Introduction
Colorectal cancer (CRC) is the third most common
malignant tumor worldwide, with more than one million newly
diagnosed cases each year (1,2). The
5-year survival rate of patients with CRC remains poor (3), albeit with a consecutive reduction in
the morbidity and mortality rates of most cancer types, including
colorectal carcinoma (4), in the
past few decades. The major treatment strategies for CRC include
surgery, radiotherapy and chemotherapy, and the primary reason for
the poor prognosis of patients with CRC is the lack of reliable
biomarkers and therapeutic targets (5). Therefore, it is of great significance
to investigate the pathogenesis of CRC to improve diagnostic
accuracy and treatment options.
Long non-coding RNAs (lncRNAs) are RNA molecules
>200 nucleotides in length (6).
lncRNAs were found to be important in tumorigenesis and to have
various modes of action, such as chromatin modification,
transcription and post-transcriptional regulation (7,8).
Emerging evidence indicates that lncRNAs may also play an important
role in the diagnosis and treatment of CRC (9). lncRNA LINC00662 participates in the
growth and metastasis of CRC by activating the ERK signaling
pathway via the miR-340-5p/claudin-8/IL22 axis (10). lncRNA LINC00858 serves as an oncogene
in CRC, altering the expression of key genes hepatocyte nuclear
factor 4α and WNK2 (11). Previous
studies have also indicated that lncRNA HLA complex group 11
(HCG11) participates in the regulation of tumor progression in
gastric cancer (12), brain glioma
(13) and prostatic cancer (14), and is closely associated with the
prognosis of patients with tumors. However, to the best of our
knowledge, no study has investigated the expression profile of
HCG11 and its role in CRC.
Epithelial-mesenchymal transition (EMT) is the
acquisition of mesenchymal features, discovered and named by
Greenburg and Hay in 1982 (15). EMT
exerts its regulatory roles via complex molecular mechanisms,
including epigenetics, posttranslational regulation and alternative
splicing, as well as the influence of microRNAs (miRs) (16). In highly invasive human tumors, tumor
cells possess significant characteristics of high malignancy for
epithelial and stromal elements, and variations in EMT have been
associated with poor prognosis (17,18). In
addition, increasing evidence indicates that EMT is also involved
in tumor cell resistance to chemotherapy (19–21).
The present study investigated the expression
profile and carcinogenesis role of HCG11 in CRC. Clinical data were
collected to analyze its prognostic value, and tumor cells were
cultured to perform functional experiments, including proliferation
and migration assays. Furthermore, a chemotherapeutic drug
resistance assay was performed by culturing resistant cell lines.
Bioinformatics predictions and validation experiments were used to
explore the mechanisms of HCG11 involved in CRC progression.
Therefore, the present study investigated the roles of HCG11 in CRC
to identify targets and biomarkers for the clinical diagnosis and
treatment of this malignancy.
Materials and methods
Patients and specimens
A total of 23 pairs of CRC and paired non-cancerous
tissues were collected at The First Affiliated Hospital of Yangtze
University (Jingzhou, China) between June 2018 and December 2019.
The mean age of patients (17 males and 6 females) was 61 years,
ranging between 44 and 73 years. None of the patients received
adjuvant treatment, including preoperative chemotherapy, prior to
radical surgery. The present study was reviewed and approved by the
medical ethics committee of The First Affiliated Hospital of
Yangtze University, and all enrolled patients provided written
informed consent. All experiments met the requirements of the
Declaration of Helsinki. The collected tissue samples were frozen
in liquid nitrogen and stored at −80°C until use. All patients were
followed up for long-term clinical data, including overall survival
(OS) and tumor prognosis.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Ambion; Thermo Fisher Scientific,
Inc.). The corresponding cDNA was obtained by reverse transcription
of miRNA, mRNA and lncRNA using the TaqMan miR kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) or ImProm-II kit
(Promega Corporation) according to the manufacturer's instructions,
respectively; qPCR was conducted using SYBR Premix EX Taq™ (Takara
Biotechnology Co., Ltd.) per the manufacturer's instructions (95°C
for 30 sec, followed by 40 cycles at 95°C for 4 sec and 60°C for 34
sec). Gene expression was quantified using the 2−ΔΔCq
method (22) with U6 and GAPDH as a
relative quantitative reference. The primer sequences were as
follows: HCG11 forward, 5′-GCTCTATGCCATCCTGCTT-3′ and reverse,
5′-TCCCATCTCCATCAACCC-3′; SOX4 forward, 5′-GCAAGATCATGGAGCAGTCG-3′
and reverse, 5′-GGGCCGGTACTTGTAGTCG-3′; GAPDH forward,
5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′;
miR-214-5p forward, 5′-ACACTCCAGCTGGGACAGCAGGCACAGAC-3′ and
reverse, 5′-CTCAACTGGTGTCGTGGA-3′; and U6 forward,
5′-GCGCGTCGTGAAGCGTTC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′.
Cell culture
Primary CRC cell lines (HCT116, SW620, SW480 and
HT-29) and HIEC-6 normal colon tissue cells were purchased from the
American Type Culture Collection. The cell lines were verified by
STR profiling before experimental use. All cells were cultured
using Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (Thermo Fisher
Scientific, Inc.), and maintained at 37°C with 5%
CO2.
Transfection
Small interfering (si)RNAs against SOX4 and HCG11
(si-SOX4 and si-HCG11, respectively), and the pcDNA3.1 vector for
HCG11 overexpression (oe-HCG11) were all purchased from Invitrogen
(Thermo Fisher Scientific, Inc.), and miR-214-5p mimics, miR-214-5p
inhibitor and the corresponding negative controls were all
purchased from Shanghai GenePharma Co., Ltd. siRNAs (30 nM) or
plasmids (1.5 µg/well) were transfected into cells at 37°C for 24 h
using Lipofectamine® 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, and cells were harvested for RT-qPCR
analysis 48 h post-transfection. The oligonucleotide sequences were
as follows: siRNA SOX4, 5′-GGACAGACGAAGAGUUUAATT-3′; siRNA HCG11,
5′-UUCUCCGAACGUGUCACGUTT-3′; siRNA negative control,
5′-GCCAGAAUGUUCCUAUUUATT-3′; miR-214-5p mimics,
5′-GGCCTGGCTGGACAGAGTTG-3′; miR-214-5p inhibitor
5′-ACAGCAGGCACAGACAGGCAG-3′; miRNA mimics negative control,
5′-CAGUACUUUUGUGUAGUACAA-3′; and miRNA inhibitor negative control
5′-GUGAGAAGUACCACCGAGACAG-3′.
Cell Counting Kit 8 (CCK-8)
Cells were seeded into a 96-well plate (6 wells per
group) at a density of ~1×104 cells/well. After
transfection, 10 µl CCK-8 reagent was added at 0, 24, 48 or 72 h,
followed by a 2-h incubation at 37°C. To determine cellular
proliferation activity, a microplate reader (iMARK; Bio-Rad
Laboratories, Inc.) was used to measure the absorbance of each well
at 450 nm. In addition, a drug sensitivity assay was conducted
using CCK-8 reagent. Transfected cells were seeded into a 96-well
plate for 24 h, and then cisplatin (Beijing Solarbio Science &
Technology Co., Ltd.) was added to the medium at a concentration of
0, 20, 40, 60, 80 or 100 µM. The absorbance at 450 m of the
corresponding wells was measured after 72 h.
Colony formation assay
Cells were seeded into a 6-well plate at a density
of ~600 cells/well. After 2 weeks of stable culture, the cells were
stained with crystal violet dye for 15 min at room temperature, and
the resulting colonies were counted. The experiment was repeated
three times.
Cell cycle analysis
Cells were collected and resuspended in DMEM with
10% FBS at a density of ~2×105 cells/ml. After digestion
with RNase A, the cells were fixed with 75% ethanol at 4°C
overnight. The cells were then stained with propidium iodide (BD
Biosciences) for 15 min in the dark, and cell cycle analysis was
conducted using a flow cytometer (FACScan; BD Biosciences) and
analyzed using the NovoExpress software v1.2 (ACEA Bioscience,
Inc.; Agilent Technologies, Inc.).
5-Ethynyl-2′-deoxyuridine (EdU)
staining
CRC cells were seeded into a 96-well plate at
1×104 cells/well for 48 h. The cells were subsequently
added to 500 µl EdU reagent (25 µM; Guangzhou RiboBio Co., Ltd.)
and incubated for 2 h, followed by 30 min of fixation in 4%
paraformaldehyde at room temperature. The cells were then
permeabilized with 0.5% Triton X-100 for 10 min, followed by Apollo
and DAPI staining of diagnostic nuclei in a dark environment for 30
min at room temperature. Finally, the stained cells were observed
and counted under a fluorescence microscope (magnification, ×200;
Olympus Corporation).
Transwell assay
Cellular migration assays were performed using an
8-mm Transwell, 24-well plate (Corning Life Sciences). CRC cells
were premixed at ~400 cells/µl in serum-free media, and 100 µl was
added to the upper chamber of the Transwell insert; ~700 µl culture
medium containing 10% fetal bovine serum was added to the lower
chamber. After incubation at 37°C for 24 h, a swab was used to
remove the cells in the upper chamber. CRC cells that had migrated
into the lower chamber were fixed with 4% paraformaldehyde at room
temperature for 15 min and stained using crystal violet dye for 20
min at room temperature. An inverted microscope (magnification,
×200; Olympus Corporation) was used to observe and count the
migrated cells. Cells in a total of six fields of view were
randomly selected for statistical analysis.
Bioinformatics analysis
The bioinformatics prediction websites starBase
(http://starbase.sysu.edu.cn/),
TargetScan (http://www.targetscan.org/) and miRDB (http://mirdb.org/index.html) were used to predict the
target genes of HCG11 and miR-214-5p. Gene Expression Profiling
Interactive Analysis (GEPIA) (23)
is an interactive web for analyzing The Cancer Genome Atlas (TCGA)
and Genotype-Tissue Expression projects online, and it was used to
assess SOX4 expression in CRC and normal tissues.
RNA pull-down assay
The complementary biotin-labeled DNA probe for HCG11
was synthesized by Shanghai GenePharma Co., Ltd. The biotin-labeled
probe was dissolved in buffer according the manufacturer's
protocol, Dynabeads™ M-280 streptavidin (Invitrogen; Thermo Fisher
Scientific, Inc.) was added, and the sample was incubated for 10
min at 24–26°C. Then, the beads were placed into CRC cell lysis
solution overnight at 4°C, and RNA fragments that specifically
bound to the beads were extracted using TRIzol® reagent.
The biotin-labeled full-length HCG11 sequence was used to
specifically pull down miRNAs for subsequent quantitative RT-qPCR
analysis.
Dual-luciferase reporter gene
assay
A dual-luciferase reporter gene assay was used to
verify the binding association of HCG11 and SOX4 to miR-214-5p. The
3′ untranslated region (UTR) containing HCG11 or SOX4, and sequence
fragments of miR-214-5p predicted binding sites, were ligated into
luciferase reporter plasmids and used to co-transfect CRC cells
together with miR-214-5p mimics or miR-214-5p inhibitor using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.); the imported mutant sequence
fragment was used as the reference group. To determine the gene
binding relationship, the dual-luciferase reporter gene system
(Promega Corporation) was used to measure luciferase activity.
Luciferase activity was measured 24 h after transfection and the
results were normalized to Renilla luciferase activity.
Western blot analysis
Cell lysis was performed using RIPA reagent (Thermo
Fisher Scientific, Inc.) containing a protease inhibitor. The BCA
method was used to determine protein concentration. For western
blot analysis, 20 µg protein/lane were separated by 10% SDS-PAGE
and transferred onto a PVDF membrane. The membrane was blocked with
5% skimmed milk at room temperature for 2 h before incubation with
primary antibodies overnight at 4°C. For specific determination of
the expression of the targeted proteins, horseradish
peroxidase-conjugated secondary antibodies (1:2,000; cat. no. 7076;
Cell Signaling Technology, Inc.) were added and incubated at room
temperature for 2 h. ECL solution (Pierce; Thermo Fisher
Scientific, Inc.) was used for visualization, and GAPDH (Cell
Signaling Technology, Inc.) was used as the internal reference. The
primary antibodies used in experiments were as follows: Vimentin
(1:1,000; cat. no. 49636; Cell Signaling Technology, Inc.);
E-cadherin (1:1,000; cat. no. 14472; Cell Signaling Technology,
Inc.); GAPDH (1:1,000; cat. no. 51332; Cell Signaling Technology,
Inc.); and SOX4 (1:500; cat. no. ab70598; Abcam).
Statistical analysis
GraphPad 7.0 software (GraphPad Software, Inc.) was
used for statistical analysis and graphical presentation. The data
are presented as the mean ± standard deviation, and the relevant
experiments were repeated at least three times. Independent-sample
t-tests and paired-sample t-tests were used for the analysis of
significant differences between two groups. Multi-group comparison
analysis was performed using one-way ANOVA validated by
Bonferroni's post-hoc test. Survival analysis of clinical
prognostic data was performed using the log rank test, and
correlation analysis was performed using Pearson's correlation
coefficient. According to the median expression of HCG11 in 23
pairs of specimens, the OS data were grouped into high and low
expression groups, and Kaplan-Meier analysis was used to assess
survival. P<0.05 was considered to indicate a statistically
significant difference.
Results
Chemotherapy inhibits the expression
of HCG11 in CRC
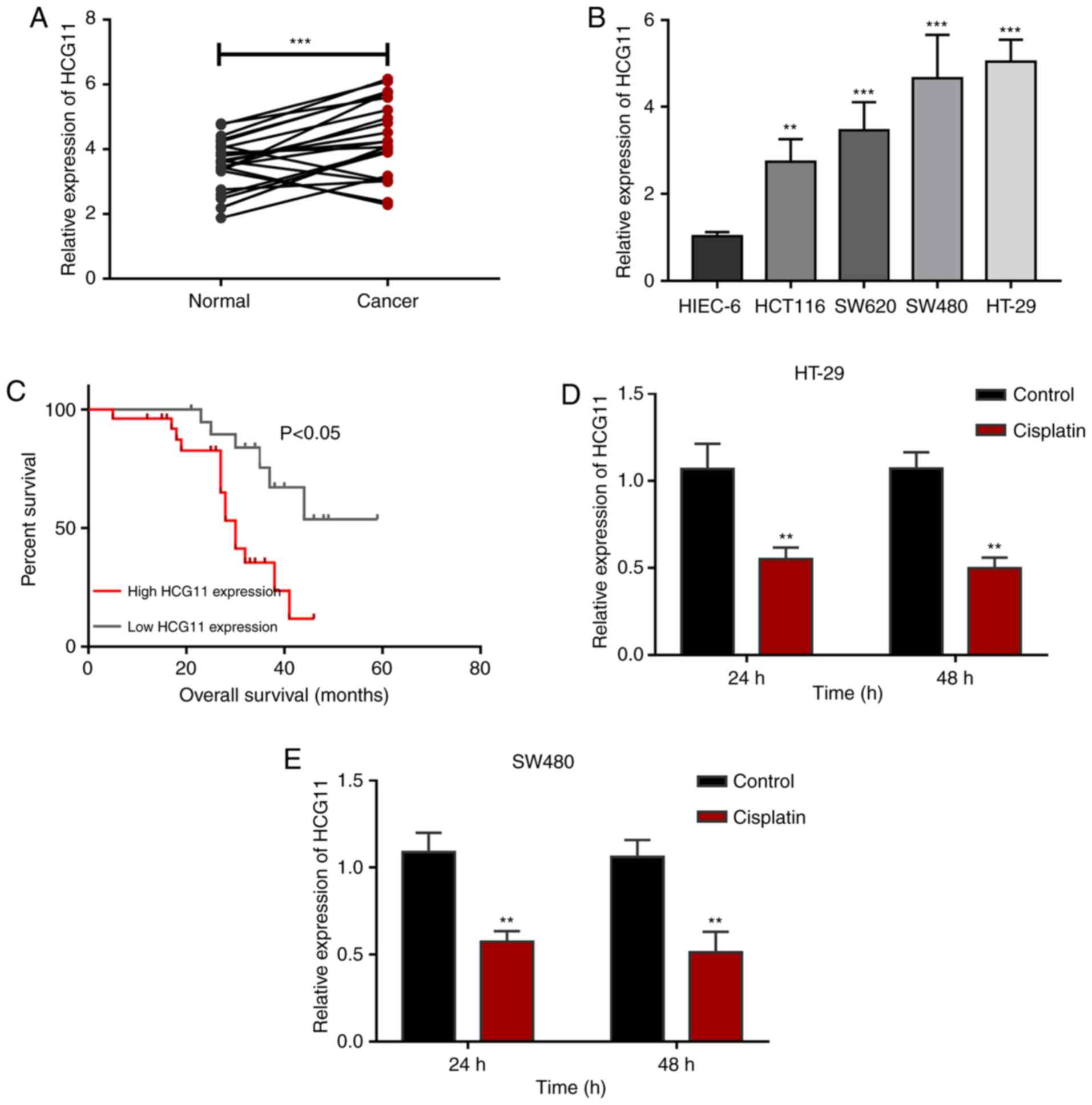
To determine the expression profiles of HCG11 in
CRC, and its potential effects on chemotherapy resistance in CRC
cells, HCG11 expression was compared in clinical samples and cell
lines. RT-qPCR analysis revealed that HCG11 expression was notably
upregulated in CRC tissues compared with normal tissues (Fig. 1A). CRC cell lines (HCT116, SW620,
SW480 and HT-29), and a normal colon cell line (HIEC-6), were
selected for HCG11 expression detection. The results showed that
HCG11 expression was consistently higher in CRC cells than in
normal colon cells (Fig. 1B).
Furthermore, the prognostic clinical data of CRC patients were
randomly collected and survival analysis was conducted to verify
the correlation between HCG11 expression and survival. The results
revealed that CRC patients that highly expressed HCG11 had
considerably lower overall survival times than low-expression
patients (Fig. 1C), suggesting the
value of HCG11 as a clinical prognostic marker in CRC. Chemotherapy
resistance is one of the key factors for the poor prognosis of CRC.
To further investigate the influence of HCG11 on the
chemotherapeutic resistance of CRC, the HT-29 and SW480 cell lines
were used to assess the response to chemotherapy after 24 and 48 h.
Changes in HCG11 expression during chemotherapy were then analyzed
via RT-qPCR in order to determine the association between
chemotherapy and HCG11. The results revealed that mean HCG11
expression in CRC cells was reduced following cisplatin treatment
(Fig. 1D and E), suggesting that
HCG11 expression was affected by tumor chemotherapy. In general,
these findings indicate that HCG11 is highly expressed in CRC and
is associated with poor prognosis. Additionally, chemotherapy had
certain regulatory effects on HCG11 expression in CRC cells,
implying that HCG11 may be involved in the resistance mechanisms of
CRC to chemotherapy.
HCG11-knockdown suppresses the
proliferation and migration of CRC cells
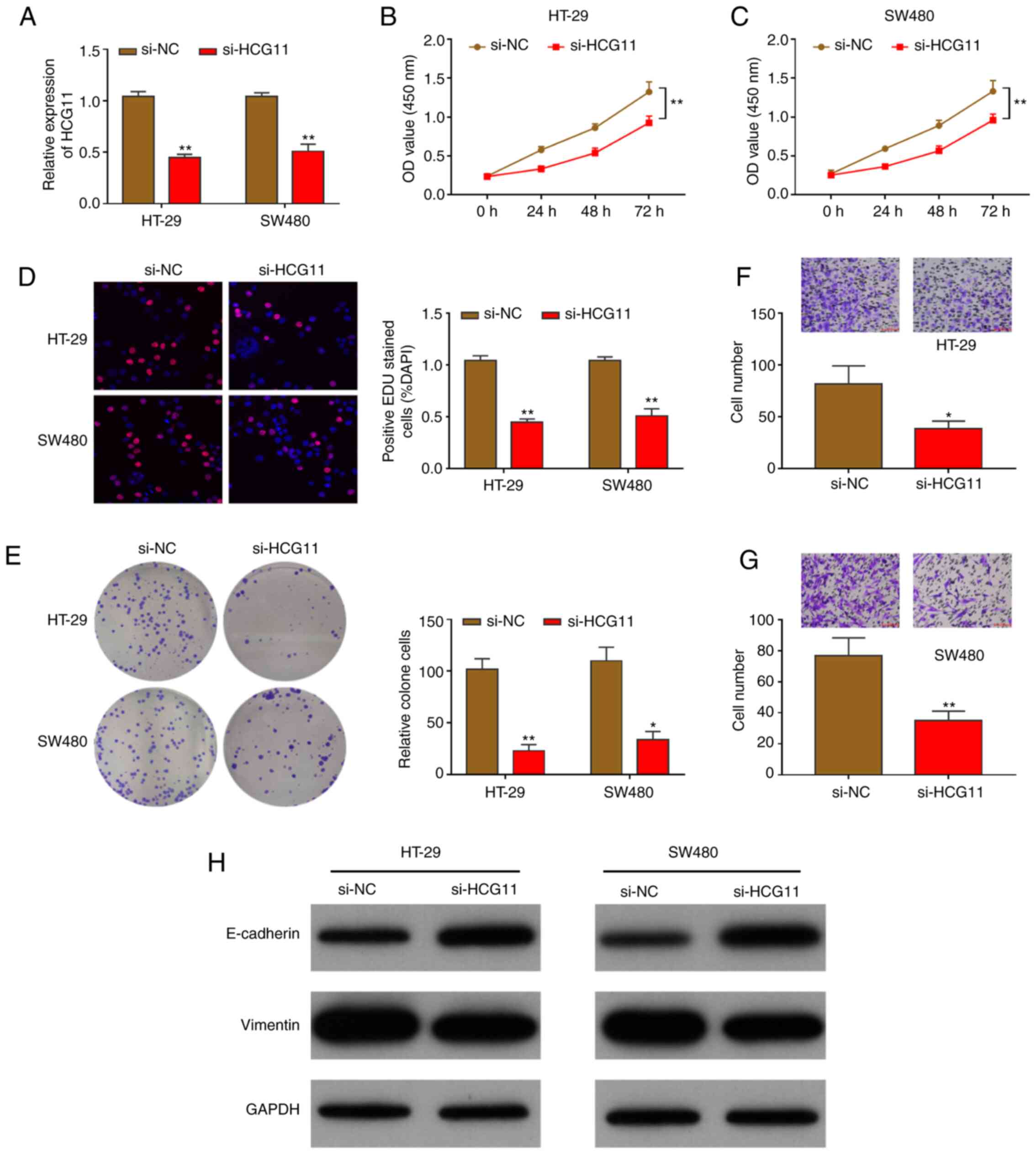
First, HCG11 expression was knocked down in CRC
cells to identify its effects on cellular malignant phenotypes,
including proliferation and migration; si-HCG11 transfection was
conducted to knockdown HCG11 expression in HT-29 and SW480 cells.
transfection efficiency was verified by RT-qPCR, which showed that
HCG11 expression was notably suppressed in HT-29 and SW480 cells
(Fig. 2A). Furthermore, the
proliferation of HT-29 and SW480 cells was assessed using the CCK-8
assay 24 h post-transfection. The results showed that the
proliferation of CRC cells was significantly attenuated after
HCG11-knockdown (Fig. 2B and C).
Furthermore, EdU staining was performed to confirm the effects of
HCG11-knockdown on CRC cell proliferation, which showed that the
ratio of EdU-positive cells was markedly decreased, indicating that
proliferation was attenuated (Fig.
2D). A colony formation assay was also performed to detect
changes in cell colony formation after HCG11-knockdown. Following 2
weeks of culture, colony formation ability was also markedly
suppressed, and the number of colonies formed was significantly
reduced (Fig. 2E). Cellular
migration was then evaluated using a Transwell assay, revealing
that HT-29 and SW480 cell migration was significantly weakened
following HCG11-knockdown. The number of migrated cells was
considerably reduced compared with that of the control group
(Fig. 2F and G).
EMT is the key mechanism mediating cellular
proliferation and migration; therefore, the expression of EMT
pathway-related proteins, such as E-cadherin and vimentin, was
determined. As shown in Fig. 2H,
HCG11-knockdown suppressed vimentin, but increased E-cadherin
protein levels, reflecting the inhibition of the intracellular EMT
pathway. These experimental results further demonstrate that
HCG11-downregulation suppressed the proliferation and migration of
CRC cells via the EMT pathway.
HCG11 induces chemotherapeutic
resistance of CRC cells to cisplatin
Based on the aforementioned findings, the influence
of HCG11 on the chemotherapeutic resistance of tumor cells was then
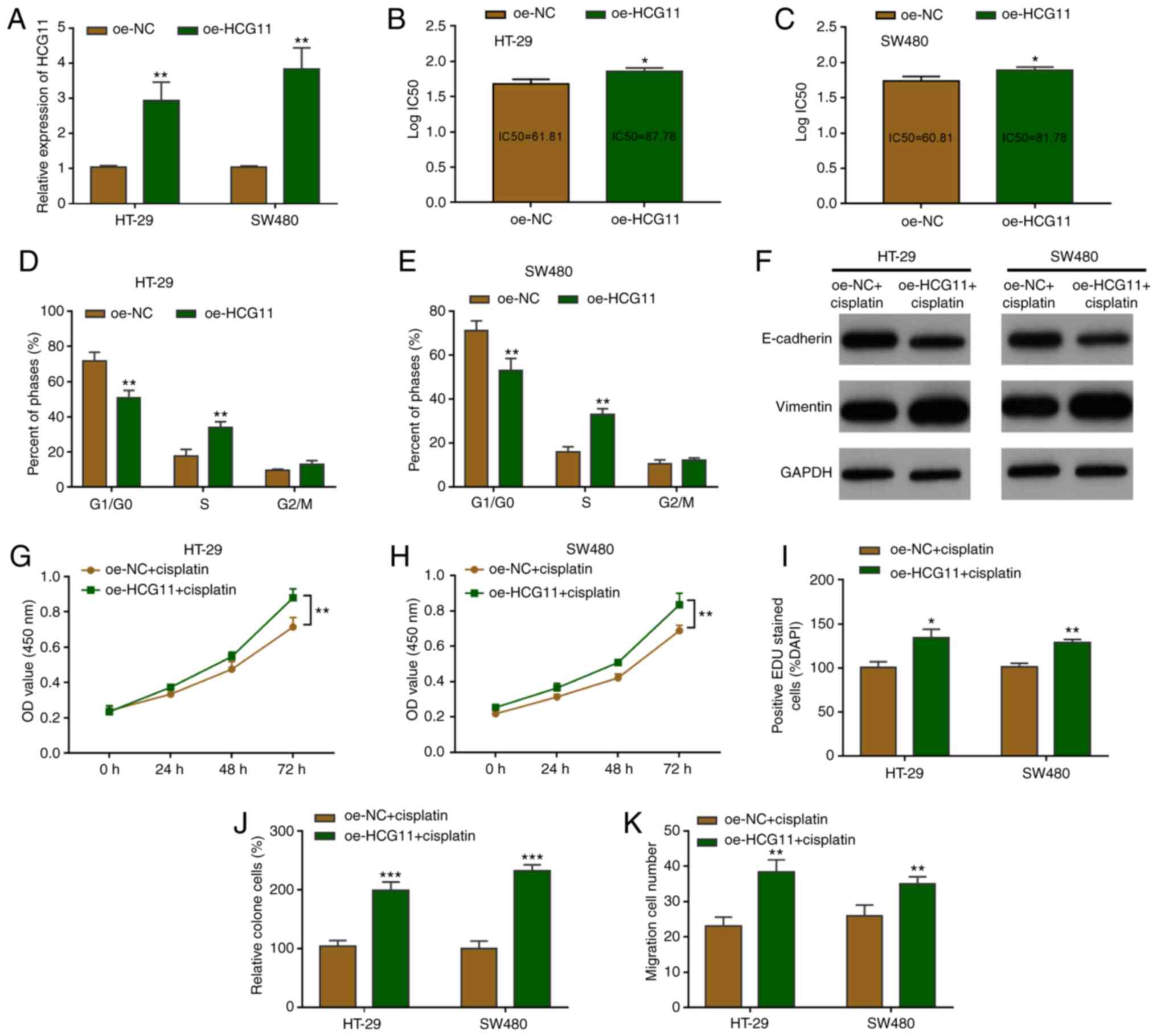
investigated. RT-qPCR demonstrated that oe-HCG11 transfection
resulted in HCG11-overexpression in HT-29 and SW480 cells (Fig. 3A). The effects of HCG11 on tumor cell
sensitivity to chemotherapy were then investigated. Cell viability
was detected 72 h post-cisplatin treatment, and
HCG11-overexpression was found to restrain chemotherapeutic
sensitivity and increase the IC50 value (Fig. 3B and C). A cell cycle assay was then
performed to identify the cell death pattern during chemotherapy.
The results showed that the overexpression of HCG11 decreased
G1/S arrest compared with the control following
cisplatin chemotherapy (20 µM) (Figs. 3D
and E, and S1A). Furthermore,
the EMT pathway was significantly inactivated by consistent
cisplatin treatment. However, HCG11-overexpression could reactivate
the EMT pathway in CRC cells (Fig.
3F). In addition, the degree of chemotherapeutic tolerance was
detected by determining cell viability, proliferation and migration
abilities. CRC cells overexpressing HCG11 exhibited increased
proliferative activity under culture with chemotherapy (Fig. 3G and H). Consistent with these
findings, cellular EdU staining post-HCG11 overexpression showed
that the proportion of EdU-positive cells was increased, suggesting
stronger cellular proliferation activity (Figs. 3I and S1B). Cell clone formation experiments
revealed stronger cisplatin resistance in CRC cells under treatment
with chemotherapy drugs, and a higher growth rate of cell clones
(Fig. 3J and S1C). Similarly, cellular migration
experiments indicated that migration was enhanced relative to the
control group, and that CRC cells overexpressing HCG11 resisted
chemotherapeutic suppression (Fig.
3K and S1D). Collectively, the
HCG11-overexpression experiments confirmed the effects of
chemotherapeutic resistance on CRC cells, which was induced by the
EMT pathway.
miR-214-5p is a target gene of HCG11
in CRC
As predicted by starBase, miR-214-5p was confirmed
to be a target gene of HCG11. The potential binding sites are shown
in Fig. 4A, and a dual-luciferase
reporter assay was used to demonstrate the binding association.
Binding site plasmids containing the HCG11 sequences were
co-transfected with miR-214-5p mimics or inhibitor and the
corresponding negative controls, and luciferase activity was then
detected. The results showed a binding association between HCG11
and miR-214-5p (Fig. 4B). An RNA
pull-down experiment was conducted to analyze gene-binding and
complex-formation ability. In HT-29 and SW480 cells, high
expression of miR-214-5p was observed in HCG11-anchored RNA, which
further confirmed their binding state in CRC cells (Fig. 4C and D). CRC cells were selected for
the targeted knockdown of HCG11 to detect changes in miR-214-5p
expression 24 h post-transfection. The upregulation of miR-214-5p
expression in tumor cells was observed. After HCG11-overexpression,
miR-214-5p expression in tumor cells was downregulated (Fig. 4E). miR-214-5p expression was also
detected in clinical tissues, and was higher in normal tissues than
in CRC tissues (Fig. 4F). Pearson's
correlation analysis revealed a significant negative correlation
between miR-214-5p and HCG11 expression in clinical tissues
(Fig. 4G). These findings indicated
that miR-214-5p bound to and suppressed the expression of HCG11 in
CRC, suggesting a potential mechanism of action for HCG11.
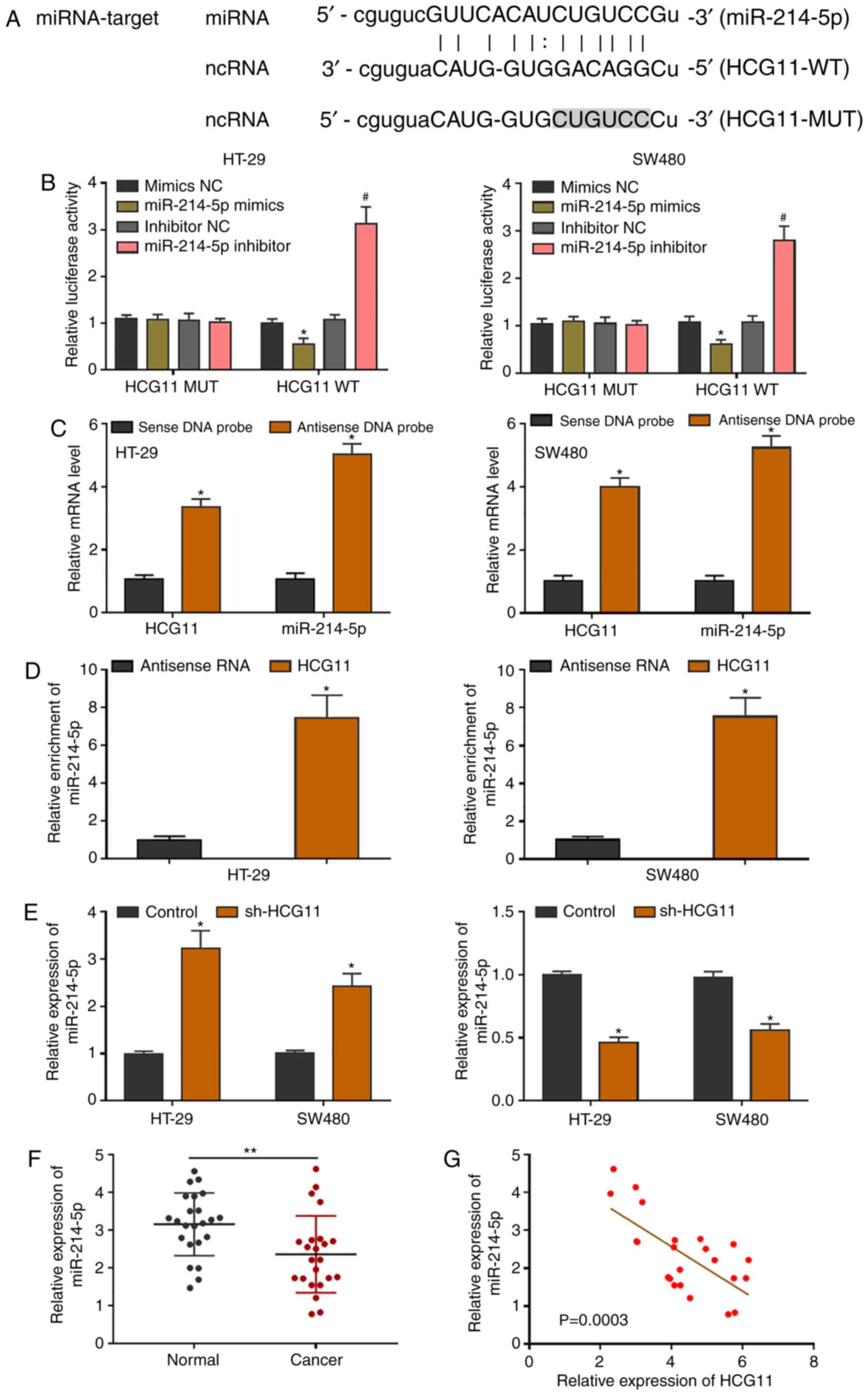 | Figure 4.miR-214-5p is a target gene of HCG11.
(A) Mutation sequence of binding sites between the 3′ untranslated
region of HCG11 and miR-214-5p, as predicted using the starBase
database. (B) Dual-luciferase reporter assay of HCG11 binding to
miR-214-5p in colorectal carcinoma cells. (C) In RNAs pulled down
by a biotin-labeled HCG11 DNA probe, rich expression of miR-214-5p
was observed, suggesting binding in colorectal carcinoma cells. (D)
Through co-incubation of the full-length, biotin-labeled HCG11
sequence and CRC cell lysis solution, rich expression of miR-214-5p
in the extracted RNA was shown by RT-qPCR. (E) miR-214-5p presented
with the opposite changes with knockdown or overexpression of HCG11
in colorectal carcinoma cells. (F) RT-qPCR analysis of miR-214-5p
downregulation in colorectal cancer tissues. (G) Negative
correlation between HCG11 and miR-214-5p expression in clinical
tumor samples was determined by Pearson's analysis. *P<0.05 and
#P<0.05 vs. control or normal tissues. HCG11, HLA
complex group 11; miR, microRNA; NC, negative control; MUT, mutant;
WT, wild-type; RT-q, reverse transcription-quantitative. |
Inhibition of miR-214-5p reverses the
effects of HCG11 on CRC cell malignancy
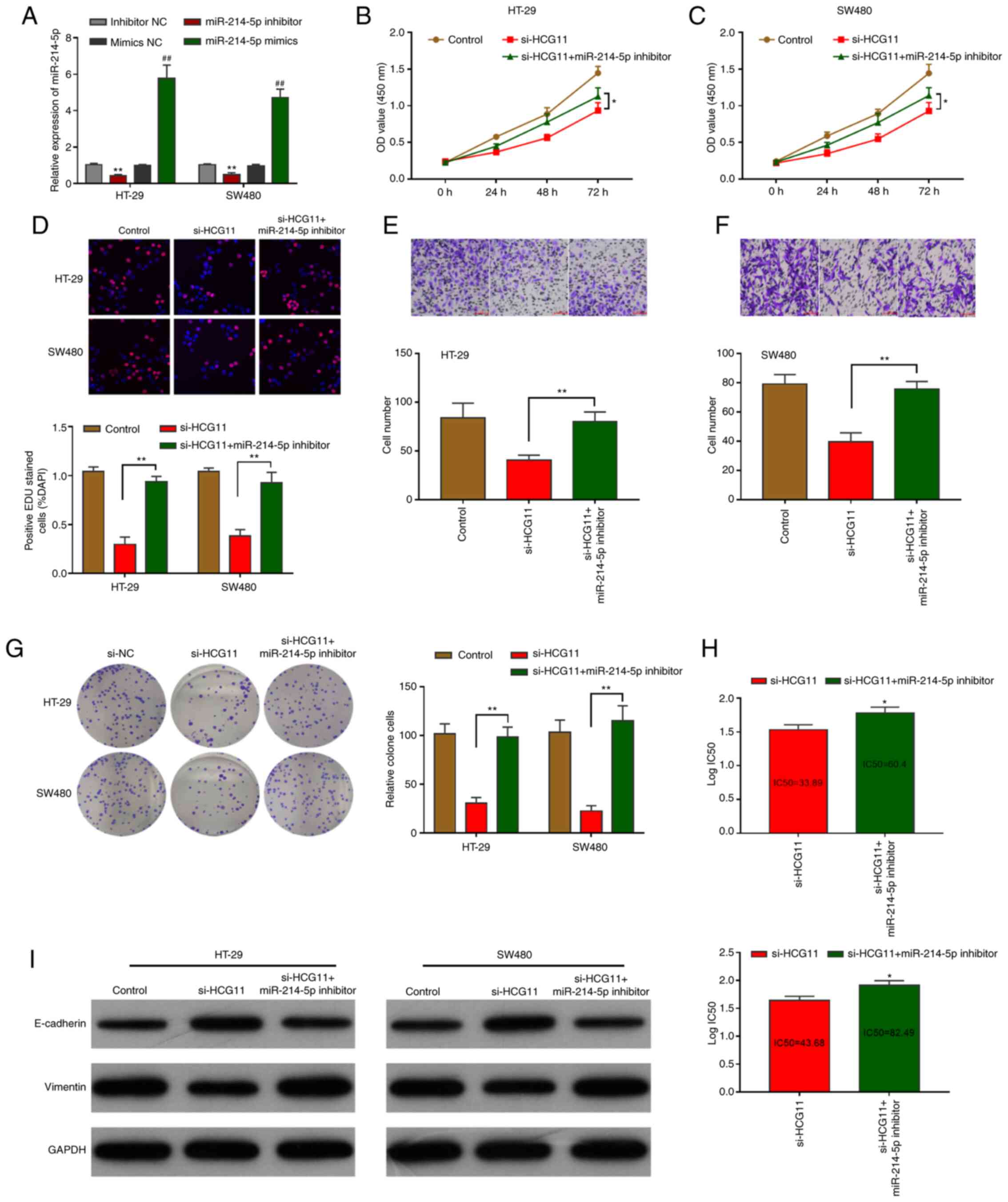
A rescue experiment was performed to determine the
involvement of miR-214-5p in the regulatory effects of HCG11 on
CRC. It was demonstrated that under normal conditions, or CRC cell
culture under chemotherapy, the HCG11-induced inhibition of
cellular proliferation and migration were recovered by m
iR-214-5p-knockdown. RT-qPCR demonstrated that the miR-214-5p
inhibitor and mimics resulted in miR-214-5p expression in HT-29 and
SW480 cells (Fig. 5A). Then, CCK-8
assays indicated that the proliferation of HT-29 and SW480 cells
was reactivated by inhibiting miR-214-5p expression (Fig. 5B and C). Furthermore, EdU experiments
indicated that miR-214-5p-knockdown restored the proliferative
activity of tumor cells (Fig. 5D).
Similarly, Transwell assays showed that miR-214-5p-knockdown
rescued the migration ability of tumor cells (Fig. 5E and F). The colony formation assay
also indicated that miR-214-5p-knockdown improved the clonal
propagation of tumor cells (Fig.
5G). Compared with si-HCG11-transfected CRC cells, CRC cells
co-transfected with si-HCG11+miR-214-5p inhibitor showed drug
resistance to cisplatin. The IC50 value of co-transfected CRC cells
was increased accordingly (Fig. 5H).
Mechanistically, the EMT pathway in CRC cells was inhibited by
si-HCG11 transfection, but was restored by si-HCG11+miR-214-5p
inhibitor co-transfection (Fig. 5I).
These results indicate that in CRC cells, miR-214-5p is involved in
the regulatory carcinogenic pathway.
SOX4 is a target of the
HCG11/miR-214-5p circuit and involved in EMT pathway mediation
It is well known that miRNAs widely regulate
cellular mRNAs, and exert corresponding regulatory roles. In CRC,
potential mRNA targets of miR-214-5p were predicted, and SOX4 was
found to be a candidate target gene (Fig. 6A). Analysis of TCGA database revealed
that SOX4 expression was higher in the CRC cohort than in normal
colon tissues (Fig. 6B). Based on
the starBase prediction results, the binding sites of miR-214-5p
and SOX4 are shown in Fig. 6C. A
dual-luciferase reporter gene assay and gene interference assay
were used to verify the binding relationship between miR-214-5p and
its target gene SOX4 in CRC, and demonstrated that miR-214-5p
suppressed SOX4 expression by binding to its 3′ UTR region
(Fig. 6D and E). To verify the
association between HCG11 and SOX4, SOX4 expression was detected in
CRC patient specimens. The results of correlation analysis revealed
a positive relationship between HCG11 and SOX4 (Fig. 6F). Furthermore, RT-qPCR revealed that
the mRNA level of SOX4 was influenced by HCG11 in CRC cells
(Fig. 6G). The transfection
efficiency of si-SOX4 as verified by RT-qPCR (Fig. 6H), which also revealed that the
HCG11/miR-214-5p loop regulates the mRNA level of SOX4 in tumor
cells (Fig. 6I). In addition,
western blotting showed that SOX4 protein expression was affected
by the HCG11/miR-214-5p axis in CRC cells (Fig. 6J). Following transfection with the
miR-214-5p inhibitor, the protein level of E-cadherin was notably
reduced, whereas the protein level of vimentin was increased.
However, this process was reversed by co-transfection with si-SOX4
and the miR-214-5p inhibitor (Fig.
6K). Collectively, these results identified SOX4 as a target of
the HCG11/miR-214-5p circuit, and confirmed that SOX4 mediates the
EMT pathway in SW480 and HT-29 cells.
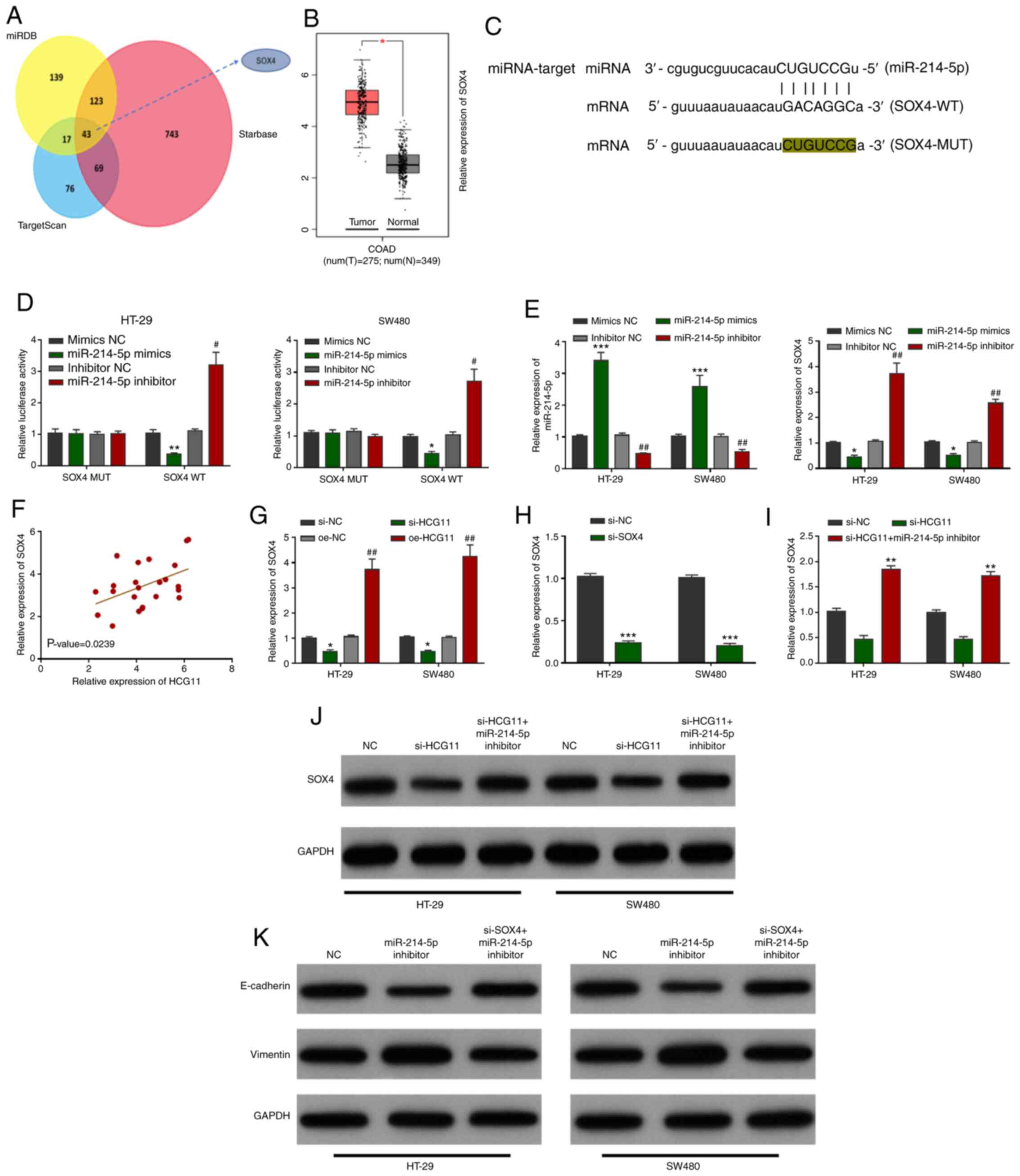 | Figure 6.SOX4 is a target of the
HCG11/miR-214-5p axis and is involved in EMT. (A) SOX4 was found to
be a downstream target gene of miR-214-5p, as predicted by Venn
diagram screening. (B) High expression of SOX4 in colorectal
carcinoma tissues as shown by The Cancer Genome Atlas database
analysis. (C) Predicted binding sites between SOX4 mRNA and
miR-214-5p. (D) Mutual binding between SOX4 and miR-214-5p was
confirmed by the dual-luciferase reporter assays. (E) Interference
of miR-214-5p expression in colorectal carcinoma cells indirectly
negatively regulated the mRNA level of SOX4. (F) HCG11 expression
was positively correlated with that of SOX4 in colorectal carcinoma
tissue samples. (G) Interference of HCG11 expression in colorectal
carcinoma cells regulated the mRNA level of SOX4 in tumor cells.
(H) Transfection efficiency of si-SOX4 as verified by RT-qPCR. (I)
RT-qPCR results showed that the HCG11/miR-214-5p loop regulated
SOX4 mRNA levels in tumor cells. (J) Western blot results revealed
that the HCG11/miR-214-5p loop regulated the protein level of SOX4
in tumor cells. (K) Western blot results revealed that miR-214-5p
regulated the level of SOX4-mediated epithelial-mesenchymal
transition. *P<0.05, **P<0.01 and ***P<0.001, and
#P<0.05 and ##P<0.01 vs. the
corresponding control. miR, microRNA; NC, negative control; HCG11,
HLA complex group 11; Si, small interfering; oe, overexpression;
RT-q, reverse transcription-quantitative; MUT, mutant; WT,
wild-type; COAD, colon adenocarcinoma. |
Discussion
The drug resistance of tumors has been widely
studied, and the resistance of most tumor types to treatment
frequently results in treatment failure and poor prognosis
(24,25). Therefore, it is necessary to
investigate the genetic features of drug resistance in tumors to
further understand the mechanisms of tumor progression, and to
identify novel therapeutic targets. The chemotherapeutic resistance
of CRC is a treatment challenge associated with patient prognosis
and recurrence (26,27). HCG11 was found to be highly expressed
in CRC tissues and to act as a marker for poor prognosis in
clinical assessments. It is also worth noting that HCG11
accelerates the progression of various human tumors, and is also
involved in the induction of tumor proliferation, migration and
other malignant abilities (14,28).
Also, the addition of chemotherapeutic drugs to the CRC cell
culture medium considerably downregulated HCG11 gene expression,
suggesting a potential association between HCG11 and CRC
chemotherapy, albeit with no reports of HCG11 involvement in any
chemotherapy. Our subsequent experiments demonstrated that in CRC,
HCG11 promoted the proliferation and migration of tumor cells,
induced the resistance of CRC cells to chemotherapy, and improved
the IC50 value of chemotherapy compounds towards CRC cells. HCG11
was also found to be involved in mediating the chemotherapeutic
sensitivity of CRC cells; however, its mechanism remains unclear.
The results of the present study indicated that high expression of
HCG11 enhanced the chemotherapy resistance and maintained the
malignant activity of CRC cells, including proliferation and
metastasis. These results imply that HCG11 is closely involved in
chemotherapeutic resistance and predicts poor prognosis in patients
with CRC.
lncRNAs are able to sponge their miRNA targets in
cells, thus limiting gene expression by exerting
post-transcriptional regulatory functions (29,30). As
predicted using bioinformatics techniques, miR-214-5p is a
potential molecular target of HCG11, and dual-luciferase and RNA
pull-down experiments verified the correlation between binding and
mutual regulation in CRC cells. In previous studies, miR-214-5p was
found to regulate constituents of intracellular signaling pathways,
including the jagged 1 gene and EMT-related genes in pancreatic
cancer (31), as well as regulating
dihydropyrimidinase-related protein 5 expression in prostatic
cancer (32), and the expression of
the target gene of rho-associated protein kinase 1 by direct
binding in bone sarcoma (33).
However, to the best of our knowledge, no study has demonstrated
tumor protection of CRC cells. Therefore, an intracellular
functional reverse experiment was performed, which revealed that
miR-214-5p markedly suppressed the carcinogenesis of HCG11-induced
CRC. Similarly, miR-214-5p-overexpression notably recovered the
inhibitory effects of HCG11 on the chemotherapeutic sensitivity of
CRC. The results of the present study indicate the important roles
of HCG11/miR-214-5p in CRC as a new cyclic regulatory
mechanism.
As a target gene of miR-214-5p, SOX4 has previously
been identified as a key oncogene in various different tumors
(34). The primary protein functions
regulated by SOX4 include the mediation of tumorigenesis and
involvement in various EMT-associated signaling pathways (35). The SOX family of protein are closely
associated with tumor progression under the tight regulation of
miRNAs and lncRNAs (36). SOX4 is
associated with the prognosis and grading of CRC, and increased
SOX4 expression has been identified as a marker of poor prognosis
(37). Data mining analysis of TCGA
revealed higher expression levels of SOX4 in CRC than in
normal cells. EMT pathways regulated by SOX4 are key signaling
pathways that mediate the chemotherapeutic resistance of tumor
cells (38,39). The present study revealed that
miR-214-5p binds to and negatively regulates SOX4
expression. Furthermore, high levels of SOX4 expression inhibited
EMT in CRC cells, while co-transfection with miR-214-5p suppressed
the EMT to protect cells from chemotherapeutic drug resistance.
This process is known as the competing endogenous RNA mechanism, in
which HCG11 promotes tumor progression in CRC cells. In summary,
the results of the present study demonstrated that lncRNA HCG11
mediated the chemotherapy resistance of CRC cells, and elucidated
the potential mechanisms of action (in the form of an
lncRNA-miRNA-mRNA axis) to further explain the causes of
chemotherapy resistance in CRC.
In conclusion, the present study demonstrated that
lncRNA HCG11 is significantly upregulated in CRC, and associated
with chemotherapeutic resistance of and poor prognosis.
HCG11-knockdown markedly reduced the proliferation and migration of
CRC cells and increased the sensitivity of tumor cells to
chemotherapy. However, HCG11 overexpression enhanced the
chemotherapy resistance of CRC cells. Therefore, these data
indicate that HCG11 might act as an oncogene in CRC, similar to
previously reported tumor studies. Further studies identified
miR-214-5p as a target gene of HCG11, which mediates EMT regulation
in tumor cells, and is a key downstream molecule for
HCG11-associated carcinogenicity. However, further studies are
required to evaluate the role of HCG11 as a therapeutic target in
CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JP and YM conceived the study idea; JX and YM
participated in the design and implementation of the experiments;
JX and JZ analysed the final data and JX wrote the manuscript. All
authors have read and approved the manuscript, and agree to be
responsible for all aspects of the study to ensure the accuracy or
completeness of any part of the work is properly investigated and
resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of the First Affiliated Hospital of Yangtze University.
All enrolled patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gu L, Xu Y, Xu W, Li M, Su H, Li C and Liu
Z: The exosome secretion inhibitor neticonazole suppresses
intestinal dysbacteriosis-induced tumorigenesis of colorectal
cancer. Invest New Drugs. 38:221–228. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fidler MM and Bray F: Global cancer
inequalities. Front Oncol. 8:2932018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hong J, Lu H, Meng X, Ryu JH, Hara Y and
Yang CS: Stability, cellular uptake, biotransformation, and efflux
of tea polyphenol (−)-epigallocatechin-3-gallate in HT-29 human
colon adenocarcinoma cells. Cancer Res. 62:7241–7246.
2002.PubMed/NCBI
|
|
4
|
Araghi M, Soerjomataram I, Bardot A,
Ferlay J, Cabasag CJ, Morrison DS, De P, Tervonen H, Walsh PM,
Bucher O, et al: Changes in colorectal cancer incidence in seven
high-income countries: A population-based study. Lancet
Gastroenterol Hepatol. 4:511–518. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsukuda K, Tanino M, Soga H, Shimizu N and
Shimizu K: A novel activating mutation of the K-ras gene in human
primary colon adenocarcinoma. Biochem Biophys Res Commun.
278:653–658. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deng H, Wang JM, Li M, Tang R, Tang K, Su
Y, Hou Y and Zhang J: Long non-coding RNAs: New biomarkers for
prognosis and diagnosis of colon cancer. Tumour Biol. Jun
23–2017.(Epub ahead of print). View Article : Google Scholar
|
|
10
|
Cheng B, Rong A, Zhou Q and Li W: LncRNA
LINC00662 promotes colon cancer tumor growth and metastasis by
competitively binding with miR-340-5p to regulate CLDN8/IL22
co-expression and activating ERK signaling pathway. J Exp Clin
Cancer Res. 39:52020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu T, Wu K and Zhang L, Zheng S, Wang X,
Zuo H, Wu X, Tao G, Jiang B and Zhang L: Long non-coding RNA
LINC00858 exerts a tumor-promoting role in colon cancer via HNF4α
and WNK2 regulation. Cell Oncol (Dordr). 43:297–310. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H, Huang H, Xu X, Wang H, Wang J,
Yao Z, Xu X, Wu Q and Xu F: LncRNA HCG11 promotes proliferation and
migration in gastric cancer via targeting miR-1276/CTNNB1 and
activating Wnt signaling pathway. Cancer Cell Int. 19:3502019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Y, Bao C, Zhang X, Lin X, Huang H and
Wang Z: Long non-coding RNA HCG11 modulates glioma progression
through cooperating with miR-496/CPEB3 axis. Cell Prolif.
52:e126152019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang YC, He WY, Dong CH, Pei L and Ma YL:
lncRNA HCG11 regulates cell progression by targeting miR-543 and
regulating AKT/mTOR pathway in prostate cancer. Cell Biol Int.
43:1453–1462. 2019. View Article : Google Scholar
|
|
15
|
Greenburg G and Hay ED: Epithelia
suspended in collagen gels can lose polarity and express
characteristics of migrating mesenchymal cells. J Cell Biol.
95:333–339. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Skrypek N, Goossens S, De Smedt E,
Vandamme N and Berx G: Epithelial-to-mesenchymal transition:
Epigenetic reprogramming driving cellular plasticity. Trends Genet.
33:943–959. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Loric S, Paradis V, Gala JL, Berteau P,
Bedossa P, Benoit G and Eschwège P: Abnormal E-cadherin expression
and prostate cell blood dissemination as markers of biological
recurrence in cancer. Eur J Cancer. 37:1475–1481. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sato M, Shames DS and Hasegawa Y: Emerging
evidence of epithelial-to-mesenchymal transition in lung
carcinogenesis. Respirology. 17:1048–1059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qiu E, Gao Y, Zhang B, Xia T, Zhang Z and
Shang G: Upregulation of cell division cycle 20 in cisplatin
resistance-induced epithelial-mesenchymal transition in
osteosarcoma cells. Am J Transl Res. 12:1309–1318. 2020.PubMed/NCBI
|
|
20
|
Ye Y, Gu J, Liu P, Wang H, Jiang L, Lei T,
Yu S, Han G and Wang Z: Long non-coding RNA SPRY4-IT1 reverses
cisplatin resistance by downregulating MPZL-1 via suppressing EMT
in NSCLC. OncoTargets Ther. 13:2783–2793. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Acikgoz E, Tatar C and Oktem G: Triptolide
inhibits CD133+/CD44+ colon cancer stem cell
growth and migration through triggering apoptosis and represses
epithelial-mesenchymal transition via downregulating expressions of
snail, slug, and twist. J Cell Biochem. 121:3313–3324. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Eberhardt W, Nasrullah U and Haeussler K:
Inhibition of Caspase-2 translation by the mRNA binding protein
HuR: A novel path of therapy resistance in colon carcinoma Cells?
Cells. 8:E7972019. View Article : Google Scholar
|
|
25
|
Paldino E, Tesori V, Casalbore P,
Gasbarrini A and Puglisi MA: Tumor initiating cells and
chemoresistance: Which is the best strategy to target colon cancer
stem cells? BioMed Res Int. 2014:8598712014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kozovska Z, Gabrisova V and Kucerova L:
Colon cancer: Cancer stem cells markers, drug resistance and
treatment. Biomed Pharmacother. 68:911–916. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uppada SB, Gowrikumar S, Ahmad R, Kumar B,
Szeglin B, Chen X, Smith JJ, Batra SK, Singh AB and Dhawan P: MASTL
induces colon cancer progression and chemoresistance by promoting
Wnt/β-catenin signaling. Mol Cancer. 17:1112018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Frankiewicz Z and Leski J: Adaptive
fiducial point detector for ECG stress testing systems. Int J
Biomed Comput. 28:127–135. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chan JJ and Tay Y: Noncoding RNA: RNA
Regulatory Networks in Cancer. Int J Mol Sci. 19:E13102018.
View Article : Google Scholar
|
|
31
|
Cao TH, Ling X, Chen C, Tang W, Hu DM and
Yin GJ: Role of miR-214-5p in the migration and invasion of
pancreatic cancer cells. Eur Rev Med Pharmacol Sci. 22:7214–7221.
2018.PubMed/NCBI
|
|
32
|
Zheng C, Guo K, Chen B, Wen Y and Xu Y:
miR-214-5p inhibits human prostate cancer proliferation and
migration through regulating CRMP5. Cancer Biomark. 26:193–202.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang M, Wang D, Zhu T and Yin R:
miR-214-5p targets ROCK1 and suppresses proliferation and invasion
of human osteosarcoma cells. Oncol Res. 25:75–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu
X, Li Z, Wei J, Liu M and Li G: Circular RNA circ-DONSON
facilitates gastric cancer growth and invasion via NURF complex
dependent activation of transcription factor SOX4. Mol Cancer.
18:452019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hanieh H, Ahmed EA, Vishnubalaj R and
Alajez NM: SOX4: Epigenetic regulation and role in tumorigenesis.
Semin Cancer Biol. 67:91–104. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grimm D, Bauer J, Wise P, Krüger M,
Simonsen U, Wehland M, Infanger M and Corydon TJ: The role of SOX
family members in solid tumours and metastasis. Semin Cancer Biol.
67:122–153. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin CM, Fang CL, Hseu YC, Chen CL, Wang
JW, Hsu SL, Tu MD, Hung ST, Tai C, Uen YH, et al Clinical
prognostic implications of transcription factor SOX4 in patients
with colon cancer, : Clinical and prognostic implications of
transcription factor SOX4 in patients with colon cancer. PLoS One.
8:e671282013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Du B and Shim JS: Targeting
epithelial-mesenchymal transition (EMT) to overcome drug resistance
in cancer. Molecules. 21:E9652016. View Article : Google Scholar
|