Introduction
Osteosarcoma (OS) is a primary malignant bone tumor
typically diagnosed in children and young adults, and patients may
suffer from severe pain or other symptoms associated with
pathological fractures (1). Distant
metastases have been reported in ~15-20% of patients diagnosed with
OS (2). Current therapies for OS
include surgical resection and combined chemotherapy, which cure
~70% of cases (3). However, the
overall 5-year survival rate for patients with OS is estimated at
20%, and the survival rates for patients with distant metastatic or
relapsed OS have not improved for the past 30 years (4,5).
Tumor metastasis is associated with the tumor
microenvironment, which plays a key role in the progression of
malignant tumors (6). The
epithelial-to-mesenchymal transition (EMT) process is a crucial
mechanism in the progression of tumor metastasis and is
characterized by the loss of cell-cell adhesions and the
acquisition of cell-matrix interactions (7). EMT is involved in the formation of
several tissues and organs during embryonic development, in
addition to tumor invasion and metastases (8).
Pleckstrin-2 (PLEK2) is a 353 amino acid protein
that is expressed in a variety of tissues (9). PLEK2 expression has been implicated in
breast cancer cell dissemination and is negatively associated with
clinical outcomes (10). Notably,
PLEK2 has been suggested as a potential gene marker for
distinguishing the CD45 subgroup between patients with melanoma and
healthy individuals (11). A
previous study demonstrated that PLEK2 expression in blood serves
as an early biomarker for melanoma diagnosis (11), and another study provided compelling
evidence that PLEK2 can promote gallbladder carcinoma cell
migration, invasion and liver metastasis by regulating the EMT
process (12). Although the role of
PLEK2 in tumor development is gradually being recognized, the
contributions of PLEK2 to OS progression have not yet been
studied.
The present study aimed to investigate the role of
PLEK2 in OS and determine its underlying molecular mechanisms. The
results may provide an avenue for prospective studies and provide a
potential therapeutic target for OS.
Materials and methods
Clinical tissues specimens
The age distribution of patients ranged from 14–22
years, and there were seven patients (all men) >18 years and
thirteen patients (all men) <18 years. A total of 20 pairs of OS
tissues and adjacent normal tissues (2 cm away from tumor) were
collected during surgical resection from the Department of
Orthopedics, The Affiliated Wuxi No. 2 People's Hospital of Nanjing
Medical University (Wuxi, China) and the Department of Orthopedics,
The First Affiliated Hospital of Nanjing Medical University
(Nanjing, China). All tissues were stored at −80°C until protein
extraction or paraffin embedding. The present study was approved by
the Ethics Committee of the Affiliated Wuxi No. 2 People's Hospital
of Nanjing Medical University (Wuxi, China; approval no.
2019-01-0202-05) and written informed consent was provided by
patients or their legal guardians prior to the study start.
Reagents and antibodies
The cell culture reagents, including culture medium,
fetal bovine serum (FBS) and penicillin-streptomycin were purchased
from Gibco; Thermo Fisher Scientific, Inc. The Cell Counting Kit-8
(CCK-8) and EdU Proliferation kits were purchased from Dojindo
Molecular Technologies, Inc., and Guangzhou RiboBio Co., Ltd.,
respectively. The agonist (insulin-like growth factor 1) and
inhibitor (LY 294002) of PI3K/AKT pathway were purchased from
MedChemExpress, Inc. The primary antibodies used were against PLEK2
(ProteinTech Group, Inc.), N-cadherin (Cell Signaling Technology,
Inc.), matrix metalloproteinase (MMP)2 (Cell Signaling Technology,
Inc.), vimentin (Cell Signaling Technology, Inc.), PI3K (Cell
Signaling Technology, Inc.), phosphorylated (p)-PI3K (Cell
Signaling Technology, Inc.), AKT (Cell Signaling Technology, Inc.),
p-AKT (Cell Signaling Technology, Inc.) and GAPDH (Jackson
ImmunoResearch Labortaories, Inc.).
Microarray data analysis
A total of three Gene Expression Omnibus (GEO)
datasets, including GSE12865 (13),
GSE14359 (14) and GSE33382
(15), were downloaded from the GEO
database (https://www.ncbi.nlm.nih.gov/geo), which compare the
mRNA levels of OS tissues with those of normal tissues. After
importing the profiles into R studio (version 3.6.0; RStudio,
Inc.), the data were normalized and probes were nominated.
Differentially expressed genes (DEGs) were identified using R
package (Limma; version 3.30.11; http://www.bioconductor.org/). Genes with |log fold
change| ≥1 were consdiered DEGs. To investigate the biological
processes that PLEK2 participates in, the correlation coefficients
between each gene and PLEK2 were calculated to perform gene set
enrichment analysis (GSEA).
Cell culture
The OS cell lines, U-2 OS, HOS, Saos-2 and 143B, and
human osteoblast hFOB 1.19 cells were purchased from the Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences. OS
cells were maintained in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% FBS and 1% penicillin-streptomycin, at 37°C
with 5% CO2. The hFOB 1.19 osteoblast cells were
maintained in DMEM/F-12 medium containing 0.3 mg/ml geneticin
(Gibco, Thermo Fisher Scientific, Inc.), 10% FBS and 1%
penicillin-streptomycin, at 33.5°C with 5% CO2.
Cell transfection and lentivirus
construction
To determine the biofunctions of PLEK2 in OS cells,
PLEK2 overexpressing U-2 OS cells and PLEK2 knockdown 143B cells
were constructed and verified for PLEK2 expression. Lentiviral
vectors carrying human PLEK2 cDNA and short hairpin RNA (shRNA)
targeting PLEK2 (sh-PLEK2, 5′-CCTGGAGAATGGGAGTGTTAA-3′) were
constructed by Shanghai GenePharma Co., Ltd. When the 2nd cells
reached 40% confluence, they were infected with vector (GenePharma
Co., Ltd.), PLEK2 (GenePharma Co., Ltd.), shRNA encoding a
non-specific control sequence (sh-NC, 5′-TTCTCCGAACGTGTCACGT-3′,
MOI=50), or sh-PLEK2 (MOI=50), using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) overnight at 37°C.
Stably transfected cell lines were obtained by treatment with 5
µg/ml puromycin (Sigma-Aldrich; Merck KGaA) for 1 week before
experimentation. PLEK2 mRNA and protein expression levels in stably
transfected cells were validated via reverse
transcription-quantitative (RT-q)PCR and western blot analyses,
respectively.
CCK-8 assay
The CCK-8 assay was performed to assess OS cell
proliferation. Stably transfected cells were seeded into 96-well
plates at a density of 1,000 cells/well and allowed to grow for
different time periods (every 12 h until 72 h). At the specified
time points, 10 µl CCK-8 reagent was added to each well. Following
incubation for 2 h, cell proliferation was detected at a
wavenlength of at 450 nm, using a spectrophotometer (Thermo Fisher
Scientific, Inc.).
EdU assay
To further investigate the effects of PLEK2 on OS
cell proliferation, the EdU assay was performed. Cells were seeded
into 96-well plates at a density of 4,000 cells/well and allowed to
grow for 24 h. Subsequently, 50 µM EdU solution was added to each
well and incubated for 2 h. Following fixation with 4%
polyformaldehyde, the cells were permeated with 0.5% Triton X-100
and stained with 1× Apollo reagent for 30 min. Nuclei were stained
with 1× Hoechst 33342 and cells were observed under a fluorescence
microscope (magnification, ×200; Carl Zeiss AG).
Colony formation assay
Stably transfected OS cells were seeded into 6-well
plates at a density of 200 cells/well and allowed to grow for 2
weeks. Following incubation, cell colonies were fixed with 4%
polyformaldehyde and visualized by staining with 0.1% crystal
violet. The number of cell colonies was counted manually.
Transwell assay
To assess the pro-metastatic effects of PLEK2 in OS
cells, the Transwell assay was performed using an insert with an
8-µm pore (Corning, Inc.). A total of 10,000 cells were seeded into
the upper chambers in 200 µl DMEM with no FBS, and 600 µl DMEM
supplemented with 10% FBS was plated in the lower chambers.
Following incubation for 24 h, cells were fixed with 4%
polyformaldehyde and stained with 0.1% crystal violet for 10 min.
Cells that migrated into the lower chambers were observed under a
light microscope (magnification, ×200), and the number of
cells/field was calculated using ImageJ software (version 2.1.4.7;
National Institutes of Health). For the invasion assay, Matrigel
was used to pretreat the Transwell inserts at 37°C for 1 h until
gel formation occurred, and the migration assay was performed using
Matrigel-coated inserts.
Wound healing assay
Stably transfected cells were seeded into 6-well
plates until they reached 100% confluence. A P200 pipette tip was
used to scratch the cell monolayers. Cells were subsequently
cultured in DMEM medium without serum to exclude the interference
of cell proliferation in migration distance. At the indicated time
points (0 and 24 h after scratching), the cell layers were observed
under a a fluorescence microscope (magnification, ×40), and the
migration distance was measured to assess cell motility.
RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (Takara, Japan), following the
manufacturer's instructions. The extracted mRNA was subsequently
reverse transcribed at 37°C for 15 min and 85°C for 5 sec using the
PrimeScript RT Reagent kit (Takara Bio, Inc.). qPCR was
subsequently performed (predenaturation at 95°C for 10 min,
denaturation at 95°C for 15 sec, renaturation at 60°C for 60 sec
and repeat for 40 cycles) using a SYBR Green PCR master mix
(Nanjing KeyGen Biotech Co., Ltd.). The following primer sequences
were used for qPCR: PLEK2 forward, 5′-GTGCTCAAGGAGGGCTTC-3′ and
reverse, 5′-GCTTGTAGTACACCAGCGTGTT-3′; and GAPDH forward,
5′-AATGGGCAGCCGTTAGGAAA-3′ and reverse, 5′-GCGCCCAATACGACCAAATC-3′.
PLEK2 expression was normalized to GAPDH and relative expression
was calculated using the 2−ΔΔCq method (16).
Western blotting
Total protein of OS cells or tissues was extracted
using RIPA lysis buffer (Beyotime Institute of Biotechnology)
containing phenylmethylsulfonyl fluoride and phosphatase inhibitor,
and protein concentration was determined using the BCA Protein
Assay kit (Beyotime Institute of Biotechnology). Following
denaturation at 95°C, 30 µg proteins were separated via 10%
SDS-PAGE, electrically transferred onto PVDF membranes and blocked
with 5% skimmed milk at room temperature for 2 h. The membranes
were incubated with primary antibodies overnight at 4°C, followed
by incubation with HRP-conjugated secondary antibodies (Jackson
ImmunoResearch Labortaories, Inc., 1:5,000, cat. no. 111-035-003)
for 2 h at room temperature. TBST (0.1% Tween-20) was used to wash
the membranes between each incubation, and the proteins were
visualized using an ECL detection kit (Tanon Science and Technology
Co., Ltd.). The expression levels of all proteins were normalized
to that of GAPDH and ImageJ software (version 2.1.4.7; National
Institutes of Health) was used for densitometry.
Immunohistochemistry
For immunohistochemistry analysis, all specimens
were embedded in paraffin and cut into 5-µm-thick sections.
Following antigen retrieval with citrate sodium and blocking in 5%
bovine serum albumin at room temperature for 2 h, the sections were
incubated with primary antibody for PLEK2 overnight at 4°C. The
sections were incubated with the corresponding HRP-conjugated
secondary antibody (Jackson ImmunoResearch Labortaories, Inc.,
1:200, cat. no. 111-035-003) for 2 h at room temperature and
visualized using 3,3′-diaminobenzidine (Beijing Solarbio Science
& Technology Co., Ltd.) for 3 min. The sections were observed
under a light microscope (magnification, ×200).
Animal experiments
All in vivo animal experiments were approved
by the Institutional Animal Care and Use Committee of the
Affiliated Wuxi No. 2 People's Hospital of Nanjing Medical
University. Mice were housed under a controlled temperature
(23±0.5°C), humidity (50±10%) and a 12:12 h light:dark cycle, food,
drinking water and litter were changed every 2 days. A total of 16
6-week-old male nude mice (18–20 g) were randomly divided into two
groups (sh-NC and sh-PLEK2, n=8) and injected with stably
transfected OS cells to perform subcutaneous tumorigenesis
analysis. Tumor volumes were examined and calculated (Volume=length
× width2 × 0.5) every 4 days after injection until mice
were euthanized at day 28 post injection. All mice were enthanized
by intraperitoneal injection of pentobarbital sodium (100 mg/kg) 4
weeks after injection (17). The
death of mice was verified 10 min after injection by loss of
movement, breath, heartbeat, corneal reflex and muscular tension.
Tumor weight was measured after the tumor node was removed, and
Ki-67 expression was detected via immunohistochemistry
analysis.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0 software (GraphPad Software, Inc.). All experiments were
performed in triplicate and data are presented as the mean ±
standard deviation. Unpaired Student's t-test was used to compare
differences between two groups, and paired Student's t-test was
used to compare differences between paired OS tissues and normal
tissues. Two-way ANOVA followed by Sidak's multiple comparisons
test were used to compare differences between multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
PLEK2 is significantly upregulated in
OS cell lines and tissues
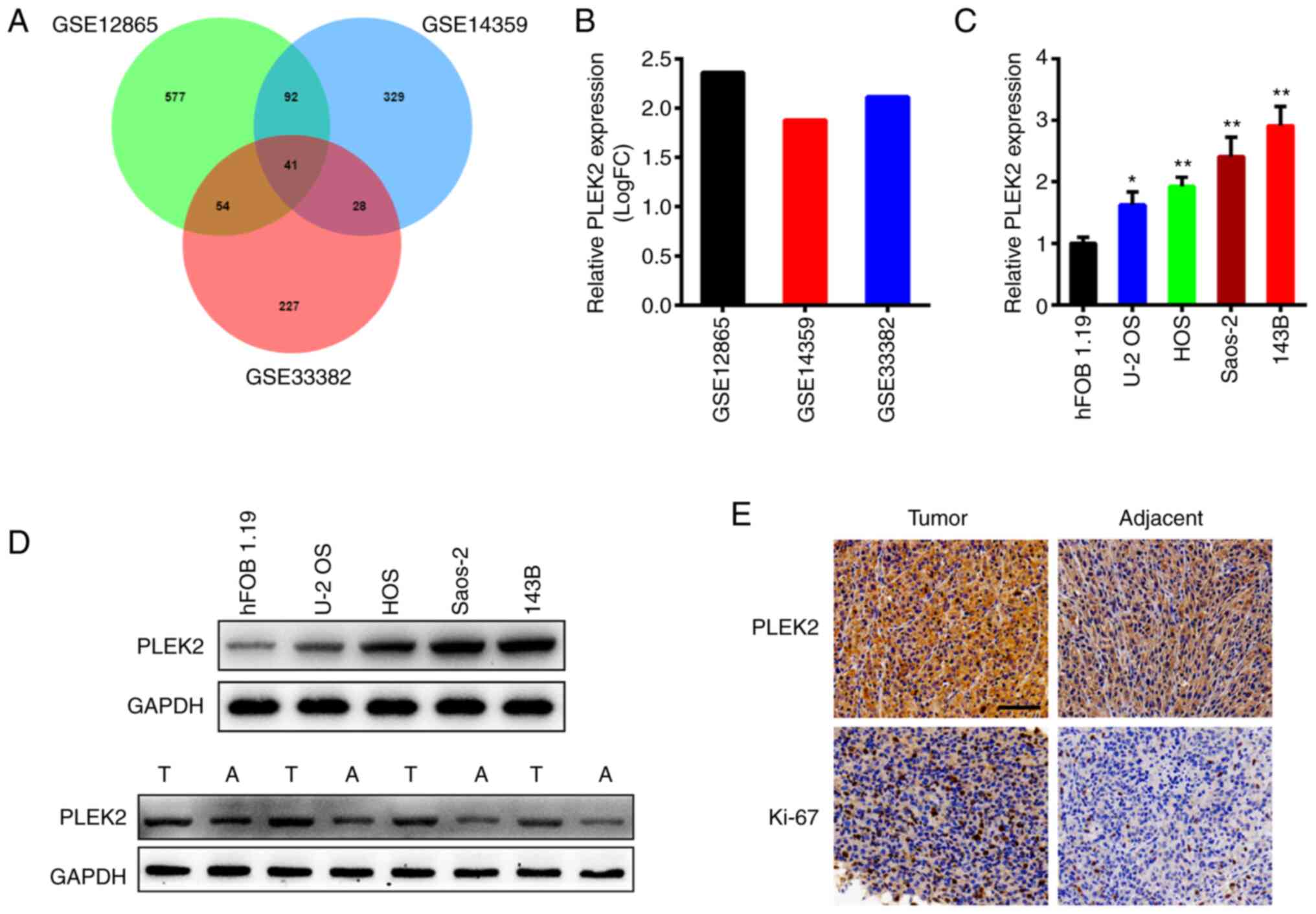
After analyzing three OS datasets using R language,
the results demonstrated that PLEK2 expression was notably higher
in OS tissues compared with adjacent normal tissues in all three
datasets (Fig. 1A), with log fold
change >1 (Fig. 1B). RT-qPCR and
western blot analyses were performed to detect PLEK2 expression in
OS cell lines and tissues. As presented in Fig. 1C (P=0.0272 for U-2 OS, P=0.0023 for
HOS, P=0.0001 for Saos-2 and 143B compared to hFOB 1.19), PLEK2
mRNA expression was significantly upregulated in OS cell lines
compared with osteoblast cells, and 143B cells exhibited the
highest PLEK2 expression, whereas U-2 OS cells exhibited the lowest
expression. Western blot analysis further demonstrated that PLEK2
protein expression was significantly higher in the OS cell lines
and tissues (Fig. 1D). In addition,
immunohistochemistry analysis demonstrated that PLEK2 was highly
expressed in OS tissues (Fig. 1E).
Taken together, these results suggest that PLEK2 expression is
upregulated in OS.
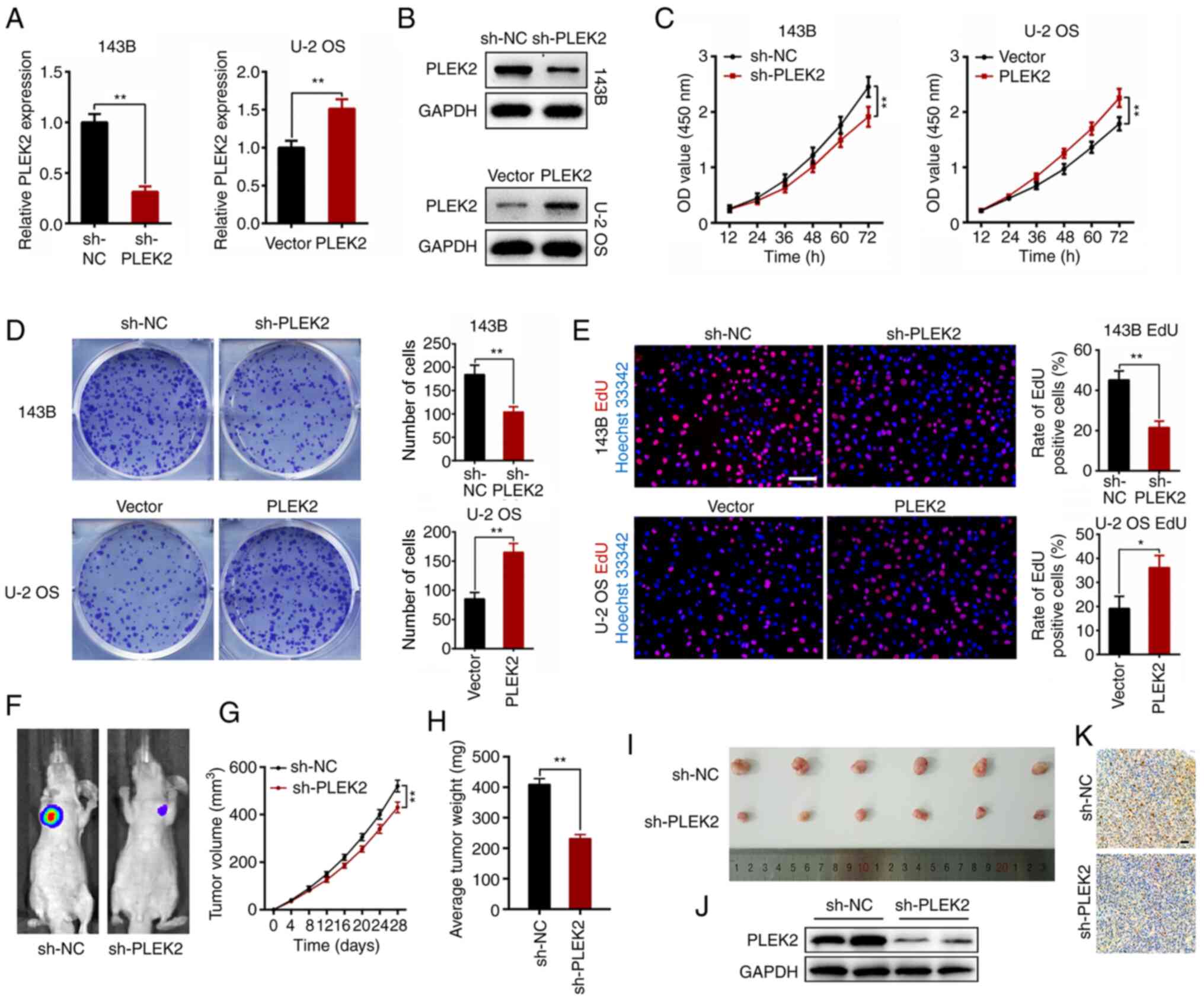
PLEK2 promotes OS proliferation in
vitro and in vivo
Given that PLEK2 expression is different in the four
OS cell lines assessed in the present study, and 143B cells
exhibited the maximum level, whereas U-2 OS exhibited the minimum
level, these cell lines were selected to comprehensively
investigate the effects of PLEK2 on OS proliferation, based on
previous studies (18–22). U-2 OS and 143B cells were transfected
with a PLEK2-coding sequence and shRNA targeting PLEK2,
respectively, and the mRNA and protein levels of PLEK2 following
transfection were detected via RT-qPCR for knockdown (P=0.0002) and
overexpression (P=0.0046; Fig. 2A)
and western blot analyses (Fig. 2B).
The CCK-8, colony formation and EdU assays were subsequently
performed. As presented in Fig. 2C,
PLEK2 knockdown significantly inhibited OS cell proliferation
(P<0.0001), whereas overexpression of PLEK2 promoted OS cell
proliferation (P<0.0001). In addition, PLEK2 knockdown
suppressed the colony formation of OS cells (P=0.0038), whereas
overexpression of PLEK2 increased the number of cell colonies
compared with the control group (P=0.0018; Fig. 2D). The results of the EdU assay
demonstrated that PLEK2 knockdown decreased the percentage of
mitotic cells (P=0.0018), the effects of which were reversed
following overexpression of PLEK2 (P=0.0153; Fig. 2E).
A subcutaneous tumor model was established to better
understand the role of PLEK2 in OS in vivo. The results
demonstrated that PLEK2 knockdown significantly decreased tumor
volume and suppressed OS growth (P<0.0001; Fig. 2F and G). Furthermore, the tumor nodes
retrieved from the PLEK2-knockdown group were much smaller compared
with the control group, with a lower average tumor weight (Fig. 2H and I). Western blotting verified
that PLEK2 expression was suppressed in the sh-PLEK2 group
(Fig. 2J). Immunohistochemistry
analysis demonstrated that PLEK2 knockdown markedly downregulated
Ki-67 expression (Fig. 2K).
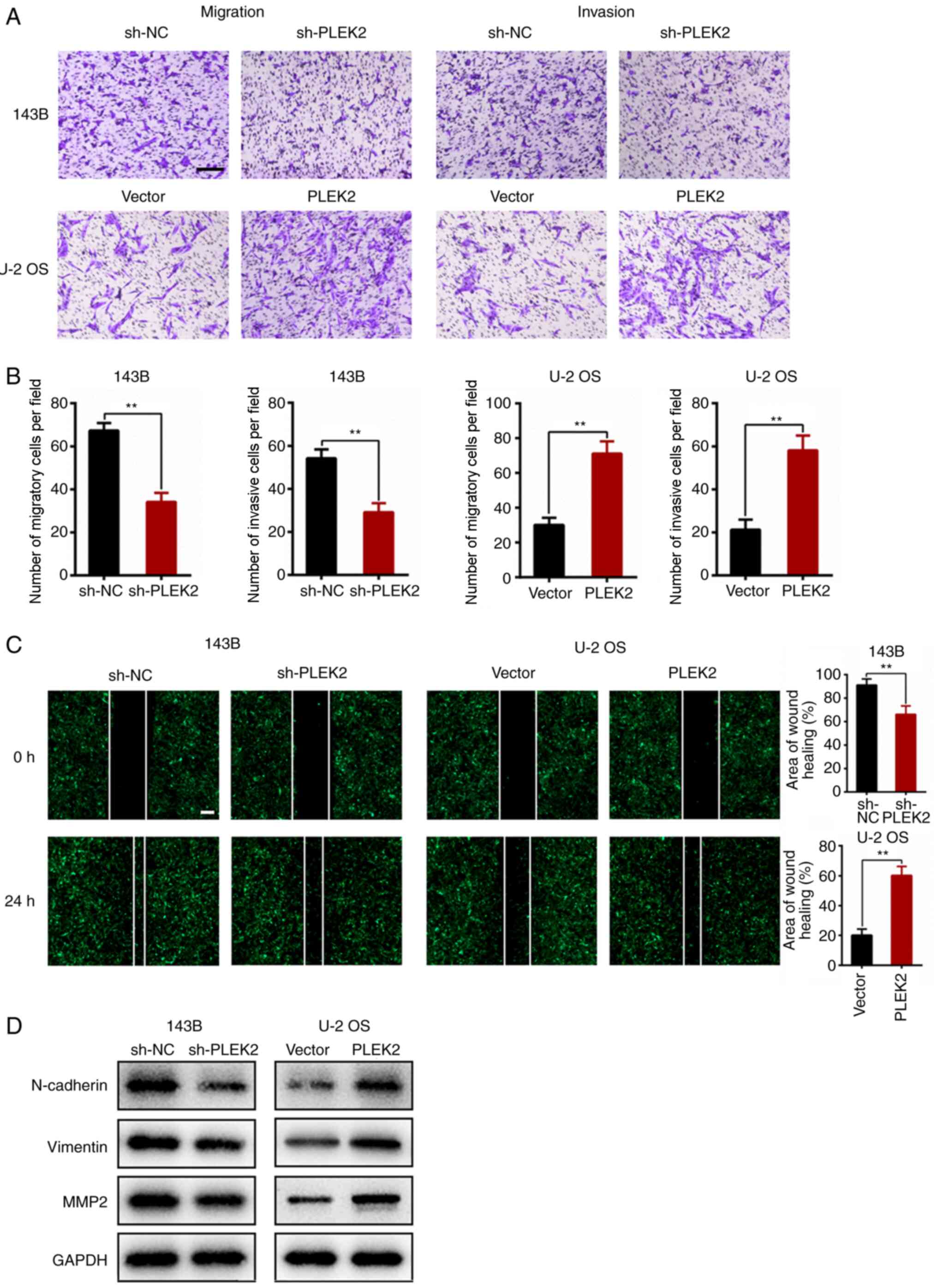
PLEK2 promotes OS metastasis in
vitro
The Transwell assay was performed to investigate the
role of PLEK2 during OS metastasis. The results demonstrated that
PLEK2 knockdown suppressed the migratory and invasive abilities of
143B (P=0.0005 for migration and P=0.0018 for invasion), whereas
overexpression of PLEK2 promoted cell migration and invasion
(P=0.001 for migration and P=0.0016 for invasion, Fig. 3A and B). In addition, the wound
healing assay indicated that PLEK2 knockdown decreased the
migration distance of 143B cells (P=0.0082), whereas overexpression
of PLEK2 enhanced motility of U-2 OS cells (P=0.0007; Fig. 3C).
The EMT process plays a crucial role in tumor
metastasis (23); thus, the present
study analyzed the expression levels of several EMT-related
proteins in OS cells following PLEK2 overexpression or depletion.
The results demonstrated that PLEK2 knockdown attenuated the
expression levels of MMP-2, N-cadherin and vimentin, the effects of
which were reversed following overexpression of PLEK2 (Fig. 3D), which suggests that PLEK2
activates the EMT process.
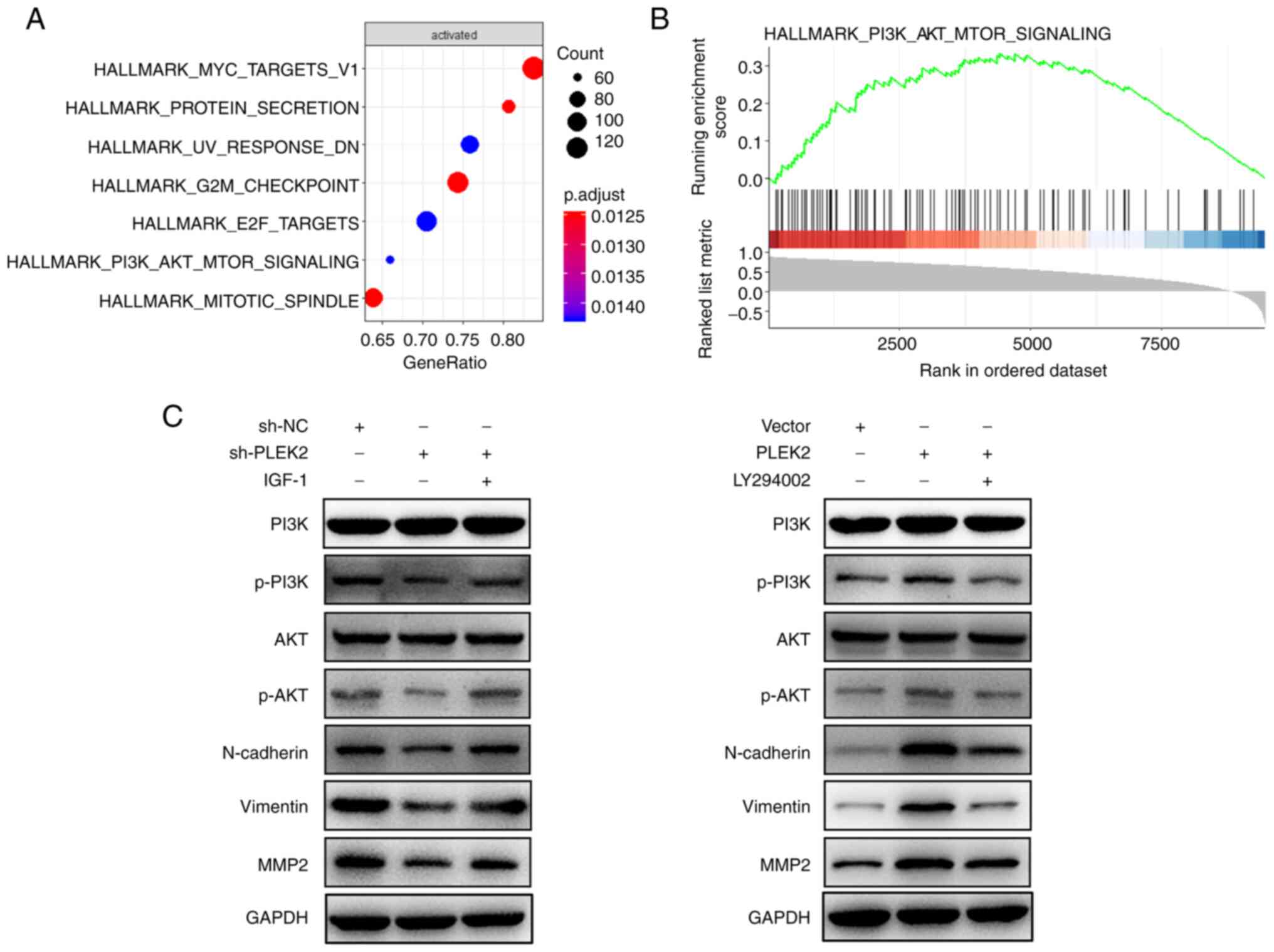
PLEK2 promotes the EMT process by
activating the PI3K/AKT/mTOR pathway
To investigate the underlying molecular mechanism by
which PLEK2 induces the EMT process, single-gene GSEA was
performed, which indicated that the PI3K/AKT/mTOR signaling pathway
was activated following overexpression of PLEK2 (Fig. 4A and B). Western blot analysis
confirmed that PLEK2 knockdown significantly suppressed activation
of the PI3K/AKT/mTOR pathway by decreasing the phosphorylation of
PI3K and AKT, which resulted in the suppression of EMT. However,
the addition of insulin-like growth factor 1, an agonist of the
PI3K/AKT pathway (24), was able to
reverse the effects of PLEK2 knockdown. In addition, overexpression
of PLEK2 activated the PI3K/AKT/mTOR pathway and EMT process, which
was reversed by LY 294002, an inhibitor of the PI3K/AKT pathway
(25) (Fig. 4C).
Discussion
OS is derived from primitive mesenchymal cells, and
mainly originates in bones, with soft tissue origination occurring
only rarely (26). OS is a
high-grade tumor that represents a primary bone malignancy and can
be fatal in children, adolescents and young adults (27). PLEK2 has been demonstrated to be
upregulated in gallbladder carcinoma tissues, which is associated
with the TNM stage and liver metastasis (12). In addition, PLEK2 expressionis
positively associated with breast cancer dissemination to the bone
marrow (10). PLEK2 induces cellular
spreading, causing the formation of large lamellipodia with ruffles
by regulating cytoskeletal rearrangement (28,29). The
movements of lamellipodia are principal mediators of the EMT
process; thus, PLEK2 is considered a dominant driver of cellular
metastasis (30,31). Based on previous studies and
bioinformatics analyses (9,11,12), the
present study investigated the role of PLEK2 in OS and demonstrated
that PLEK2 expression was upregulated in OS tissues and cell
lines.
The EMT process is closely associated with cancer
progression and metastasis. The molecular changes that occur in
epithelial cancer cells are characteristic of EMT, including the
acquisition of mesenchymal qualities and the loss of epithelial
features (7,23). These primary features enhance cancer
cell migration and invasion, and increase antitumoral therapy
resistance (32). In addition,
upregulation of N-cadherin and vimentin, and downregulation of
E-cadherin are significant EMT signals (33). In the present study, E-cadherin
expression decreased, while vimentin and N-cadherin expression
levels increased following overexpression of PLEK2, which promoted
tumor progression and metastasis. According to the in vitro
Transwell and wound healing assays, the OS cell metastasis capacity
was also promoted following overexpression of PLEK2, suggesting
that PLEK2 is a likely risk factor for OS progression.
Environmental signals regulate movement and shape
cells through large protein assemblies at the plasma membrane that
interact with the actin cytoskeleton (34). This regulatory process involves
proteins downstream from PI3K that directly control actin
polymerization (35,36). These sets of proteins regulate cell
protrusions and are important for changing the cellular shape and
facilitating cellular spread (37).
The PI3K/AKT/mTOR pathway is an important regulator of cell
survival during cellular stress conditions (38). Tumors typically occur in a stressful
environment, indicating that the PI3K/AKT/mTOR pathway plays a
crucial role in cancer progression. Somatic mutations and gains and
losses in key genes are among the abundant genetic alterations that
can affect these pathways in a variety of solid and hematological
tumors (39,40). Activation of the PI3K/AKT/mTOR
pathway disrupts cell proliferation and survival regulation,
providing tumor cells with a competitive growth advantage and
inducing angiogenesis, metastatic competence and therapy resistance
(41). Thus, the present study
investigated whether PLEK2 activates the PI3K/AKT/mTOR pathway. The
results demonstrated that PLEK2 knockdown decreased the expression
levels of phosphorylated PI3K, AKT and EMT-related proteins, the
effects of which were reversed following treatment with a
PI3K/AKT/mTOR pathway agonist. Overexpression of PLEK2 reversed
these effects in OS cells, suggesting that PLEK2 promotes OS
metastasis by inducing the EMT process, mediated by activation of
the PI3K/AKT pathway. A limitation of the present study is the lack
of follow-up data, which may be helpful to verify the role of PLEK2
in OS.
In conclusion, the results of the present study
demonstrated that PLEK2 expression was upregulated in OS, and
played a crucial role in OS progression. Furthermore, PLEK2
promoted the EMT process by activating the PI3K/AKT/mTOR signaling
pathway. Taken together, these results suggest that PLEK2 mediates
the promotion of tumorigenesis and metastasis, both in vitro
and in vivo. PLEK2 promotes the EMT process by targeting the
PI3K/AKT/mTOR signaling pathway, promoting metastasis and OS
progression. Thus, these results suggest that the inhibition of
PLEK2 may represent a potential therapeutic target for OS.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Six Talent
Peaks Project in Jiangsu Province (grant no. WSW-099).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YL, SY, FW, ZZ, WX, JX, LQ and YG contributed to
data analysis, drafting and revised the manuscript for important
intellecutual content. YL and YG confirmed the authenticity of all
the raw data. All authors have read and approved the final version
of the manuscript, and agree to be accountable for all aspects of
the work.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Wuxi No. 2 People's Hospital of Nanjing
Medical University (Wuxi, China; approval no. 2019-01-0202-05) and
written informed consent was provided by patients or their legal
guardians prior to the study start. All in vivo animal
experiments were approved by the Institutional Animal Care and Use
Committee of the Affiliated Wuxi No. 2 People's Hospital of Nanjing
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wittig JC, Bickels J, Priebat D, Jelinek
J, Kellar-Graney K, Shmookler B and Malawer MM: Osteosarcoma: A
multidisciplinary approach to diagnosis and treatment. Am Fam
Physician. 65:1123–1132. 2002.PubMed/NCBI
|
|
2
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Link MP, Goorin AM, Miser AW, Green AA,
Pratt CB, Belasco JB, Pritchard J, Malpas JS, Baker AR, Kirkpatrick
JA, et al: The effect of adjuvant chemotherapy on relapse-free
survival in patients with osteosarcoma of the extremity. N Engl J
Med. 314:1600–1606. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meyers PA, Healey JH, Chou AJ, Wexler LH,
Merola PR, Morris CD, Laquaglia MP, Kellick MG, Abramson SJ and
Gorlick R: Addition of pamidronate to chemotherapy for the
treatment of osteosarcoma. Cancer. 117:1736–1744. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taki M, Abiko K, Ukita M, Murakami R,
Yamanoi K, Yamaguchi K, Hamanishi J, Baba T, Matsumura N and Mandai
M: Tumor immune microenvironment during epithelial-mesenchymal
transition. Clin Cancer Res. Apr 7–2021.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu MH, Bauman EM, Roll RL, Yeilding N and
Abrams CS: Pleckstrin 2, a widely expressed paralog of pleckstrin
involved in actin rearrangement. J Biol Chem. 274:21515–21518.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Naume B, Zhao X, Synnestvedt M, Borgen E,
Russnes HG, Lingjaerde OC, Strømberg M, Wiedswang G, Kvalheim G,
Kåresen R, et al: Presence of bone marrow micrometastasis is
associated with different recurrence risk within molecular subtypes
of breast cancer. Mol Oncol. 1:160–171. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo Y, Robinson S, Fujita J, Siconolfi L,
Magidson J, Edwards CK, Wassmann K, Storm K, Norris DA,
Bankaitis-Davis D, et al: Transcriptome profiling of whole blood
cells identifies PLEK2 and C1QB in human melanoma. PLoS One.
6:e209712011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen H, He M, Lin R, Zhan M, Xu S, Huang
X, Xu C, Chen W, Yao Y, Mohan M and Wang J: PLEK2 promotes
gallbladder cancer invasion and metastasis through EGFR/CCL2
pathway. J Exp Clin Cancer Res. 38:2472019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sadikovic B, Yoshimoto M, Chilton-MacNeill
S, Thorner P, Squire JA and Zielenska M: Identification of
interactive networks of gene expression associated with
osteosarcoma oncogenesis by integrated molecular profiling. Hum Mol
Genet. 18:1962–1975. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fritsche-Guenther R, Noske A, Ungethüm U,
Kuban RJ, Schlag PM, Tunn PU, Karle J, Krenn V, Dietel M and Sers
C: De novo expression of EphA2 in osteosarcoma modulates activation
of the mitogenic signalling pathway. Histopathology. 57:836–850.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuijjer ML, Rydbeck H, Kresse SH, Buddingh
EP, Lid AB, Roelofs H, Bürger H, Myklebost O, Hogendoorn PC,
Meza-Zepeda LA and Cleton-Jansen AM: Identification of osteosarcoma
driver genes by integrative analysis of copy number and gene
expression data. Genes Chromosomes Cancer. 51:696–706. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Laferriere CA and Pang DS: Review of
intraperitoneal injection of sodium pentobarbital as a method of
euthanasia in laboratory rodents. J Am Assoc Lab Anim Sci.
59:254–263. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ge X, Liu W, Zhao W, Feng S, Duan A, Ji C,
Shen K, Liu W, Zhou J, Jiang D, et al: Exosomal transfer of LCP1
promotes osteosarcoma cell tumorigenesis and metastasis by
activating the JAK2/STAT3 signaling pathway. Mol Ther Nucleic
Acids. 21:900–915. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kong Y, Wu R, Zhang S, Zhao M, Wu H, Lu Q,
Fu S and Su Y: Wilms' tumor 1-associating protein contributes to
psoriasis by promoting keratinocytes proliferation via regulating
cyclinA2 and CDK2. Int Immunopharmacol. 88:1069182020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin L, Xiao J, Shi L, Chen W, Ge Y, Jiang
M, Li Z, Fan H, Yang L and Xu Z: STRA6 exerts oncogenic role in
gastric tumorigenesis by acting as a crucial target of miR-873. J
Exp Clin Cancer Res. 38:4522019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu W, Jiang D, Gong F, Huang Y, Luo Y,
Rong Y, Wang J, Ge X, Ji C, Fan J and Cai W: miR-210-5p promotes
epithelial-mesenchymal transition by inhibiting PIK3R5 thereby
activating oncogenic autophagy in osteosarcoma cells. Cell Death
Dis. 11:932020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen T, Wang W, Zhou W, Coleman I, Cai Q,
Dong B, Ittmann MM, Creighton CJ, Bian Y, Meng Y, et al: MAPK4
promotes prostate cancer by concerted activation of androgen
receptor and AKT. J Clin Invest. 131:e1354652021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bakir B, Chiarella AM, Pitarresi JR and
Rustgi AK: EMT, MET, plasticity, and tumor metastasis. Trends Cell
Biol. 30:764–776. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng HC, Chang TK, Su WC, Tsai HL and
Wang JY: Narrative review of the influence of diabetes mellitus and
hyperglycemia on colorectal cancer risk and oncological outcomes.
Transl Oncol. 14:1010892021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang J, Xu Y, Ren H, Wudu M, Wang Q, Song
X, Su H, Jiang X, Jiang L and Qiu X: MKRN2 inhibits migration and
invasion of non-small-cell lung cancer by negatively regulating the
PI3K/Akt pathway. J Exp Clin Cancer Res. 37:1892018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim EH, Kim MS, Lee KH, Sai S, Jeong YK,
Koh JS and Kong CB: Effect of low- and high-linear energy transfer
radiation on in vitro and orthotopic in vivo models of osteosarcoma
by activation of caspase-3 and −9. Int J Oncol. 51:1124–1134. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Biermann JS, Adkins DR, Agulnik M,
Benjamin RS, Brigman B, Butrynski JE, Cheong D, Chow W, Curry WT,
Frassica DA, et al: Bone cancer. J Natl Compr Canc Netw.
11:688–723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bach TL, Kerr WT, Wang Y, Bauman EM, Kine
P, Whiteman EL, Morgan RS, Williamson EK, Ostap EM, Burkhardt JK,
et al: PI3K regulates pleckstrin-2 in T-cell cytoskeletal
reorganization. Blood. 109:1147–1155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hamaguchi N, Ihara S, Ohdaira T, Nagano H,
Iwamatsu A, Tachikawa H and Fukui Y: Pleckstrin-2 selectively
interacts with phosphatidylinositol 3-kinase lipid products and
regulates actin organization and cell spreading. Biochem Biophys
Res Commun. 361:270–275. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ombrato L and Malanchi I: The EMT
universe: Space between cancer cell dissemination and metastasis
initiation. Crit Rev Oncog. 19:349–361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Smith BN and Bhowmick NA: Role of EMT in
metastasis and therapy resistance. J Clin Med. 5:172016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu Y, Sarkissyan M and Vadgama JV:
Epithelial-mesenchymal transition and breast cancer. J Clin Med.
5:132016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chaki SP, Barhoumi R, Berginski ME,
Sreenivasappa H, Trache A, Gomez SM and Rivera GM: Nck enables
directional cell migration through the coordination of polarized
membrane protrusion with adhesion dynamics. J Cell Sci.
126:1637–1649. 2013.PubMed/NCBI
|
|
35
|
Lafuente EM, van Puijenbroek AA, Krause M,
Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA,
Gertler FB and Boussiotis VA: RIAM, an Ena/VASP and profilin
ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion.
Dev Cell. 7:585–595. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Krause M, Leslie JD, Stewart M, Lafuente
EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H,
Way M, et al: Lamellipodin, an Ena/VASP ligand, is implicated in
the regulation of lamellipodial dynamics. Dev Cell. 7:571–583.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tscharntke M, Pofahl R, Krieg T and Haase
I: Ras-induced spreading and wound closure in human epidermal
keratinocytes. FASEB J. 19:1836–1838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Datta SR, Brunet A and Greenberg ME:
Cellular survival: A play in three Akts. Genes Dev. 13:2905–2927.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ediriweera MK, Tennekoon KH and Samarakoon
SR: Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer:
Biological and therapeutic significance. Semin Cancer Biol.
59:147–160. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hoxhaj G and Manning BD: The PI3K-AKT
network at the interface of oncogenic signalling and cancer
metabolism. Nat Rev Cancer. 20:74–88. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Porta C, Paglino C and Mosca A: Targeting
PI3K/Akt/mTOR signaling in cancer. Front Oncol. 4:642014.
View Article : Google Scholar : PubMed/NCBI
|