45,46).
Whether other epigenetic modifications, such as histone
deacetylation or chromatin remodeling, affects the expression or
function of TMC8 requires further study.
The relationship between TMC8 and lymphocytes is not
completely clear. Research by Lazarczyk et al (25) showed that TMC8 is also involved in
the maintenance of lymphocyte zinc homeostasis. Moreover, mutations
in the TMC8 gene result in an excess of zinc ions, which in turn
blocks the activation and proliferation of T cells. Crequer et
al (26) studied the lymphocytes
of three adult patients with EV and TMC8 mutations and found that
the number of CD4+ and CD8+ T cells and the
response to stimulation were normal. However, the number of memory
CD4+ T cells and effector memory CD8+ T cells
increased significantly. This finding suggests that patients with
TMC8 dysfunction have mild T cell dysfunction. The significant
positive correlation between TMC8 and T cells is due to its high
expression in T cells in the current study, which also shows that
it can be used as an effective indicator of T cell infiltration and
function. However, different subpopulations of lymphocytes exert
different effects (47,48), and whether the expression of TMC8 can
reflect the enrichment of specific subpopulations has not yet been
reported to the authors' knowledge. Therefore, the present study
further explored the role of TMC8 in immune network through
correlation analysis.
Various immune deconvolution methods (49) can be used to estimate the abundance
of immune infiltrates, including TIMER, CIBERSORT, quanTIseq,
xCell, MCPCounter and EPIC methods. To avoid bias, multiple
algorithms were used for cross-validation in the present study. The
results showed that different algorithms had almost consistent
results. The primary CD4+ T cells differentiate into Th1
cells, Th2 cells, Th7 cells, T reg cells and follicular helper T
cells under the action of different cytokines and transcription
factors. Among them, the body's antitumor and anti-virus effect is
dominated by Th1-mediated cellular immunity. IFN-γ secreted by Th1
can significantly promote the expression level of MHC class I
molecules in tumor cells, thereby activating the function of
specific CD8+ T lymphocytes. IFN-γ can also increase the
phagocytic activity of macrophages and inhibit the formation of
tumor blood vessels (47,50). Previous research also shows that the
Th1 cell immune response plays a very important role in controlling
and eliminating HPV infection and determines the outcome of HPV
infection (51,52). In the current study, the expression
of TMC8 and Th1 infiltration were significantly positively
correlated, suggesting that TMC8-upregulation is an important
reference indicator of Th1 cell function. The expression of TMC8 in
patients with HPV-positive HNSC was significantly upregulated
compared with that of patients who were HPV-negative, suggesting
that TMC8 may play the role of a HPV barrier through Th1 cells.
Tfh cells are key Th cells that can activate naive B
cells to differentiate into plasma cells to produce antibodies and
produce a humoral immune response (53). However, the direct effect of Tfh on
tumors has not yet been clarified. At present, Tfh is closely
associated with the etiology in angioimmunoblastic T-cell lymphoma
(AITL) (54), peripheral T-cell
lymphomas with follicular growth pattern (PTCL-F) (55) and B cell lymphoma 6 (BCL-6) (53). Th2 stimulates B cell proliferation by
secreting interleukin (IL)-4, which mediates the humoral immune
response (56). The current study
showed that there was a significant positive correlation between
TMC8 expression and Tfh, Th2 infiltration and B cell enrichment,
suggesting that TMC8 may further regulate B cell activation through
Tfh and Th2.
Th17 cells have opposite functions at different
stages of the tumor. In the microenvironment of solid tumors, it
exerts anticancer effects by secreting IL-17 (57,58).
However, as the tumor progresses, the infiltration of T reg cells
will induce Th1 to secrete a large amount of IL-10 and promote
tumor angiogenesis. The results of the present study indicated that
in advanced HNSC (grades G3 and G4), TMC8 expression was
upregulated, while TMC8 and Th17 were significantly co-expressed.
This is consistent with the change of Th17 function, suggesting
that TMC8 plays a complex role in the tumor microenvironment.
In addition to lymphocytes, TMC8 was also found to
be significantly positively correlated with infiltration of
mesenchymal cells, including macrophages and DCs. Macrophages
account for >50% of the tumor stroma (59). M1 and M2 macrophages play opposite
roles in the tumor microenvironment (60), but TMC8 was upregulated in both types
of macrophages, indicating that TMC8 is not involved in macrophage
polarization. To further clarify the possible function of TMC8
expression in immunity, previously reported gene markers were
compared. Co-expression of markers for TAMs, M1 phenotype and M2
phenotype with TMC8 remained consistent with the results of the
previous algorithm results. DC markers also showed a significant
correlation with TMC8 expression, indicating a close relationship
between TMC8 and DC penetration. The combination of DC and T cells
can secrete IL-12 and IL-18 to activate T cell proliferation,
induce CTL production and the Th1 type immune response, which is
conducive to tumor clearance (61).
The co-expression of TMC8 and marker genes of specific immune cells
further verified the results of the different immune
algorithms.
Tumors of different cancer types may share
underlying similarities (62). Thus,
pan-cancer analysis of large-scale datasets has the potential to
improve disease modeling by exploiting these similarities. In
addition to HNSC, the results of the Kaplan-Meier database also
showed that in bladder cancer, cervical squamous cell carcinoma,
pheochromocytoma and paraganglioma, thymoma and sarcoma,
TMC8-upregulation and improved OS time were significantly
correlated. It was hypothesized that TMC8 was responsible for the
resistance to HPV infection, thereby increasing cancer
susceptibility. In the present study, TMC8-overexpression was
associated with the improved prognosis of a variety of cancer
types; however, the mechanism of how these different types of
cancer are associated with HPV infection still needs further
confirmation. In pathogen-associated human malignancies, up to 35%
are caused by HPV and the carcinogenic potential of different HPV
species varies widely (63). It is
well known that cervical cancer and HNSC have also been shown to be
associated with HPV infection (64).
Studies have reported that mutations in TMC6 and TMC8 increases the
risk of developing cervical cancer from persistent HPV infection
(24,65). Liang et al (36) reported that the common genetic
variations in TMC8 are associated with the etiology of high-risk
HPV infection and HNSC. These results show that the TMC8 gene is a
key component of human keratinocyte HPV barrier that may affect the
biological behavior of cells through immune-mediated pathways and
ultimately affect disease prognosis. However, the opposite outcome
was observed in other cancer types. Yamada et al (66) suggested that TMC8 is one of the
numerous downstream genes of microRNA-144-5p/oncogenic syndecan-3
axes, which are associated with a poor prognosis of renal clear
cell carcinoma. In addition, Lu et al (67). Reported that a higher level of TMC8
expression is associated with a poorer prognosis for hepatocellular
carcinoma.
The present study had certain limitations. The
correlation strength between TMC8 and the infiltration level of
some immune cells was only weak to moderate. Further research,
including deep sequencing, is needed to identify the full spectrum
of variability and any functional variants of TMC8. It is also
necessary to further explore the specific molecular mechanism of
TMC8 in HNS carcinoma cells.
In summary, the present study reported that
variations in the level of TMC8 expression correlated with HNSC
prognosis. High levels of TMC8 expression were associated with
improved OS time, which suggested that TMC8 expression could
predict tumor prognosis. Furthermore, the findings demonstrated
that the extent of immune cell infiltration and the diversity of
immune marker expression were correlated with TMC8 expression in
HNSC. Therefore, these results provided insight into the potential
function of TMC8 in tumor immunology and its potential as a
biomarker for HNSC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by Shenzhen Healthcare
Research Project (grant no. SZLY2018022), the Sanming Project of
Medicine in Shenzhen (grant no. SZSM 201512036) and Shenzhen Fund
for Guangdong Provincial High-level Clinical Key Specialties (grant
no. SZGSP008).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. Additional datasets generated
and/or analyzed during the current study are available in the
following repositories: The Cancer Genome Atlas (https://portal.gdc.cancer.gov), Gene Expression
Profiling Interactive Analysis (http://gepia.cancer-pku.cn/index.html), Oncomine
database (https://www.oncomine.org/resource/login.html) Tumor
Immune Estimation Resource (https://cistrome.shinyapps.io/timer/), cBioPortal
(https://www.cbioportal.org) and
Kaplan-Meier plotter (http://kmplot.com/analysis/).
Authors' contributions
HYY and YHS designed the research and reviewed the
writing. BL analyzed the data and prepared the original draft. SJW
and YDY performed the statistical calculations and experiments. All
authors read and approved the manuscript. BL and YHS confirmed the
authenticity of all raw data.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Peking University Shenzhen Hospital (Shenzhen, China). Signed
informed consent were obtained from the patients and/or
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sanderson RJ and Ironside JA: Squamous
cell carcinomas of the head and neck. BMJ. 325:822–827. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chaturvedi AK, Anderson WF,
Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, Rosenberg PS,
Bray F and Gillison ML: Worldwide trends in incidence rates for
oral cavity and oropharyngeal cancers. J Clin Oncol. 31:4550–4559.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leemans CR, Snijders PJF and Brakenhoff
RH: The molecular landscape of head and neck cancer. Nat Rev
Cancer. 18:269–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oliva M, Spreafico A, Taberna M, Alemany
L, Coburn B, Mesia R and Siu LL: Immune biomarkers of response to
immune-checkpoint inhibitors in head and neck squamous cell
carcinoma. Ann Oncol. 30:57–67. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hira-Miyazawa M, Nakamura H, Hirai M,
Kobayashi Y, Kitahara H, Bou-Gharios G and Kawashiri S: Regulation
of programmed-death ligand in the human head and neck squamous cell
carcinoma microenvironment is mediated through matrix
metalloproteinase-mediated proteolytic cleavage. Int J Oncol.
52:379–388. 2018.PubMed/NCBI
|
|
7
|
Zhou C, Ye M, Ni S, Li Q, Ye D, Li J, Shen
Z and Deng H: DNA methylation biomarkers for head and neck squamous
cell carcinoma. Epigenetics. 13:398–409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allameh A, Moazeni-Roodi A, Harirchi I,
Ravanshad M, Motiee-Langroudi M, Garajei A, Hamidavi A and
Mesbah-Namin SA: Promoter DNA methylation and mRNA expression level
of p16 gene in oral squamous cell carcinoma: Correlation with
Clinicopathological Characteristics. Pathol Oncol Res.
25:1535–1543. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fomenkov A, Zangen R, Huang YP, Osada M,
Guo Z, Fomenkov T, Trink B, Sidransky D and Ratovitski EA: RACK1
and stratifin target DeltaNp63alpha for a proteasome degradation in
head and neck squamous cell carcinoma cells upon DNA damage. Cell
Cycle. 3:1285–1295. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mirza AH, Thomas G, Ottensmeier CH and
King EV: Importance of the immune system in head and neck cancer.
Head Neck. 41:2789–2800. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rossa C Jr and D'silva NJ: Immune-relevant
aspects of murine models of head and neck cancer. Oncogene.
38:3973–3988. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pardoll DM: The blockade of immune
checkpoints in cancer immunotherapy. Nat Rev Cancer. 12:252–264.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bardhan K, Anagnostou T and Boussiotis VA:
The PD1: PD-L1/2 pathway from discovery to clinical implementation.
Front Immunol. 7:5502016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wollenberg B: PD-1 antibodies in
head-and-neck cancer. Lancet. 393:108–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kansy BA, Concha-Benavente F, Srivastava
RM, Jie HB, Shayan G, Lei Y, Moskovitz J, Moy J, Li J, Brandau S,
et al: PD-1 Status in CD8+T cells associates with survival and
Anti-PD-1 therapeutic outcomes in head and neck cancer. Cancer Res.
77:6353–6364. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bauml J, Seiwert TY, Pfister DG, Worden F,
Liu SV, Gilbert J, Saba NF, Weiss J, Wirth L, Sukari A, et al:
Pembrolizumab for platinum- and cetuximab-refractory head and neck
cancer: Results from a single-arm, phase ii study. J Clin Oncol.
35:1542–1549. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pai SI, Zandberg DP and Strome SE: The
role of antagonists of the PD-1: PD-L1/PD-L2 axis in head and neck
cancer treatment. Oral Oncol. 61:152–158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
De Felice F, Tombolini M, Abate G, Salerno
F, Bulzonetti N, Tombolini V and Musio D: Prognostic significance
of the neutrophil/lymphocyte ratio in patients with non-human
papilloma virus-related oropharyngeal cancer: A retrospective
cohort study. Oncology. 96:8–13. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang SH, Waldron JN, Milosevic M, Shen X,
Ringash J, Su J, Tong L, Perez-Ordonez B, Weinreb I, Bayley AJ, et
al: Prognostic value of pretreatment circulating neutrophils,
monocytes, and lymphocytes in oropharyngeal cancer stratified by
human papillomavirus status. Cancer. 121:545–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chew EY, Hartman CM, Richardson PA,
Zevallos JP, Sikora AG, Kramer JR and Chiao EY: Risk factors for
oropharynx cancer in a cohort of HIV-infected veterans. Oral Oncol.
68:60–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Faraji F, Fung N, Zaidi M, Gourin CC,
Eisele DW, Rooper LM and Fakhry C: Tumor-infiltrating lymphocyte
quantification stratifies early-stage human papillomavirus
oropharynx cancer prognosis. Laryngoscope. 130:930–938. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Patel T, Morrison LK, Rady P and Tyring S:
Epidermodysplasia verruciformis and susceptibility to HPV. Dis
Markers. 29:199–206. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Madeleine MM, Carter JJ, Johnson LG, Wipf
GC, Davis C, Berg D, Nelson K, Daling JR, Schwartz SM and Galloway
DA: Risk of squamous cell skin cancer after organ transplant
associated with antibodies to cutaneous papillomaviruses,
polyomaviruses, and TMC6/8 (EVER1/2) variants. Cancer Med.
3:1440–1447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Castro FA, Ivansson EL, Schmitt M,
Juko-Pecirep I, Kjellberg L, Hildesheim A, Gyllensten UB and
Pawlita M: Contribution of TMC6 and TMC8 (EVER1 and EVER2) variants
to cervical cancer susceptibility. Int J Cancer. 130:349–355. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lazarczyk M, Dalard C, Hayder M, Dupre L,
Pignolet B, Majewski S, Vuillier F, Favre M and Liblau RS: EVER
proteins, key elements of the natural anti-human papillomavirus
barrier, are regulated upon T-cell activation. PLoS One.
7:e399952012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Crequer A, Picard C, Pedergnana V, Lim A,
Zhang SY, Abel L, Majewski S, Casanova JL, Jablonska S, Orth G and
Jouanguy E: EVER2 deficiency is associated with mild T-cell
abnormalities. J Clin Immunol. 33:14–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Colevas AD, Yom SS, Pfister DG, Spencer S,
Adelstein D, Adkins D, Brizel DM, Burtness B, Busse PM, Caudell JJ,
et al: NCCN Guidelines Insights: Head and neck cancers, version
1.2018. J Natl Compr Canc Netw. 16:479–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna: 2014
|
|
31
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen B, Khodadoust MS, Liu CL, Newman AM
and Alizadeh AA: Profiling tumor infiltrating immune cells with
CIBERSORT. Methods Mol Biol. 1711:243–259. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Aran D, Hu Z and Butte AJ: xCell:
Digitally portraying the tissue cellular heterogeneity landscape.
Genome Biol. 18:2202017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nagy A, Lanczky A, Menyhart O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang C, Kelsey KT, Mcclean MD,
Christensen BC, Marsit CJ, Karagas MR, Waterboer T, Pawlita M and
Nelson HH: A coding variant in TMC8 (EVER2) is associated with high
risk HPV infection and head and neck cancer risk. PLoS One.
10:e01237162015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Antonsson A, Law MH, Neale RE, Coman WB,
Pryor DI; Study of Digestive Health (SDH), ; Porceddu SV and
Whiteman DC: Variants of EVER1 and EVER2 (TMC6 and TMC8) and human
papillomavirus status in patients with mucosal squamous cell
carcinoma of the head and neck. Cancer Causes Control. 27:809–815.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jablonska S and Majewski S:
Epidermodysplasia verruciformis: immunological and clinical
aspects. Curr Top Microbiol Immunol. 186:157–175. 1994.PubMed/NCBI
|
|
39
|
Majewski S, Jablonska S and Orth G:
Epidermodysplasia verruciformis. Immunological and nonimmunological
surveillance mechanisms: role in tumor progression. Clin Dermatol.
15:321–334. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Patel AS, Karagas MR, Pawlita M, Waterboer
T and Nelson HH: Cutaneous human papillomavirus infection, the
EVER2 gene and incidence of squamous cell carcinoma: A case-control
study. Int J Cancer. 122:2377–2379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Antonsson A, Law MH, Neale RE, Coman WB,
Pryor DI; Study of Digestive Health (SDH), ; Porceddu SV and
Whiteman DC: Erratum to: Variants of EVER1 and EVER2 (TMC6 and
TMC8) and human papillomavirus status in patients with mucosal
squamous cell carcinoma of the head and neck. Cancer Causes
Control. 27:9512016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Henderson NC, Rieder F and Wynn TA:
Fibrosis: From mechanisms to medicines. Nature. 587:555–66. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Su AI, Wiltshire T, Batalov S, Lapp H,
Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al:
A gene atlas of the mouse and human protein-encoding
transcriptomes. Proc Natl Acad Sci USA. 101:6062–6067. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Morgan AE, Davies TJ and Mc Auley MT: The
role of DNA methylation in ageing and cancer. Proc Nutr Soc.
77:412–422. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chatterjee S and Ahituv N: Gene regulatory
elements, major drivers of human disease. Annu Rev Genomics Hum
Genet. 18:45–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y
and Ren J: Regulatory network of miRNA on its target: Coordination
between transcriptional and post-transcriptional regulation of gene
expression. Cell Mol Life Sci. 76:441–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Amaya-Uribe L, Rojas M, Azizi G, Anaya JM
and Gershwin ME: Primary immunodeficiency and autoimmunity: A
comprehensive review. J Autoimmun. 99:52–72. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Iwasaki A and Medzhitov R: Control of
adaptive immunity by the innate immune system. Nat Immunol.
16:343–353. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hammerbacher J and Snyder A: Informatics
for cancer immunotherapy. Ann Oncol. 28 (Suppl-12):xii56–xii73.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kumar BV, Connors TJ and Farber DL: Human
T cell development, localization, and function throughout life.
Immunity. 48:202–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Park H, Li Z, Yang XO, Chang SH, Nurieva
R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q and Dong C: A distinct
lineage of CD4 T cells regulates tissue inflammation by producing
interleukin 17. Nat Immunol. 6:1133–1141. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Scott M, Nakagawa M and Moscicki A: B.
Cell-mediated immune response to human papillomavirus infection.
Clin Diagn Lab Immunol. 8:209–20. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jogdand GM, Mohanty S and Devadas S:
Regulators of Tfh cell differentiation. Front Immunol. 7:5202016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Renand A, Milpied P, Rossignol J, Bruneau
J, Lemonnier F, Dussiot M, Coulon S and Hermine O: Neuropilin-1
expression characterizes T follicular helper (Tfh) cells activated
during B cell differentiation in human secondary lymphoid organs.
PLoS One. 8:e855892013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Huang Y, Moreau A, Dupuis J, Streubel B,
Petit B, Le Gouill S, Martin-Garcia N, Copie-Bergman C, Gaillard F,
Qubaja M, et al: Peripheral T-cell lymphomas with a follicular
growth pattern are derived from follicular helper T cells (TFH) and
may show overlapping features with angioimmunoblastic T-cell
lymphomas. Am J Surg Pathol. 33:682–690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Schmitt N, Bentebibel SE and Ueno H:
Phenotype and functions of memory Tfh cells in human blood. Trends
Immunol. 35:436–442. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yasuda K, Takeuchi Y and Hirota K: The
pathogenicity of Th17 cells in autoimmune diseases. Semin
Immunopathol. 41:283–297. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chang SH: T helper 17 (Th17) cells and
interleukin-17 (IL-17) in cancer. Arch Pharm Res. 42:549–559. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Goswami KK, Ghosh T, Ghosh S, Sarkar M,
Bose A and Baral R: Tumor promoting role of anti-tumor macrophages
in tumor microenvironment. Cell Immunol. 316:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Murray PJ: Macrophage Polarization. Annu
Rev Physiol. 79:541–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lefebvre JL, Sanes JR and Kay JN:
Development of dendritic form and function. Annu Rev Cell Dev Biol.
31:741–777. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cooper LA, Demicco EG, Saltz JH, Powell
RT, Rao A and Lazar AJ: PanCancer insights from the cancer genome
atlas: The pathologist's perspective. J Pathol. 244:512–524. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dunne EF and Park IU: HPV and
HPV-associated diseases. Infect Dis Clin North Am. 27:765–78. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Goodman A: HPV testing as a screen for
cervical cancer. BMJ. 350:h23722015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang SS, Gonzalez P, Yu K, Porras C, Li Q,
Safaeian M, Rodriguez AC, Sherman ME, Bratti C, Schiffman M, et al:
Common genetic variants and risk for HPV persistence and
progression to cervical cancer. PLoS One. 5:e86672010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yamada Y, Arai T, Kojima S, Sugawara S,
Kato M, Okato A, Yamazaki K, Naya Y, Ichikawa T and Seki N:
Regulation of antitumor miR-144-5p targets oncogenes: Direct
regulation of syndecan-3 and its clinical significance. Cancer Sci.
109:2919–2936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lu P, Ding Q, Ding S, Fan Y, Li X, Tian D
and Liu M: Transmembrane channel-like protein 8 as a potential
biomarker for poor prognosis of hepatocellular carcinoma. Mol Clin
Oncol. 7:244–248. 2017.PubMed/NCBI
|