Introduction
Serine threonine tyrosine kinase 1 (STYK1), also
known as novel oncogene with kinase domain (NOK), belongs to the
receptor protein tyrosine kinases (RPTKs) subfamily (1); it has been demonstrated to be a potent
oncogene that enhances cell proliferation in vitro, and
drives both tumorigenesis and metastasis in animal model systems
(2). Aberrant STYK1/NOK expression
has been identified in a wide range of cancer types, including
lung, ovarian, breast, colorectal, prostate and renal cell cancer
(3–8). Notably, cells overexpressing STYK1/NOK
exhibit a similar metabolic profile compared with cancer cells,
namely functions in aerobic glycolysis or the Warburg effect, which
is reflected in augmented glucose uptake and lactate production,
upregulation of key glycolytic enzymes and regulators, impaired
electron transport and mitochondrial oxidative phosphorylation
(OXPHOS) (9).
As a consequence of aerobic glycolysis, cancer cells
become heavily dependent on both glycolysis and glucose uptake. In
order to incorporate sufficient amounts of glucose, cells increase
the expression levels of different glucose transporters (GLUTs). At
present, 14 types of human GLUTs encoded by different genes have
been identified. Although their substrate specificity and tissue
distribution are different, these GLUTs have common sequence
characteristics and are highly conserved in numerous species, such
as mice and rats (10). According to
the differences in extracellular structure, these GLUTs can be
classified into three categories: Class I (GLUT1-4), class II
(GLUT5, 7, 9 and 11) and class III (GLUT6, 8, 10, 12 and 13)
(11). Class I GLUTs were discovered
first and studied in depth. Among them, GLUT1 and GLUT3 are widely
distributed in the plasma membrane of all tissues and cells, and
are responsible for maintaining the basic level of glucose uptake
under normal physiological conditions (10–12).
GLUT2 is mainly present in certain tissues with high glucose
concentrations, such as those in the intestine and liver (13). GLUT4 is highly expressed in
insulin-sensitive tissues, including brown and white fat, skeletal
muscle and the myocardium (10). The
newly discovered GLUT14 has 95% sequence homology with GLUT3 and is
only present in the testis; its role in glucose transport remains
unclear.
Dysfunctions of certain GLUTs are closely associated
with various diseases. Accumulating data have indicated that most
tumor tissues have an abnormal GLUT expression profile compared
with normal tissues, which is crucial for maintaining the
proliferation, metastasis and survival of cancer cells under
hypoxia (14,15). In recent years, an increasing number
of researchers have paid attention to the structural
characteristics, the expression and regulation, and the clinical
application of the main GLUTs in terms of their role as malignant
tumor markers (16–18). However, most reports have focused on
a specific type of tumor cell, and there are relatively few studies
on the role of GLUTs in carcinogenic RPTKs, including
STYK1/NOK-mediated malignant transformation and tumorigenesis
(19–21).
The present study focused on the most significant
class I GLUTs (GLUT1-4) and provided evidence for the functional
involvement of the GLUT3 transporter in STYK1/NOK-mediated
metabolic reprogramming and cell proliferation characteristics.
Materials and methods
Cell lines and reagents
The murine NIH-3T3 fibroblast cell line was obtained
from the China Infrastructure of Cell Line Resources, Institute of
Basic Medical Sciences, Chinese Academy of Medical Sciences.
NIH-3T3 cells were grown in DMEM supplemented with 10% FBS (both
Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. The
following antibodies were used in the present study: Anti-STYK1
(cat. no. 18028-1-AP), anti-GLUT1 (cat. no. 21829-1-AP), anti-GLUT2
(cat. no. 20436-1-AP), anti-GLUT4 (cat. no. 21048-1-AP),
anti-hexokinase (HK)1 (cat. no. 19662-1-AP), anti-platelet
phosphofructokinase (PFKP) (cat. no. 13389-1-AP) and anti-pyruvate
kinase (PKM)1 (cat. no. 15821-1-AP), all from ProteinTech Group,
Inc.; anti-GLUT3 (cat. no. ab191071) and anti-pyruvate
dehydrogenase α1 (PDHA1) (cat. no. ab168379), all from Abcam, Inc.;
anti-β-actin (cat. no. 4970) and anti-β-tubulin (cat. no. 2146),
both from Cell Signaling Technology, Inc.; and HRP-conjugated
secondary antibody (cat. no. TA130003) from OriGene Technologies,
Inc. MTT, DMSO and G418 were purchased from Sigma-Aldrich; Merck
KGaA.
Plasmid construction and transient
transfection
The pcDNA3.0 and pcDNA3.0-STYK1/NOK plasmids were
constructed previously (9). For
construction of pSilencer-small interfering RNA (si/siRNA) GLUT3,
the single-stranded oligonucleotides
(5′-AGCTTAAGTAGCTAAGTCGGTTGAAACTCGAGTTTCAACCGACTTAGCTACTTG-3′ and
5′-GATCCAAGTAGCTAAGTCGGTTGAAACTCGAGTTTCAACCGACTTAGCTACTTA-3′) were
annealed to double strands before being subcloned into the
HindIII and BamHI sites of pSilencer 4.1-CMV neo to
form the pSilencer-siGLUT3 construct. For construction of
pSilencer-small interfering RNA (si/siRNA) control, the
single-stranded oligonucleotides
(5′-AGCTTAATGGATCAATGGCTGGTAAACTCGAGTTTACCAGCCATTGATCCATTG-3′ and
5′-GATCCAATGGATCAATGGCTGGTAAACTCGAGTTTACCAGCCATTGATCCATTA-3′) were
annealed to double strands before being subcloned into the
HindIII and BamHI sites of pSilencer 4.1-CMV neo to
form the pSilencer-siCtrl construct. All enzymes and reagents used
for plasmid construction were purchased from Ambion, Inc. For
transient transfection, NIH-3T3 cells at ~80% confluence were
transiently transfected with 4 µg plasmid DNA using
Lipofectamine® 2000 transfection reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Construction of the NIH-3T3 stable
cells
Plasmids pcDNA3.0 or pcDNA3.0-STYK1/NOK (4 µg) were
transfected into NIH-3T3 cells. After 24 h of transfection, the
cell culture medium was replaced with fresh medium containing 600
µg/ml G418. After 2 weeks of screening, cell colonies were picked
up and expanded in a 24-well tissue culture plate. Finally, reverse
transcription-quantitative PCR (RT-qPCR) and western blotting were
performed to detect STYK1/NOK expression. NIH-3T3-pcDNA3.0 stable
cells were represented as ‘vehicle’ and NIH-3T3-pcDNA3.0-STYK1/NOK
stable cells were represented as ‘STYK1/NOK’.
RT-PCR and RT-qPCR
Total RNA was extracted from the cultured cells
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.). RT-PCR was performed using the PrimeScript one-step RT-PCR
kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocols. The operating conditions for RT-PCR were
as follows: 50°C for 35 min for reverse transcription and 94°C for
5 min for denaturation. The PCR conditions were: 94°C for 30 sec,
50°C for 30 sec and 72°C for 50 sec, repeated for 20–30 cycles; the
reaction was extended at 72°C for 10 min before the reaction
product was stored at 4°C. RT-qPCR was performed using the One Step
SYBR PrimeScript RT-PCR kit II (Perfect Real Time; Takara
Biotechnology Co., Ltd.). The operating conditions for RT-qPCR
were: 42°C for 5 min and 95°C for 10 sec; 95°C for 5 sec and 60°C
for 30 sec (repeated for 40 cycles). The dissociation of the
reaction products was from 55°C to 95°C as the temperature
increased at a rate of 0.2°C per 10 sec. The gene expression levels
of β-actin were used as an internal control. The RT-qPCR results
were calculated using the 2−ΔΔCq method (22). All primers were designed using the
PrimerBank online program (https://pga.mgh.harvard.edu/primerbank/). Primer
sequences are listed in Table I.
 | Table I.Primers used in RT-PCR/RT-qPCR
analysis. |
Table I.
Primers used in RT-PCR/RT-qPCR
analysis.
| Gene name | GenBank ID | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Size of product,
bp |
|---|
| β-actin | BC009275 |
CAGCCTCGTCCCGTAGACA |
CGCTCCTGGAAGATGGTGAT | 161 |
| STYK1/NOK | KP729000 |
TCACCTAGAGAGCTGCGCTT |
CGTAGTCTGGGACGTCGTATG | 181 |
| GLUT1 | NM_011400 |
GATCCCAGCAGCAAGAAGGT |
AGAGACCAAAGCGTGGTGAG | 197 |
| GLUT2 | NM_031197 |
TGAGTTCCTTCCAGTTCGGC |
TGTAAGTGGGGTGTCTGTGC | 152 |
| GLUT3 | NM_011401 |
CAGGAATCTTCAAGGACGCGG |
CGAAATCGTCATGAAAACGGAGC | 179 |
| GLUT4 | NM_009204 |
ATTCTGCTGCCCTTCTGTCC |
GGAGCTGGAGCAAGGACATT | 184 |
| HK1 | NM_001146100 |
GAGTCTGAGGTCTACGACACC |
CCCACGGGTAATTTCTTGTCC | 131 |
| PFKP | NM_019703 |
CGCCTATCCGAAGTACCTGGA |
CCCCGTGTAGATTCCCATGC | 130 |
| PKM | NM_011099 |
GCCGCCTGGACATTGACTC |
CCATGAGAGAAATTCAGCCGAG | 145 |
Western blot analysis
Western blotting was performed as described
previously (23). Cells were
collected and lysed in RIPA buffer (Beijing Solarbio Science &
Technology Co., Ltd.) containing 0.1 M PMSF, protease and
phosphatase inhibitor cocktail for 30 min on ice. The lysates were
centrifuged at 12,000 × g at 4°C for 15 min, and the supernatant
was collected. Protein concentration was determined by the BCA
method. Cell lysates (20 µg) were separated using a 10% gel by
SDS-PAGE and transferred to a nitrocellulose membrane (Cytiva) at
200 mA for 1.5 h. The membrane was blocked with 5% BSA (Beijing
Solarbio Science & Technology Co., Ltd.) for 1 h at room
temperature, probed with a primary antibody (dilution, 1:1,000) at
4°C overnight and an appropriate secondary antibody (dilution,
1:5,000) for 1 h at room temperature. Finally, the proteins were
visualized using EasySee Western Blot kit (Beijing TransGen Biotech
Co., Ltd., Beijing, China), and imaged and quantified using
ChemiDoc MP Imaging system (Image Lab Software, version 4.1;
Bio-Rad Laboratories Co., Ltd.).
Glucose consumption and lactate
production assays
For the glucose consumption assay, cells were seeded
into a 24-well culture plate in DMEM supplemented with 10% FBS.
After 48 h, 100 µl culture medium was obtained from each well for
the determination of glucose content using a glucose assay kit
(cat. no. BC2490; Beijing Solarbio Science & Technology Co.,
Ltd.) according to the manufacturer's instructions. For the lactate
production assay, cells were seeded into a 6-well culture plate in
normal growth medium. After 48 h, 1×106 cells were
collected from each well for the determination of lactate content
using a lactate assay kit (cat. no. BC2230; Beijing Solarbio
Science & Technology Co., Ltd.) according to the manufacturer's
instructions.
Cell proliferation assay using
MTT
For the proliferation assay, cells were seeded into
96-well plates at a density of 5,000 cells per well. After
incubation for 24, 48 and 72 h, cell proliferation was assessed by
the addition of MTT at a final concentration of 0.5 mg/ml for 1–4
h. After removing the culture medium, the cells in each well were
re-suspended with 150 µl DMSO and then shaken for 10 min at 37°C to
fully dissolve the crystals. The reaction products were measured at
an optical density of 490 nm with a spectrophotometer.
Cell migration assay
Cell migration was analyzed using 8.0-µm
Transwell® Inserts (Corning Life Sciences). Briefly,
1×105 cells were seeded into the upper chamber
supplemented with 100 µl serum-free medium, while 650 µl DMEM
supplemented with 10% FBS was added to the lower chamber as the
inducer. Following 24 h of incubation with 5% CO2 at
37°C, non-migrated cells in the upper chamber were removed using a
cotton swab, while migrated cells in the lower chamber were stained
with 0.1% crystal violet for 10 min at room temperature. Finally,
the cells were washed with PBS and counted under an inverted
fluorescence microscope (magnification, ×200; Olympus Corporation)
using the image processing program ImageJ_v1.8.0 software (National
Institutes of Health). Images of five random fields of view were
captured for each group for analysis.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 8 software (GraphPad Software Inc.). Student's
t-test or one-way analysis of variance (ANOVA) followed by Tukey's
post-hoc test was conducted to account for the comparison of two
groups and multiple groups, respectively. Data are presented as the
mean ± SD. P<0.05 was considered to indicate a statistically
significant difference.
Results
Effects of STYK1/NOK on the expression
of GLUT1-4 participating in the glycolysis process
As aforementioned, GLUTs are required for glucose to
enter cells on most occasions. Class I GLUTs (GLUT1-4) were
discovered first and may serve an important role in this process.
Our previous study revealed that STYK1/NOK can promote glucose
uptake in NIH-3T3 cells (9), which
is likely due to the enhanced expression of GLUTs. To investigate
the expression profile of these four GLUTs in STYK1/NOK-mediated
aerobic glycolysis in the present study, NIH-3T3 cells stably
expressing STYK1/NOK were first generated. Cells transfected with
pcDNA3.0 or pcDNA3.0-STYK1/NOK were subsequently selected using
G418.
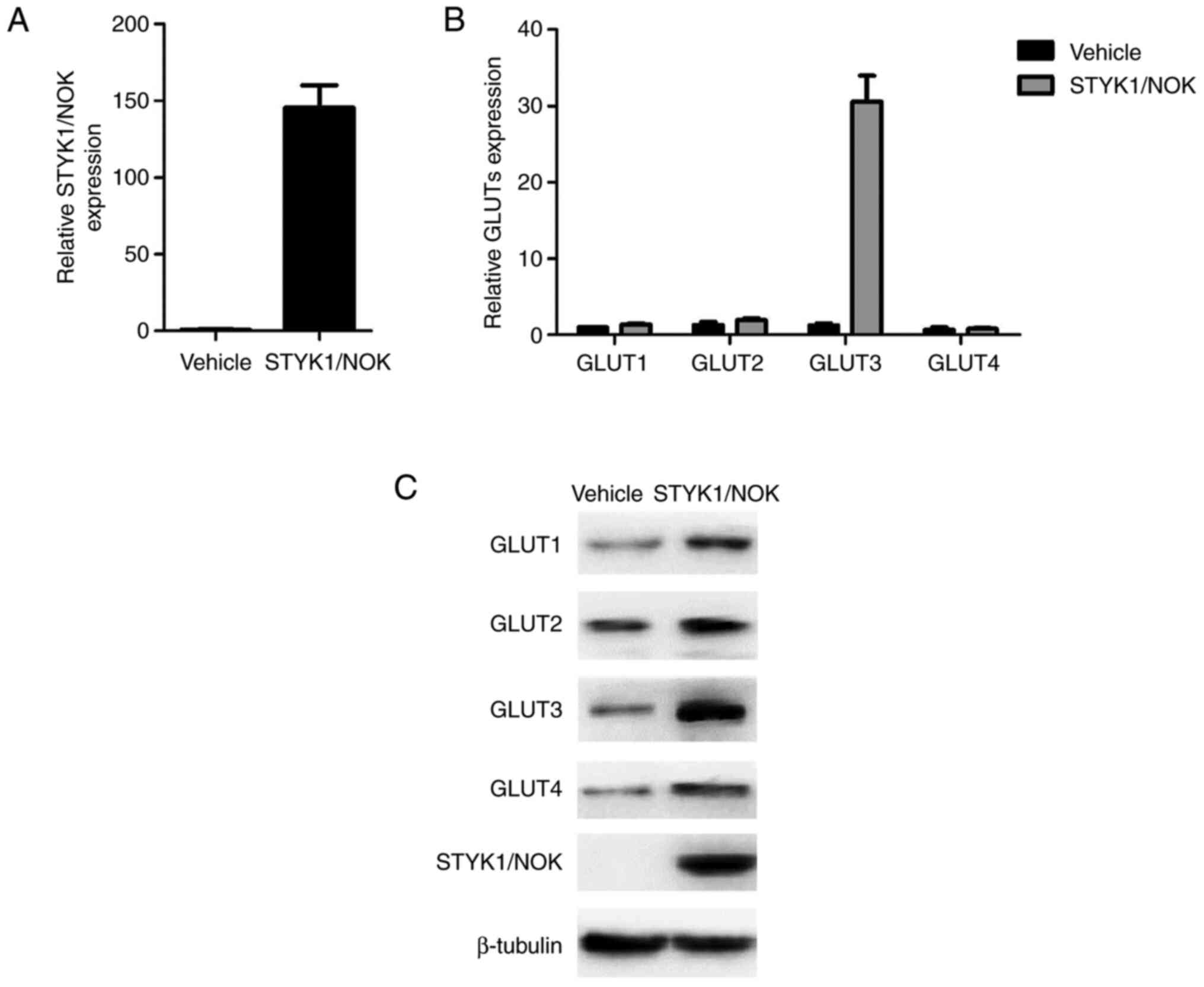
Following successful construction, total RNA and
proteins were extracted from both the NIH-3T3-pcDNA3.0 and
NIH-3T3-pcDNA3.0-STYK1/NOK stable cells. Subsequently, the mRNA and
protein expression levels of STYK1/NOK and GLUT1-4 were detected
using RT-qPCR (Fig. 1A and B) or
western blotting (Fig. 1C),
respectively. Fig. 1B shows that the
mRNA expression levels of GLUT1-4, particularly GLUT3, were
upregulated to varying degrees. Additionally, the results of
western blot analysis were consistent with those of RT-qPCR
(Fig. 1C). These data indicated that
GLUT3 may have a greater impact on cell biological effects driven
by STYK1/NOK. In the present study, GLUT3 was selected as the
target transporter for subsequent experiments.
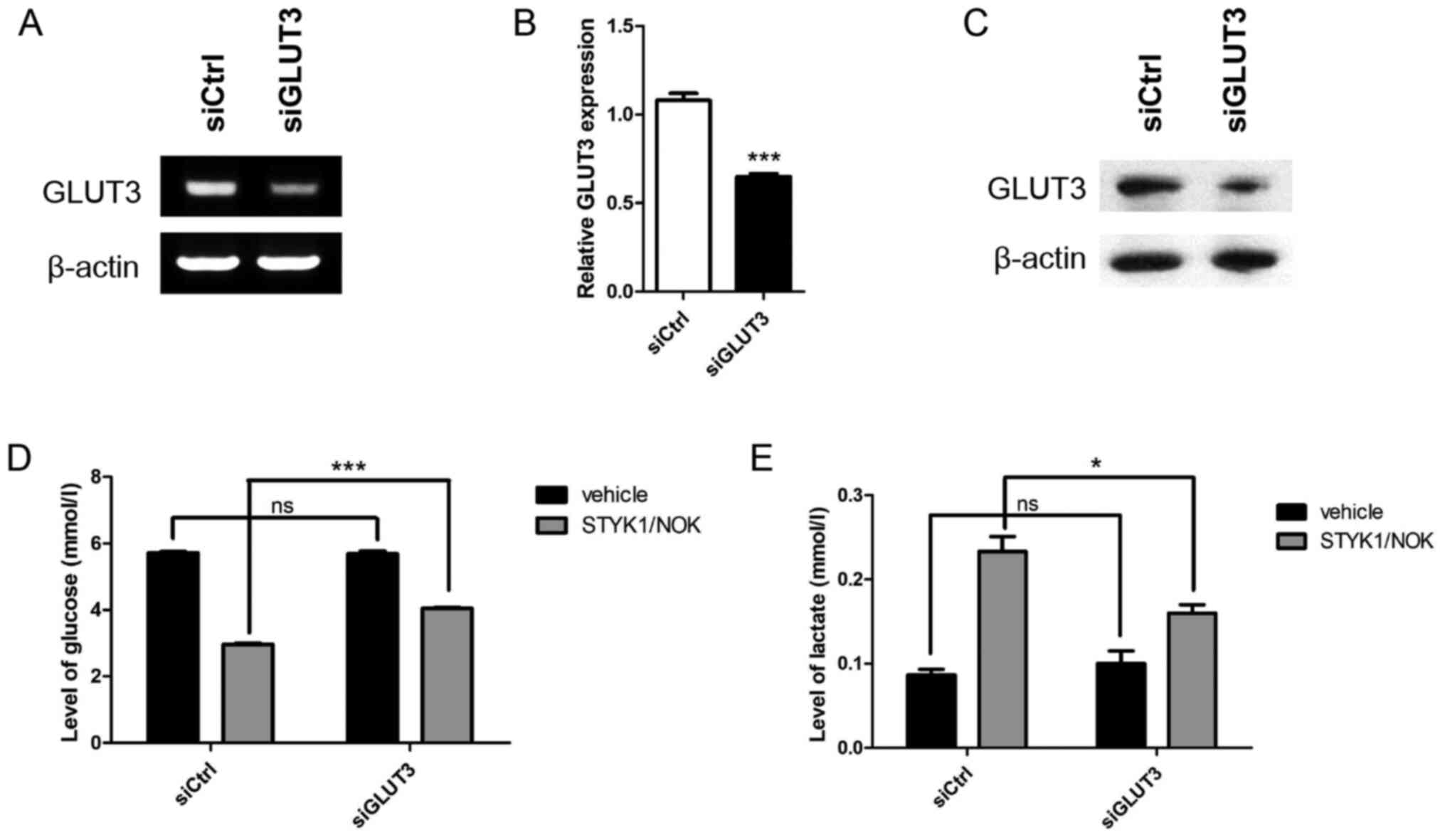
GLUT3 silencing reduces
STYK1/NOK-mediated aerobic glycolysis
To gain insights into the functional involvement of
GLUT3 in STYK1/NOK-induced aerobic glycolysis, siCtrl or siGLUT3
was transiently transfected into NIH-3T3 stable cells. GLUT3
knockdown was assessed by RT-PCR (Fig.
2A), RT-qPCR (Fig. 2B) and
western blotting (Fig. 2C). To
examine the metabolic influence of the downregulation of GLUT3, the
present study subsequently analyzed the glucose uptake capacity of
control and GLUT3-silenced cells by measuring the content of
glucose in the culture medium. Notably, although STYK1/NOK
overexpression promoted glucose consumption in both groups compared
with the vehicle, the increase in STYK1/NOK-mediated glucose uptake
was significantly reduced in the siGLUT3 group compared with the
siCtrl group (Fig. 2D).
Additionally, GLUT3-silenced NIH-3T3-STYK1/NOK stable cells
exhibited reduced levels of lactate compared with non-interfering
NIH-3T3-STYK1/NOK stable cells (Fig.
2E). Overall, these results suggested that metabolic
reprogramming induced by STYK1/NOK in NIH-3T3 cells was impaired as
a consequence of GLUT3 downregulation.
GLUT3 knockdown weakens the effects of
STYK1/NOK on the expression levels of key glycolytic enzymes
For further verification, the present study analyzed
the expression levels of three rate-limiting enzymes, HK, PFKP and
PKM, in the glycolysis signaling pathway, as well as PDHA1, which
is one of the subunits of the pyruvate dehydrogenase complex (PDC),
which catalyzes the conversion of pyruvate to acetyl-CoA (24). Fig.
3A-C shows that the mRNA expression levels of the three
glycolytic enzymes were all upregulated whether or not endogenous
GLUT3 was knocked down, yet the degrees of upregulation of
glycolytic enzymes in NIH-3T3-STYK1/NOK stable cells were
significantly decreased after GLUT3 silencing. Western blot
analysis revealed similar results, with the exception that the
enhanced expression levels of glycolytic enzymes mediated by
STYK1/NOK almost disappeared following GLUT3 knockdown (Fig. 3D).
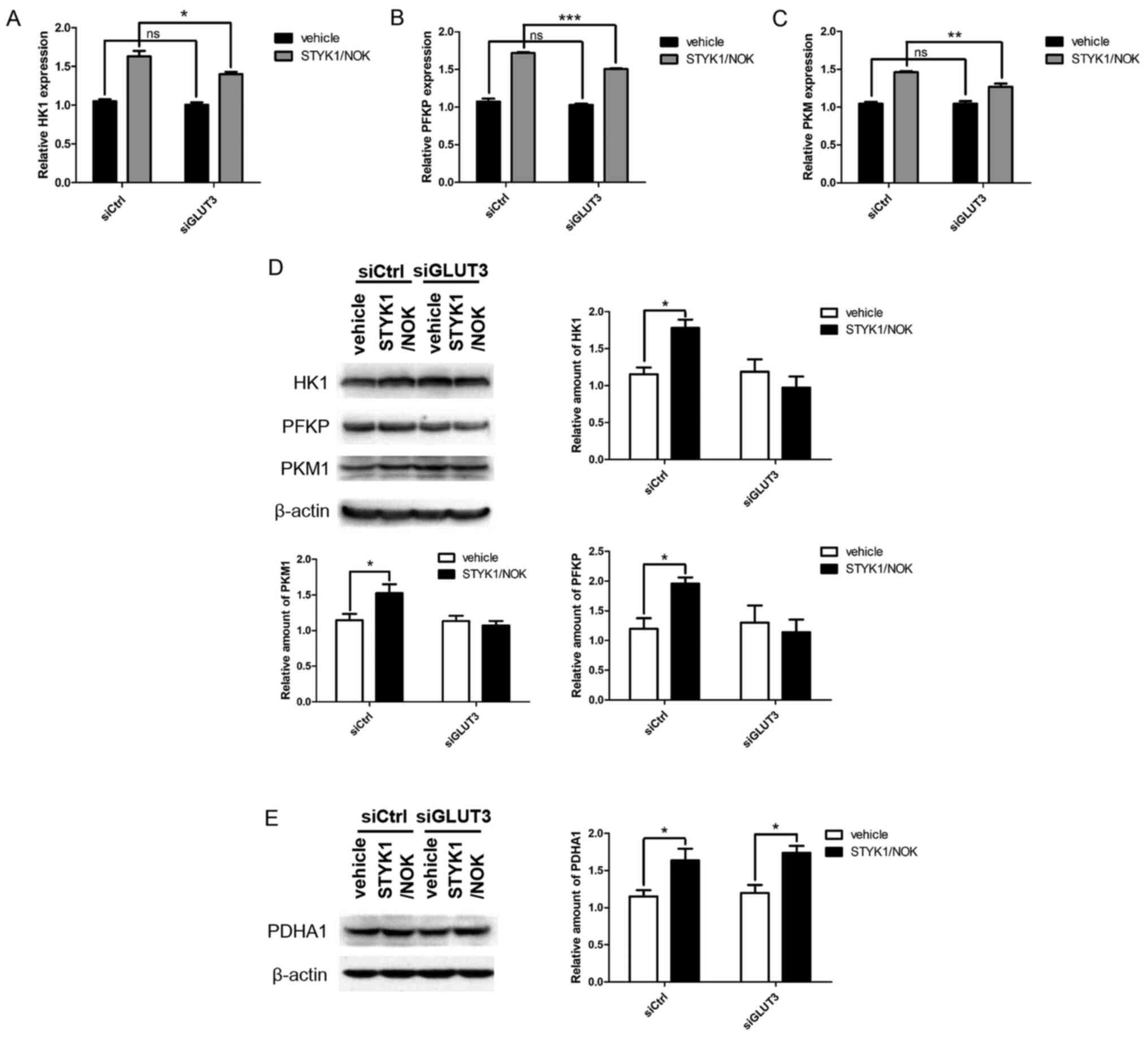 | Figure 3.GLUT3 knockdown weakens the effects of
STYK1/NOK on key enzymes involved in glycolysis. NIH-3T3 stable
cells described as aforementioned were transiently transfected with
either siCtrl or siGLUT3 for 48 h. Subsequently, total RNA was
extracted and subjected to RT-qPCR analysis using primers specific
for (A) HK1, (B) PFKP and (C) PKM. Each value is presented as the
mean ± SD of three independent experiments, which has been adjusted
based on β-actin expression. (D and E) Cell lysates were prepared
and subjected to western blot analysis. The reaction products were
probed using anti-HK1, anti-PFKP, anti-PKM1 and anti-PDHA1. β-actin
was used as a loading control. The statistical histograms represent
the relative amounts of HK1, PFKP, PKM1 and PDHA1, which were
quantitated based on the expression levels of β-actin from three
independent assays using the Image J program. *P<0.05;
**P<0.01; ***P<0.001. ns, no significance; GLUT, glucose
transporter; STYK1, serine threonine tyrosine kinase 1; NOK, novel
oncogene with kinase domain; si/siRNA, small interfering RNA;
siCtrl, control siRNA; siGLUT3, GLUT3 siRNA; HK1, hexokinase 1;
PFKP, platelet phosphofructokinase; PKM, pyruvate kinase; PDHA1,
pyruvate dehydrogenase α1; STYK1, serine threonine tyrosine kinase
1; NOK, novel oncogene with kinase domain. |
In contrast to the clear changes in the expression
levels of glycolytic enzymes, no marked difference in PDHA1
upregulation mediated by STYK1/NOK was identified between the
siCtrl and siGLUT3 groups (Fig. 3E),
suggesting that GLUT3 may mainly affect STYK1/NOK-controlled
aerobic glycolysis, but not the mitochondrial tricarboxylic acid
(TCA) cycle.
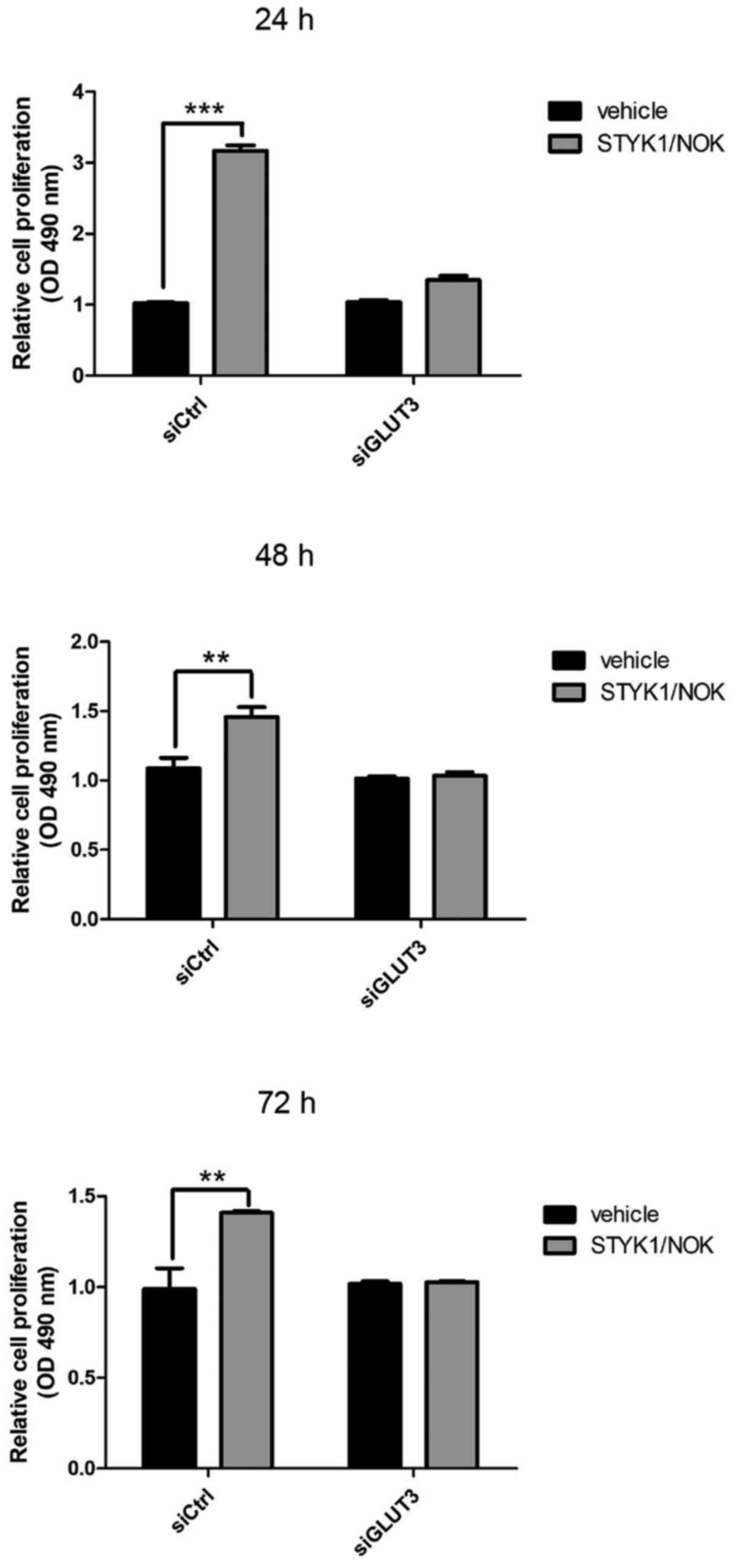
Loss of GLUT3 diminishes cell
proliferation and migration driven by STYK1/NOK
Changes in the glucose metabolism pattern may affect
cell proliferation. To investigate the influence of GLUT3
downregulation on NIH-3T3 cell properties, the present study
analyzed the proliferation rate of GLUT3-silenced and control
NIH-3T3 stable cells using an MTT assay. Fig. 4 shows that STYK1/NOK overexpression
markedly promoted cell proliferation in the siCtrl group at several
different time points (24, 48 and 72 h), which was in agreement
with previous studies. However, this increase in cell proliferation
caused by STYK1/NOK was markedly reduced following GLUT3
knockdown.
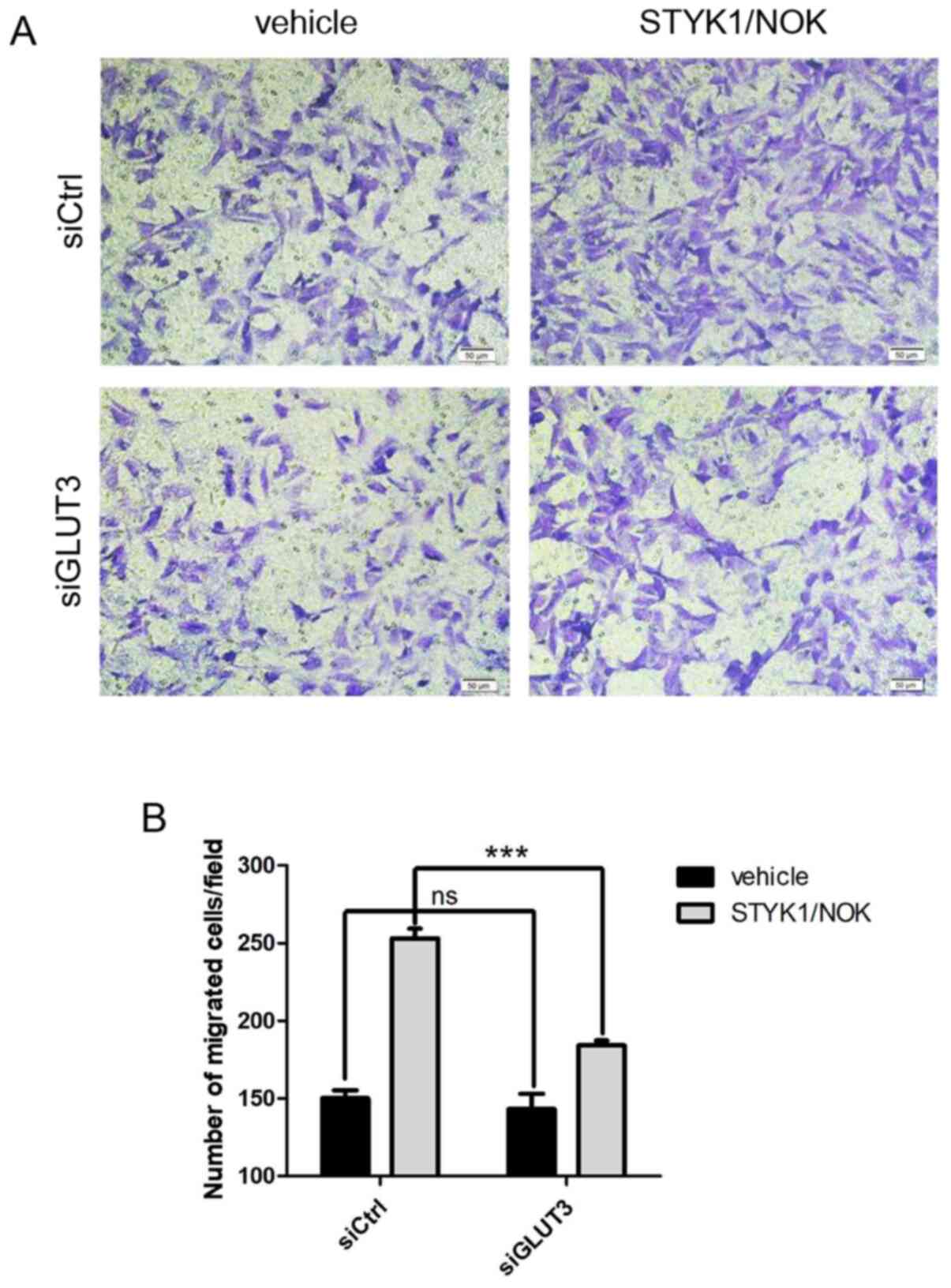
The malignant proliferation of cells is often
accompanied by an enhanced migratory ability. The present study
subsequently investigated whether loss of GLUT3 might also
influence the augmented cell migration caused by STYK1/NOK. A
Transwell chamber-based cell migration assay was performed using
NIH-3T3 stable cells. As shown in Fig.
5, GLUT3 knockdown diminished NIH-3T3-STYK1/NOK stable cell
migration compared with that in the control group. Collectively,
these findings revealed that GLUT3 may be crucial to
STYK1/NOK-mediated malignant transformation and tumorigenesis.
Discussion
Aerobic glycolysis, also referred to as the Warburg
effect, is a highly conserved phenomenon observed in cancer cells
and rapidly proliferating cells. According to Warburg's hypothesis,
cells preferentially transform glucose to lactate even in the
presence of sufficient oxygen (25–27). In
cancer cells, this phenotype is mainly driven by the activation of
oncogenes or loss of tumor suppressor genes (28). For instance, the Warburg effect can
be induced by oncogenic RPTKs, in which a constitutively active
form of RPTKs is usually required (29,30).
STYK1/NOK represents a typical example of RPTKs, which directly
accelerates aerobic glycolysis.
As a consequence of this metabolic reprogramming,
cells depend on a high glucose uptake to produce sufficient amounts
of ATP to maintain their elevated proliferation rate. Therefore,
cancer cells or rapidly proliferating cells exhibit high levels of
GLUTs. Among these, class I GLUTs (GLUT1-4) are considered to serve
a prominent role in this process due to their ubiquitous expression
(10), whereas the involvement of
class I GLUTs in the STYK1/NOK-induced Warburg effect remains
largely unexplored. To the best of our knowledge, the present study
was the first to describe the importance of the GLUT3 transporter
in glucose metabolism and cell biological activities in
STYK1/NOK-overexpressing NIH-3T3 cells. The data revealed that
GLUT3 knockdown led to an impaired capability of basal glucose
uptake and lactate production, demonstrating that STYK1/NOK-induced
aerobic glycolysis depended on the involvement of GLUT3 to a
certain extent. Notably, no apparent effect of GLUT3 downregulation
on PDHA1, which is one of the subunits of PDC that catalyzes the
conversion of pyruvate to acetyl-CoA, was observed in the present
study. This suggested that the influence of GLUT3 on glucose
metabolism in STYK1/NOK-overexpressing cells is mainly located
upstream of the metabolic pathway, whereas the TCA cycle in
mitochondria may not be limited by the downregulation of GLUT3. Our
previous study revealed that STYK1/NOK not only markedly enhanced
aerobic glycolysis, but also markedly inhibited the process of
electron transport and OXPHOS in mitochondria (9). Based on this, a profound study on the
mitochondrial function, including the TCA cycle, electron transport
and OXPHOS, will be performed in the future to comprehensively
analyze the role of GLUT3 in STYK1/NOK-mediated metabolic
reprogramming. Additionally, whether other types of GLUTs serve
similar or different roles in the biological events caused by
STYK1/NOK should be investigated further.
Abnormal energy metabolism is often coupled with
changes in cell biological characteristics. For rapidly
proliferating cells or cancer cells, more energy and intermediate
products can be obtained by reprogramming the metabolic process to
meet their own proliferation requirements. Once this metabolic
pattern is disturbed, cell proliferation characteristics may also
be affected. The present study demonstrated that knockdown of GLUT3
diminished cell proliferation and migration driven by STYK1/NOK,
demonstrating that GLUT3 may be crucial to STYK1/NOK-mediated
malignant transformation and tumorigenesis. Notably, a considerable
number of studies have suggested that STYK1/NOK can activate
various proliferation-related signaling pathways or molecules
(31–33). A number of the signaling pathways
have been reported to strictly regulate the expression and
subcellular distribution of GLUTs, for instance, PI3K/Akt/mTOR,
hypoxia-inducible factor-1, c-myc and tumor suppressor protein p53
(19,34–36),
which provides clues for the further exploration of the upstream
molecular mechanism of GLUT3 in STYK1/NOK-overexpressing cells.
This is the field of research that will be the focus of our work
and studies in the future.
Overall, the data presented in the current study
support the notion that tumor cells are highly dependent on GLUTs.
To the best of our knowledge, the present study was the first to
reveal that GLUT3 downregulation impaired STYK1/NOK-induced
metabolic reprogramming, cell proliferation and migration.
Therefore, the present study provided insights clarifying the
modulatory effects of STYK1/NOK in cell energy metabolism, and for
the future development of pharmacological approaches aimed at
targeting GLUT3.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Hebei Province (grant no. H2019208216), the
Scientific Research Foundation for PhD (grant no. 81/1181286) and
the Natural Science Foundation of Hebei Province (grant no.
H2020208002).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WS was involved in the conception and design of the
present study, performed the experiments and wrote the manuscript.
YF and YW were involved in data analysis and interpretation. WS and
YF confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ye X, Ji C, Huang Q, Cheng C, Tang R, Xu
J, Zeng L, Dai J, Wu Q, Gu S, et al: Isolation and characterization
of a human putative receptor protein kinase cDNA STYK1. Mol Biol
Rep. 30:91–96. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu L, Yu XZ, Li TS, Song LX, Chen PL, Suo
TL, Li YH, Wang SD, Chen Y, Ren YM, et al: A novel protein tyrosine
kinase NOK that shares homology with platelet-derived growth
factor/fibroblast growth factor receptors induces tumorigenesis and
metastasis in nude mice. Cancer Res. 64:3491–3499. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cao Q, Chen M, Li Z, Huang W, Jin Y, Ye X
and Tong M: High novel oncogene with kinasedomain (NOK) gene
expression is associated with the progression of renal cell
carcinoma. Clin Lab. 62:179–186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen P, Li WM, Lu Q, Wang J, Yan XL, Zhang
ZP and Li XF: Clinicopathologic features and prognostic
implications of NOK/STYK1 protein expression in non-small cell lung
cancer. BMC Cancer. 14:4022014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jackson KA, Oprea G, Handy J and Kimbro
KS: Aberrant STYK1 expression in ovarian cancer tissues and cell
lines. J Ovarian Res. 2:152009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moriai R, Kobayashi D, Amachika T, Tsuji N
and Watanabe N: Diagnostic relevance of overexpressed NOK mRNA in
breast cancer. Anticancer Res. 26((6c)): 4969–4973. 2006.PubMed/NCBI
|
|
7
|
Orang AV, Safaralizadeh R, Hosseinpour
Feizi MA and Somi MH: Diagnostic relevance of overexpressed serine
threonine tyrosine kinase/novel oncogene with kinase domain
(STYK1/NOK) mRNA in colorectal cancer. Asian Pac J Cancer Prev.
15:6685–6689. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chung S, Tamura K, Furihata M, Uemura M,
Daigo Y, Nasu Y, Miki T, Shuin T, Fujioka T, Nakamura Y and
Nakagawa H: Overexpression of the potential kinase
serine/threonine/tyrosine kinase 1 (STYK 1) in castrationresistant
prostate cancer. Cancer Sci. 100:2109–2114. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi WY, Yang X, Huang B, Shen WH and Liu
L: NOK mediates glycolysis and nuclear PDC associated histone
acetylation. Front Biosci (Landmark Ed). 22:1792–1804. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thorens B and Mueckler M: Glucose
transporters in the 21st Century. Am J Physiol Endocrinol Metab.
298:E141–E145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barrona CC, Bilan PJ, Tsakiridis T and
Tsiani E: Facilitative glucose transporters: Implications for
cancer detection, prognosis and treatment. Metabolism. 65:124–139.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ganapathy V, Thangaraju M and Prasad PD:
Nutrient transporters in cancer: Relevance to Warburg hypothesis
and beyond. Pharmacol Ther. 121:29–40. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao FQ and Keating AF: Functional
properties and genomics of glucose transporters. Curr Genomics.
8:113–128. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi Y, Liu S, Ahmad S and Gao Q: Targeting
key Transporters in tumor glycolysis as a novel anticancer
strategy. Curr Top Med Chem. 18:454–466. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao M and Zhang Z: Glucose transporter
regulation in cancer: A profile and the loops. Crit Rev Eukaryot
Gene Expr. 26:223–238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alexandra C and Al-Hasani H: Glucose
transporters in adipose tissue, liver, and skeletal muscle in
metabolic health and disease. Pflugers Arch. 472:1273–1298. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reckzeh ES and Waldmann H: Development of
glucose transporter (GLUT) inhibitors. European J Org Chem.
2020:2321–2329. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Groves AM, Shastry M, Rodriguez-Justo M,
Malhotra A, Endozo R, Davidson T, Kelleher T, Miles KA, Ell PJ and
Keshtgar MR: 18F-FDG PET and biomarkers for tumour
angiogenesis in early breast cancer. Eur J Nucl Med Mol Imaging.
38:46–52. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Makinoshima H, Takita M, Saruwatari K,
Umemura S, Obata Y, Ishii G, Matsumoto S, Sugiyama E, Ochiai A, Abe
R, et al: Signaling through the phosphatidylinositol 3-Kinase
(PI3K)/mammalian target of rapamycin (mTOR) axis is responsible for
aerobic glycolysis mediated by glucose transporter in epidermal
growth factor receptor (EGFR)-mutated lung adenocarcinoma. J Biol
Chem. 290:17495–17504. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu H, Ertay A, Peng P, Li J, Liu D, Xiong
H, Zou Y, Qiu H, Hancock D, Yuan X, et al: SGLT1 is required for
the survival of triple-negative breast cancer cells via
potentiation of EGFR activity. Mol Oncol. 13:1874–1886. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Whiteman EL, Chen JJ and Birnbaum MJ:
Platelet-derived growth factor (PDGF) stimulates glucose transport
in 3T3-L1 adipocytes overexpressing PDGF receptor by a pathway
independent of insulin receptor substrates. Endocrinology.
144:3811–3820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhuo Z, Hu J, Yang X, Chen M, Lei X, Deng
L, Yao N, Peng Q, Chen Z, Ye W and Zhang D: Ailanthone inhibits
Huh7 cancer cell growth via cell cycle arrest and apoptosis in
vitro and in vivo. Sci Rep. 5:161852015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bose S and Le A: Glucose metabolism in
cancer. Adv Exp Med Biol. 1063:3–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.PubMed/NCBI
|
|
26
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Warburg O and Minami S: Versuche an
uberlebendem carcinom-gewebe. Klin Wochenschr. 2:776–777. 1923.
View Article : Google Scholar
|
|
28
|
Levine AJ and Puzio-Kuter AM: The control
of the metabolic switch in cancers by oncogenes and tumor
suppressor genes. Science. 330:1340–1344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hitosugi T, Kang S, Vander Heiden MG,
Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et
al: Tyrosine phosphorylation inhibits PKM2 to promote the Warburg
effect and tumor growth. Sci Signal. 2:ra732009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ji H, Lee JH, Wang Y, Pang Y, Zhang T, Xia
Y, Zhong L, Lyu J and Lu Z: EGFR phosphorylates FAM129B to promote
Ras activation. Proc Natl Acad Sci USA. 113:644–649. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li YH, Wang YY, Zhong S, Rong ZL, Ren YM,
Li ZY, Zhang SP, Chang ZJ and Liu L: Transmembrane helix of novel
oncogene with kinase-domain (NOK) influences its oligomerization
and limits the activation of RAS/MAPK signaling. Mol Cells.
27:39–45. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li J, Wu F, Sheng F, Li YJ, Jin D, Ding X
and Zhang S: NOK/STYK1 interacts with GSK-3β and mediates Ser9
phosphorylation through activated Akt. FEBS Lett. 586:3787–3792.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang Z, Ma N, Xiong YL, Wang L, Li WM,
Lai YY, Zhang CX, Zhang ZP, Li XF and Zhao JB: Aberrantly high
expression of NOK/STYK1 is tightly associated with the activation
of the AKT/GSK3β/N-cadherin pathway in non-small cell lung cancer.
Onco Targets Ther. 12:10299–10309. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song K, Li M, Xu XJ, Xuan L, Huang GN,
Song XL and Liu QF: HIF-1α and GLUT1 gene expression is associated
with chemoresistance of acute myeloid leukemia. Asian Pac J Cancer
Prev. 15:1823–1829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miller DM, Thomas SD, Islam A, Muench D
and Sedoris K: c-Myc and cancer metabolism. Clin Cancer Res.
18:5546–5553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ramos H, Calheiros J, Almeida J,
Barcherini V, Santos S, Carvalho ATP, Santos MMM and Saraiva L:
SLMP53-1 inhibits tumor cell growth through regulation of glucose
metabolism and angiogenesis in a P53-Dependent manner. Int J Mol
Sci. 21:5962020. View Article : Google Scholar : PubMed/NCBI
|