Introduction
Cervical cancer, one of the most common
gynaecological tumours, is the second-most common cancer among
women and the fourth leading cause of tumour-associated deaths in
females worldwide (1). Statistics
indicate that over half a million new cervical cancer cases occur
every year worldwide, and 85% of these occur in developing and
underdeveloped countries (2).
Extensive cervical cancer screening has drastically decreased the
morbidity from cervical cancer in the past decade. However,
cervical cancer still substantially influences women's health, and
will remain an important public health issue for several decades
(3). Currently, the primary
treatments for cervical cancer are a radical hysterectomy with
pelvic lymph node dissection, chemotherapy, and radiotherapy
(4). To date, despite improvements
in the clinical outcomes of cervical cancer following first-line
treatments, its prognosis remains unsatisfactory (5). Furthermore, distant metastasis and
tumour recurrence make cervical cancer a highly malignant and
deadly human cancer (6). Therefore,
investigating the molecular mechanisms underlying cervical cancer
pathogenesis is urgently required in order to develop effective
targeted treatments.
The expression profiles and roles of long non-coding
RNAs (lncRNAs) in human cancers are a research focus in the
oncology field (7). lncRNAs are a
subcategory of transcripts that are >200 nucleotides in length
(8). They have no protein-coding
ability but participate in regulating gene expression at both the
transcriptional and post-transcriptional levels (9). lncRNAs have been extensively studied
and identified as novel modulators during carcinogenesis and cancer
progression (10–12). The aberrant expression of lncRNAs has
been widely discovered in human cancers, manifesting a close
correlation with aggressive biological events (13–15).
Several studies have demonstrated the crucial regulatory actions of
lncRNAs in modulating pathological conditions during cervical
carcinogenesis and cancer progression (16–18).
MicroRNAs (miRNAs/miRs) are another group of
single-stranded RNA molecules with 17–23 nucleotides. They lack
protein-coding ability (19) but
directly interact with the 3′-untranslated regions (3′-UTRs) of
their target genes and subsequently trigger mRNA degradation and/or
translational repression (20).
Recently, several miRNAs have been confirmed to be dysregulated and
perform important regulatory actions towards cervical cancer
oncogenesis by exerting tumour-inhibiting or tumour-facilitating
roles (21,22). Recently, a novel regulatory
mechanism, called the competing endogenous RNA (ceRNA) theory, was
uncovered, based on which lncRNAs can decoy certain miRNAs and thus
indirectly affect the miRNA's target genes (23). Therefore, therapies specifically
against lncRNAs or miRNAs may be promising therapeutic techniques
for cervical cancer.
A novel lncRNA, called USP30 antisense RNA 1
(USP30-AS1), has been studied in bladder urothelial carcinoma
(24). Utilizing The Cancer Genome
Atlas (TCGA) database, it was revealed that USP30-AS1 was one of
the most dysregulated lncRNAs in cervical squamous cell carcinoma
and endocervical adenocarcinoma (CESC), suggesting that USP30-AS1
may exert important roles during cervical cancer progression.
Additionally, the detailed role of USP30-AS1 in cervical cancer
remains unknown. Therefore, the present study aimed to determine
the potential functions of USP30-AS1 in cervical cancer and uncover
its underlying molecular events.
Materials and methods
Human samples
Tumour tissues and matched adjacent normal tissues
were obtained from 56 patients with cervical cancer (age range,
41–73 years) at The First People's Hospital of Chongqing Liangjiang
New Area (Chongqing, China). Patients with International Federation
of Gynaecology and Obstetrics stage (25) I–II and III–IV were 22 and 34,
respectively. Immediately after tissue excision, samples were
frozen in liquid nitrogen and stored in liquid nitrogen (−196°C)
until further use. The follow-up lasted for 60 months. Patients
that had received radiochemotherapy were excluded. Patients with
other types of human cancer were also excluded from the study. All
experimental procedures were approved by the Human Ethics Approval
Committee of The First People's Hospital of Chongqing Liangjiang
New Area (approval no. ECAC-TFHCQLJ.20150601). Furthermore, written
informed consent was obtained from all patients before the
study.
TCGA program
TCGA dataset of CESC (TCGA-CESC) (26) was downloaded from TGCA (https://tcga-data.nci.nih.gov/tcga/), and used in
the expression analysis of USP30-AS1 and miR-299-3p. The dataset
included 306 CESC tissues and 3 normal cervical tissues. In
addition, the clinicopathological features of these cohorts were
obtained, and the correlation between USP30-AS1/miR-299-3p
expression and clinicopathological features in CESC was also
assessed. The survival analysis of PTP4A1 in TCGA-CESC cohorts was
implemented using The Human Protein Atlas (https://www.proteinatlas.org/).
Cell culture
The normal human cervical epithelial cell line
Ect1/E6E7 was acquired from the American Type Culture Collection
(ATCC). It was cultured in keratinocyte serum-free medium (cat. no.
17005-042; Gibco; Thermo Fisher Scientific, Inc.) supplemented with
0.1 ng/ml human recombinant epidermal growth factor, 0.05 mg/ml
bovine pituitary extract, and 0.4 mM calcium chloride. The three
human cervical cancer cell lines, SiHa (ATCC), HeLa (ATCC) and
CaSki (Cell Bank of the Chinese Academy of Sciences), were cultured
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.).
Another cervical cancer cell line, C-33A (Cell Bank of the Chinese
Academy of Sciences), was maintained in minimum essential medium
(Gibco; Thermo Fisher Scientific, Inc.). All culture media were
supplemented with 10% foetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Gibco;
Thermo Fisher Scientific, Inc.). Cells were cultured at 37°C in
humidified air with 5% CO2.
Transfection experiments
Small interfering (si)RNAs against USP30-AS1
(si-USP30-AS1s) were designed to knockdown USP30-AS1 expression
using scrambled control (si-NC) as the control. All siRNAs were
constructed by Shanghai GenePharma Co., Ltd. The si-USP30-AS1
sequences were as follows: si-USP30-AS1#1,
5′-CTGCTATAATTAGTTATTATTGT-3′; si-USP30-AS1#2,
5′-AGCCAACAAACTTATTGTTTACT-3′; and si-USP30-AS1#3,
5′-TTCATTATTTACATTTAATATTC-3′. The si-NC sequence was
5′-CACGATAAGACAATGTATTT-3′. miRNA-299-3p (miR-299-3p) mimic, miRNA
scrambled control (miR-NC), miR-299-3p inhibitor (anti-miR-299-3p),
and miRNA inhibitor control (anti-miR-NC) were also synthesised by
Shanghai GenePharma Co., Ltd. The miR-299-3p mimics sequence was
5′-UUCGCCAAAUGGUAGGGUGUAU-3′ and the miR-NC sequence was
5′-UUGUACUACACAAAAGUACUG-3′. The anti-miR-299-3p inhibitor sequence
was 5′-AAGCGGUUUACCAUCCCACAUA-3′ and the anti-miR-NC sequence was
5′-ACUACUGAGUGACAGUAGA-3′. The protein tyrosine phosphatase type
IVA (PTP4A1)-overexpressing vector pcDNA3.1-PTP4A1 (pc-PTP4A1;
GeneChem Co., Ltd.) was produced by cloning PTP4A1 sequences into
pcDNA3.1. Cells were placed into 6-well plates and grown to 60–70%
confluency, followed by transfection with siRNA (100 pmol), miRNA
oligonucleotides (100 pmol) or plasmids (4 µg) using Lipofectamine™
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). All
transfections were performed at room temperature. Reverse
transcription-quantitative PCR (RT-qPCR), flow cytometric analysis,
and Transwell migration and invasion assays and western blotting
were executed at 48 h post-transfection. After 24 h culture at
37°C, Cell Counting kit-8 (CCK-8) assay was performed.
RT-qPCR
Total RNA was isolated from tissues or cultured
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). To quantify USP30-AS1 and PTP4A1 expression,
total RNA was reverse transcribed to complementary DNA (cDNA) using
the PrimeScript™ RT Reagent kit with gDNA Eraser (both from Takara
Biotechnology Co., Ltd.). The thermocycling conditions were as
follows: 37°C for 15 min, and 85°C for 5 sec. PCR amplification was
performed using TB Green® Premix Ex Taq™ II (Takara
Biotechnology Co., Ltd.). The thermocycling conditions were as
follows: 95°C for 30 sec, 95°C for 5 sec, 60°C for 30 sec, for 40
cycles. GAPDH functioned as an internal control for USP30-AS1 and
PTP4A1. To detect miR-299-3p expression, cDNA was obtained from
total RNA via RT using the miRcute miRNA First-Strand cDNA
Synthesis kit (Tiangen Biotech Co., Ltd.). The thermocycling
conditions were as follows: 42°C for 60 min, and 95°C for 3 min.
The miRcute miRNA qPCR Detection kit SYBR Green (Tiangen Biotech
Co., Ltd.) was used for qPCR. The thermocycling conditions were as
follows: 95°C for 15 min, 94°C for 20 sec, 60°C for 34 sec, for 45
cycles. The miRNA expression was normalized to that of U6 small
nuclear RNA. The relative gene expression was calculated using the
2−ΔΔCq method (27). The
following primer sequences were used: USP30-AS1 forward,
5′-GTCTCCCCAGGTCTGTGCTTAA-3′ and reverse,
5′-GTATTTTTTCCTTATGCTGCCAAA-3′; PTP4A1 forward,
5′-ACCAATGCGACCTTAAACAAATT-3′ and reverse,
5′-CTTCTTTCTCCACAAGAGTAGTGTCAT-3′; GAPDH forward,
5′-ACCTGACCTGCCGTCTAGAAAA-3′ and reverse,
5′-TTGAAGTCAGAGGAGACCACCTG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; miR-299-3p forward,
5′-TCGGCAGGUUCGCCAAAUG-3′ and reverse, 5′-CACTCAACTGGTGTCGTGGA-3′;
miR-127-5p forward, 5′-TCGGCAGGCUGAAGCUCAG-3′ and reverse,
5′-CACTCAACTGGTGTCGTGGA-3′; miR-186-3p forward,
5′-TCGGCAGGGGGUUUUUUAAGU-3′ and reverse,
5′-CACTCAACTGGTGTCGTGGA-3′; miR-204-5p forward,
5′-TCGGCAGGUUCCCUUUGUCA-3′ and reverse, 5′-CACTCAACTGGTGTCGTGGA-3′;
miR-379-3p forward, 5′-TCGGCAGGUCAAUCACCUGG-3′ and reverse,
5′-CACTCAACTGGTGTCGTGGA-3′; and miR-411-3p forward,
5′-TCGGCAGGCCAAUCACCUGG-3′ and reverse,
5′-CACTCAACTGGTGTCGTGGA-3′.
Subcellular fractionation
Cervical cancer cells in the logarithmic growth
phase were digested with 0.25% (w/v) trypsin and centrifuged at
1,000 × g for 5 min. The cytoplasmic and nuclear fractions were
separated using the Cytoplasmic and Nuclear RNA Purification kit
(Norgen Biotek Corp.). The relative abundances of USP30-AS1, GAPDH
and U6 in both fractions were determined using RT-qPCR. GAPDH and
U6 were used as the cytoplasmic and nuclear controls,
respectively.
CCK-8 assay
Infected cervical cancer cells were harvested and
used to prepare a single-cell suspension. Each well of the 96-well
plates was covered with 100 µl cell suspension containing 2,000
cells. The CCK-8 assay was performed at 0, 1, 2 and 3 days after
incubation at 37°C with 5% CO2. A 10-µl volume of CCK-8
reagent (Beyotime Institute of Biotechnology) was directly added to
each well. After 2 h, the optical density was measured at 450 nm
(OD450) using a microplate spectrophotometer.
Flow cytometric analysis of cell
apoptosis
Cervical cancer cells transfected with the
aforementioned molecular products were cultivated at 37°C with 5%
CO2 for 48 h before cell apoptosis detection using the
Annexin V-FITC Apoptosis Detection kit (Beyotime Institute of
Biotechnology). Cells were digested with 0.25% (w/v) trypsin,
centrifuged in 1,000 × g at room temperature for 5 min and rinsed
with ice-cold phosphate-buffered saline, followed by staining with
5 µl of Annexin V-FITC and 10 µl of propidium iodide. After 20 min
of incubation at 20–25°C in the dark, apoptotic cells were
identified using a flow cytometer (BD Biosciences).
Transwell migration and invasion
assays
For the migration assay, a single-cell suspension
was prepared from infected cervical cancer cells using serum-free
culture medium. The apical chambers of 24-well
Transwell® inserts (pore size, 8 µm; BD Biosciences)
were filled with 200 µl cell suspension containing 5×104
cells, whereas 600 µl of 20% FBS-containing culture medium was
added into the lower chamber. After 24 h culture at 37°C, the
inserts were immersed in 4% paraformaldehyde at room temperature
for 30 min and washed twice with phosphate-buffered saline. Crystal
violet staining was performed at room temperature for 30 min. After
extensive washing, the non-migrated cells were cleaned from the
upper face of the inserts with a cotton swab, whereas migrated
cells were imaged and counted in five visual fields using a light
microscope (Olympus Corporation; ×200 magnification). The cell
invasion assay was performed in a similar manner, with the
exception that the inserts were precoated with Matrigel (BD
Biosciences).
Mouse tumour model
Short hairpin RNA (shRNA) sequences targeting
USP30-AS1 and negative control shRNA (sh-NC) designed and
synthesized by Shanghai GenePharma Co., Ltd., were inserted into a
lentiviral vector. The vectors alongside psAX2 and pMD2G vectors
were injected into 293T cells (Cell Bank of the Chinese Academy of
Sciences). The lentiviral supernatant was collected after 48 h of
cultivation and used to infect HeLa cells. Finally, the stably
transfected cells were filtered using puromycin (Sigma-Aldrich;
Merck KGaA).
A total of six female BALB/c nude mice (20 g; age,
4–6 weeks) were purchased from HFK Bioscience (http://www.hfkbio.com). The animals were housed under
specific pathogen-free conditions at 25°C and 50% humidity, with a
10:14 light/dark cycle and ad libitum access to food and
water. The flank of mice were subcutaneously injected with
2×106 HeLa cells stably transfected with sh-USP30-AS1 or
sh-NC. The condition of the mice was monitored once every 2 days,
and the width and length of the subcutaneous xenografts were
measured weekly. The recorded data were then used to determine the
tumour volume using the following formula: Volume=1/2× length ×
width2. At 4 weeks after cell injection, all mice were
euthanized by means of cervical dislocation. Solid tumours were
excised and weighed. Protocols involving animals were approved by
the Ethical Guidelines for Animal Care and Use Committee of The
First People's Hospital of Chongqing Liangjiang New Area
(Chongqing, China; approval no. EGACUC-TFHCQLJ.20181205). The
humane endpoints used in the present study included: A tumour
diameter >15 mm, tumour ulceration, abnormal feeding, weight
loss or presence of cachexia.
Bioinformatics analysis
The potential miRNA targets of USP30-AS1 were
predicted using miRDB (http://mirdb.org/). The binding interaction between
PTP4A1 and miR-299-3p was predicted using TargetScan (http://www.targetscan.org/vert_60/) and
miRDB.
Dual-luciferase reporter assay
USP30-AS1 sequences containing the wild-type (WT) or
mutant (MUT) binding sites for miR-299-3p were constructed by
Shanghai GenePharma Co., Ltd., and cloned into the pmirGLO
luciferase vector (Promega Corporation) to obtain WT-USP30-AS1 and
MUT-USP30-AS1 reporter vectors. The recombinant vectors, WT-PTP4A1
and MUT-PTP4A1, were designed and produced using the same method.
The resulting vectors together with miR-299-3p mimic or miR-NC were
transfected into cervical cancer cells seeded into 6-well plates at
a density of 6×105 cells/well. Transfection experiments
were performed using Lipofectamine 2000. After 48 h of incubation,
the injected cells were collected and measured using the
Dual-luciferase Reporter Assay kit (Promega Corporation).
RNA immunoprecipitation (RIP)
Cervical cancer cells were harvested and treated
using the EZ-Magna RIP RNA-binding Protein Immunoprecipitation kit
(cat. no. 03-110; EMD Millipore), which was performed according to
manufacturer's protocol. Cells were lysed in RIP cell lysis buffer
that came from the kit, and the supernatant was collected after
centrifugation at 1,000 × g and 4°C for 10 min. The cell extract
(100 µl) was cultured, and magnetic beads conjugated with human
anti-AGO2 antibody (5 µl) or normal mouse IgG (5 µl) (both from
cat. no. 03-110; EMD Millipore) were added. Cells were incubated
for 12 h and kept at 4°C. Finally, magnetic beads were collected
and treated with proteinase K to remove proteins. The relative
enrichments of USP30-AS1, miR-299-3p and PTP4A1 in
immunoprecipitated RNA were quantified via RT-qPCR.
Western blotting
The total protein was extracted from cultured cells
by cultivating the cultured cells with RIPA lysis buffer (Beyotime
Institute of Biotechnology). An enhanced BCA Protein Assay kit
(Beyotime Institute of Biotechnology) was used to measure protein
concentrations. A total of 30 µg protein were separated via
SDS-PAGE (10% gel). The separated proteins were then transferred
onto polyvinylidene difluoride membranes and blocked with 5%
non-fat dried milk at room temperature for 2 h. After overnight
incubation at 4°C with primary antibodies targeting PTP4A1 (1:1,000
dilution; cat. no. ab121185) or GAPDH (1:1,000 dilution; cat. no.
ab128915) (both from Abcam), horseradish peroxidase
(HRP)-conjugated IgG secondary antibody (1:5,000 dilution; cat. no.
ab205718; Abcam) was added, and cultivated at room temperature for
1 h. Ultimately, target protein development was achieved using
Immobilon® ECL Ultra Western HRP Substrate (EMD
Millipore). The densitometry was implemented utilizing Quantity One
software (4.62 version; Bio Rad Laboratories, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS 19.0
software (SPSS, Inc.). All experiments were performed in triplicate
and data are presented as the mean ± standard error. Paired
Student's t-test was used to compare the levels of genes
(USP30-AS1, miR-299-3p and PTP4A1) between cervical cancer tissues
and normal tissues, while unpaired Student's t-test was used to
compare all other differences between two groups. One-way analysis
of variance followed by Tukey's post hoc test was used to compare
differences between multiple groups. The gene correlation analysis
was performed using Pearson's correlation analysis. Survival curves
were generated using the Kaplan-Meier method and compared using the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
USP30-AS1 depletion inhibits cervical
cancer cell proliferation, migration and invasion, and promotes
cell apoptosis in vitro
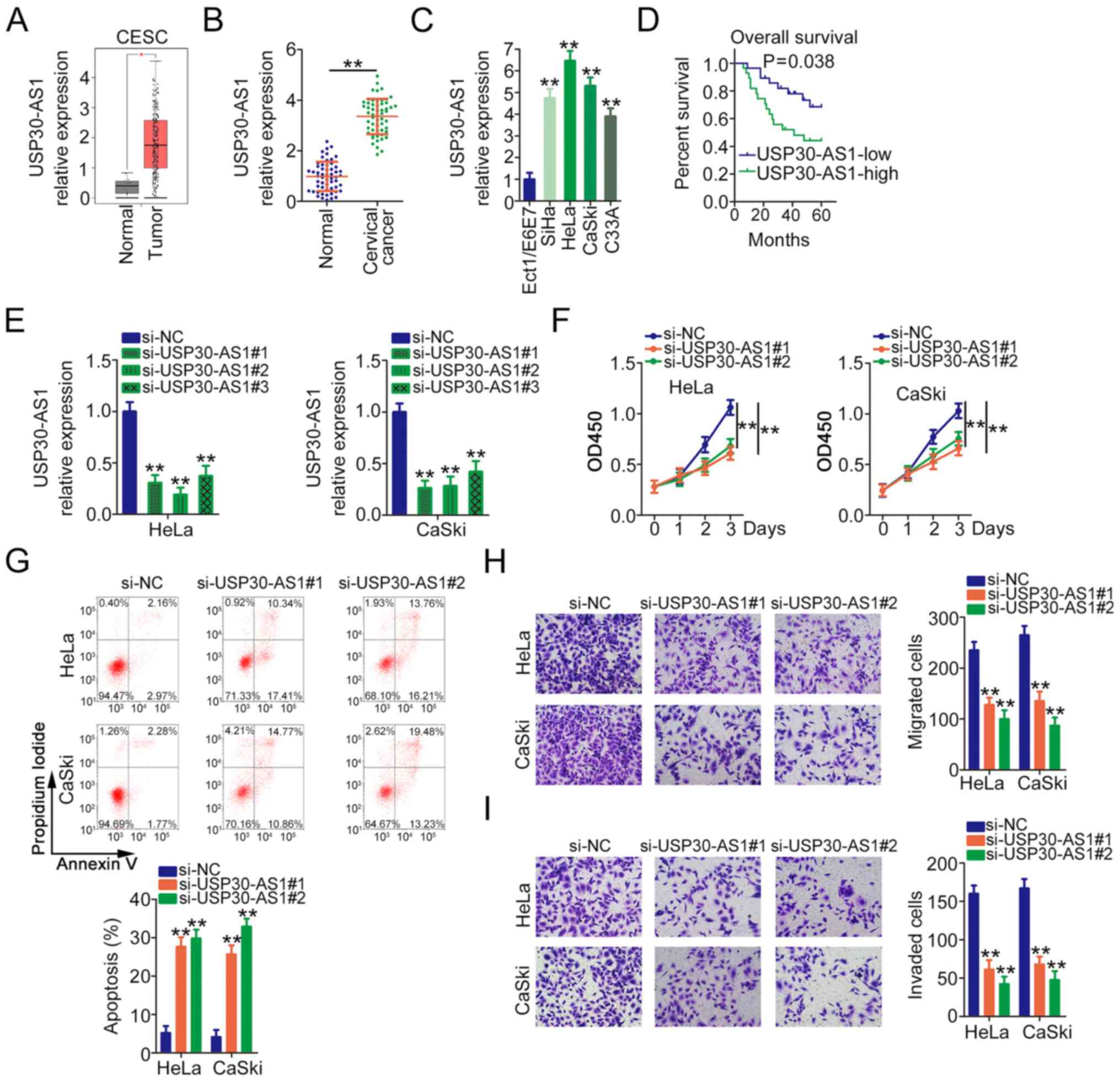
First, USP30-AS1 expression in CESC was determined
using TCGA database. A marked increase in USP30-AS1 expression was
identified in CESC tissues when compared with normal tissues
(Fig. 1A). Also, the correlation
between USP30-AS1 expression and clinicopathological features in
CESC was analysed employing TCGA database. The results are
presented in Fig. S1A and B. Then,
56 pairs of cervical cancer tissue and adjacent normal tissues were
collected to determine USP30-AS1 expression. USP30-AS1 was
upregulated in cervical cancer tissues when compared with matched
normal tissues (Fig. 1B).
Furthermore, compared with Ect1/E6E7 cells, USP30-AS1 levels were
increased in the tested cervical cancer cell lines (Fig. 1C). Based on the median value of
USP30-AS1, all patients with cervical cancer were classified into
two groups: USP30-AS1-high (n=28) and USP30-AS1-low (n=28)
expression groups. The Kaplan-Meier method alongside the log-rank
test was applied for the survival analysis. Patients with high
USP30-AS1 expression levels had shorter overall survival times than
those with low USP30-AS1 expression (Fig. 1D).
Among the four cervical cancer cell lines, HeLa and
CaSki cells exhibited the highest USP30-AS1 expression levels, as
evidenced by RT-qPCR. Therefore, these two cell lines were selected
for subsequent functional experiments. To address the effects of
USP30-AS1 in cervical cancer, USP30-AS1-targeting siRNAs were
synthesized to knock down endogenous USP30-AS1 expression, with
si-NC as the control. Fig. 1E
depicts the efficiency of siRNAs in decreasing USP30-AS1
expression. There were two siRNAs, si-USP30-AS1#1 and
si-USP30-AS1#2, used to avoid off-target effects. USP30-AS1
knockdown significantly inhibited HeLa and CaSki cell proliferation
(Fig. 1F). Furthermore, a notable
increase in cell apoptosis was observed in USP30-AS1-deficient HeLa
and CaSki cells (Fig. 1G). Transwell
migration and invasion assays revealed that the absence of
USP30-AS1 hindered the migratory (Fig.
1H) and invasive (Fig. 1I)
abilities of HeLa and CaSki cells. Taken together, these results
suggest that USP30-AS1 functions as a carcinogenic lncRNA in
cervical cancer.
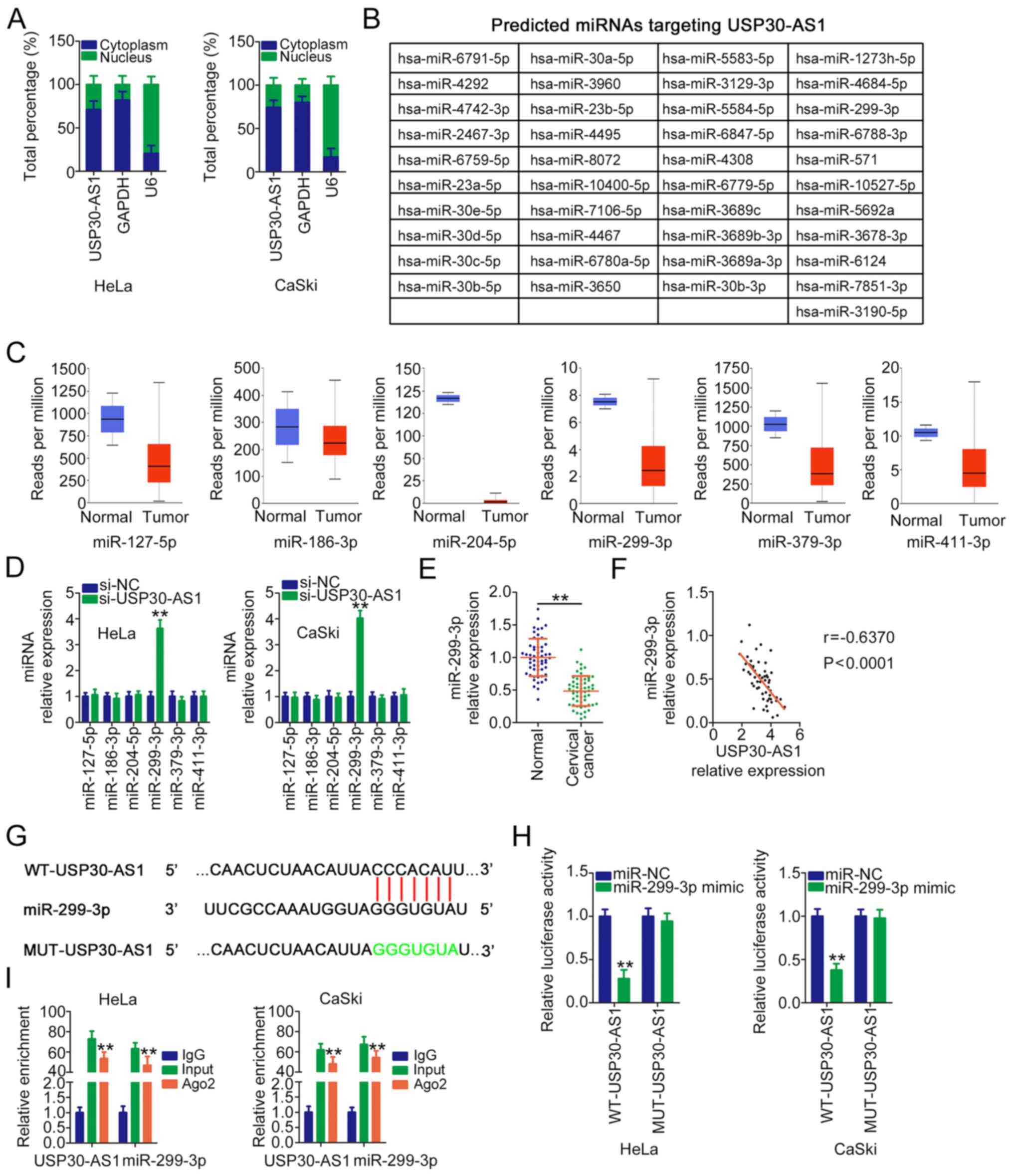
USP30-AS1 directly binds to miR-299-3p
and functions as a miR-299-3p sponge in cervical cancer
In order to investigate the carcinogenic mechanism
of USP30-AS1 in cervical cancer, the subcellular distribution of
USP30-AS1 was examined via a subcellular fractionation assay.
USP30-AS1 was mostly localized in the cytoplasm of HeLa and CaSki
cells (Fig. 2A). Several studies
have reported that cytoplasmic lncRNAs can function as molecular
sponges for certain miRNAs and further regulate gene expression. By
searching the miRDB, a total of 41 miRNAs (Fig. 2B) were predicted as potential
downstream targets of USP30-AS1. These miRNAs were analysed using
TCGA database to screen miRNAs that are expressed at low levels in
cervical cancer. A total of six miRNAs (miR-127-5p, miR-186-3p,
miR-204-5p, miR-299-3p, miR-379-3p and miR-411-3p) were
downregulated in CESC (Fig. 2C);
therefore, they were selected for further investigation. Next,
RT-qPCR was performed to measure the expression levels of these
candidate miRNAs in cervical cancer cells following USP30-AS1
silencing. Transfection with si-USP30-AS1 resulted in markedly high
expression levels of miR-299-3p in HeLa and CaSki cells (Fig. 2D). Furthermore, miR-299-3p was
expressed at low levels in cervical cancer tissues (Fig. 2E) and exhibited an inverse expression
correlation with USP30-AS1 (Fig.
2F). Furthermore, the correlation between miR-299-3p expression
and clinicopathological features in CESC was analysed via TCGA
database. The results are presented in Fig. S1C-E. Low miR-299-3p expression was
significantly associated with tumour stage in patients with
CESC.
Luciferase reporter and RIP assays were performed to
verify the functional interaction between USP30-AS1 and miR-299-3p
(Fig. 2G). The luciferase reporter
assay revealed that the luciferase activity of WT-USP30-AS1 was
alleviated by the miR-299-3p mimic. However, exogenous miR-299-3p
expression did not affect MUT-USP30-AS1 activity (Fig. 2H). Furthermore, the RIP assay
revealed that both USP30-AS1 and miR-299-3p had a tendency to be
present in Ago2-rich magnetic beads (Fig. 2I). Taken together, the results
suggest that USP30-AS1 is an endogenous miR-299-3p sponge in
cervical cancer.
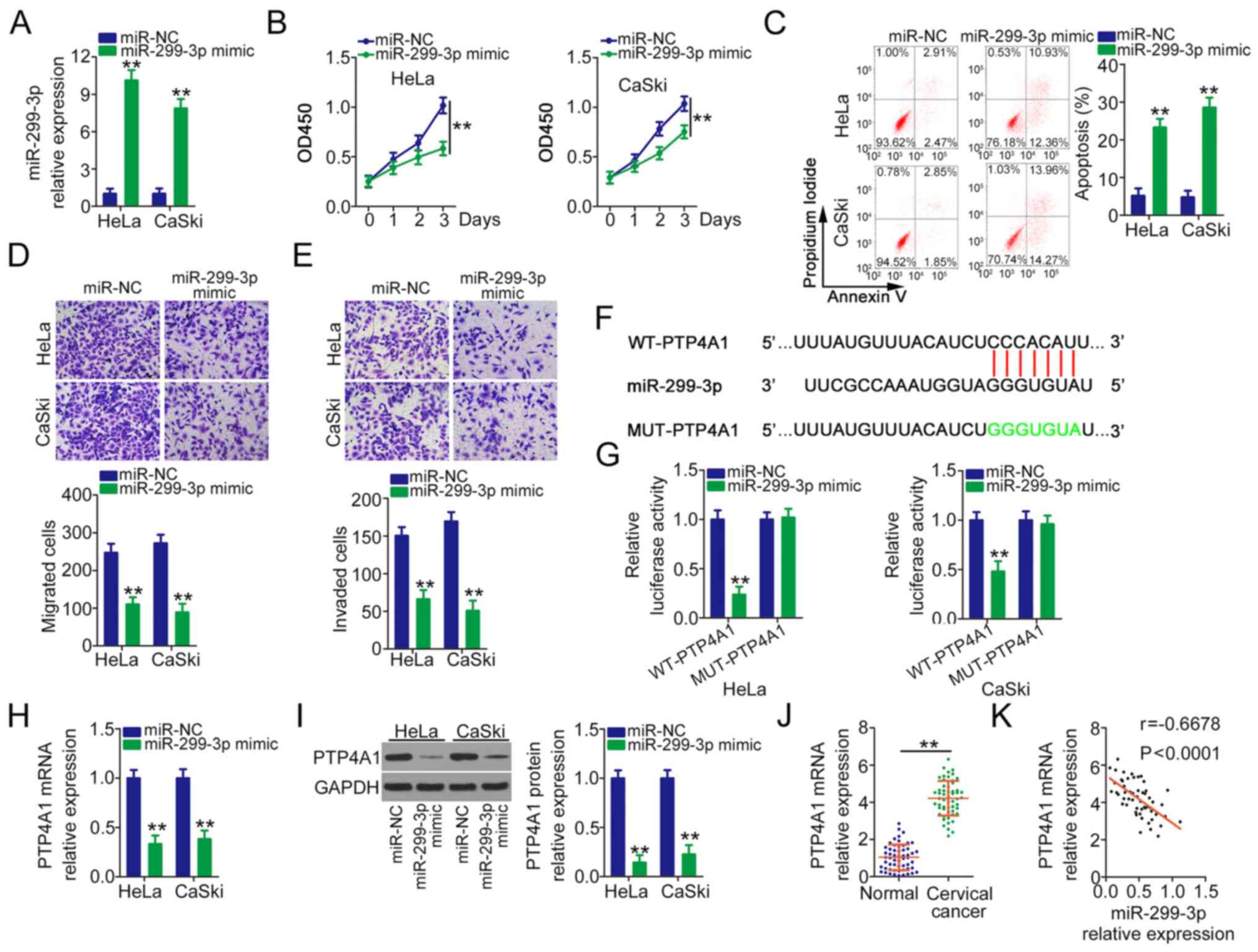
miR-299-3p plays an anti-oncogenic
role and directly targets PTP4A1 in cervical cancer
In order to detect the role of miR-299-3p in
cervical cancer, miR-299-3p was overexpressed in HeLa and CaSki
cells following transfection with miR-299-3p mimic (Fig. 3A). Overexpressed miR-299-3p evidently
lowered the proliferation ability (Fig.
3B) and facilitated the apoptosis (Fig. 3C) of HeLa and CaSki cells.
Furthermore, the migration (Fig. 3D)
and invasion (Fig. 3E) of HeLa and
CaSki cells were obviously impaired after miR-299-3p mimic
injection. Bioinformatics prediction indicated a targeting
relationship between miR-299-3p and PTP4A1 (Fig. 3F). Based on the binding sequences, WT
and MUT luciferase reporter vectors were designed, synthesized and
cotransfected with miR-299-3p mimic or miR-NC into HeLa and CaSki
cells. The reinforced expression of miR-299-3p considerably lowered
WT-PTP4A1 activity, whereas miR-299-3p mimic transfection did not
inhibit the luciferase activity of PTP4A1 when the binding
sequences were mutated (Fig. 3G).
Furthermore, PTP4A1 mRNA (Fig. 3H)
and protein (Fig. 3I) expression
levels were downregulated by miR-299-3p upregulation in HeLa and
CaSki cells. In addition, PTP4A1 overexpression in cervical cancer
tissues (Fig. 3J) was negatively
correlated with miR-299-3p expression (Fig. 3K). Through TCGA database, it was
revealed that high expression of PTP4A1 shows worse prognosis in
cervical cancer in TCGA database (Fig.
S1F). In general, miR-299-3p directly targets PTP4A1 and
suppresses malignant behaviour in cervical cancer.
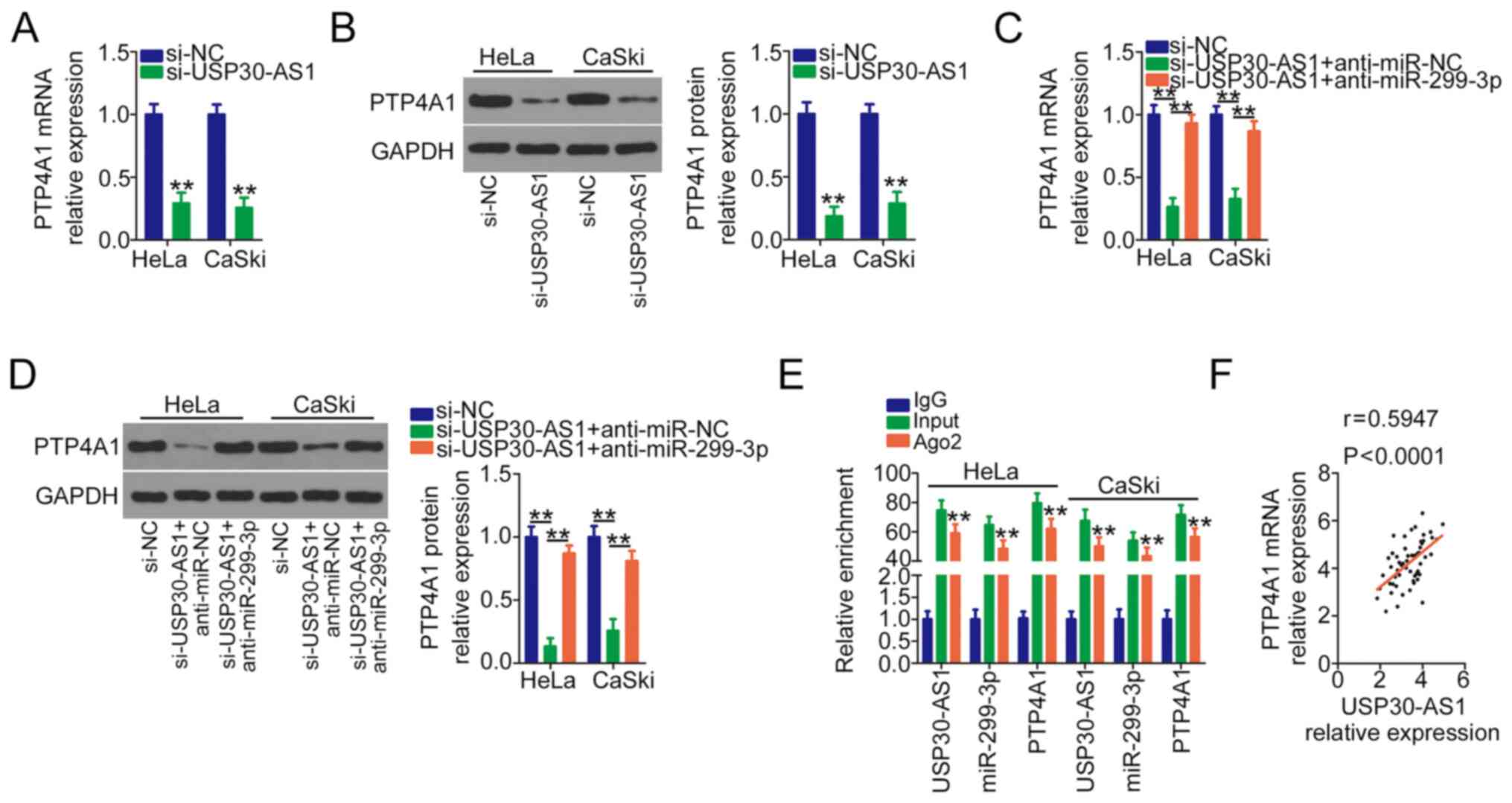
USP30-AS1 competitively sponges
miR-299-3p to positively regulate PTP4A1 expression in cervical
cancer
The present study attempted to further understand
the relationship among USP30-AS1, miR-299-3p and PTP4A1 in cervical
cancer. HeLa and CaSki cells were transfected with si-USP30-AS1
alone or cotransfected with anti-miR-299-3p or anti-miR-NC. PTP4A1
mRNA and protein levels were assayed by performing RT-qPCR and
western blotting, respectively. USP30-AS1 interference markedly
decreased PTP4A1 expression at both the mRNA (Fig. 4A) or protein (Fig. 4B) levels, whereas anti-miR-299-3p
cotransfection reversed its regulatory actions on PTP4A1 expression
(Fig. 4C and D). Furthermore, a RIP
assay was performed to assess the potentially endogenous
interaction among USP30-AS1, miR-299-3p and PTP4A1. The results
revealed that all three molecules were substantially enriched by
the anti-Ago2 antibody (Fig. 4E).
Furthermore, a positive correlation was observed between USP30-AS1
and PTP4A1 expression in cervical cancer tissues (Fig. 4F). In short, USP30-AS1 functions as a
ceRNA for miR-299-3p and thereby upregulates PTP4A1 in cervical
cancer.
Increased output of miR-299-3p/PTP4A1
neutralises the actions of si-USP30-AS1 on cervical cancer
cells
As the present study demonstrated that USP30-AS1
functions as a ceRNA to positively regulate PTP4A1 expression by
sequestering miR-299-3p in cervical cancer, rescue experiments were
performed to understand whether the tumour-promoting roles of
USP30-AS1 were mediated by the miR-299-3p/PTP4A1 axis. Initially,
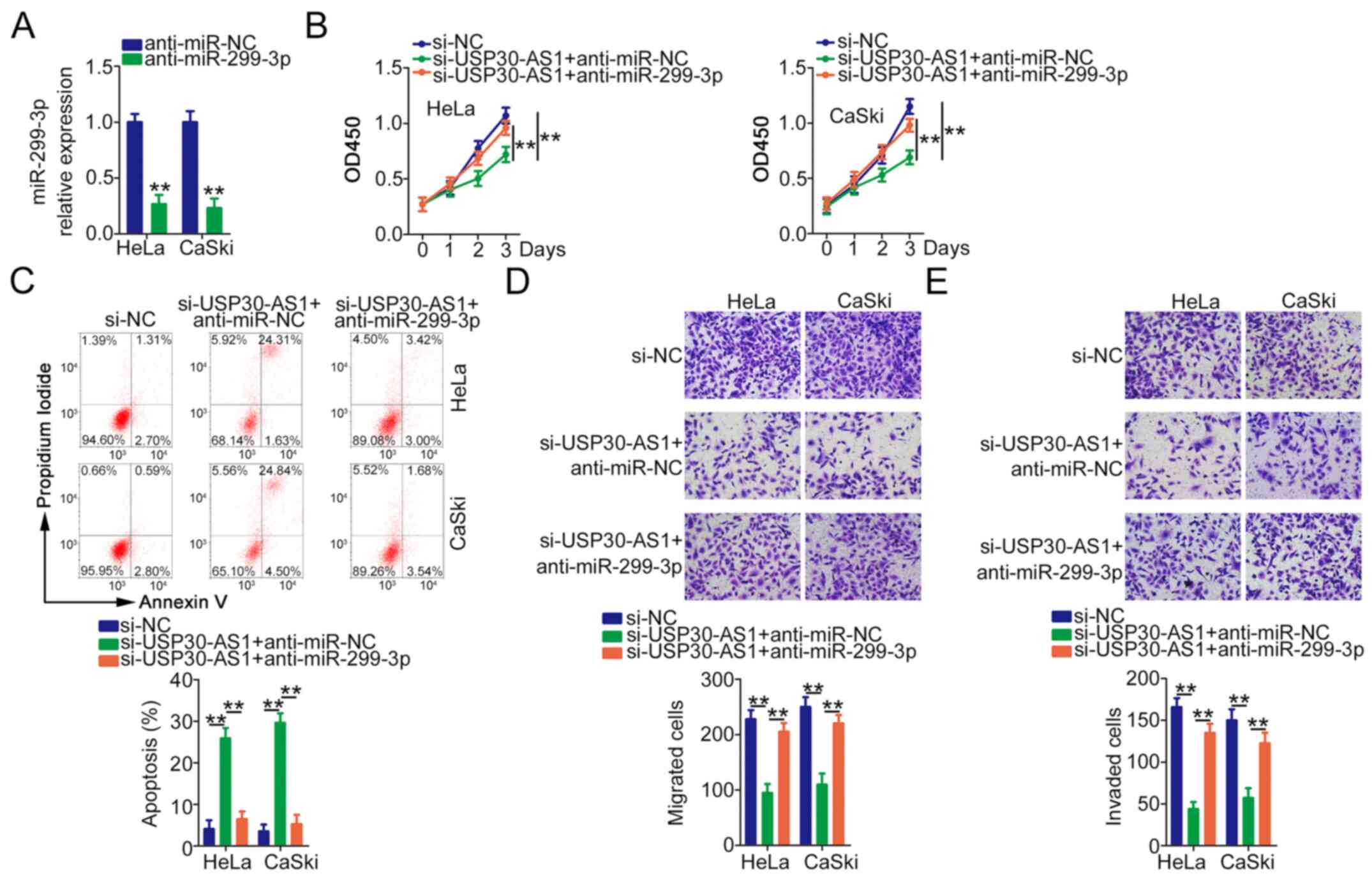
RT-qPCR verified the efficiency of anti-miR-299-3p in decreasing
miR-299-3p expression (Fig. 5A).
HeLa and CaSki cells were cotransfected with si-USP30-AS1 alongside
anti-miR-299-3p or anti-miR-NC, followed by functional experiments.
miR-299-3p downregulation abrogated the antiproliferative (Fig. 5B) and proapoptotic (Fig. 5C) effects of si-USP30-AS1 in HeLa and
CaSki cells. Similarly, anti-miR-299-3p markedly rescued the
migration (Fig. 5D) and invasion
(Fig. 5E) abilities of HeLa and
CaSki cells hindered by USP30-AS1 deficiency.
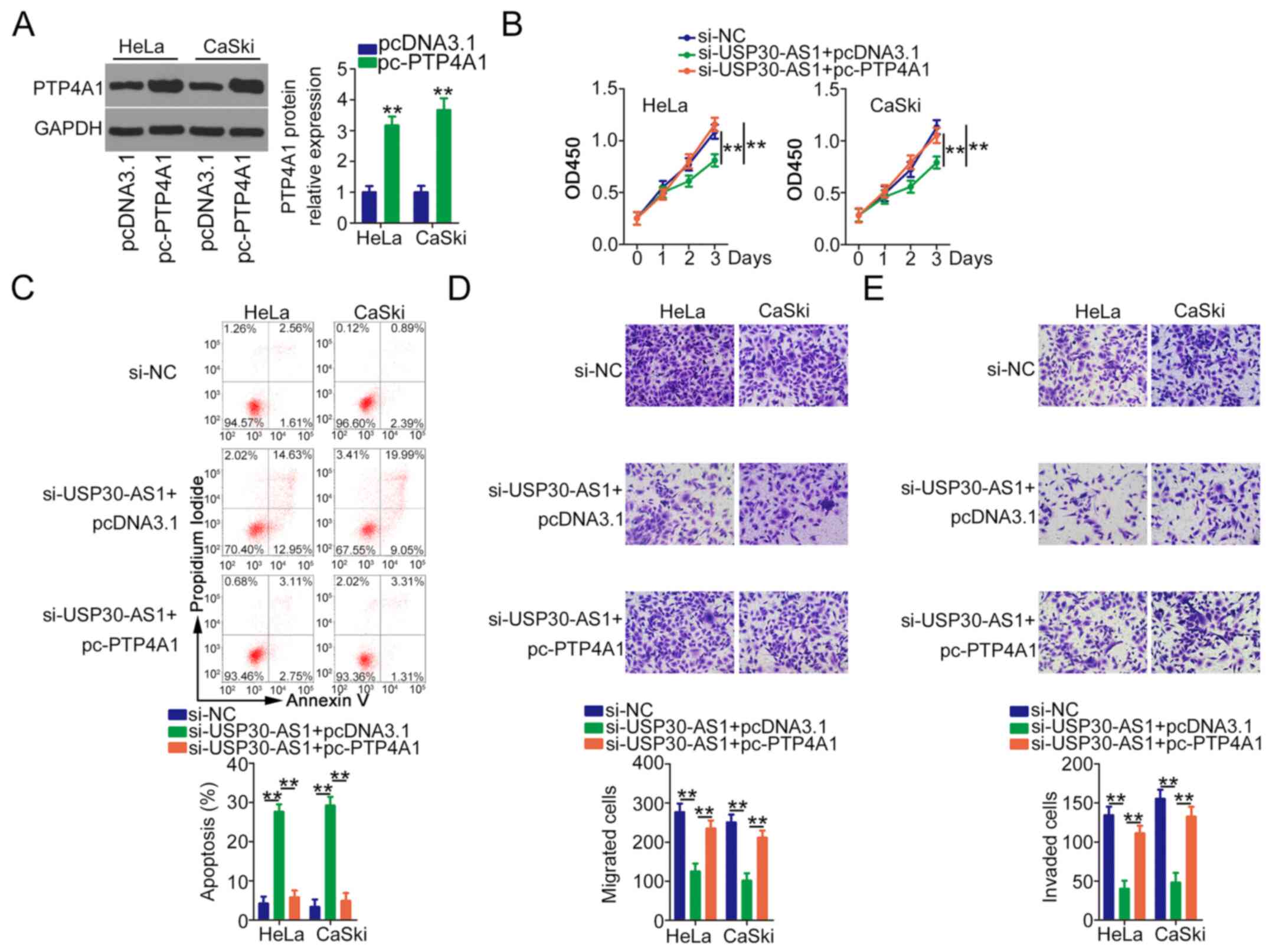
Western blotting revealed that transfection of
pc-PTP4A1 induced PTP4A1 overexpression in HeLa and CaSki cells
(Fig. 6A). pc-PTP4A1 or pcDNA3.1
along with si-USP30-AS1 was transfected into HeLa and CaSki cells.
In HeLa and CaSki cells, USP30-AS1 loss suppressed cell
proliferation (Fig. 6B) and
increased cell apoptosis (Fig. 6C),
whereas PTP4A1 overexpression abolished these effects. Furthermore,
cotransfection with pc-PTP4A1 reversed the inhibition of the
migration (Fig. 6D) and invasion
(Fig. 6E) of HeLa and CaSki cells by
si-USP30-AS1. Taken together, these results suggest that the ceRNA
mechanism-based USP30-AS1/miR-299-3p/PTP4A1 network can promote
cervical cancer cell malignant potential.
Depletion of USP30-AS1 restrains
tumour growth in vivo
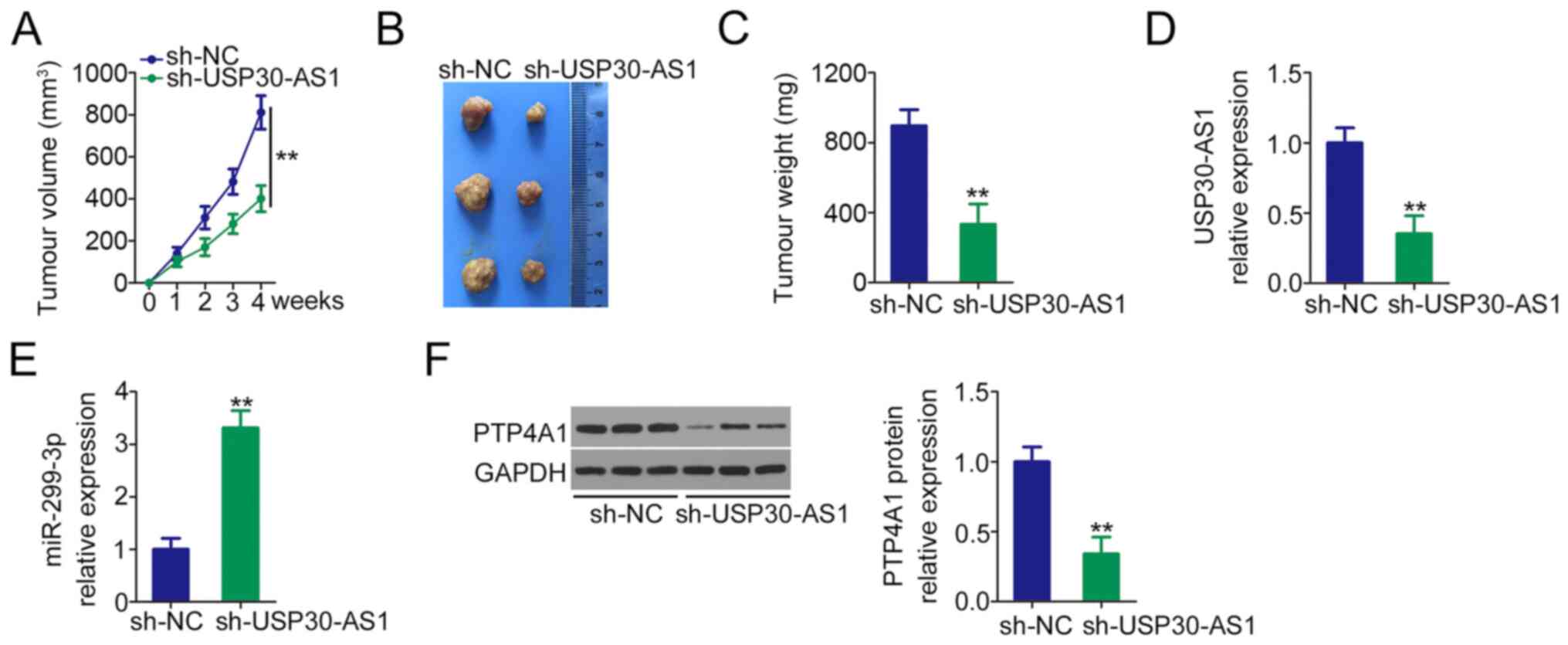
To analyse the effects of USP30-AS1 on tumour growth
in vivo, HeLa cells stably expressing sh-USP30-AS1 were
constructed and subcutaneously injected into nude mice to construct
a mouse tumour model. The volume (Fig.
7A and B) and weight (Fig. 7C)
of the subcutaneous tumours in the sh-USP30-AS1 group were
evidently smaller than those in the sh-NC group. Additionally, the
expression analyses revealed that USP30-AS1 was downregulated
(Fig. 7D) but miR-299-3p was
upregulated (Fig. 7E) in the tumours
in the USP30-AS1-silenced group. Furthermore, the tumours in the
sh-USP30-AS1 group had decreased PTP4A1 protein expression compared
with the sh-NC group (Fig. 7F).
Therefore, USP30-AS1 interference decreases the tumour growth of
cervical cancer cells in vivo.
Discussion
Recently, a substantial amount of research has
investigated the roles of lncRNAs in cervical cancer, and
satisfactory results have been obtained (28–30).
Several lncRNAs are differentially expressed in cervical cancer,
which affect several aggressive properties of the tumour (31). Considering the importance of lncRNAs,
they have huge potential for exploitation as targets for the
diagnosis, prognosis and treatment of cancer. The ENCODE database
reported that the human genome possesses 33,829 lncRNAs (32); however, the regulatory activities of
most lncRNAs remain unknown, including USP30-AS1. Therefore, the
present study determined whether USP30-AS1 is implicated in
cervical cancer malignancy and investigated its relevant underlying
molecular mechanisms.
USP30-AS1 is upregulated in bladder urothelial
carcinoma and is associated with cell autophagy (24). Furthermore, lncRNAs have been
verified as independent prognostic factors for bladder urothelial
carcinoma (24). To date, neither
the expression status nor the detailed role of USP30-AS1 have been
elucidated in cervical cancer. In the present study, increased
USP30-AS1 expression was observed in both cervical cancer tissues
and cell lines. Furthermore, patients with cervical cancer that
exhibit high USP30-AS1 expression levels had shorter overall
survival than those with low USP30-AS1 expression levels. In
vitro and in vivo experiments revealed that USP30-AS1
downregulation induced cell apoptosis; suppressed cell
proliferation, migration and invasion in vitro; and impeded
tumour growth in vivo. All these results offer a new
theoretical and experimental basis for developing USP30-AS1 as a
diagnostic and prognostic biomarker as well as a therapeutic target
for cervical cancer.
In terms of the working mechanism, lncRNAs can
compete for miRNAs through the same miRNA recognition element,
which lowers the negative regulatory actions of miRNAs on target
genes and indirectly modulates target mRNA levels (33). To investigate whether USP30-AS1 acts
as a miRNA sponge, the exact distribution of USP30-AS1 in cervical
cancer cells was determined via a subcellular fractionation assay.
USP30-AS1 was identified as a cytoplasmic lncRNA in cervical cancer
cells. Then, using bioinformatics analyses, the binding site
between USP30-AS1 and miR-299-3p was determined, and subsequent
mechanistic studies corroborated that USP30-AS1 functions as an
endogenous sponge for miR-299-3p in cervical cancer. Furthermore,
PTP4A1 was validated as the downstream target of miR-299-3p, and
miR-299-3p could lower PTP4A1 expression in cervical cancer cells
by base pairing with its 3′-UTR. Notably, further investigation
determined the effects of USP30-AS1 on PTP4A1 expression in
cervical cancer cells. The results of the present study suggest
that PTP4A1 is indirectly regulated by USP30-AS1 by decoying
miR-299-3p. Taken together, these observations unveil a new ceRNA
network involving USP30-AS1, miR-299-3p and PTP4A1.
miR-299-3p dysregulation has been reported in
several types of human cancer (34–36). In
cervical cancer, miR-299-3p is weakly expressed and plays an
anti-oncogenic role during cancer progression by inhibiting cell
proliferation, colony formation and invasion (37), which is in accordance with the
results of the present study. Mechanistically, PTP4A1, a prenylated
protein tyrosine phosphatase, was identified as the direct target
of miR-299-3p in cervical cancer. PTP4A1, also known as a
phosphatase of the regenerating liver, is upregulated in patients
with cervical cancer and is implicated in the control of multiple
malignant behaviours (38,39). Post-transcriptional regulation of
PTP4A1 by miRNAs has been investigated in several types of human
cancer. For example, miR-1271 directly targets PTP4A1 and decreases
its expression in hepatocellular carcinoma, subsequently resulting
in the inhibition of tumour metastasis and the
epithelial-to-mesenchymal transition (40). Nevertheless, the lncRNA-mediated
modulation of PTP4A1 is still far from being fully addressed in
cervical cancer. To the best of our knowledge, the presented
reported for the first time that PTP4A1 could be regulated by
USP30-AS1 at the post-transcriptional level in cervical cancer.
USP30-AS1, a type of ceRNA, competes for miR-299-3p in cervical
cancer and thereby reverses the suppressive effects of miR-299-3p
on PTP4A1, resulting in PTP4A1 overexpression. In addition, rescue
experiments confirmed that miR-299-3p interventions or exogenous
PTP4A1 counteracted the cancer-inhibiting actions of USP30-AS1
deficiency in cervical cancer cells. Taken together, the
miR-299-3p/PTP4A1 axis mediates the central roles of USP30-AS1 in
cervical cancer and forms the USP30-AS1/miR-299-3p/PTP4A1
pathway.
The present study had some limitations; it lacked
the recurrence monitoring of all patients that participated in this
research. This will be the focus of future studies.
In conclusion, to the best of our knowledge, the
present study demonstrated for the first time that high USP30-AS1
expression is closely associated with worse overall survival
outcomes in patients with cervical cancer. USP30-AS1 functions as a
ceRNA and upregulates PTP4A1 by decoying miR-299-3p, thereby
aggravating the oncogenic potential of cervical cancer cells both
in vitro and in vivo. The USP30-AS1/miR-299-3p/PTP4A1
network may be an important molecular marker for cervical cancer
progression and may be researched as a new strategy for targeted
therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MC and LZ designed the present study. MC, YC, HC and
LZ performed the experiments. MC and LZ confirm the authenticity of
all the raw data. LZ analyzed the data. MC drafted the initial
manuscript and revised the manuscript for important intellectual
content. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by the
Human Ethics Approval Committee of The First People's Hospital of
Chongqing Liangjiang New Area (approval no. ECAC-TFHCQLJ.20150601).
Furthermore, written informed consent was obtained from all
patients before the study. Protocols involving animals were
approved by the Ethical Guidelines for Animal Care and Use
Committee of The First People's Hospital of Chongqing Liangjiang
New Area (approval no. EGACUC-TFHCQLJ.20181205).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abbas KM, van Zandvoort K, Brisson M and
Jit M: Effects of updated demography, disability weights, and
cervical cancer burden on estimates of human papillomavirus
vaccination impact at the global, regional, and national levels: A
PRIME modelling study. Lancet Glob Health. 8:e536–e544. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tsikouras P, Zervoudis S, Manav B, Tomara
E, Iatrakis G, Romanidis C, Bothou A and Galazios G: Cervical
cancer: Screening, diagnosis and staging. J BUON. 21:320–325.
2016.PubMed/NCBI
|
|
4
|
Nuchpramool P and Hanprasertpong J:
Preoperative neutrophil-lymphocyte ratio and platelet-lymphocyte
ratio are not clinically useful in predicting prognosis in early
stage cervical cancer. Surg Res Pract. 2018:91629212018.PubMed/NCBI
|
|
5
|
Venkatas J and Singh M: Cervical cancer: A
meta-analysis, therapy and future of nanomedicine.
Ecancermedicalscience. 14:11112020.PubMed/NCBI
|
|
6
|
Small W Jr, Bacon MA, Bajaj A, Chuang LT,
Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR,
Viswanathan AN and Gaffney DK: Cervical cancer: A global health
crisis. Cancer. 123:2404–2412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kornienko AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC Biol. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
DiStefano JK: The emerging role of long
noncoding RNAs in human disease. Methods Mol Biol. 1706:91–110.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang H, Wang Y, Liu X and Li Y: Progress
of long noncoding RNAs in anti-tumor resistance. Pathol Res Pract.
216:1532152020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Teppan J, Barth DA, Prinz F, Jonas K,
Pichler M and Klec C: Involvement of long non-coding RNAs (lncRNAs)
in tumor angiogenesis. Noncoding RNA. 6:422020.PubMed/NCBI
|
|
15
|
Ye M, Zhang J, Wei M, Liu B and Dong K:
Emerging role of long noncoding RNA-encoded micropeptides in
cancer. Cancer Cell Int. 20:5062020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi D, Zhang C and Liu X: Long noncoding
RNAs in cervical cancer. J Cancer Res Ther. 14:745–753. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Aalijahan H and Ghorbian S: Long
non-coding RNAs and cervical cancer. Exp Mol Pathol. 106:7–16.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun W, Shen NM and Fu SL: Involvement of
lncRNA-mediated signaling pathway in the development of cervical
cancer. Eur Rev Med Pharmacol Sci. 23:3672–3687. 2019.PubMed/NCBI
|
|
19
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen S, Zhang S, Liu P, Wang J and Du H:
Potential role of microRNAs in the treatment and diagnosis of
cervical cancer. Cancer Genet. 248-249:25–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miao J, Regenstein JM, Xu D, Zhou D, Li H,
Zhang H, Li C, Qiu J and Chen X: The roles of microRNA in human
cervical cancer. Arch Biochem Biophys. 690:1084802020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao D, Wang Y, Li D and Wang L:
Reconstruction and analysis of the differentially expressed
IncRNA-miRNA-mRNA Network based on competitive endogenous RNA in
hepatocellular carcinoma. Crit Rev Eukaryot Gene Expr. 29:539–549.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun Z, Jing C, Xiao C and Li T: An
autophagy-related long non-coding RNA prognostic signature
accurately predicts survival outcomes in bladder urothelial
carcinoma patients. Aging. 12:15624–15637. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saleh M, Virarkar M, Javadi S, Elsherif
SB, de Castro Faria S and Bhosale P: Cervical cancer: 2018 revised
international federation of gynecology and obstetrics staging
system and the role of imaging. AJR Am J Roentgenol. 214:1182–1195.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cancer Genome Atlas Research Network;
Albert Einstein College of Medicine; Analytical Biological
Services; Barretos Cancer Hospital; Baylor College of Medicine;
Beckman Research Institute of City of Hope; Buck Institute for
Research on Aging; Canada's Michael Smith Genome Sciences Centre;
Harvard Medical School; Helen F. Graham Cancer Center &
Research Institute at Christiana Care Health Services et al, .
Integrated genomic and molecular characterization of cervical
cancer. Nature. 543:378–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Wang D, Wu D, Zhang D and Sun M:
Long noncoding RNA KCNMB2-AS1 stabilized by
N6-methyladenosine modification promotes cervical cancer
growth through acting as a competing endogenous RNA. Cell
Transplant. 29:9636897209643822020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Min H and He W: Long non-coding RNA
ARAP1-AS1 promotes the proliferation and migration in cervical
cancer through epigenetic regulation of DUSP5. Cancer Biol Ther.
21:907–914. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang AH, Zhao JM, Du J, Pang QX and Wang
MQ: Long noncoding RNA LUCAT1 promotes cervical cancer cell
proliferation and invasion by upregulating MTA1. Eur Rev Med
Pharmacol Sci. 24:86232020.PubMed/NCBI
|
|
31
|
Taheri M and Ghafouri-Fard S: Long
Non-coding RNA signature in cervical cancer. Klin Onkol.
31:403–408. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bu D, Yu K, Sun S, Xie C, Skogerbø G, Miao
R, Xiao H, Liao Q, Luo H, Zhao G, et al: NONCODE v3.0: Integrative
annotation of long noncoding RNAs. Nucleic Acids Res. 40:D210–D215.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Braga EA, Fridman MV, Moscovtsev AA,
Filippova EA, Dmitriev AA and Kushlinskii NE: LncRNAs in ovarian
cancer progression, metastasis, and main pathways: ceRNA and
alternative mechanisms. Int J Mol Sci. 21:88552020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng D, Dai Y, Wang S and Xing X:
MicroRNA-299-3p promotes the sensibility of lung cancer to
doxorubicin through directly targeting ABCE1. Int J Clin Exp
Pathol. 8:10072–10081. 2015.PubMed/NCBI
|
|
35
|
Wang JY, Jiang JB, Li Y, Wang YL and Dai
Y: MicroRNA-299-3p suppresses proliferation and invasion by
targeting VEGFA in human colon carcinoma. Biomed Pharmacother.
93:1047–1054. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao R, Liu Q and Lou C: MicroRNA-299-3p
regulates proliferation, migration and invasion of human ovarian
cancer cells by modulating the expression of OCT4. Arch Biochem
Biophys. 651:21–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu Y, Zhao JD and Yang H: MiR-299-3p
inhibits proliferation and invasion of cervical cancer cell via
targeting TCF4. Eur Rev Med Pharmacol Sci. 23:5621–5627.
2019.PubMed/NCBI
|
|
38
|
Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang
X, Li X, Wu D, Wang D, Yang Z and Zhang S: Circular RNA circNRIP1
promotes migration and invasion in cervical cancer by sponging
miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death
Dis. 11:3992020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dong J, Sui L, Wang Q, Chen M and Sun H:
MicroRNA-26a inhibits cell proliferation and invasion of cervical
cancer cells by targeting protein tyrosine phosphatase type IVA 1.
Mol Med Rep. 10:1426–1432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li C, Jiang Y, Miao R, Qu K, Zhang J and
Liu C: MicroRNA-1271 functions as a metastasis and
epithelial-mesenchymal transition inhibitor in human HCC by
targeting the PTP4A1/c-Src axis. Int J Oncol. 52:536–546.
2018.PubMed/NCBI
|