Introduction
Non-small cell lung cancer (NSCLC) accounts for ~85%
of all diagnosed cases of lung cancer (1). Lung adenocarcinoma (LUAD) is the most
common subtype of NSCLC, and its morbidity and mortality rates have
increased to 0.057 and 85%, respectively, in China in 2017
(2,3). Advancements in science and technology
have allowed the development of novel therapeutic strategies for
patients with lung cancer. For example, Han et al (4) discovered that pemetrexed plus
carboplatin combined with gefitinib prolongs the survival time of
patients with LUAD who harbor sensitive EGFR mutations.
Furthermore, Zhuang et al (5)
demonstrated that combination treatment of nadroparin with
radiotherapy induces stronger synergistic antitumor effects in LUAD
A549 cells. However, recent studies suggest that current treatment
strategies can be further improved (6–8).
Despite advancements in surgery, molecular subtyping
and targeted therapy, the prognosis of patients with LUAD remains
relatively poor (9). Patients with
LUAD often relapse and develop metastases following surgery,
chemotherapy and radiotherapy (10).
Due to its malignant characteristics, the 5-year survival rate of
patients with early stage LUAD is 50–70% (11). Patients with advanced LUAD are often
resistant to conventional chemotherapies or targeted therapeutic
drugs (11). Furthermore,
high-potency anticancer drugs remain ineffective for long-term use,
and cancer cells become resistant to anticancer drugs as the
mutation rate rapidly increases (12). Despite consistent improvements to the
surgical methods for treating cancer, and regular updates to
chemotherapy drugs, the complete removal of residual cancer cells
remains difficult, and thus, the risk of recurrence remains
relatively high (13). The
recurrence of cancer proves problematic to subsequent treatment
strategies, and the rate of deterioration increases (13). Furthermore, the molecular mechanisms
underlying recurrence have not been fully elucidated. A previous
study reported that 85/289 patients with stage I and II LUAD
developed distant recurrence within 5 years (14), suggesting that cancer recurrence
holds important clinical value.
With the development of precision medicine, several
biomarkers have been demonstrated to be positively associated with
biological events. For example, Krishnamurthy et al
(15) reported that cyclin-dependent
kinase inhibitor 2A (CDKN2A) expression is a biomarker of aging,
with high CDKN2A expression being associated with advanced aging in
rodent tissues. In addition, β-amyloid 1–42 in cerebrospinal fluid
(CSF) has been verified as a biomarker for Alzheimer's disease in
the autopsy cohort of CSF samples, with a high sensitivity for
detection of 96.4% (16). These
biomarkers are not just limited to proteins, since mRNAs can also
act as biomarkers. Ji et al (17) demonstrated that microRNA-208 is a
useful indicator of myocardial injury. Notable progress has been
made in the discovery and verification of tumor biomarkers,
particularly in the discovery of molecular markers associated with
the clinical effects of tumor therapy. In 2011, human epididymis
protein 4 was approved by the Food and Drug Administration to
monitor the recurrence or disease progression of epithelial ovarian
cancer in conjunction with CA125 (18). Huang et al (19) reported that abnormalities of
amplified in breast cancer 1 (AIB1) are significantly associated
with prognostic significance in urothelial carcinoma, and high AIB1
expression is associated with increased hazard ratios for the
5-year cause-specific survival rate (80.6 vs. 55.8% for high and
low AIB1 expression, respectively; P=0.008) and 5-year overall
survival rate (78.1 vs. 54.8% for high and low AIB1 expression,
respectively; P=0.006). The homeobox B13/IL17 receptor B biomarker
predicts the recurrence risk in estrogen receptor-positive and
lymph node-negative patients with breast cancer (20). Previous studies have identified
several biomarkers that are currently applied in clinical settings
(21,22).
The present study performed bioinformatics analysis
to assess a robust sequence of data, and several biomarkers were
verified using clinical samples of LUAD. Further evaluations were
performed to verify whether cell-free UPK2 mRNA may be used as a
potential biomarker for postoperative recurrence in patients with
early stage LUAD.
Materials and methods
Data analysis
The datasets used in the present study were
downloaded from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga;
Project ID, TCGA-LUAD). The transcriptome data of 24 relapsed
patients, including 14 men and 10 women, was screened.
Additionally, the transcriptome data of paracancerous tissues from
53 patients (33 men and 20 women) was included. The soft
connectivity function in the weighted gene co-expression network
analysis (WGCNA; v1.69; http://cran.r-project.org/web/packages/WGCNA/index.html)
package (23) was used to assess the
effects of different power values on the co-expression network and
co-expression modules in the scale independence and average
connectivity by Pearson's correlation analysis. The ‘randomly
selected gene’ parameter was set to 5,000, and all other parameters
were set to the default values. The expression values were
summarized using the collapse rows function in the WGCNA package,
and cluster analysis was subsequently performed using flashClust
v1.01-2 (https://cran.r-project.org/web/packages/flashClust/index.html)
(24). The interaction/association
of each module was visualized using heat maps. In addition, Gene
Ontology (GO) analysis of the gene modules of interest was
performed using the Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.ncifcrf.gov/summary.jsp). The
differential expression of genes was defined as
log(fold-change)|<0.6|; false discovery rate <0.05. Cytoscape
v3.11 was used to construct co-expression network (https://cytoscape.org/download.html).
Kaplan-Meier survival analysis of the corresponding genes was
performed using the Oncomine database (https://www.oncomine.org/resource/login.html) and the
P-values were calculated using the log-rank test.
Patient recruitment
Blood samples were collected from 132 patients with
LUAD who underwent excision surgery at the First Affiliated
Hospital of the Second Military Medical University (Shanghai,
China) between February 2006 and March 2010. All patients received
systematic treatment following surgery, including optimal local
treatments, such as radiochemotherapy and targeted therapy. Disease
recurrence was based on chest X-ray or CT imaging. The present
study was performed in accordance with the Declaration of Helsinki
and Good Clinical Practice, and was approved by the Ethics
Committee of the First Affiliated Hospital of the Second Military
Medical University. All patients provided oral informed consent for
the use of their blood for scientific purposes.
Sample collection and processing
Blood samples (15 ml) from 105 patients with early
stage LUAD of the 132 patients (pathological stage I, II and IIIA)
were initially collected 90 days after surgery. The second, third
and fourth samples were collected 180 days, 1 year and 2 years
after surgery, respectively. Blood samples were no longer collected
if disease recurrence was detected during repeated examinations.
The samples were collected in 5-ml heparin anticoagulation tubes
and immediately used to extract free mRNA according to the
manufacturer's protocol (Qiagen, Inc.; cat. no. 72022).
Free mRNA was reverse transcribed using the reverse
transcription (RT) kit according to the manufacturer's protocol
(Takara Bio, Inc.; cat. no. 639522). A quarter of the RT product
was mixed with a pre-formulated 2X SYBR Green PCR mix (Roche
Diagnostics; cat. no. 06924204001) containing uroplakin 2 (UPK2)
RT-quantitative (q)PCR primers (UPK2 forward,
5′-CACTGAGTCCAGCAGAGAGATC-3′ and reverse,
5′-ACAGAGAGCAGCACCGTGATGA-3′; GAPDH forward,
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3′) and was subsequently supplemented
with distilled water to reach a final volume of 20 µl.
Amplification was performed to obtain the dissolution curve, using
the following thermocycling conditions: 94°C for 5 min, followed by
40 cycles at 94°C for 2 min, 60°C for 1 min and 72°C for 2 min, and
a final extension step at 72°C for 2 min. Relative UPK2 expression
was calculated using the 2−ΔΔCq method (25) and normalized to the internal
reference gene GAPDH.
Statistical analysis
The serum expression levels of UPK2 were detected in
triplicate. GraphPad Prism 8.0 (GraphPad Software Inc.) was used
for statistical analysis. The data are presented as the mean ± SD.
The difference between two groups was analyzed by unpaired
Student's t-test. The difference between paired samples (before and
after relapse) was analyzed by paired Student's t-test. P<0.05
was considered to indicate a statistically significant difference.
The receiver operating characteristic (ROC) curve of LUAD
recurrence was assessed using the pROC package v1.16.2 (https://cran.r-project.org/web/packages/pROC/index.html)
(26) within R software, and the
area under the curve (AUC) was calculated. The survival curve was
plotted using the ggsurvplot package v0.4.8 (https://cran.r-project.org/web/packages/survminer/readme/README.html)
within R language and the log-rank test was used to obtain the
P-value.
Results
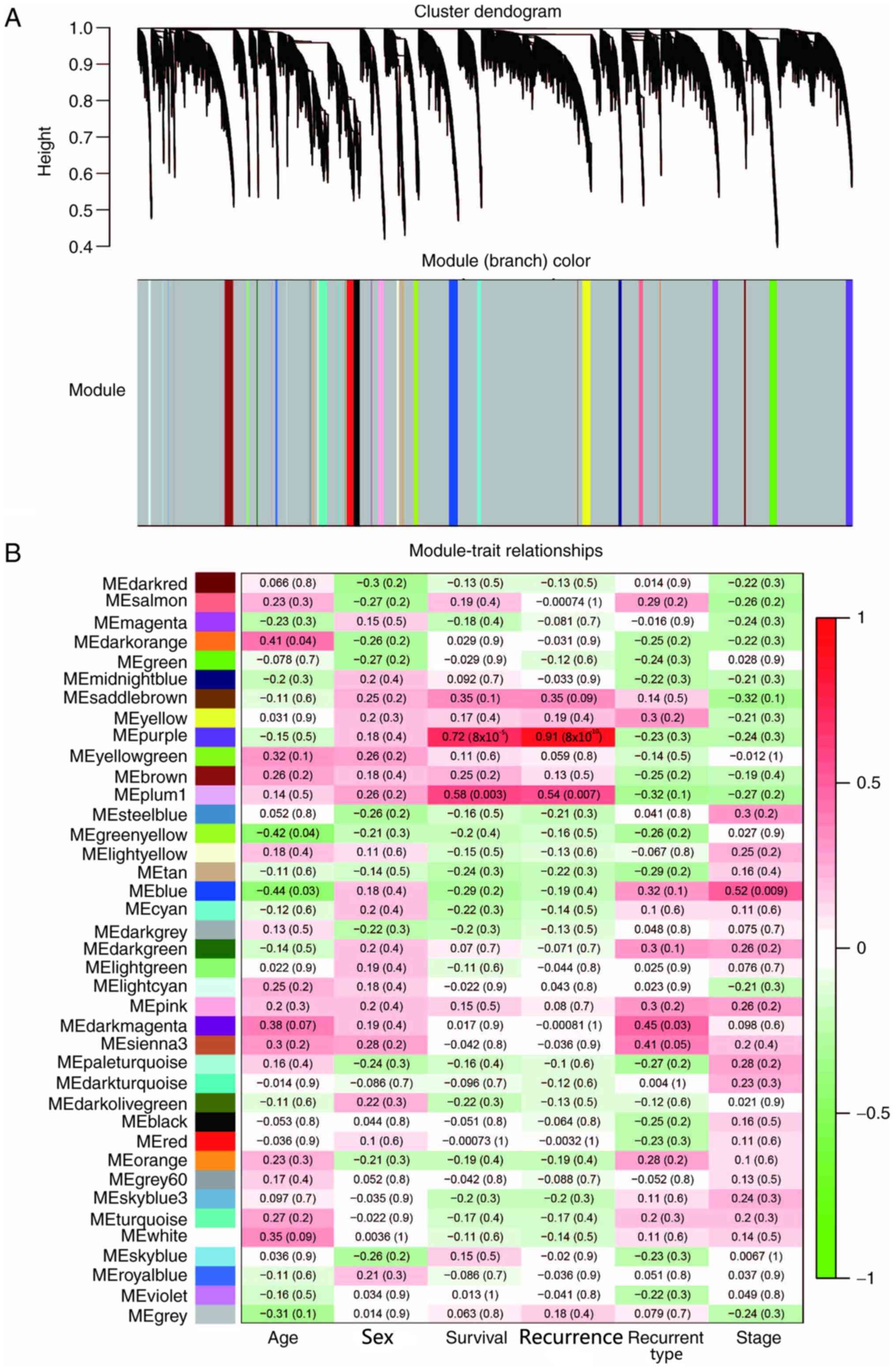
Specific co-expression modules
associated with LUAD recurrence
In order to establish a co-expressed gene network
associated with postoperative recurrence of LUAD, WGCNA was used to
assess the gene expression profile data from patients with
recurrent LUAD within the TCGA database. The transcriptome data of
24 relapsed patients, including 14 men and 10 women, were screened.
The association between genes was calculated using WGCNA, and the
gene expression data of patients with recurrent LUAD were
classified into 39 gene modules using unsupervised average linkage
hierarchical clustering and were labelled in a heat map with
different colors (Fig. 1A). Gene
modules of different colors contained mutually exclusive
co-expressed genes. Genes that could not be classified into a
specific module were incorporated into gray modules. WGCNA can
determine the association between gene modules and a series of
phenotypes (23); therefore, this
method was used to assess the association between the specific gene
modules of patients with postoperative recurrent early stage LUAD
and a series of phenotypes, including age, sex, survival,
recurrence, recurrence type and pathological stage (27). Without any phenotypic or genetic
preferences for module partitioning, the results demonstrated that
the purple module was significantly associated with survival and
recurrence, with Pearson's correlation coefficients >0.7
(Fig. 1B). Overall, the present
results suggested that these genes and their co-expression patterns
may be associated with the recurrence of LUAD.
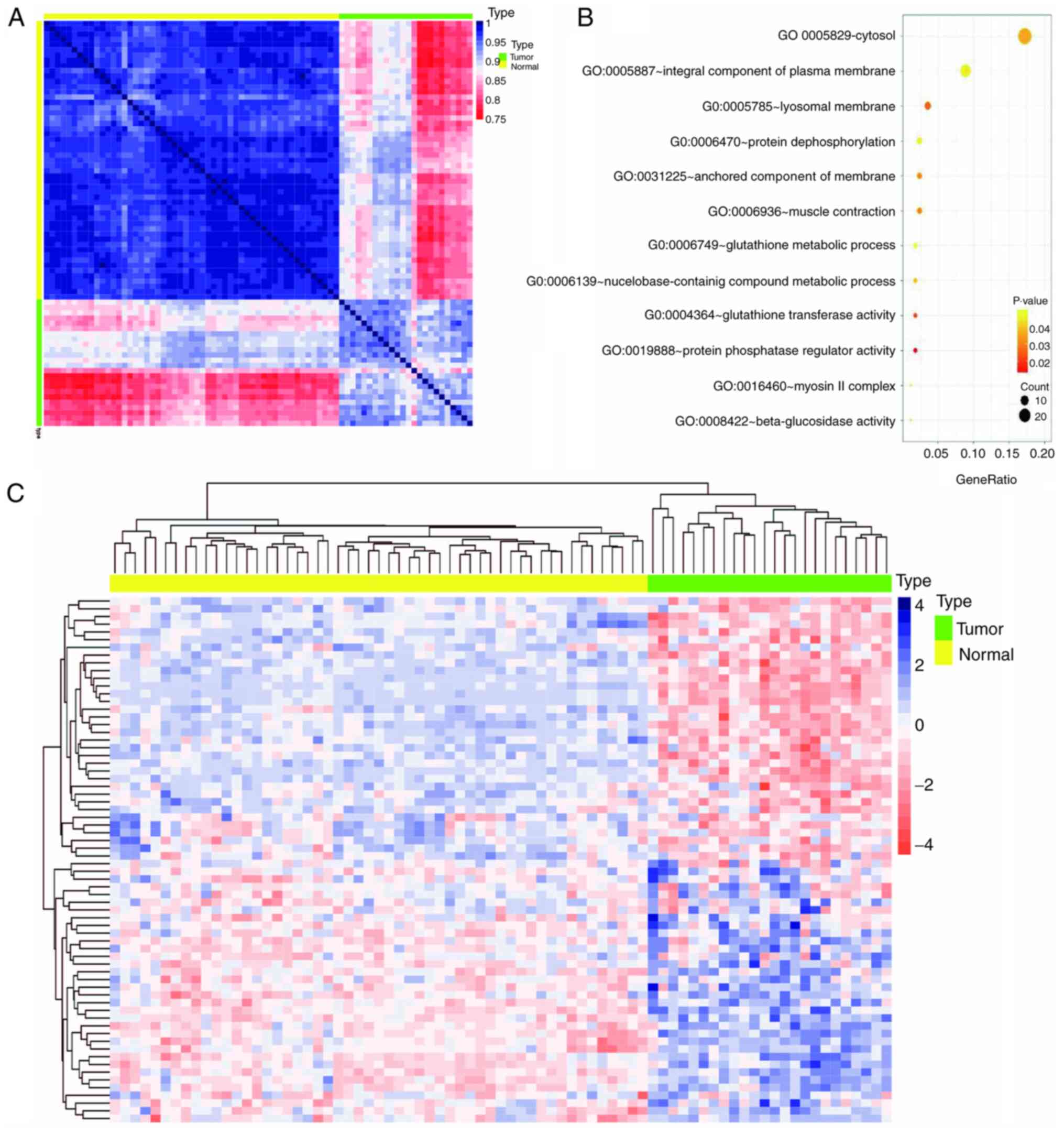
Biological insights from the purple
module
WGCNA classifies co-expressed genes of patient
samples into specific modules associated with a series of traits
regulated by the same mechanism (23). The purple module was identified as
the most relevant to postoperative recurrence. In order to verify
the association between the co-expressed genes within the purple
module and LUAD, a heat map containing the gene expression levels
of 24 recurrent tumor tissues and 53 paracancerous tissues from
TCGA was constructed. The results demonstrated a marked difference
in the expression pattern of the purple module between
paracancerous and recurrent tumor tissues (Fig. 2A). However, the expression pattern of
the purple module genes in patients with recurrent tumor tissues
was not as consistent compared with paracancerous tissues; the
expression pattern in tumor tissues exhibited three expression
patterns of light red, light blue and deep red, suggesting
different mechanisms of relapse (Fig.
2A). Of the 117 genes in the purple module, 68 genes exhibited
significant differences (data not shown) between recurrent tumor
tissues and paracancerous tissues [log(fold-change)|<0.6|; false
discovery rate <0.05]. These 68 genes were used to construct an
expression heat map of the two types of tissues (Fig. 2C), which demonstrated that the
expression patterns of the 68 genes were more uniform than that of
the heat map constructed using 117 genes. The biological function
of the 117 genes in the purple module was further analyzed via GO
analysis using DAVID, with the most significant GO term being
‘cytosol’ (P=0.0356; Fig. 2B).
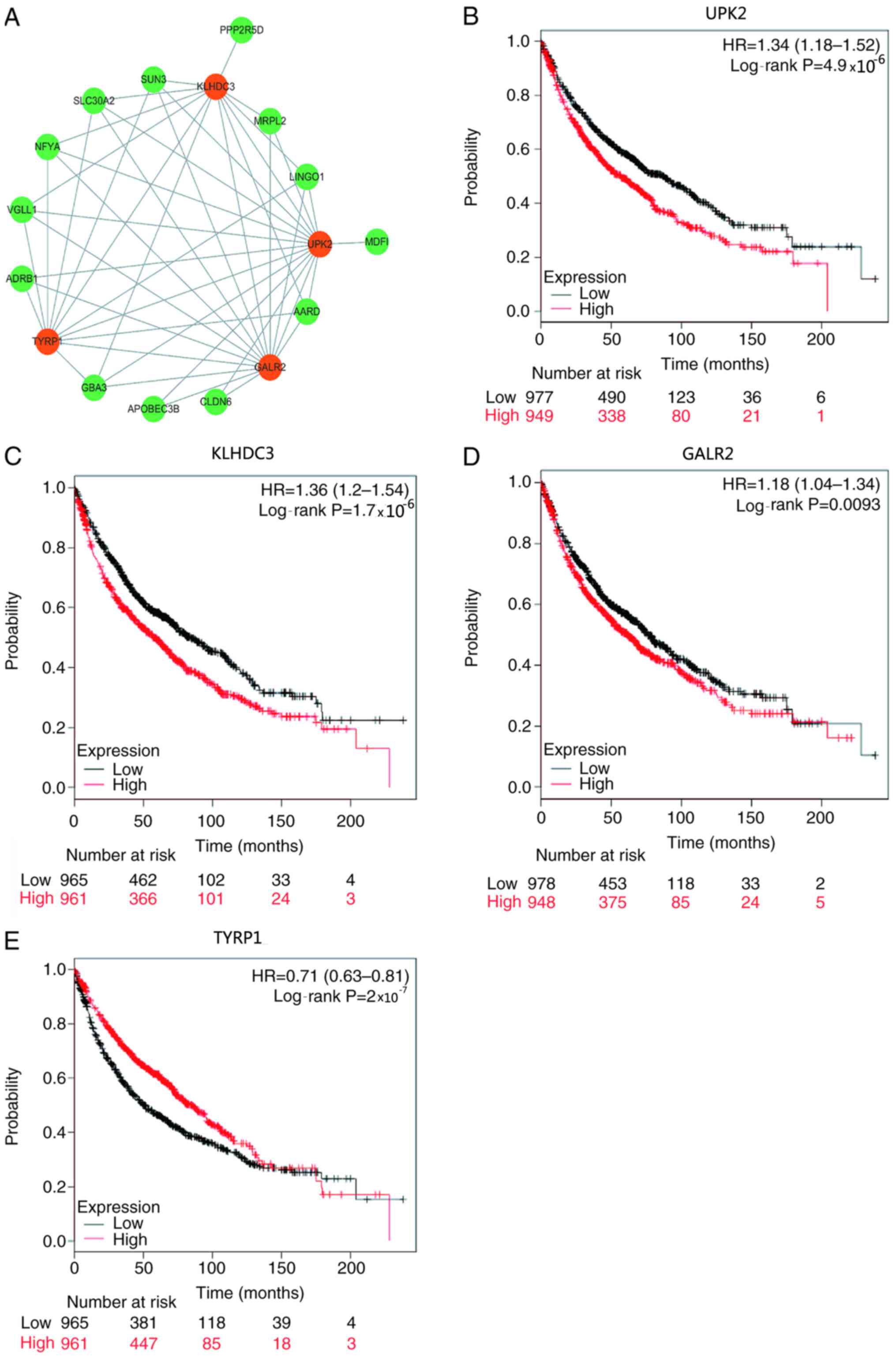
Key genes associated with LUAD
recurrence in the purple module
Gene significance is closely associated with gene
connectivity, which means that nodes with higher connectivity in
the co-expression network serve an important role in the process of
performing biological functions (28). Therefore, a co-expression network of
genes for LUAD recurrence was constructed, identifying 47 edges and
17 nodes (power=8; Fig. 3A). The
results demonstrated that there were four genes [UPK2, kelch domain
containing 3 (KLHDC3), galanin receptor 2 (GALR2) and
tyrosinase-related protein 1 (TYRP1)] in the co-expression network
with more nodes linked appearing in the purple module, which were
significantly associated with survival and recurrence (Table I and Fig.
3B-E). Among these genes, high expression levels of UPK2,
KLHDC3 and GALR2, and low expression levels of TYRP1 were
associated with a poor prognosis (Table
I and Fig. 3B-E). The function
of these four genes in LUAD was further analyzed using clinical
data downloaded from the Oncomine database. The results
demonstrated that patients with low expression levels of UPK2,
KLHDC3 and GALR2 had significantly improved survival outcomes than
those with high expression levels of UPK2, KLHDC3 and GALR2
(P=4.9×10−5, P=1.7×10−5 and P=0.0093,
respectively; Fig. 3B-D).
Conversely, patients with high TYRP1 expression had a significantly
improved prognosis than those with low TYRP1 expression
(P=2×10−7; Fig. 3E).
 | Table I.UPK2, KLHDC3, GALR2 and TYRP1
expression are highly associated with survival and recurrence. |
Table I.
UPK2, KLHDC3, GALR2 and TYRP1
expression are highly associated with survival and recurrence.
| Gene | Nodes no.
(power=8) | Probes name used by
oncomine database | P-value |
|---|
| UPK2 | 15 | 207862_at |
4.9×10−06 |
| GALR2 | 14 | 211226_at | 0.0093 |
| KLHDC3 | 12 | 208784_s_at |
1.7×10−06 |
| TYRP1 | 11 | 205694_at |
2×10−07 |
Clinical characteristics of patients
with early stage LUAD who received UPK2 free mRNA plasma
testing
Previous studies have reported that free mRNA in
plasma has the potential to act as a tumor marker (29,30).
Table II presents the demographic
information and clinical characteristics of the 105 patients with
early stage LUAD who met the study criteria out of 132 patients. Of
these LUAD patients, 58 were men (55%) and 47 were women (45%),
with a median age of 58 years (age range, 39–83 years). The
pathological stage of most patients was stage I or II (83%), whilst
the remaining patients were at stage IIIa (17%). Following surgery,
43 patients received adjuvant therapy (41%), including 35 patients
receiving radiotherapy, 4 patients receiving adjuvant chemotherapy
and 4 patients receiving both radiotherapy and chemotherapy
(Table II).
 | Table II.Demographic information and clinical
characteristics of 105 patients with lung adenocarcinoma with
(n=35) or without (n=70) recurrence. |
Table II.
Demographic information and clinical
characteristics of 105 patients with lung adenocarcinoma with
(n=35) or without (n=70) recurrence.
|
Characteristics | All | Recurrence | No recurrence |
|---|
| Sex, n (%) |
|
|
|
|
Male | 58 (55) | 23 (66) | 35 (50) |
|
Female | 47 (45) | 12 (34) | 35 (50) |
| Median age (range),
years | 58 (39–83) | 59 (39–80) | 58 (42–83) |
| Mortality, n
(%) |
|
|
|
|
Dead | 74 (70) | 26 (74) | 48 (69) |
|
Alive | 23 (22) | 7 (20) | 16 (23) |
|
Unknown | 8 (8) | 2 (6) | 6 (9) |
| Smoking status, n
(%) |
|
|
|
|
Former | 70 (67) | 22 (63) | 48 (69) |
|
Active | 25 (24) | 11 (31) | 14 (20) |
|
Never | 8 (8) | 2 (6) | 6 (9) |
|
Unknown | 2 (2) | 0 (0) | 2 (3) |
| Stage at diagnosis,
n (%) |
|
|
|
| I | 65 (62) | 21 (60) | 44 (63) |
| II | 22 (21) | 10 (29) | 12 (17) |
|
IIIA | 18 (17) | 4 (11) | 14 (20) |
| Treatment, n
(%) |
|
|
|
| No
adjuvant treatment | 62 (59) | 16 (46) | 46 (66) |
|
Adjuvant treatment | 43 (41) | 19 (54) | 24 (34) |
|
Adjuvant chemotherapy | 4 (4) | 2 (6) | 2 (3) |
|
Radiation | 35 (33) | 15 (43) | 20 (27) |
|
Radiation and
chemotherapy | 4 (4) | 2 (6) | 2 (3) |
| Mean uroplakin 2
expression (range) | 0.20
(0.05–0.80) | 0.28
(0.08–0.61) | 0.16
(0.05–0.80) |
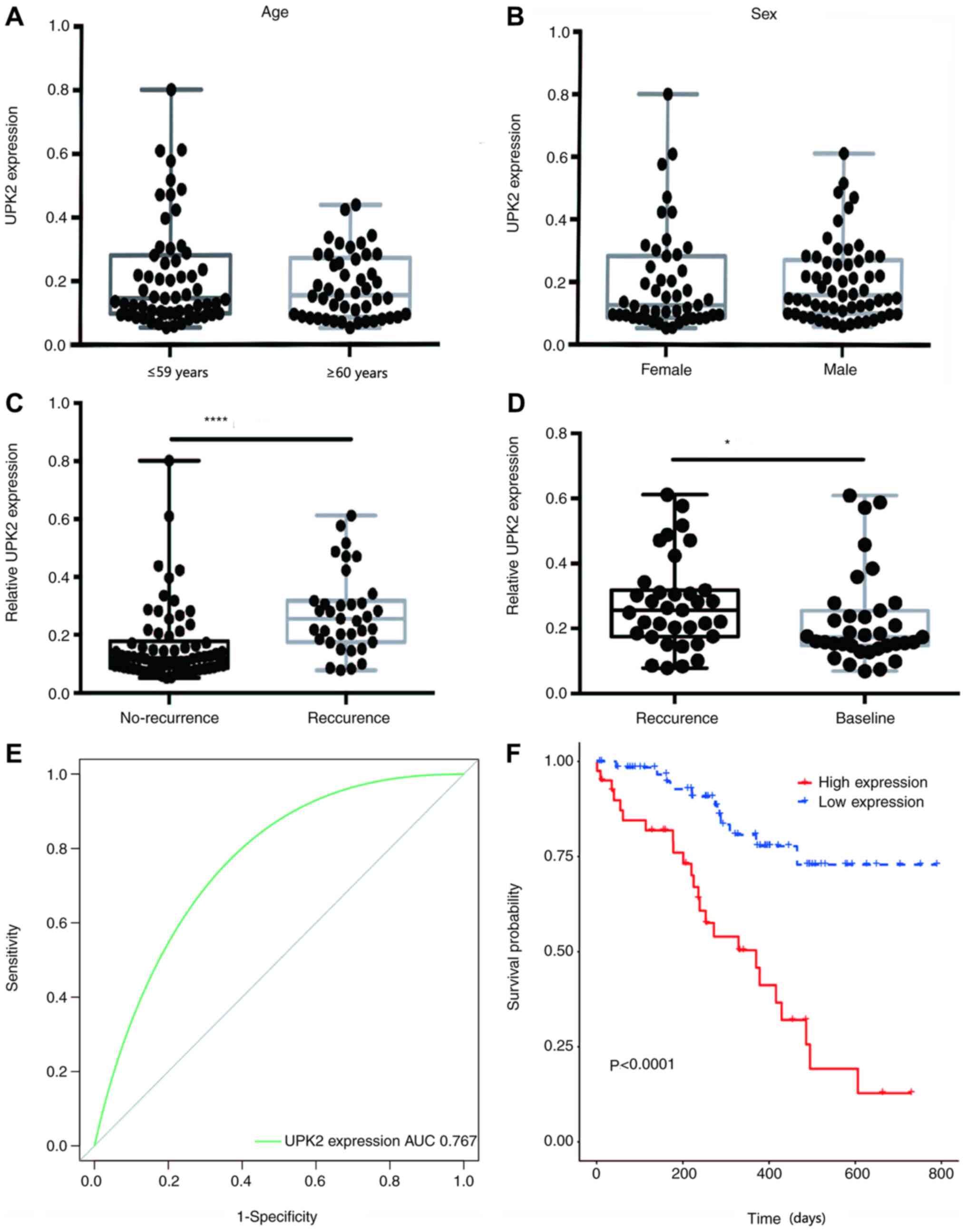
Diagnostic performance of UPK2
Patient blood samples were collected and assessed
for free UPK2 expression. Imaging examination was performed every
three months from the time of the first repeated examination, which
was 90 days after surgery. If the imaging examination indicated
that the patient had relapsed, they were classified into the
relapsed group (35 patients), while 70 patients did not relapse,
and the level of UPK2 mRNA detected was recorded. If the patient
had no recurrence during the 3-year follow-up period, then the mean
value of multiple testing was recorded as the relative expression
level of UPK2. The results demonstrated no significant differences
in UPK2 expression between patients with LUAD of different ages and
sex (Fig. 4A and B). Notably,
non-relapsed patients exhibited low UPK2 expression. The mean
expression level of UPK2 for relapsed patients was measured after
recurrence was detected via imaging. The mean UPK2 expression
relative to GADPH in relapsed patients was 0.2763, while the mean
UPK2 expression in non-relapsed patients was 0.1623, which was
significantly lower than that of relapsed patients (P<0.0001;
Fig. 4C). Notably, for relapsed
patients, UPK2 expression at relapse was significantly higher than
that before relapse (P=0.0351; Fig.
4D). In addition, the ROC curve was plotted and the AUC was
calculated to determine whether plasma UPK2 expression may be used
to distinguish between relapsed and non-relapsed patients (Fig. 4E). The results demonstrated that when
plasma UPK2 expression was used alone as a diagnostic biomarker,
the AUC was 0.767 with a 95% CI of 0.675–0.858. Furthermore,
patients with LUAD were divided into two groups, namely the high
(≥0.1623) and low (<0.1623) UPK2 expression groups, and their
survival curves were plotted. The results indicated that patients
with high plasma UPK2 mRNA expression had a poorer survival outcome
than those with low plasma UPK2 mRNA expression (Fig. 4F).
Discussion
Lung cancer has become a global health problem due
to the high morbidity and mortality rates (0.057 and 85%,
respectively) in China in 2017 (2,3). LUAD is
the main subtype of NSCLC (31–33).
Despite recent advancements in cancer treatment, the 5-year
survival rate of patients with LUAD remains relatively low (~15%)
(34,35). With the advent of precision medicine
concepts, molecular biomarkers and molecular drug targets have
become hotspots in cancer research, improving the long-term
outcomes for patients with different types of cancer. For example,
patients with lung cancer with EGFR mutations can benefit from the
treatment of tyrosine kinase inhibitors, such as gefitinib and
erlotinib (36). Other potential
biomarkers are predominantly oncogene-driven mutations, including
ALK translocation and ROS1 gene rearrangement (37,38).
Thus, it remains critical to identify and validate clinically
relevant and effective prognostic markers for LUAD to complement
existing molecular biomarkers and further guide treatment
decisions.
UPK2 is a highly specific marker of bladder
transitional cell carcinoma. UPK2 mRNA expression was initially
detected in blood samples from two patients with metastatic bladder
cancer who did not receive chemotherapy and 1 out of 8 patients
with metastatic bladder cancer who received chemotherapy (39). However, it was not detected in
patients with non-metastatic bladder cancer or in the normal
control group, indicating that detection of UPK2 in the peripheral
blood is associated with metastasis of bladder cancer (39). Therefore, assessment of UPK2
specificity and sensitivity may be a potential means of detecting
bladder cancer metastasis, staging and monitoring chemotherapy
response. Lotan et al (40)
assessed 11 immunohistochemical markers at the primary sites of
several micropapillary carcinomas and demonstrated that urinary
tract proteins can be used as markers to identify urinary
mesothelial invasive micropapillary carcinoma. Li et al
(41) reported that UPK2 is
expressed in 63% of plasma cell samples, which is significantly
higher than UPK3 expression (6%), and further suggested that UPK2
is a valuable marker and should be included in immunohistochemical
markers to facilitate the differential diagnosis of tumors with
plasmacytoid features. Furthermore, Matuszewski et al
(42) demonstrated that the
concentration of UPK2 in urine decreased with the progression of
bladder cancer, which further confirms the diagnostic value of UPK2
concentration in plasma and urine for bladder cancer. Tian et
al (43) assessed UPK2
expression via bladder tissue microarray and reported that UPK2 is
highly specific (100%) and can be used as a marker to identify
urothelial lineage tumors and to help distinguish between bladder
and prostate cancers, or can be used in combination with GATA3 as a
potential marker for metastatic breast cancer. Hoang et al
(44) demonstrated that the positive
rate of UPK2, GATA3 and p40 antibody combined testing was 94.2%
(97/103) in invasive urothelial carcinoma, indicating that
combination of these three antibodies has a high sensitivity to the
differential diagnosis of invasive urothelial carcinoma. However,
the combination testing of UPK2, GATA3 and p40 is negative in LUAD,
colon adenocarcinoma and renal cell carcinoma (44). Furthermore, the expression and role
of UPK2 in LUAD recurrence has not been fully investigated.
The results of the present study demonstrated that
UPK2 expression was significantly increased in patients with LUAD
recurrence, and the prognosis of patients with high UPK2 expression
was poor. These different expression levels of UPK2 before and
after LUAD recurrence are consistent with the aforementioned
findings on bladder cancer, suggesting that the differential
expression of UPK2 in patients with LUAD recurrence may have
clinical implications (39).
Enrichment analysis demonstrated that the function of UPK2 was
predominantly associated with ‘cytosol’. Blood samples were
collected from 105 patients with LUAD, of which 35 had LUAD
recurrence and 70 patients had no recurrence. RT-qPCR analysis
demonstrated that UPK2 mRNA expression in the blood of relapsed
patients was significantly higher than that of patients without
recurrence, indicating that the difference in UPK2 expression was
significantly associated with the recurrence of LUAD. Therefore,
the present results suggested that UPK2 may be used as a biomarker
to detect recurrence instead of using imaging techniques, since the
detection of free UPK2 mRNA is easier to perform and less invasive.
Prospective studies should further investigate UPK2 expression in
patients with LUAD, with different stages and lymph node
metastasis, and should further validate the clinicopathological
characteristics associated with UPK2 expression, in order to
develop novel therapeutic strategies for patients with LUAD.
Acknowledgements
Not applicable.
Funding
This study was funded by National Natural Science
Foundation of China (grant no. 81672890).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ performed the experiments, prepared the figures
and wrote the manuscript. QL and BL performed clinical sample
collection and preparation. HL and CW performed the bioinformatics
analysis. CL and HJ interpreted the data, drafted and revised the
manuscript, and gave final approval of the version to be published.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee and
Institutional Review Board of the First Affiliated Hospital of the
Second Military Medical University (approval no. CHEC2020-021). All
patients provided oral informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LUAD
|
lung adenocarcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
WGCNA
|
weighted gene co-expression network
analysis
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
ULPK2
|
uroplakin 2
|
|
KLHDC3
|
kelch domain containing 3
|
|
GALR2
|
galanin receptor 2
|
|
TYRP1
|
tyrosinase-related protein 1
|
|
GO
|
Gene Ontology
|
References
|
1
|
Zhu Y, Xing P, Wang S, Ma D, Mu Y, Li X,
Xu Z and Li J: Evaluation of calculating carboplatin dosage in
carboplatin-pemetrexed therapy as the first-line therapy for
Chinese patients with advanced lung adenocarcinoma. Thorac Cancer.
9:400–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu AN, Qu HJ, Yu CY and Sun P: Knockdown
of LINC01614 inhibits lung adenocarcinoma cell progression by
up-regulating miR-217 and down-regulating FOXP1. J Cell Mol Med.
22:4034–4044. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou X, Zhang P, Luo W, Zhang L, Hu R, Sun
Y and Jiang H: Ketamine induces apoptosis in lung adenocarcinoma
cells by regulating the expression of CD69. Cancer Med. 7:788–795.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han B, Jin B, Chu T, Niu Y, Dong Y, Xu J,
Gu A, Zhong H, Wang H, Zhang X, et al: Combination of chemotherapy
and gefitinib as first-line treatment for patients with advanced
lung adenocarcinoma and sensitive EGFR mutations: A randomized
controlled trial. Int J Cancer. 141:1249–1256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhuang X, Qiao T, Xu G, Yuan S, Zhang Q
and Chen X: Combination of nadroparin with radiotherapy results in
powerful synergistic antitumor effects in lung adenocarcinoma A549
cells. Oncol Rep. 36:2200–2206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Noronha V, Zanwar S, Joshi A, Patil VM,
Mahajan A, Janu A, Agarwal JP, Bhargava P, Kapoor A and Prabhash K:
Practice patterns and outcomes for pemetrexed plus platinum doublet
as neoadjuvant chemotherapy in adenocarcinomas of lung: Looking
beyond the usual paradigm. Clin Oncol (R Coll Radiol). 30:23–29.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marini KD, Croucher DR, McCloy RA,
Vaghjiani V, Gonzalez-Rajal A, Hastings JF, Chin V, Szczepny A,
Kostyrko K, Marquez C, et al: Inhibition of activin signaling in
lung adenocarcinoma increases the therapeutic index of platinum
chemotherapy. Sci Transl Med. 10:eaat35042018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tao H, Meng Q, Li M, Shi L, Tang J and Liu
Z: Outcomes of bevacizumab combined with chemotherapy in lung
adenocarcinoma-induced malignant pleural effusion. Thorac Cancer.
9:298–304. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu K, Zhang X, Li F, Xiao D, Hou Y, Zhu S,
Liu D, Ye X, Ye M, Yang J, et al: Frequent alterations in
cytoskeleton remodelling genes in primary and metastatic lung
adenocarcinomas. Nat Commun. 6:101312015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Xu W, Zhan P, Xu T, Jin J, Miu Y,
Zhou Z, Zhu Q, Wan B, Xi G, et al: Overexpression of geranylgeranyl
diphosphate synthase contributes to tumour metastasis and
correlates with poor prognosis of lung adenocarcinoma. J Cell Mol
Med. 22:2177–2189. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao S, Guo W, Li J, Yu W, Guo T, Deng W
and Gu C: High expression of Y-box-binding protein 1 correlates
with poor prognosis and early recurrence in patients with small
invasive lung adenocarcinoma. Onco Targets Ther. 9:2683–2692. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mansoori B, Mohammadi A, Davudian S,
Shirjang S and Baradaran B: The different mechanisms of cancer drug
resistance: A brief review. Adv Pharm Bull. 7:339–348. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang C, Leighl NB, Wu YL and Zhong WZ:
Emerging therapies for non-small cell lung cancer. J Hematol Oncol.
12:452019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aramini B, Casali C, Stefani A, Bettelli
S, Wagner S, Sangale Z, Hughes E, Lanchbury JS, Maiorana A and
Morandi U: Prediction of distant recurrence in resected stage I and
II lung adenocarcinoma. Lung Cancer. 101:82–87. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krishnamurthy J, Torrice C, Ramsey MR,
Kovalev GI, Al-Regaiey K, Su L and Sharpless NE: Ink4a/Arf
expression is a biomarker of aging. J Clin Invest. 114:1299–1307.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shaw LM, Vanderstichele H, Knapik-Czajka
M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A,
Lewczuk P, et al: Cerebrospinal fluid biomarker signature in
Alzheimer's disease neuroimaging initiative subjects. Ann Neurol.
65:403–413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ji X, Takahashi R, Hiura Y, Hirokawa G,
Fukushima Y and Iwai N: Plasma miR-208 as a biomarker of myocardial
injury. Clin Chem. 55:1944–1949. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Plotti F, Capriglione S, Terranova C,
Montera R, Aloisi A, Damiani P, Muzii L, Scaletta G,
Benedetti-Panici P and Angioli R: Does HE4 have a role as biomarker
in the recurrence of ovarian cancer? Tumour Biol. 33:2117–2123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang Y, Cen J, Wei J, Chen Z, Fang Y,
Feng Z, Lu J, Liang Y, Luo J, Mo C and Chen W: Impact of AIB1
expression on the prognosis of upper tract urothelial carcinoma
after radical nephroureterectomy. Cancer Biomark. 25:151–160. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sgroi DC, Carney E, Zarrella E, Steffel L,
Binns SN, Finkelstein DM, Szymonifka J, Bhan AK, Shepherd LE, Zhang
Y, et al: Prediction of late disease recurrence and extended
adjuvant letrozole benefit by the HOXB13/IL17BR biomarker. J Natl
Cancer Inst. 105:1036–1042. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ni KW and Sun GZ: The identification of
key biomarkers in patients with lung adenocarcinoma based on
bioinformatics. Math Biosci Eng. 16:7671–7687. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li R, Yang YE, Yin YH, Zhang MY, Li H and
Qu YQ: Methylation and transcriptome analysis reveal lung
adenocarcinoma-specific diagnostic biomarkers. J Transl Med.
17:3242019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Langfelder P and Horvath S: Fast R
functions for robust correlations and hierarchical clustering. J
Stat Softw. 46:i112012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu L, Tao G, Zhu L, Wang G, Li Z, Ye J and
Chen Q: Prediction of pathologic stage in non-small cell lung
cancer using machine learning algorithm based on CT image feature
analysis. BMC Cancer. 19:4642019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mähler N, Wang J, Terebieniec BK,
Ingvarsson PK, Street NR and Hvidsten TR: Gene co-expression
network connectivity is an important determinant of selective
constraint. PLoS Genet. 13:e10064022017. View Article : Google Scholar
|
|
29
|
Miura N, Hasegawa J and Shiota G: Serum
messenger RNA as a biomarker and its clinical usefulness in
malignancies. Clin Med Oncol. 2:511–527. 2008.PubMed/NCBI
|
|
30
|
Tani N, Ichikawa D, Ikoma D, Tomita H, Sai
S, Ikoma H, Okamoto K, Ochiai T, Ueda Y, Otsuji E, et al:
Circulating cell-free mRNA in plasma as a tumor marker for patients
with primary and recurrent gastric cancer. Anticancer Res.
27:1207–1212. 2007.PubMed/NCBI
|
|
31
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Youlden DR, Cramb SM and Baade PD: The
international epidemiology of lung cancer: Geographical
distribution and secular trends. J Thorac Oncol. 3:819–831. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rosell R, Bivona TG and Karachaliou N:
Genetics and biomarkers in personalisation of lung cancer
treatment. Lancet. 382:720–731. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hung JJ, Yeh YC, Jeng WJ, Wu KJ, Huang BS,
Wu YC, Chou TY and Hsu WH: Predictive value of the international
association for the study of lung cancer/American Thoracic
Society/European Respiratory Society classification of lung
adenocarcinoma in tumor recurrence and patient survival. J Clin
Oncol. 32:2357–2364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Subramaniam S, Thakur RK, Yadav VK, Nanda
R, Chowdhury S and Agrawal A: Lung cancer biomarkers: State of the
art. J Carcinog. 12:32013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Remon J, Moran T, Reguart N, Majem M,
Carcereny E and Lianes P: Beyond EGFR TKI in EGFR-mutant non-small
cell lung cancer patients: Main challenges still to be overcome.
Cancer Treat Rev. 40:723–729. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, et al: Identification of the transforming EML4-ALK
fusion gene in non-small-cell lung cancer. Nature. 448:561–566.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bergethon K, Shaw AT, Ou SH, Katayama R,
Lovly CM, McDonald NT, Massion PP, Siwak-Tapp C, Gonzalez A, Fang
R, et al: ROS1 rearrangements define a unique molecular class of
lung cancers. J Clin Oncol. 30:863–870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li SM, Zhang ZT, Chan S, McLenan O, Dixon
C, Taneja S, Lepor H, Sun TT and Wu XR: Detection of circulating
uroplakin-positive cells in patients with transitional cell
carcinoma of the bladder. J Urol. 162((3 Pt 1)): 931–935. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lotan TL, Ye H, Melamed J, Wu XR, Shih IM
and Epstein JI: Immunohistochemical panel to identify the primary
site of invasive micropapillary carcinoma. Am J Surg Pathol.
33:1037–1041. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li W, Liang Y, Deavers MT, Kamat AM, Matin
SF, Dinney CP, Czerniak B and Guo CC: Uroplakin II is a more
sensitive immunohistochemical marker than uroplakin III in
urothelial carcinoma and its variants. Am J Clin Pathol.
142:864–871. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Matuszewski M, Szymanska B, Dlugosz A,
Malkiewicz B, Dembowski J and Piwowar A: Preliminary evaluation of
the diagnostic usefulness of uroplakin 2 with an assessment of the
antioxidant potential of patients with bladder cancer. Biomed Res
Int. 2018:86932972018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tian W, Guner G, Miyamoto H,
Cimino-Mathews A, Gonzalez-Roibon N, Argani P, Li X, Sharma R,
Subhawong AP, Rezaei K, et al: Utility of uroplakin II expression
as a marker of urothelial carcinoma. Hum Pathol. 46:58–64. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hoang LL, Tacha D, Bremer RE, Haas TS and
Cheng L: Uroplakin II (UPII), GATA3, and p40 are highly sensitive
markers for the differential diagnosis of invasive urothelial
carcinoma. Appl Immunohistochem Mol Morphol. 23:711–716. 2015.
View Article : Google Scholar : PubMed/NCBI
|