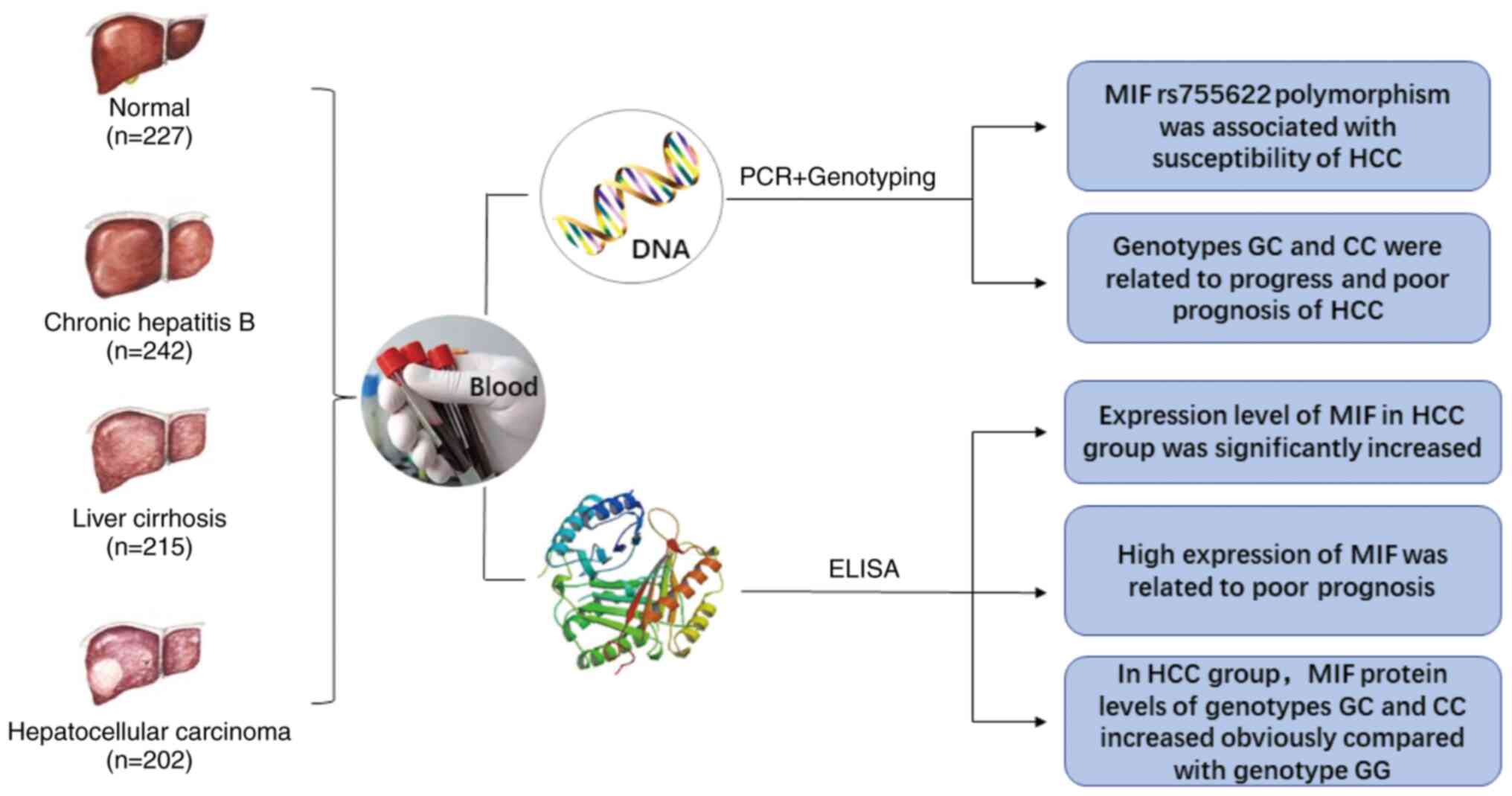
Introduction
Hepatocellular carcinoma (HCC), one of the most
common malignancies in the world (1). According to statistics, there are
~700,000 new cases in the world every year, and Chinese patients
account for as much as 54% of these cases (2). It develops mostly on the basis of viral
hepatitis and cirrhosis, which is characterized by insidious onset,
poor prognosis and the large consumption of medical resources
(1). The etiology and pathogenesis
of HCC are not completely clear, and it is considered to be
associated with the hepatitis virus, contaminated food and water,
poisons, parasites and genetic factors (1). Importantly, genetic factors are an
important basis of HCC. Studies have confirmed that single
nucleotide polymorphisms (SNPs) located in promoter regions and
coding regions of critical genes can affect susceptibility to HCC
(3,4).
Macrophage migration inhibitory factor (MIF), a
cytokine with multiple biological activities, plays an important
regulatory role in the endocrine-immune network of the human body.
It can inhibit the migration of macrophages and promote the
accumulation, proliferation, activation and secretion of
macrophages at the site of inflammation, thus playing a critical
role in the pathogenesis of diseases such as inflammation,
autoimmune diseases and tumors (5).
The gene encoding MIF in humans is located on the long arm of
chromosome 22 (22q11.2), covering a length of 1 kb. There are
several polymorphic loci in the MIF gene. In previous years,
numerous studies (6,7) have found that polymorphisms in the MIF
gene and the expression levels of MIF are closely associated with
the susceptibility, progression, prognosis and drug resistance of a
number of diseases, for example, sepsis, autoimmune diseases,
cancer, metabolic disorders such as type 2 diabetes and
obesity.
To date, the association between MIF polymorphisms
and their expression levels in HCC has rarely been demonstrated.
The present study aimed to analyze the association of MIF gene SNPs
with HCC and detected MIF protein expression levels in peripheral
blood from patients with HCC in the Chinese Han population
(Fig. 1). Since most HCC cases are
due to hepatitis and cirrhosis, patients with chronic hepatitis B
(CHB) and liver cirrhosis (LC) were used as controls.
Currently, HCC is a refractory disease, the present
study will help to explore new predictive/prognostic indicators and
therapeutic targets for HCC.
Materials and methods
Patients and controls
From February 2014 to January 2018, 886 participants
were enrolled in the First Affiliated Hospital of Guangxi Medical
University, including 202 patients with HCC (HCC group), 242
patients with CHB (CHB group), 215 patients with LC (LC group) and
227 healthy volunteers (normal group). Inclusion criteria included:
i) Patients living in China in the long-term, ii) the last three
generations of immediate family members being Han, with no
marriages to individuals of other nationalities and iii) patients
with HCC, CHB and LC meeting the relative diagnosis norms and
guidelines (8–10). Exclusion criteria included: i)
Participants with other neoplastic diseases or hepatic diseases and
ii) participants with genetic relationships to each other. Tumor,
node and metastasis (TNM) staging was judged according to the
Barcelona Clinic Liver Cancer strategy (11). All patients with HCC underwent
α-fetoprotein (AFP) testing and imaging examinations (CT and MRI).
Image classification (including lump type, nodular type and diffuse
type) was carried out through imaging examination (12) and the presence or absence of lymph
node metastasis and distant metastasis was determined by
independent pathologists and imaging experts in the First
Affiliated Hospital of Guangxi Medical University (Nanning, China)
according to the Diagnosis and staging of hepatocellular carcinoma
(HCC): Current guidelines (12).
Written informed consent was obtained from all
participants. The Ethics Committee of The First Affiliated Hospital
of Guangxi Medical University approved the study (approval no.
2014-KY-E-049). The privacy of all participants was protected
consistently.
DNA extraction, PCR and
genotyping
In total, 4 ml of peripheral blood was collected
from the median elbow vein of every participant and stored at
−80°C. DNA was extracted using the Genomic DNA Extraction kit
(Tiangen Biotechnology Co., Ltd.) according to the manufacturer's
instructions (centrifugal column method). DNA was added to 50–100
µl Tris-ethylenediamine tetraacetic acid buffer (TE) for
rehydration, and the concentration and purity of DNA was assessed
before storage at −80°C.
The primers used to amplify the MIF gene (the
sequences included rs755622, rs1007888 and rs2096525) were designed
and synthesized by Takara Biotechnology Co., Ltd. The sequences
were as follows: MIF rs755622 Forward: 5′-GGCGACTAACATCGGTGA-3′ and
reverse: 5′-GCAGAAGGACCAGGAGAC-3′, MIF rs1007888 forward:
5′-TTAGGGAGGGGTAAGAAC-3′ and reverse: 5′-GAAGCCCATGTAAAAGAA-3′, MIF
rs2096525 forward: 5′-GGTGCCCACCGGACGAGGGAT-3′ and reverse:
5′-GTCGGGCCCCGAACGTCCACT-3′.
PCR reactions were performed in a thermocycler
(Veriti; Applied Biosystems; Thermo Fisher Scientific, Inc.). The
total reaction system was 20 µl, including 1 µl genomic DNA, 1 µl
Tks Gflex DNA polymerase, 0.5 µl forward primer, 0.5 µl reverse
primer, 10 µl 2X Gflex PCR buffer and up to 20 µl with
double-distilled water. PCRs for MIF rs755622 were performed under
the following conditions: Initial denaturation at 95°C for 3 min,
followed by amplification for three cycles (denaturation at 95°C
for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for
30 sec), followed by three cycles (denaturing at 95°C for 30 sec,
annealing at 58°C for 30 sec and extension at 72°C for 30 sec),
followed by 25 cycles (denaturing at 95°C for 30 sec, annealing at
55°C for 30 sec and extension at 72°C for 30 sec), followed by five
cycles (denaturing at 95°C for 30 sec, annealing at 53°C for 30 sec
and extension at 72°C for 30 sec) with a final extension at 72°C
for 7 min. The PCR for MIF rs1007888 was performed under the
following conditions: Initial denaturation at 95°C for 4 min,
followed by amplification for 35 cycles (denaturation at 94°C for
30 sec, annealing at 55°C for 30 sec and extension at 72°C for 30
sec) and a final extension at 72°C for 7 min. PCR for MIF rs2096525
was performed under the following conditions: Initial denaturation
at 95°C for 5 min, followed by amplification for 40 cycles
(denaturation at 95°C for 30 sec, annealing at 65°C for 30 sec and
extension at 72°C for 30 sec) with a final extension at 72°C for 7
min.
To confirm the genotypes for each sample, PCR
products with forward primers were sent to GenScript Biotechnology
Co., Ltd. for DNA sequencing. The sequencer used was an Applied
Biosystems 3730×l DNA Analyzer (Thermo Fisher Scientific,
Inc.).
Protein expression of MIF detection
via ELISA
The expression levels of MIF protein in the
peripheral blood from all participants were tested with Human MIF
ELISA kits (cat. no. EK0813; Boster Biological Technology Co.,
Ltd.). The specific method of ELISA followed the manufacturer's
instructions. Assays were performed in triplicate for each sample.
With 50% of the maximum MIF concentration as the cut-off point,
patients with HCC with survival information were divided into the
high expression group and the low expression group for subsequent
survival analysis.
Patient follow-up
Clinical follow-up after diagnosis was performed
every 3 months during the first two years, and then every 6 months
until the patient died. The content of the follow-up included
asking about the patient's symptoms, physical examination, testing
of AFP, CT or MRI of the upper abdomen. Liver function, HBV-DNA, or
HCV-RNA tests were performed when necessary. Telephone follow-up
was performed when the patients did not come to the hospital for
follow-up visits on time.
Statistical analysis
Normally distributed measurement data were
represented as mean ± SD. The Hardy-Weinberg equilibrium was
evaluated with the Haploview 4.2 software (Broad Institute). The
minimum sample size and statistical power were calculated by the
PASS 11 software (NCSS, Inc.). Allele and genotype frequencies were
compared between the groups using the Pearson's χ2 test
and Bonferroni's correction. The measurement data of multiple
groups were compared with each other using one-way and two-way
ANOVA and post hoc Bonferroni's correction. Overall survival time
(from diagnosis to the last follow-up or death) was analyzed using
the Kaplan-Meier method and compared using the log-rank test.
P<0.05 was considered to indicate a statistically significant
difference. SPSS version 20.0 (IBM Corp) was used to perform all
statistical analyses.
Results
Characteristics of the study
population
The characteristics of the study population are
shown in Table I. The four groups of
subjects are balanced and comparable in terms of age and sex. The
infection rate of hepatitis B virus in the LC group was 69.3%,
while that in the HCC group was 74.8%. The infection rate of
hepatitis C virus in the LC group was 4.2%, while the infection
rate of hepatitis C virus in the HCC group was 5.4%. Alcoholic
liver disease accounted for 28.8% in the LC group and 25.2% in the
HCC group.
 | Table I.Characteristics of the study
population. |
Table I.
Characteristics of the study
population.
|
Characteristics | Normal, n=227 | CHB, n=242 | LC, n=215 | HCC, n=202 |
|---|
| Age, years | 51.2±14.23 | 49.8±13.54 | 50.5±15.14 | 50.7±13.96 |
| Sex, n (%) |
|
Male | 166 (73.1) | 180 (74.4) | 162 (75.3) | 151 (74.8) |
|
Female | 61 (26.9) | 62 (25.6) | 53 (24.7) | 51 (25.2) |
| HBV infection, n
(%) |
|
Yes | 0 (0) | 242 (100.0) | 149 (69.3) | 151 (74.8) |
| No | 227 (100.0) | 0 (0) | 66 (30.7) | 51 (25.2) |
| HCV infection, n
(%) |
|
Yes | 0 (0) | 0 (0) | 9 (4.2) | 11 (5.4) |
| No | 227 (100.0) | 242 (100.0) | 206 (95.8) | 191 (94.6) |
| Alcoholic liver
disease, n (%) |
|
Yes | 0 (0) | 0 (0) | 62 (28.8) | 51 (25.2) |
| No | 227 (100.0) | 242 (100.0) | 153 (71.2) | 151 (74.8) |
Association of MIF polymorphisms with
the risk of CHB, LC and HCC
The genotype distributions of the two polymorphisms
among patients with HCC, CHB and LC, as well as healthy volunteers,
were all consistent with Hardy-Weinberg equilibrium (all
P>0.05).
After adjusting for age and sex, it was observed
that the MIF gene rs755622polymorphism was associated with an
increased susceptibility to HCC (P=0.001), and that allele C may be
risk factors for HCC (P<0.001; Table
II). However, the MIF rs1007888 and rs2096525 polymorphisms
were not associated with susceptibility to HCC (P>0.05; Table II). Moreover, MIF rs755622,
rs1007888 and rs2096525 polymorphisms were not associated with
susceptibility to CHB or LC (Tables
III and IV).
 | Table II.Macrophage migration inhibitory
factor gene polymorphisms in 202 patients with HCC and 227 normal
volunteers. |
Table II.
Macrophage migration inhibitory
factor gene polymorphisms in 202 patients with HCC and 227 normal
volunteers.
| SNPs | Genotype | HCC, n (%) | Normal, n (%) | χ2 | P-value |
|---|
| rs755622 | GG | 109 (54.0) | 158 (69.6) | 13.167 | 0.001 |
|
| GC | 76
(37.6) | 62
(27.3) |
|
|
|
| CC | 17
(8.4) | 7
(3.1) |
|
|
|
| Allele G | 294 (72.8) | 378 (83.3) | 13.848 | <0.001 |
|
| Allele C | 110 (27.2) | 76
(16.7) |
|
|
| rs1007888 | AA | 48
(23.8) | 66
(29.1) | 1.685 | 0.431 |
|
| AG | 109 (54.0) | 117 (51.5) |
|
|
|
| GG | 45
(22.2) | 44
(19.4) |
|
|
|
| Allele A | 205 (50.7) | 249 (54.8) | 1.445 | 0.229 |
|
| Allele G | 199 (49.3) | 205 (45.2) |
|
|
| rs2096525 | CC | 145 (71.8) | 144 (63.4) | 3.391 | 0.183 |
|
| CT | 49
(24.2) | 71
(31.3) |
|
|
|
| TT | 8
(4.0) | 12
(5.3) |
|
|
|
| Allele C | 339 (84.0) | 359 (79.1) | 3.296 | 0.069 |
|
| Allele T | 65
(16.0) | 95
(20.9) |
|
|
 | Table III.Macrophage migration inhibitory
factor gene polymorphisms in 242 patients with CHB and 227 normal
volunteers. |
Table III.
Macrophage migration inhibitory
factor gene polymorphisms in 242 patients with CHB and 227 normal
volunteers.
| SNPs | Genotype | CHB, n (%) | Normal, n (%) | χ2 | P-value |
|---|
| rs755622 | GG | 161 (66.5) | 158 (69.6) | 0.688 | 0.709 |
|
| GC | 71
(29.3) | 62
(27.3) |
|
|
|
| CC | 10
(4.2) | 7
(3.1) |
|
|
|
| Allele G | 393 (81.2) | 378 (83.3) | 0.680 | 0.409 |
|
| Allele C | 91
(18.8) | 76
(16.7) |
|
|
| rs1007888 | AA | 57
(23.6) | 66
(29.1) | 3.389 | 0.184 |
|
| AG | 123 (50.8) | 117 (51.5) |
|
|
|
| GG | 62
(25.6) | 44
(19.4) |
|
|
|
| Allele A | 237 (49.0) | 249 (54.8) | 3.243 | 0.072 |
|
| Allele G | 247 (51.0) | 205 (45.2) |
|
|
| rs2096525 | CC | 167 (69.0) | 144 (63.4) | 1.669 | 0.434 |
|
| CT | 65
(26.9) | 71
(31.3) |
|
|
|
| TT | 10 (4.1) | 12
(5.3) |
|
|
|
| Allele C | 399 (82.4) | 359 (79.1) | 1.709 | 0.191 |
|
| Allele T | 85
(17.6) | 95
(20.9) |
|
|
 | Table IV.Macrophage migration inhibitory
factor gene polymorphisms in 215 patients with liver cirrhosis and
227 normal volunteers. |
Table IV.
Macrophage migration inhibitory
factor gene polymorphisms in 215 patients with liver cirrhosis and
227 normal volunteers.
| SNPs | Genotype | LC | Normal | χ2 | P-value |
|---|
| rs755622 | GG | 139 (64.7) | 158 (69.6) | 1.334 | 0.513 |
|
| GC | 67
(31.2) | 62
(27.3) |
|
|
|
| CC | 9
(4.2) | 7
(3.1) |
|
|
|
| Allele G | 345 (80.2) | 378 (83.3) | 1.359 | 0.244 |
|
| Allele C | 85
(19.8) | 76
(16.7) |
|
|
| rs1007888 | AA | 52
(24.2) | 66
(29.1) | 1.962 | 0.375 |
|
| AG | 112 (52.1) | 117 (51.5) |
|
|
|
| GG | 51
(23.7) | 44
(19.4) |
|
|
|
| Allele A | 216 (50.2) | 249 (54.8) | 1.885 | 0.170 |
|
| Allele G | 214 (49.8) | 205 (45.2) |
|
|
| rs2096525 | CC | 152 (70.7) | 144 (63.4) | 2.633 | 0.268 |
|
| CT | 54
(25.1) | 71
(31.3) |
|
|
|
| TT | 9
(4.2) | 12
(5.3) |
|
|
|
| Allele C | 358 (83.3) | 359 (79.1) | 2.519 | 0.112 |
|
| Allele T | 72
(16.7) | 95
(20.9) |
|
|
Association of MIF rs755622
polymorphism with clinical parameters in HCC
There was a significant association between the MIF
rs755622 polymorphism and TNM stage, lymph node metastasis and
distant metastasis of HCC (all P<0.05; Table V). However, no association was found
between the MIF rs755622 polymorphism and AFP levels or imaging
classification in patients with HCC (Table V).
 | Table V.Macrophage migration inhibitory
factor rs755622 polymorphism in relation to clinical parameters in
patients with hepatocellular carcinoma. |
Table V.
Macrophage migration inhibitory
factor rs755622 polymorphism in relation to clinical parameters in
patients with hepatocellular carcinoma.
|
| Genotype, n
(%) |
|
|
|---|
|
|
|
|
|
|---|
| Clinical
variable | GG | GC | CC | χ2 | P-value |
|---|
| AFP, ng/ml |
|
|
| 0.669 | 0.716 |
|
>400 | 75 (55.1) | 51 (37.5) | 10 (7.4) |
|
|
|
<400 | 34 (51.5) | 25 (37.9) | 7 (10.6) |
|
|
| Imaging
classification |
|
|
| 0.391 | 0.822 |
| Lump
type | 72 (54.5) | 48 (36.4) | 12 (9.1) |
|
|
| Nodular
type +diffuse type | 37 (52.9) | 28 (40.0) | 5 (7.1) |
|
|
| TNM stage |
|
|
| 11.663 | 0.003 |
|
I+II | 53 (68.8) | 21 (27.3) | 3 (3.9) |
|
|
|
III+IV | 56 (44.8) | 55 (44.0) | 14 (11.2) |
|
|
| Lymph node
metastasis |
|
|
| 11.054 | 0.004 |
|
Yes | 81 (48.8) | 68 (41.0) | 17 (10.2) |
|
|
| No | 28 (77.8) | 8 (22.2) | 0 (0) |
|
|
| Distant
metastasis |
|
|
| 12.017 | 0.002 |
|
Yes | 15 (34.1) | 21 (47.7) | 8 (18.2) |
|
|
| No | 94 (59.5) | 55 (34.8) | 9 (5.7) |
|
|
MIF rs755622 polymorphism and
prognosis
Next, survival analysis for patients in the HCC
group was performed. At the end of the follow-up, survival
information was available from 163 patients (genotype distribution:
88 GG, 61 GC and 14 CC). The dropout rate was 19.3%. The reason for
the dropout rate of 19.3% was data not being available. Allele C of
MIF rs755622 was significantly related to the susceptibility of HCC
(P<0.001); in addition, the proportions of GC and CC genotypes
in the HCC group were 37.6 and 8.4%, respectively, which were
significantly higher compared with the proportions of GC and CC
genotypes in the normal group (27.3 and 3.1%). Hence, GC and CC
genotypes were risk factors for the occurrence of HCC and were
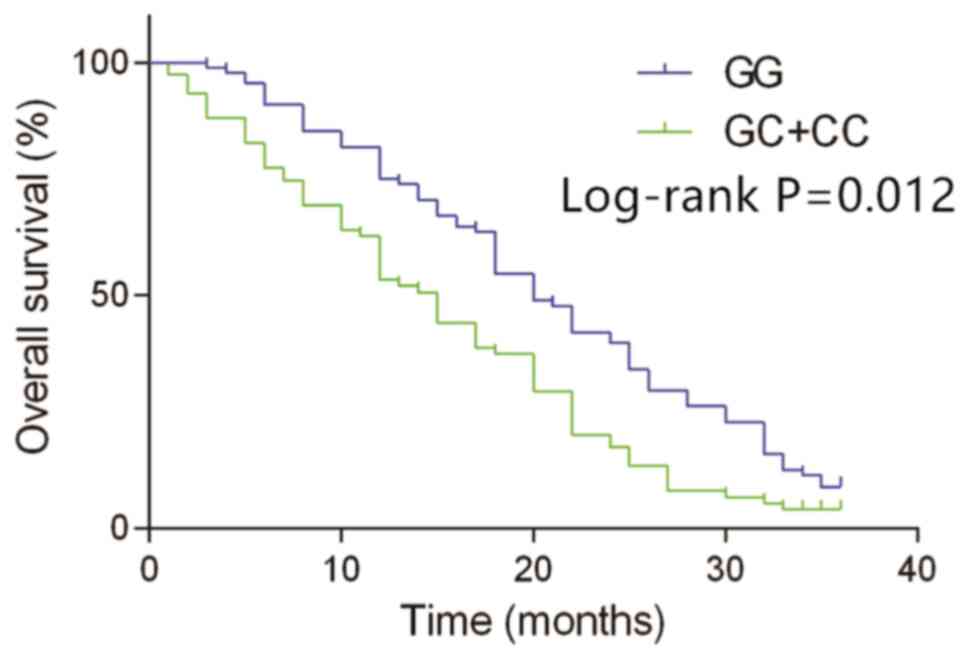
analyzed together. According to a Kaplan-Meier analysis, the
overall survival in patients with the GC and CC genotypes of MIF
rs755622 was much shorter compared with those with the GG genotype
(median overall survival time, 15.7 months vs. 20.2 months;
log-rank P=0.012; Fig. 2). These
results indicated that the GC and CC genotypes may be an indicator
of poor prognosis in patients with HCC.
Expression levels of MIF in patients
with CHB, LC and HCC
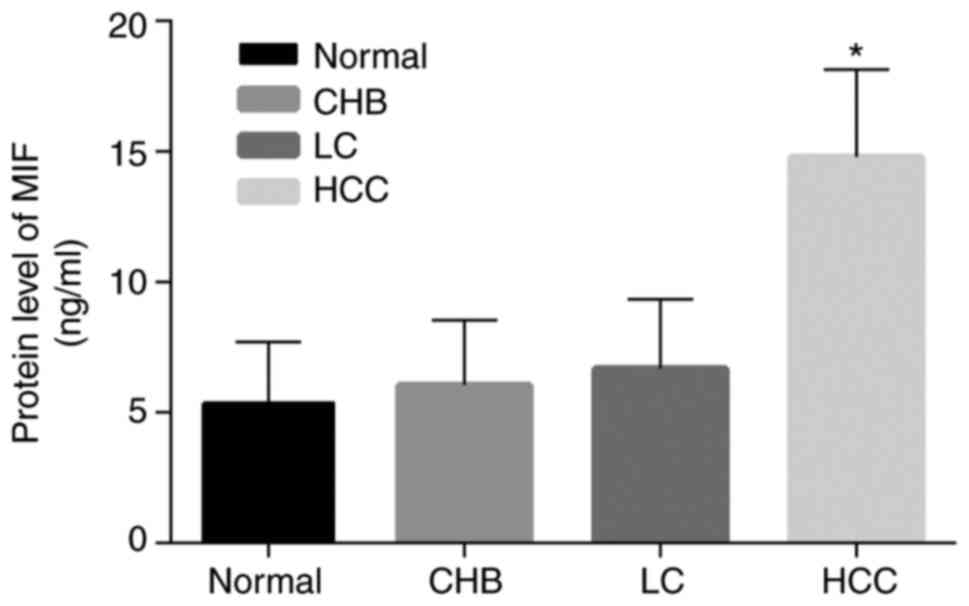
ELISA showed that, the expression of MIF in the HCC
group was significantly increased compared with the normal, CHB and
LC groups; however, the expression levels of MIF in the CHB and LC
groups were not remarkably increased compared with the normal group
(P>0.05; Fig. 3). These data
revealed that MIF in peripheral blood may be involved in the
pathogenesis of HCC.
MIF expression level and
prognosis
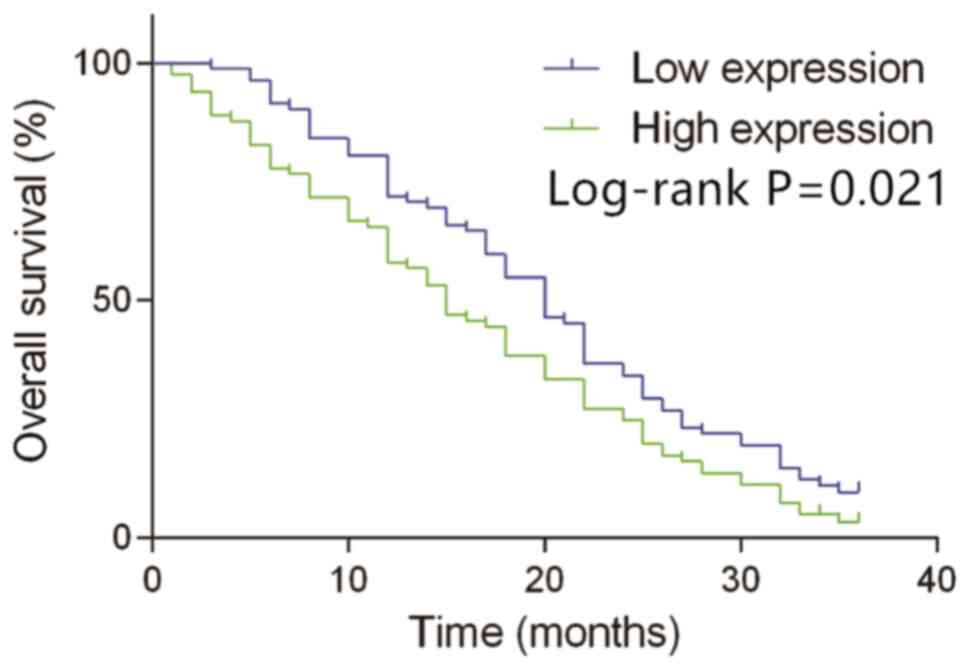
With a median concentration of MIF in peripheral
blood (14.80 ng/ml) as the cut-off point, 163 patients with HCC
with survival information were divided into the high expression
group (≥14.80 ng/ml, n=81) and the low expression group (<14.80
ng/ml, n=82). According to Kaplan-Meier analysis, the overall
survival time in patients with the high expression of MIF was much
shorter compared with those with the low expression of MIF (median
overall survival time, 15.5 months vs. 20.0 months; log-rank
P=0.021; Fig. 4). These data
suggested that the high expression of MIF may be an indicator of
poor prognosis.
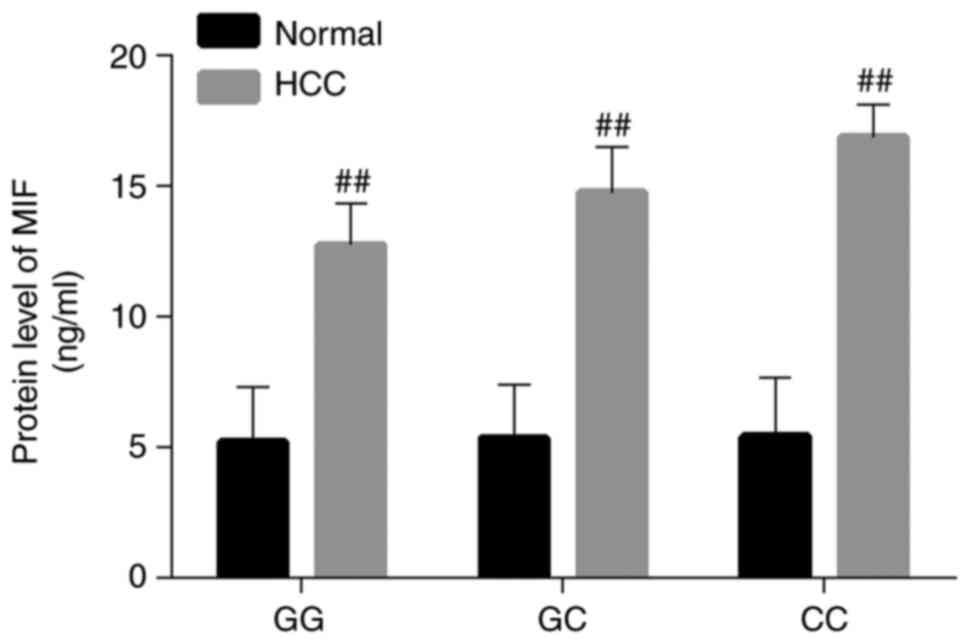
Expression levels of MIF in patients
with HCC patients with different genotypes
The MIF protein levels for all genotypes in the HCC
group increased significantly compared with the normal group
(P<0.01; Fig. 5). In the HCC
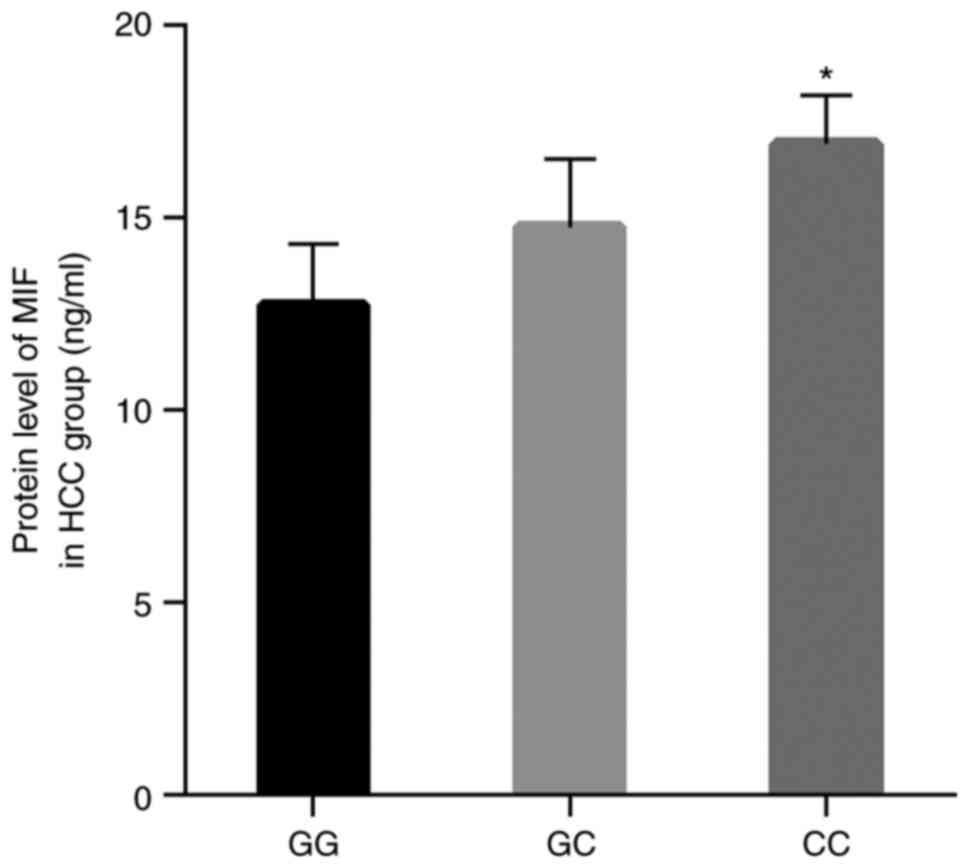
group, the MIF protein levels for the genotypes GC and CC were
significantly increased compared with the genotype GG, especially
for genotype CC (P<0.05; Fig. 6).
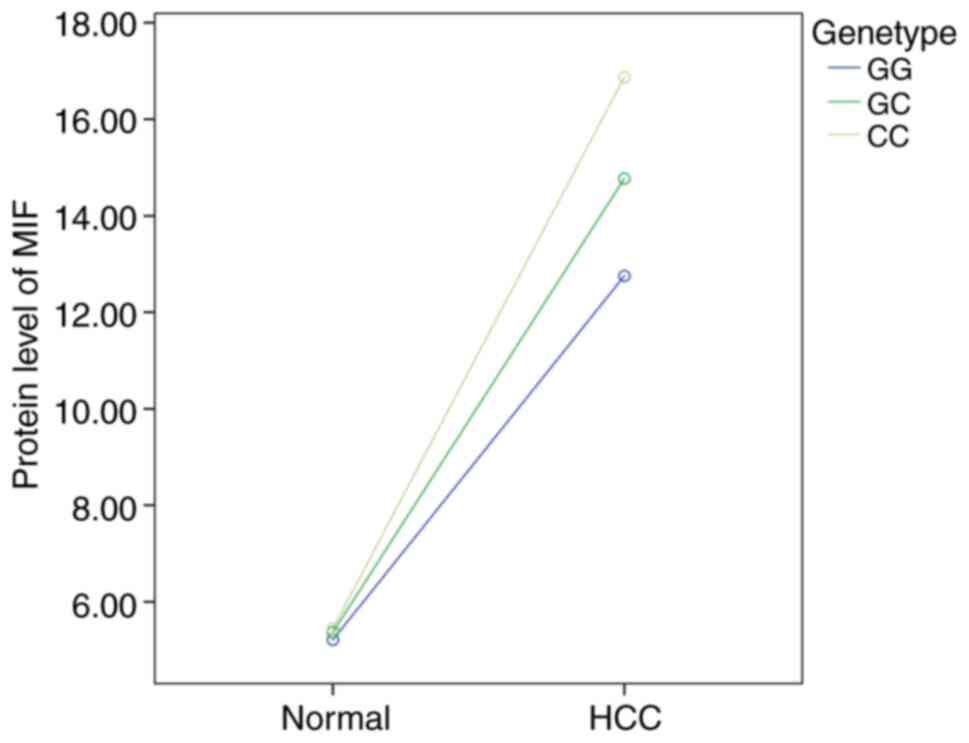
Disease status and genotype are synergistic (Fig. 7). These results confirmed that
genotypes GC and CC of MIF rs755622 may contribute to its
expression in peripheral blood.
Discussion
MIF, a protein with a molecular weight of 12.5 kDa,
is composed of 115 amino acids, and is an open-ended hollow
structure consisting of three monomers (each containing two
anti-parallel α helices and six β sheets) (13). The homology of the MIF gene in all
mammals is ~90%, suggesting that the MIF protein may have important
biological functions (14). The
human MIF gene contains two introns and three exons, and the mRNA
is ~0.8 kb in length (6). MIF is
widely expressed in various tissues and cells, such as anterior
pituitary cells, activated T lymphocytes, mononuclear macrophages,
the liver, kidneys and spleen (14).
In the past, MIF was considered an inflammatory mediator (15), but recent studies (16–18) have
demonstrated that MIF is closely related to the occurrence and
progression of tumors.
A number of studies have demonstrated that MIF plays
a crucial role in the pathogenesis of HCC through multiple
mechanisms. Firstly, MIF promotes the formation of blood vessels in
HCC. Macrophages usually accumulate in the interstitial tissue
adjacent to the infiltrating area of the tumor. The interaction of
tumor cells with macrophages results in the production of
extracellular matrix-degrading enzymes and promotes tumor
angiogenesis (5,19). Secondly, MIF promotes the
proliferation of HCC cells. MIF can upregulate the extracellular
signal-regulated kinase (ERK) and mitogen-activated protein kinase
(MAPK) through signaling pathways. Signals are transmitted to the
nucleus after the phosphorylation of ERK/MAPK, thereby causing the
proliferation and differentiation of tumor cells (20,21).
Thirdly, MIF promotes the invasion and migration of HCC cells. MIF
increases the adhesion and migration of cancer cells through the
Rho signaling transduction pathway, which can indirectly promote
the invasion and migration of cancer cells by inducing the
production of MMP-9 and IL-8 (22,23).
Fourthly, MIF affects P53; the P53 gene is one of the most
important genes that determine tumor occurrence in the human body,
such as esophagus cancer, breast cancer, hepatocellular carcinoma
and so on (24–26). MIF leads to tumorigenesis and
progression by affecting the function of P53 or cooperating with
functional mutant p53 (27,28).
The present study reported that the MIF rs755622
polymorphism is associated with susceptibility and metastasis of
HCC, which is an indicator of poor prognosis. Ramireddy et
al (29) found that the presence
of the MIF rs755662 polymorphism was associated with
susceptibility, patient age and TNM stages of colorectal cancer in
the Taiwanese population. A previous study (30) in Chinese women showed that the MIF
rs755622 polymorphism increases breast cancer susceptibility, and
that individuals with the GC and CC genotypes have a significantly
increased risk. The study by Ding et al (31) suggested that the MIF rs755662
polymorphism may be associated with an increased incidence of
prostate cancer and may be associated with higher Gleason scores,
higher clinical stages and overexpression of serological
prostate-specific antigen. A previous meta-analysis (32) included 15 studies in Asian and
Caucasian populations and indicated that the MIF rs755662
polymorphism may be an independent risk factor for gastrointestinal
cancer and hematological malignancy susceptibility. Yuan et
al (33) reported that MIF gene
polymorphisms may be associated with the surgical prognosis of HCC,
such as differentiation grade, TNM stage, survival rate,
recurrence, metastasis and average survival time. These findings
are consistent with our results.
MIF is highly expressed in a variety of tumors,
precancerous lesions and tumor metastasis tissue, especially in HCC
(34). Wang et al (35) found that the expression levels of MIF
in the serum from patients with HCC are higher compared with the
levels from healthy volunteers, and the expression levels of MIF in
tissues are also higher compared with those in the adjacent
non-tumor liver tissues. Zhao et al (36) reported that the intratumoral MIF
expression level of HCC was positively correlated with plasma MIF
levels, and that plasma MIF had an improved diagnostic value
compared with AFP. Furthermore, plasma MIF levels demonstrated a
significant association with overall and tumor-free survival time
in patients with HCC. Han and Zhang (37) deemed that MIF is important for the
progression and prognosis of HCC, which can be used as a biomarker
for HCC diagnosis. The present study showed that MIF expression
levels in the peripheral blood from patients with HCC were
significantly higher compared with those from patients with CHB and
LC or healthy volunteers, and that high levels of MIF in peripheral
blood from patients with HCC may be an indicator of poor prognosis,
and the results of the present study were consistent with the
aforementioned previous studies.
In addition, the present study also reported that
MIF rs755622 polymorphism contribute to MIF expression. The
rs755622 polymorphism is located in the promoter region of the MIF
gene. Changes in this locus, from G to C, affect the transcription
and translation of the MIF gene, thereby upregulating the
expression levels of MIF and ultimately determining the
susceptibility and prognosis of multiple diseases, such as
arthritis, autoimmune disease and cancer (38). MIF is not just only about the gene
polymorphism, it is also about the state of the disease. It is
considered that the genetic polymorphism and disease state jointly
determine the expression of MIF, and the latter may be the main
factor, and the former may be the secondary factor. At present, HCC
is still a refractory disease, but MIF is expected to improve HCC
therapies through correlation with predictive/prognostic factors in
the future (39). MIF may be a
potential diagnostic/prognostic indicator for HCC.
Most cases of HCC are based on chronic viral
hepatitis and cirrhosis (1);
therefore, the present study used CHB and LC as controls. The
results showed that the MIF rs755622 polymorphism is only
associated with HCC but not with CHB and LC. Nonetheless,
conflicting data have previously been reported, showing that the
MIF rs755622 polymorphism was associated with CHB and HBV-induced
liver cirrhosis (40). The
discrepancy may lie in the small sample size in the study by Zhang
et al (40) and ethnic or
regional differences in genetic polymorphisms.
However, there were some limitations of the present
study. In the present study, the sample size was small, and the
study was conducted only in the Chinese Han population. In
addition, the present study only detected the expression level of
MIF in the peripheral blood of subjects and did not detect the
expression level of MIF in the liver tissue. Hence, the results of
the present study have yet to be further validated in studies with
larger cohorts and other ethnic groups, as well as more in-depth
experimental tissue and cellular studies.
In summary, the MIF rs755622 polymorphism is
associated with susceptibility to HCC in the Chinese Han
population, and is related to metastasis or poor prognosis of HCC,
which may be due to the MIF rs755622 polymorphism upregulating its
expression in peripheral blood.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Sciences Funds of China (No.81860104 and 81860369).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LFQ and XPL conceived of the study. JMQ, QEZ, JQZ
and CQY performed the experiments. LFQ analyzed the data and
drafted the manuscript. LFQ, QEZ and JQZ ensure the authenticity of
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Guangxi Medical
University (Nanning, China). The patients provided written informed
consent for the use of their data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kulik L and El-Serag HB: Epidemiology and
management of hepatocellular carcinoma. Gastroenterology.
156:477–491.e1. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pal LR and Moult J: Genetic basis of
common human disease: Insight into the role of missense SNPs from
genome-wide association studies. J Mol Biol. 427:2271–2289. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nie S, Wang H and Wang X: Research
progress on polymorphism of susceptibility genes in hepatocellular
carcinoma. Int J Genet. 41:512–517. 2018.
|
|
5
|
Nobre CC, de Araújo JM, Fernandes TA,
Cobucci RN, Lanza DC, Andrade VS and Fernandes JV: Macrophage
migration inhibitory factor (MIF): Biological activities and
relation with cancer. Pathol Oncol Res. 23:235–244. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grieb G, Merk M, Bernhagen J and Bucala R:
Macrophage migration inhibitory factor (MIF): A promising
biomarker. Drug News Perspect. 23:257–264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Illescas O, Gomez-Verjan JC,
Garcia-Velazquez L, Govezensky T and Rodriguez-Sosa M: Macrophage
migration inhibitory factor-173 G/C polymorphism: A global
meta-analysis across the disease spectrum. Front Genet. 9:552018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fukui H, Saito H, Ueno Y, Uto H, Obara K,
Sakaida I, Shibuya A, Seike M, Nagoshi S, Segawa M, et al:
Evidence-based clinical practice guidelines for liver cirrhosis
2015. J Gastroenterol. 51:629–650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hou J, Wang G, Wang F, Cheng J, Ren H,
Zhuang H, Sun J, Li L, Li J, Meng Q, et al: Guideline of prevention
and treatment for chronic hepatitis B (2015 Update). J Clin Transl
Hepatol. 5:297–318. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou J, Sun H, Wang Z, Cong W, Wang J,
Zeng M, Zhou W, Bie P, Liu L, Wen T, et al: Guidelines for the
diagnosis and treatment of hepatocellular carcinoma (2019 edition).
Liver Cancer. 9:682–720. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Forner A, Reig ME, de Lope CR and Bruix J:
Current strategy for staging and treatment: The BCLC update and
future prospects. Semin Liver Dis. 30:61–74. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ayuso C, Rimola J, Vilana R, Burrel M,
Darnell A, García-Criado Á, Bianchi L, Belmonte E, Caparroz C,
Barrufet M, et al: Diagnosis and staging of hepatocellular
carcinoma (HCC): Current guidelines. Eur J Radiol. 101:72–81. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meza-Romero R, Benedek G, Leng L, Bucala R
and Vandenbark AA: Predicted structure of MIF/CD74 and RTL1000/CD74
complexes. Metab Brain Dis. 31:249–255. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jankauskas SS, Wong DWL, Bucala R, Djudjaj
S and Boor P: Evolving complexity of MIF signaling. Cell Signal.
57:76–88. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Conroy H. Mawhinney L and Donnelly SC:
Inflammation and cancer: Macrophage migration inhibitory factor
(MIF)-the potential missing link. QJM. 103:831–836. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jäger B, Klatt D, Plappert L, Golpon H,
Lienenklaus S, Barbosa PD, Schambach A and Prasse A: CXCR4/MIF axis
amplifies tumor growth and epithelial-mesenchymal interaction in
non-small cell lung cancer. Cell Signal. 73:1096722020. View Article : Google Scholar
|
|
17
|
Guda MR, Rashid MA, Asuthkar S, Jalasutram
A, Caniglia JL, Tsung AJ and Velpula KK: Pleiotropic role of
macrophage migration inhibitory factor in cancer. Am J Cancer Res.
9:2760–2773. 2019.PubMed/NCBI
|
|
18
|
Penticuff JC, Woolbright BL, Sielecki TM,
Weir SJ and Taylor JA: MIF family proteins in genitourinary cancer:
Tumorigenic roles and therapeutic potential. Nat Rev Urol.
16:318–328. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hira E, Ono T, Dhar DK, El-Assal ON,
Hishikawa Y, Yamanoi A and Nagasue N: Overexpression of macrophage
migration inhibitory factor induces angiogenesis and deteriorates
prognosis after radical resection for hepatocellular carcinoma.
Cancer. 103:588–598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li QT, Feng YM, Ke ZH, Qiu MJ and Xiong
ZF: KCNN4 promotes invasion and metastasis through the MAPK/ERK
pathway in hepatocellular carcinoma. J Investig Med. 68:68–74.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Songlin M, Fei X, Long Y and Yang L: The
effect of miRNA-451 on the proliferation and invasion capacity of
human hepatocellular carcinoma cell lines via its regulation on
COX-2 and other cytokines. Oncol Progress. 16:690–693. 2018.
|
|
22
|
Sun B, Nishihira J, Yoshiki T, Kondo M,
Sato Y, Sasaki F and Todo S: Macrophage migration inhibitory factor
promotes tumor invasion and metastasis via the Rho-dependent
pathway. Clin Cancer Res. 11:1050–1058. 2005.PubMed/NCBI
|
|
23
|
Pei XJ, Wu TT, Li B, Tian XY, Li Z and
Yang QX: Increased expression of macrophage migration inhibitory
factor and DJ-1 contribute to cell invasion and metastasis of
nasopharyngeal carcinoma. Int J Med Sci. 11:106–115. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu J, Zhang C, Hu W and Feng Z: Tumor
suppressor p53 and its mutants in cancer metabolism. Cancer Lett.
356:197–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mathumai K: Treating p53 mutant
aggregation-associated cancer. Cancers. 10:1542018. View Article : Google Scholar
|
|
26
|
Ubby I, Krueger C, Rosato R, Qian W, Chang
J and Sabapathy K: Cancer therapeutic targeting using
mutant-p53-specific siRNAs. Oncogene. 38:3415–3427. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fukaya R, Ohta S, Yaguchi T, Matsuzaki Y,
Sugihara E, Okano H, Saya H, Kawakami Y, Kawase T, Yoshida K and
Toda M: MIF maintains the tumorigenic capacity of brain
tumor-initiating cells by directly inhibiting p53. Cancer Res.
76:2813–2823. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kong F, Xuan D, Kong X, Du Y, Li L, Zhu H,
Wang Y, Xie D, Guha S, Li Z, et al: ZFPM2-AS1, a Novel lncRNA,
attenuates the p53 pathway and promotes gastric carcinogenesis by
stabilizing MIF. Oncogene. 37:5982–5996. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ramireddy L, Chen WT, Peng CT, Hu RM, Ke
TW, Chiang HC, Chang SC, Tsai FJ and Lo WY: Association between
genetic polymorphism of the MIF gene and colorectal cancer in
Taiwan. J Clin Lab Anal. 29:268–274. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin S, Wang M, Liu X, Zhu W, Guo Y, Dai Z,
Yang P, Tian T, Dai C, Zheng Y, et al: Association of genetic
polymorphisms in MIF with breast cancer risk in Chinese women. Clin
Exp Med. 17:395–401. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ding GX, Zhou SQ, Xu Z, Feng NH, Song NH,
Wang XJ, Yang J, Zhang W, Wu HF and Hua LX: The association between
MIF-173 G>C polymorphism and prostate cancer in southern
Chinese. J Surg Oncol. 100:106–110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiang T, Bing Z, Tong Q, Liu S, Peng S,
Yang X and Fan H: The MIF-173G/C gene polymorphism increase
gastrointestinal cancer and hematological malignancy risk: Evidence
from a meta-analysis and FPRP test. Int J Clin Exp Med.
8:15949–15957. 2015.PubMed/NCBI
|
|
33
|
Yuan T, Tang C, Chen M, Deng S and Chen P:
Influence of the human MIF promoter polymorphism on hepatocellular
carcinoma prognosis. Genet Mol Res. 12:6629–6635. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
O'Reilly C, Doroudian M, Mawhinney L and
Donnelly SC: Targeting MIF in cancer: Therapeutic strategies,
current developments, and future opportunities. Med Res Rev.
36:440–460. 2016. View Article : Google Scholar
|
|
35
|
Wang D, Luo L, Chen W, Chen LZ, Zeng WT,
Li W and Huang XH: Significance of the vascular endothelial growth
factor and the macrophage migration inhibitory factor in the
progression of hepatocellular carcinoma. Oncol Rep. 31:1199–1204.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao YM, Wang L, Dai Z, Wang DD, Hei ZY,
Zhang N, Fu XT, Wang XL, Zhang SC, Qin LX, et al: Validity of
plasma macrophage migration inhibitory factor for diagnosis and
prognosis of hepatocellular carcinoma. Int J Cancer. 129:2463–2472.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Han Y and Zhang C: Macrophage migration
inhibitory factor plays a pivotal role in hepatocellular carcinoma
and may be a noninvasive imaging target. Medical Hypotheses.
75:530–532. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Oscar I, Gomez-Verjan JC, Lizbeth GV,
Tzipe G and Miriam RS: Macrophage migration inhibitory factor-173
G/C polymorphism: A global meta-analysis across the disease
spectrum. Front Genet. 9:552018. View Article : Google Scholar
|
|
39
|
Gnoni A, Santini D, Scartozzi M, Russo A,
Licchetta A, Palmieri V, Lupo L, Faloppi L, Palasciano G, Memeo V,
et al: Hepatocellular carcinoma treatment over sorafenib:
Epigenetics, microRNAs and microenvironment. Is there a light at
the end of the tunnel? Expert Opin Ther Targets. 19:1623–1635.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang K, Pan X, Shu X, Cao H, Chen L, Zou
Y, Deng H, Li G and Xu Q: Relationship between MIF-173 G/C
polymorphism and susceptibility to chronic hepatitis B and
HBV-induced liver cirrhosis. Cell Immunol. 282:113–116. 2013.
View Article : Google Scholar : PubMed/NCBI
|