Introduction
Colorectal cancer (CRC) is the world's fourth most
lethal malignancy, causing ~900,000 mortalities every year
(1–4). It is the third most common malignancy
in males after prostate and lung cancer, and the second most common
malignancy in females after breast cancer (5). Over the last 20 years, the incidence of
CRC has been rapidly rising in those aged <50 years (2,4). Despite
advances in systemic therapy, many patients with metastatic CRC
succumb to their disease (6).
Therefore, there is an urgent requirement for more effective
therapeutic strategies for CRC. It is of great significance to
identify biomarkers affecting cell proliferation and migration, and
to determine the regulatory mechanisms in CRC cells.
Long non-coding RNAs (lncRNAs) are non-coding
transcripts that are >200 nucleotides long and have previously
been identified as one of the largest and most diverse RNA families
(7,8). lncRNAs are modulators involved in a
variety of biological processes (9–11). They
regulate gene expression via different mechanisms in the cytoplasm
or nucleus (11,12). Additionally, RNA-binding proteins
(RBPs) exert vital effects on RNA processing and metabolism,
including mRNA localization, mRNA stability control and translation
control (13,14). The abnormal expression and mutation
of RBP genes affect different steps of RNA processing, thereby
changing the function of the target gene (13).
RNA-protein interactions create vital checkpoints
during the regulation of gene expression at the RNA level (15). lncRNAs are associated with a variety
of cellular functions, most of which involve interactions with one
or more RBPs (16). RBPs usually
bind to numerous different RNAs. Furthermore, RBPs bind to target
transcripts and alter their translation or stability (17). lncRNAs interact with specific RBPs to
enhance mRNA stability in the cytoplasm (18,19). It
has been revealed that lncRNA long intergenic non-protein coding
RNA 857 (LINC00857) serves as a tumor promotor in esophageal
adenocarcinoma (20), hepatocellular
carcinoma (HCC) (21) and bladder
cancer (22); however, its role and
mechanism in CRC cells remain elusive.
Thus, the aim of the present study was to focus on
the role and regulatory mechanism of LINC00857 in CRC cells in
order to provide potential novel insight into CRC therapy.
Materials and methods
Bioinformatics analysis
The interactions between LINC00857, solute carrier
family 7 member 5 (SLC7A5) and YTH domain containing 1 (YTHDC1)
were predicted using the starBase database (http://starbase.sysu.edu.cn/). Additionally, the
extent of binding between LINC00857 and YTHDC1 was predicted though
the RNA-Protein Interaction Prediction (RPISeq) website (http://pridb.gdcb.iastate.edu/RPISeq/).
Patient sample collection
Forty CRC specimens and adjacent non-tumor tissue
samples were obtained from 20 male and 20 female patients (age
range, 32–73 years; mean age, 51.4±6.1 years) who did not receive
therapy before undergoing surgery at Chenzhou No. 1 People's
Hospital. The tissue samples were harvested via surgical resection
or colonoscopy. Adjacent non-tumor tissue samples were obtained
from ≥1 cm away from the tumor tissues. Only histologically
confirmed new CRC cases that had not previously been diagnosed for
cancer were included in the present study. Tumor location (left- or
right-sided CRC) was histologically confirmed. The histological
grade was assessed according to World Health Organization criteria
(23). Tumors were staged according
to the tumor-node-metastasis staging system of the American Joint
Committee on Cancer (7th edition) (24). All patients provided written informed
consent and the study was approved by the Ethics Committee of the
Chenzhou No. 1 People's Hospital (Chenzhou, China). All of the
methods included in the present study were performed rigidly
according to the approved guidelines. Immediately after surgical
resection, the tissue samples were frozen in liquid nitrogen and
stored at −80°C.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from the tissue samples and cells (CRC
cell lines, SW480, SW620, LoVo and LS174T cells and human normal
colonic epithelial cell line, FHC) was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The RNA quality and concentration were determined using the
NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Inc.)
at absorbances of 260 and 280 nm. Total RNA was digested using
DNase I (Thermo Fisher Scientific Inc.) for 90 min at 37°C and then
reverse transcribed into complementary DNA (cDNA) using a reverse
transcription kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's instructions. The cDNA template (3 µl; corresponding
to ~150 ng of the extracted total RNA) was prepared prior to each
PCR reaction. SYBR® Premix Ex Taq™ (Takara Biotechnology
Co., Ltd.) was used for RT-qPCR analysis, which was performed on an
Applied Biosystems™ 7500 Real-Time PCR System. The relative
quantification was calculated according to the 2−ΔΔCq
method as previously described (25). The results were normalized to GAPDH
expression. Primer sequences used in the present study are
presented in Table SI and the
following thermocycling conditions were used: Initial denaturation
at 95°C for 3 min, followed by 40 cycles at 95°C for 5 sec, 60°C
for 30 sec and at 72°C for 45 sec, before final extension at 72°C
for 3 min.
Cell culture and transfection
CRC cell lines, SW480, SW620, LoVo and LS174T cells
and a human normal colonic epithelial cell line, FHC were purchased
from the American Type Culture Collection. All CRC cells were
cultured in Gibco Dulbecco's modified Eagle's medium (DMEM; Thermo
Fisher Scientific, Inc.) supplemented with Gibco 10% fetal bovine
serum (FBS; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 mg/ml streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) in a humidified atmosphere at 37°C with 5%
CO2.
Short hairpin (sh)-RNAs for LINC00857
(sh-LINC00857#1/2) and YTHDC1 (sh-YTHDC1) and a negative control
(sh-NC) were transfected into CRC cells. Additionally, the sequence
of LINC00857 was synthesized and subcloned into a pcDNA3.1 plasmid
(Invitrogen; Thermo Fisher Scientific, Inc.) to overexpress
LINC00857 with an empty pcDNA3.1 vector serving as the NC. The
sequence of YTHDC1 was synthesized and subcloned into a pcDNA3.1
plasmid to overexpress YTHDC1 with an empty pcDNA3.1 vector serving
as the NC. The sequence of SLC7A5 was synthesized and subcloned
into a pcDNA3.1 plasmid to overexpress SLC7A5 with an empty
pcDNA3.1 vector serving as the NC. SW480 and SW620 cells
(1×105 cells/well) were seeded in 24-well plates and 500
µl DMEM was added to each well. When the cells reached 40–60%
confluence, the abovementioned vectors were transfected into cells
at a final concentration of 50 nM using Lipofectamine®
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C
with 5% CO2 according to the manufacturer's
instructions. Cells were harvested using TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.) and transferred to 1.5-ml Eppendorf tubes
for mixture and stored at −80°C for further analysis 48 h
post-transfection. Finally, RT-qPCR was performed to evaluate the
transfection efficiency. The plasmid sequences used in the present
study are presented in Table
SI.
Cell counting kit-8 (CCK-8) assay
Following transfection with sh-LINC00857#1/2 or
sh-NC, SW480 and SW620 cells (1×103 cells/well) were
plated into 96-well plates. Cell viability was detected on days 0–2
and 3 using a CCK-8 assay (Dojindo Molecular Technologies, Inc.)
according to the manufacturer's instructions. Optical density was
assessed at a wavelength of 450 nm using a microplate reader
(Thermo Fisher Scientific, Inc.).
Colony formation assay
For colony formation assays, SW480- and
SW620-transfected cells (1×103) were placed in 6-well
plates and cultivated in proper medium supplemented with 10% FBS
for 14 days and, during this period, the medium was replaced every
4 days. After 14 days, the cells were fixed with methanol and
stained with 0.1% crystal violet (MilliporeSigma) for 30 min at
room temperature. Images of the visible colonies were obtained and
the number of colonies was counted by a light microscope (OLYMPUS
IX-71; Olympus Corporation) (magnification, ×10) after 48 h.
5-Ethynyl-2-deoxyuridine (EdU)
assay
EdU experiments were conducted with an EdU
labeling/detection kit (Guangzhou RiboBio Co., Ltd.) according to
the manufacturer's instructions. The transfected cells were placed
in 50 µM of EdU staining medium and maintained for an additional 2
h at room temperature. After washing the cells three times with 0.5
g/ml phosphate-buffered saline (PBS), the nuclei were stained with
DAPI (Invitrogen; Thermo Fisher Scientific, Inc.) for 10 min in the
dark room at room temperature. Finally, fluorescence microscopy was
utilized to observe and count EdU-positive cells. The percentage of
EdU-positive cells was calculated from five random fields in three
wells.
Flow cytometry analysis
Cell apoptosis was assessed via flow cytometry.
SW480 and SW620 cells were subjected to double staining with
fluorescein isothiocyanate-Annexin V and propidium iodide
(Becton-Dickinson and Company) according to the manufacturer's
instructions. The cells were detected via flow cytometry using a
FACScan® (BD Biosciences) equipped with CellQuest Pro
software v5.1 (BD Biosciences).
Wound healing assay
Following transfection, SW480 and SW620
(2×105) cells seeded in 6-well plates were subjected to
serum starvation for 4 h and cultured to 100% confluence.
Thereafter, a wound was simulated by scratching a straight line in
the cell monolayer using a sterile 200-µl pipette tip. After gently
scraping the scratched monolayer cells twice with serum-free medium
(Gibco; Thermo Fisher Scientific Inc.), the wound was healed in
complete medium for 24 h. After the wound was formed, the wound
widths at 0 and 24 h were visualized using an inverted microscope.
The percentage of wound closure was assessed for cell
migration.
Western blot analysis
Total proteins were extracted from cells using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology) including protease inhibitors (Beyotime Institute of
Biotechnology) and protein concentrations were evaluated using a
bicinchoninic acid Protein Assay Kit (Beyotime Institute of
Biotechnology). The protein lysates were separated using 10%
SDS-polyacrylamide gel electrophoresis and equal quantities (5 µg
per sample) of separated proteins were transferred to 0.22-µm
nitrocellulose membranes (MilliporeSigma). After blocking with 5%
non-fat milk for 1 h at room temperature, the membranes were
incubated with specific primary antibodies and treated with
specific primary antibodies overnight at 4°C. GAPDH (cat. no.
ab8245; 1:500) served as the internal control. The primary
antibodies (Abcam) were as follows: Anti-E-cadherin (cat. no.
ab1416; 1:50), anti-N-cadherin (cat. no. ab98952; 1:500), anti-MMP2
(cat. no. ab92536; 1:1,000), anti-MMP9 (cat. no. ab76003; 1:1,000)
and anti-SLC7A5 (cat. no. ab99419; 1:1,000). After washing three
times with PBS, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit antibodies [cat. no. sc-2357
(1:5,000) Santa Cruz Biotechnology, Inc.] for another 2 h at room
temperature. The proteins were visualized using an enhanced
chemiluminescence detection kit (Amersham; Cytiva) and the Odyssey
Infrared Imaging system version 2.1 (LI-COR Biosciences).
Subcellular fractionation
Cytoplasmic and nuclear extracts were extracted from
SW480 or SW620 cells using NE-PER™ kit (cat. no. 78833; Thermo
Fisher Scientific, Inc.). RT-qPCR analysis was performed to detect
the RNAs isolated from the nucleus or cytoplasm. Expression levels
of U6, the nuclear control and GAPDH, the cytoplasmic control were
evaluated.
RNA immunoprecipitation (RIP)
assay
The RIP assay was conducted using a Magna RIP™
RNA-Binding Protein Immunoprecipitation Kit (MilliporeSigma)
according to the manufacturer's instructions. CRC cells at 80–90%
confluency were scraped off and lysed in complete RIP lysis buffer.
Whole cell extracts (100 µl) were cultivated with magnetic beads
for 6 h at 4°C and the beads were conjugated with anti-UPF1 RNA
helicase and ATPase (UPF1), anti-heterogeneous nuclear
ribonucleoprotein C (HNRNPC), anti-YTHDC1 or control IgG. After the
magnetic beads were washed three times, Proteinase K (Thermo Fisher
Scientific Inc.) was applied to remove proteins from the complexes.
Purified RNAs were subjected to RT-qPCR analysis.
In vitro transcription and RNA
pull-down assay
T7 RNA polymerase (Thermo Fisher Scientific Inc.)
was utilized to transcribe LINC00857 in vitro (YTHDC1), a
RNeasy Plus Mini Kit (Qiagen Sciences, Inc.) was utilized to purify
LINC00857 (YTHDC1) and RNase-free DNase I (Qiagen Sciences, Inc.)
was utilized to treat LINC00857 (YTHDC1). The transcribed LINC00857
(YTHDC1) was labeled with Biotin RNA Labeling Mix (Thermo Fisher
Scientific Inc.) and the RNA pull-down assays were performed with a
Pierce™ Magnetic RNA-Protein Pull-Down Kit (Pierce; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
RNA stability assay
Following transfection, CRC cells were treated with
1 µg/ml actinomycin D. Next, the cells were collected at different
time points (0, 3, 6 and 9 h) and RNA was extracted with TRIzol™
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Finally,
RT-qPCR analysis was performed to evaluate the mRNA levels.
Statistical analysis
Each experiment was repeated three times. Both
paired and unpaired Student's t-test were used to analyze
differences between two groups. Paired Student's t-test was used
for RT-qPCR analysis to evaluate gene expression in CRC tissues and
paired adjacent non-tumor tissue samples. One-way analysis of
variance followed by Tukey's post hoc test was used to analyze the
differences between three groups. All statistical analyses were
performed with SPSS v.20.0 software (IBM Corp.) and the data are
presented as the means ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Significantly upregulated LINC00857 in
CRC cells promotes CRC cell proliferative and migratory abilities
in vitro
According to previous studies, LINC00857 expression
is upregulated in certain types of cancer (20–22).
Therefore, to verify the abnormal expression of LINC00857 during
CRC progression, RT-qPCR analysis was performed with CRC and
adjacent healthy tissue samples, as well as with four CRC cell
lines (SW480, SW620, LoVo and LS174T) and a human normal colonic
epithelial cell line (FHC). The results indicated that LINC00857
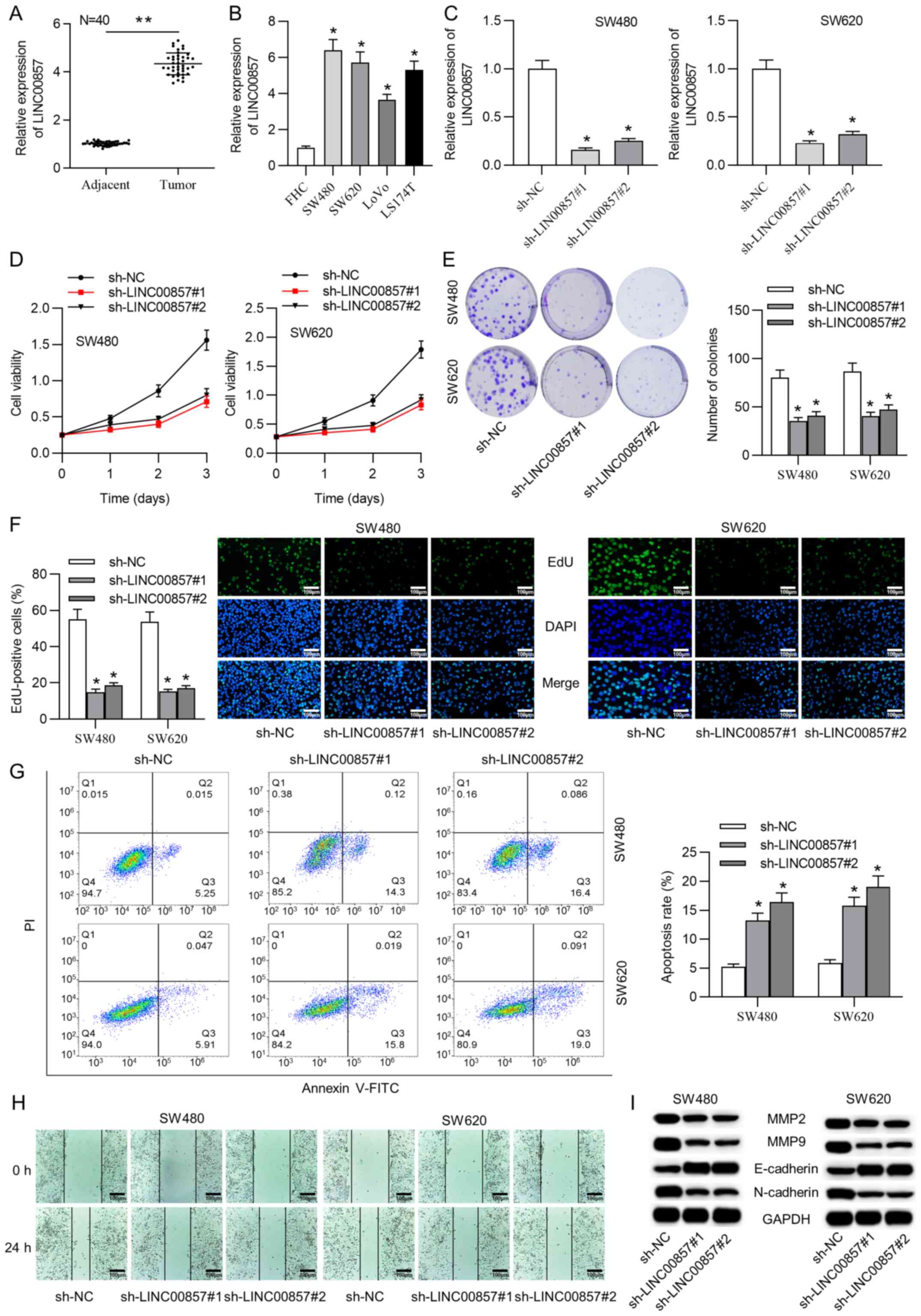
was significantly upregulated in CRC tissue samples (Fig. 1A). Consistently, LINC00857 expression
was higher in CRC cells than in healthy cells (Fig. 1B). Additionally, the RT-qPCR results
demonstrated a close association between LINC00857 and the
right-sided CRC samples (Fig. S1A).
There was a statistically significant association between high
LINC00857 expression and the right-sided CRC samples (Table I). Furthermore, LINC00857 expression
was observed to be upregulated in advanced stage CRC tissue samples
(Fig. S1B). In addition, higher
LINC00857 expression was closely associated with larger tumor size,
more advanced stage and poor histological grade (Table I).
 | Table I.Association between LINC00857
expression and clinical features of 40 colorectal cancer tissue
samples. |
Table I.
Association between LINC00857
expression and clinical features of 40 colorectal cancer tissue
samples.
|
|
| LINC00857
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | Patients, n | High, n (%) | Low, n (%) | P-value |
|---|
| Localization |
|
|
| P<0.001 |
|
Left | 11 | 2 (18.2) | 9 (81.8) |
|
|
Right | 29 | 24 (82.8) | 5 (17.2) |
|
| Tumor size
(cm) |
|
|
| P<0.01 |
| ≤2 | 14 | 4 (28.6) | 10 (71.4) |
|
|
>2 | 26 | 21 (80.8) | 5 (19.2) |
|
| Clinical T
stage |
|
|
| P<0.001 |
|
T1-T2 | 13 | 2 (15.4) | 11 (84.6) |
|
|
T3-T4 | 27 | 20 (74.1) | 7 (25.9) |
|
| Histological
grade |
|
|
| P<0.01 |
| Well
differentiated | 5 | 1 (20.0) | 4 (80.0) |
|
|
Moderately differentiated | 15 | 4 (26.7) | 11 (73.3) |
|
| Poorly
differentiated | 20 | 17 (85.0) | 3 (15.0) |
|
To investigate the role of LINC00857 in CRC cells,
SW480 and SW620 cells were transfected with sh-LINC00857#1/2 or
sh-NC vectors to silence LINC00857 and a series of loss-of-function
experiments were subsequently performed. The transfection
efficiency was analyzed by RT-qPCR and the results indicated that
LINC00857 expression was significantly decreased in the SW480 and
SW620 cells (Fig. 1C). Furthermore,
the CCK-8 assay revealed that cell viability was impaired by
LINC00857 downregulation (Fig. 1D).
Moreover, the colony formation assay demonstrated a large decrease
in the number of colonies (Fig. 1E).
Additionally, the EdU staining results illustrated that the
percentage of EdU-positive cells in the sh-LINC00857#1/2-mediated
group was diminished (Fig. 1F).
These findings cumulatively indicate that silencing LINC00857
suppressed the proliferative ability of CRC cells.
Subsequently, to investigate whether apoptosis
regulation was propelled by LINC00857 silencing, flow cytometric
analysis was conducted. The results demonstrated that LINC00857
knockdown induced CRC cell apoptosis (Fig. 1G). Additionally, to estimate the
effect of LINC00857 knockdown on CRC cell migratory ability, a
wound healing assay was performed. The number of migrated CRC cells
was decreased following LINC00857 depletion (Fig. 1H). To validate the function of
LINC00857 silencing on CRC cell migration, western blot analyses
were conducted to evaluate the MMP2, MMP9, E-cadherin and
N-cadherin levels. The results showed that the protein levels of
MMP2 and MMP9 were markedly decreased after LINC00857 silencing.
Simultaneously, the E-cadherin protein level was upregulated and
the N-cadherin protein level was downregulated in the
sh-LINC00857#1/2 groups (Fig.
1I).
LINC00857 interacts with RNA-binding
protein YTHDC1
The localization of lncRNAs within the cell
primarily determines their molecular functions (26). Therefore, to clarify the potential
mechanism of LINC00857 in CRC cells, subcellular localization was
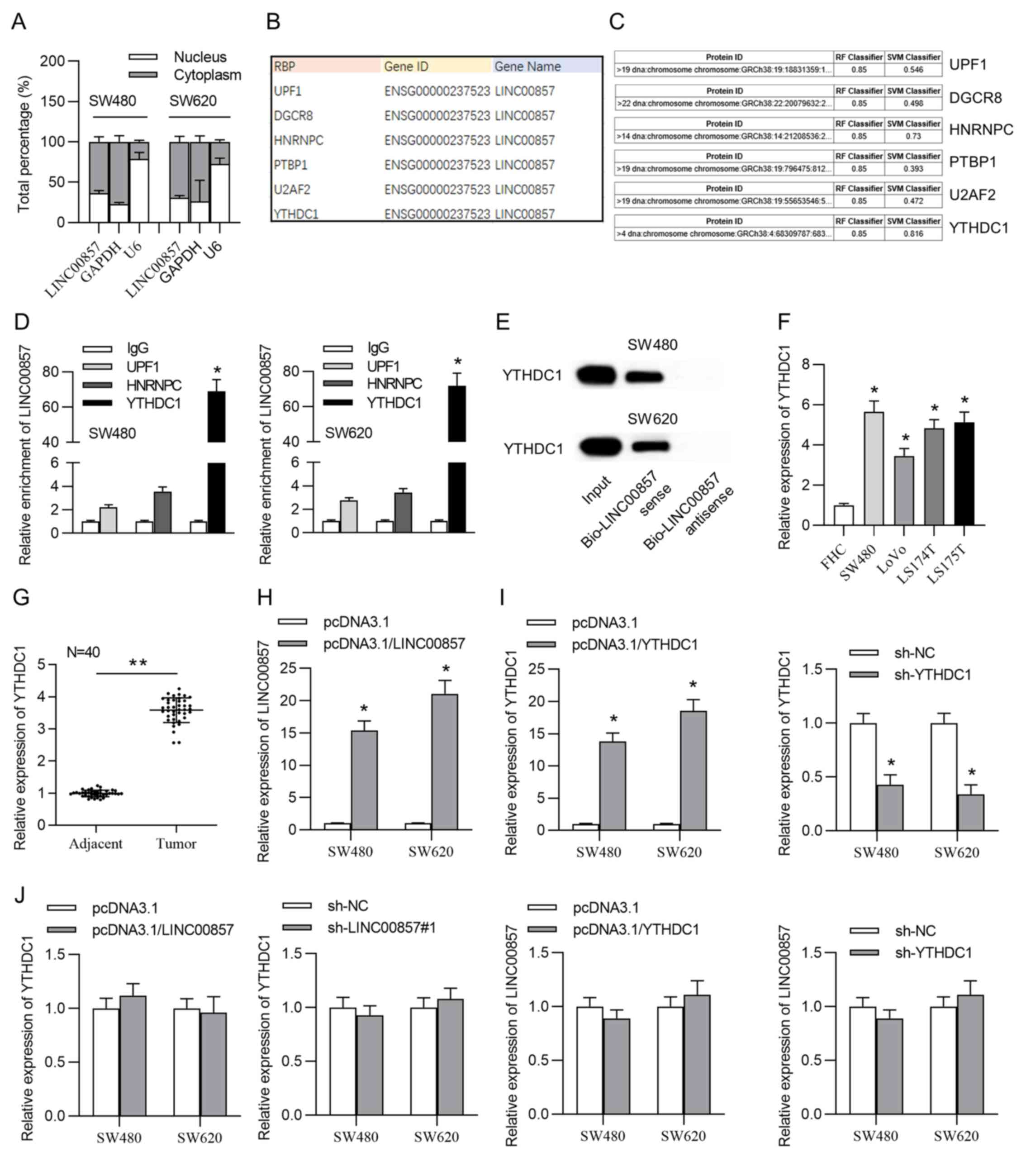
conducted. The results indicated LINC00857 was primarily
distributed in the cytoplasm of CRC cells (Fig. 2A) indicating that LINC00857
post-transcriptionally regulated gene expression. Cytoplasmic
lncRNAs cooperate with RBPs in different types of cancer (27). Therefore, it was hypothesized that
LINC00857 interacts with RBPs to regulate CRC cells. Six potential
RBPs were predicted using the starBase database (Fig. 2B). The interaction between LINC00857
and these candidate RBPs was evaluated by RPISeq. The Random Forest
classifier and Support Vector Machine classifier scores were
>0.5, indicating that LINC00857 may bind to UPF1, YTHDC1 and
HNRNPC (Fig. 2C). To determine the
interaction between these RBPs and LINC00857 an RIP assay was
performed. LINC00857 presented a significant enrichment conjugated
with anti-YTHDC1 rather than anti-UPF1 or anti-HNRNPC (Fig. 2D). Similarly, high binding affinity
between LINC00857 and YTHDC1 was verified via western blotting
performed following the RNA pull-down assay. The results
demonstrated significant enrichment in pull-down products by
biotinylated LINC00857 sense and no enrichment was observed in
pull-down products by biotinylated LINC00857 antisense by blotting
using the anti-YTHDC1 antibody (Fig.
2E). Furthermore, according to RT-qPCR results, YTHDC1 was
upregulated in CRC tissue samples and cells (Fig. 2F and G).
LINC00857 was transfected with the
pcDNA3.1/LINC00857 plasmid and RT-qPCR revealed that LINC00857
expression was significantly increased in the CRC cells (Fig. 2H). RT-qPCR was also used to examine
the transfection efficiency of pcDNA3.1/YTHDC1 and sh-YTHDC1. The
results revealed that YTHDC1 was upregulated in the pcDNA3.1/YTHDC1
group and downregulated in the sh-YTHDC1 group (Fig. 2I). YTHDC1 expression was not altered
upon LINC00857 silencing or elevation and LINC00857 expression was
not changed upon YTHDC1 upregulation or downregulation (Fig. 2J). These findings indicated that
YTHDC1 and LINC00857 cannot mutually affect gene expression,
verifying that they were bound together.
LINC00857 increases SLC7A5 stability
by recruiting YTHDC1
Previous studies reported that RBPs interact with
mRNAs and increase mRNA stability (28–30). The
present study hypothesized whether YTHDC1 had a similar regulatory
function on its target in CRC cells. First, 10 potential target
mRNAs were predicted using the starBase database (Fig. 3A). Among all the putative mRNAs,
SLC7A5, plectin (PLEC) and zinc finger homeobox 3 (ZFHX3) were
successfully confirmed with western blotting of complexes pulled
down by biotinylated YTHDC1 (Fig.
3B). RT-qPCR results demonstrated that SLC7A5 expression
decreased under YTHDC1 downregulation, and PLECs and ZFHX3 were not
decreased (Fig. 3C). SLC7A5 was
therefore identified as a downstream target of YTHDC1 and selected
for subsequent experiments. Subsequently, SLC7A5 expression in CRC
tissue samples and cells was evaluated using RT-qPCR. SLC7A5 was
found to be significantly upregulated in CRC tissue samples and
cells (Fig. 3D and E). Furthermore,
the RIP assay with SW480 and SW620 cells showed that the
interaction of YTHDC1-SLC7A5 was suppressed under LINC00857
deficiency (Fig. 3F). Furthermore,
mRNA and protein expression levels of SLC7A5 decreased as a result
of LINC00857 knockdown (Fig.
3G).
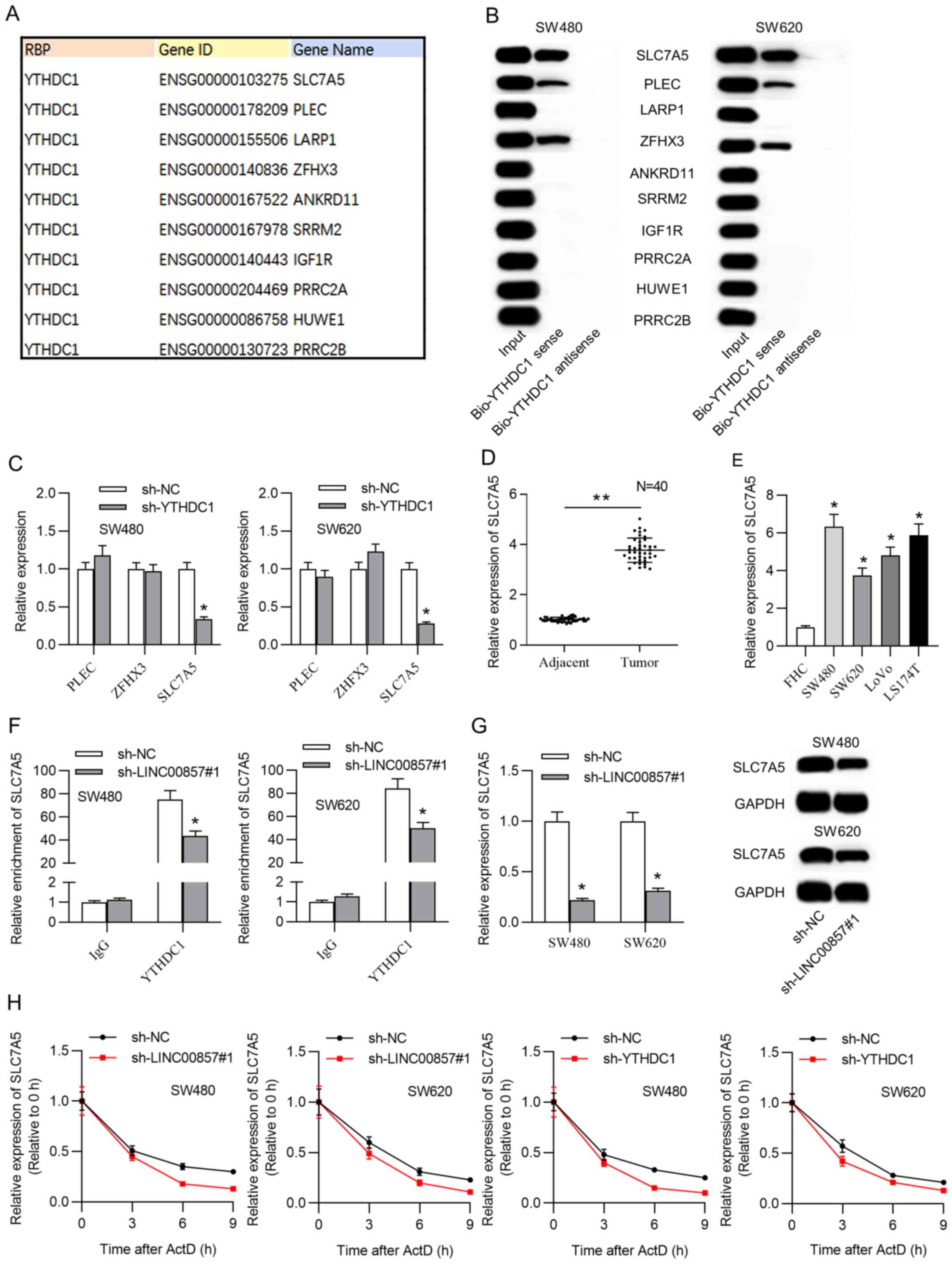 | Figure 3.SLC7A5 was a potential downstream
target of LINC00857-YTHDC1. (A) Putative targets of YTHDC1 were
acquired from the starBase database. (B) Western blot analysis
verified the identity of the mRNAs pulled down by biotinylated
YTHDC1. (C) The expression of the candidate mRNAs after YTHDC1
knockdown was evaluated via RT-qPCR analysis. (D) RT-qPCR detected
SLC7A5 levels in CRC and adjacent tissue samples. **P<0.01. (E)
YTHDC1 expression was analyzed via RT-qPCR in CRC cell lines and a
control cell line. *P<0.05 vs. FHC. (F) RIP experiments verified
SLC7A5 enrichment in YTHDC1 RIP under LINC00857 silencing in CRC
cells. *P<0.05 vs. sh-NC. (G) SLC7A5 mRNA and protein levels
were confirmed after LINC00857 knockdown in CRC cells through
RT-qPCR and western blot analysis. *P<0.05 vs. sh-NC. (H) RNA
stability assay assessed the degradation rate of SLC7A5 mRNA in CRC
cells upon LINC00857 or YTHDC1 knockdown. SLC7A5, solute carrier
family 7 member 5; LINC00857, long intergenic non-protein coding
RNA 857; YTHDC1, YTH domain containing 1; RT-qPCR, reverse
transcription-quantitative PCR; CRC, colorectal cancer; RIP, RNA
immunoprecipitation; sh, short hairpin; NC, negative control; PLEC,
plectin; ZFHX3, zinc finger homeobox 3; ActD, actinomycin D. |
These findings further supported LINC00857-YTHDC1
binding and their coregulation of SLC7A5 expression. Additionally,
to further probe whether LINC00857-YTHDC1 regulates SLC7A5 mRNA
stability, SW480 and SW620 cells were treated with actinomycin D
and the decay of pre-existing mRNA was evaluated. The results
revealed that silencing LINC00857 or YTHDC1 repressed the SLC7A5
mRNA half-life (Fig. 3H).
LINC00857 enhances CRC cell
proliferative and migratory abilities by upregulating SLC7A5
stability
To probe SLC7A5 involvement in LINC00857-mediated
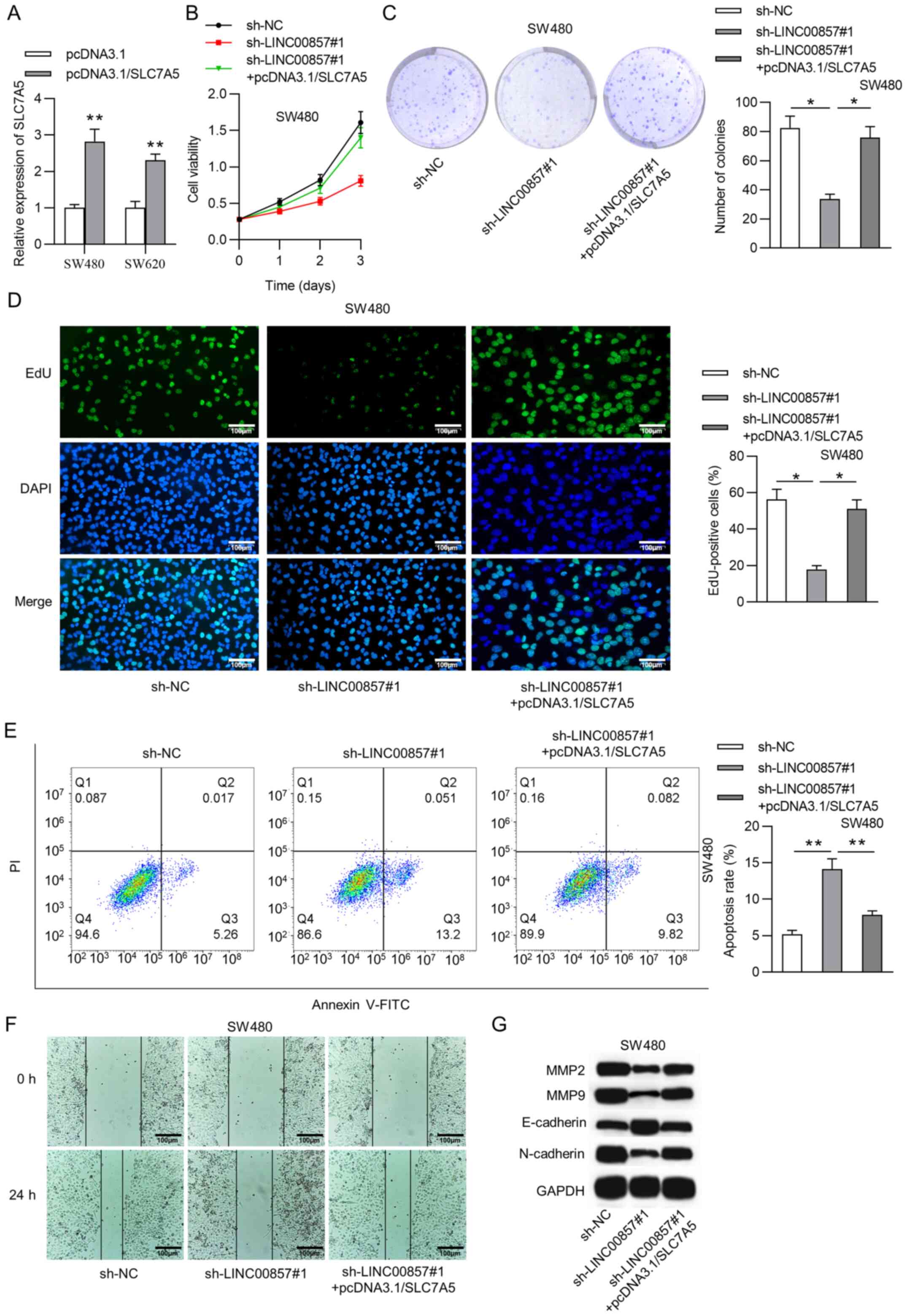
CRC cell proliferation and migration, rescue assays were performed.
CRC cells were co-transfected with sh-LINC00857#1 and
pcDNA3.1/SLC7A5. RT-qPCR analysis indicated that SLC7A5 expression
was significantly increased in CRC cells transfected with
pcDNA3.1/SLC7A5 (Fig. 4A). The CCK-8
assay indicated that upregulated SLC7A5 had the opposite effect on
CRC cell viability suppressed by LINC00857 silencing (Fig. 4B). Additionally, the proliferation
assay results revealed that SLC7A5 overexpression attenuated the
decrease in CRC cell proliferative ability under LINC00857
knockdown (Fig. 4C and D). The
proportion of apoptotic CRC cells was significantly decreased after
co-transfection (Fig. 4E).
Furthermore, according to the wound healing assay, CRC cell
migration was repressed by LINC00857 depletion, but was recovered
under SLC7A5 upregulation (Fig. 4F).
Similarly, the western blot analysis revealed that pcDNA3.1/SLC7A5
counteracted the inhibitory effect of LINC00857 silencing on the
levels of MMP2, MMP9, E-cadherin and N-cadherin in CRC cells
(Fig. 4G).
Discussion
It has been previously reported that LINC00857 has
an oncogenic effect on certain types of cancer, such as esophageal
adenocarcinoma (20) and HCC
(21). In the present study,
LINC00857 was significantly upregulated in CRC tissue samples and
cells. Additionally, a series of loss-of-function experiments
further validated that LINC00857 knockdown repressed CRC cell
proliferative and migratory abilities and the
epithelial-mesenchymal transition (EMT), as well as promoted cell
apoptosis in vitro. These data suggested that LINC00857
acted as an oncogene by promoting cell proliferation, migration and
EMT during CRC progression.
YTHD-containing proteins participate in numerous RNA
processes, such as mRNA splicing, nuclear export, translation and
decay in post-transcriptional regulation (31). In eukaryotes, YTHD-containing
proteins comprise five functional genes, including YTHDC1 (31). One of the marked effects of YTHDC1 is
RNA nuclear export (32). It has
previously been demonstrated that the upregulation of YTHDC1 acts
as a diagnostic marker, therapeutic target or oncogene in colon
cancer (33), HCC (34) and pancreatic adenocarcinoma (35). Additionally, its downregulation
indicates a favorable prognosis in patients with head and neck
squamous cell carcinoma (36).
Notably, YTHDC1 is abundantly expressed in colon adenocarcinoma
tissues (37). The localization of
lncRNAs within the cell is the primary determinant of their
molecular functions (26).
In the present study, the cytoplasmic localization
of LINC00857 in CRC cells indicated the post-transcriptional effect
of LINC00857 on gene expression. Furthermore, RBPs exert crucial
effects by post-transcriptionally regulating gene expression
(28), including pre-mRNA splicing,
mRNA stabilization, mRNA localization and translation (17,38). The
present study hypothesized the potential regulatory pattern of
LINC00857 in which LINC00857 interacted with RBPs to stabilize
mRNA. The bioinformatics analysis demonstrated that YTHDC1
potentially bound to LINC00857. Subsequent mechanistic experiments
verified the binding ability between lncRNA LINC00857 and RBP
YTHDC1 in CRC cells. In addition, YTHDC1 expression in CRC tissue
samples and cells was identified to be significantly higher than
that in normal CRC tissue samples and cells. LINC00857 and YTHDC1
did not mutually affect expression in CRC cells. These findings
indicated that LINC00857 interacted with YTHDC1 in CRC cells.
Systematically, SLC7A5 mRNA was demonstrated to
interact with YTHDC1 in CRC cells. SLC7A5 (also termed LAT1),
characterized as an amino acid transporter, has been revealed to be
associated with various types of cancer. For example, SLC7A5 serves
as a novel biomarker for high-grade malignancy in prostate cancer
(39). Another study demonstrated
that positive expression of SLC7A5 predicts poor prognosis in
patients with non-small cell lung cancer (40). Furthermore, it has been reported that
RBPs post-transcriptionally interact with target mRNAs, thereby
strengthening mRNA stability (28–30). In
the present study, SLC7A5 was upregulated in CRC tissue samples and
cells. Mechanistically, LINC00857 and YTHDC1 increased SLC7A5 mRNA
stability in CRC cells. LINC00857 was observed to cooperate with
YTHDC1 to stabilize SLC7A5 in CRC cells. Additionally, rescue
assays further verified that the decreased cell proliferation and
migration under LINC00857 knockdown was rescued by upregulation of
SLC7A5.
However, the present study had several limitations,
such as that the results were not validated in vivo. Animal
experiments should be performed to further verify in vitro
results. In addition, LINC00857 has been reported to competitively
bind with miRNAs to regulate mRNA (41). Therefore, other mechanisms mediated
by LINC00857 in CRC cells should be explored in the future.
Thus, the present study confirmed that LINC00857,
YTHDC1 and SLC7A5 were upregulated in CRC tissue samples and cells.
Furthermore, LINC00857 increased CRC cell proliferative and
migratory abilities in vitro. The present study hypothesized
that LINC00857 abundantly bound to YTHDC1 in CRC cells and that the
LINC00857-YTHDC1 complex increased SLC7A5 stability in CRC cells.
The present findings may provide novel insight into lncRNA
regulatory characteristics in CRC and a novel direction for CRC
treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ST conceived and designed the experiments. ST, QL,
and MX performed the experiments. ST and MX analyzed the data. ST
and MX drafted the manuscript. ST and MX confirm the authenticity
of all the raw data. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent and
the study was approved by the Ethics Committee of the Chenzhou No.
1 People's Hospital (approval no. 2019-039; Chenzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel SG and Ahnen DJ: Colorectal cancer
in the young. Curr Gastroenterol Rep. 20:152018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weitz J, Koch M, Debus J, Höhler T, Galle
PR and Büchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Connell LC, Mota JM, Braghiroli MI and
Hoff PM: The rising incidence of younger patients with colorectal
cancer: Questions about screening, biology, and treatment. Curr
Treat Options Oncol. 18:232017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Castells A: Hereditary forms of colorectal
cancer. Gastroenterol Hepatol. 39 (Suppl 1):S62–S67. 2016.(In
English, Spanish). View Article : Google Scholar
|
|
6
|
Wrobel P and Ahmed S: Current status of
immunotherapy in metastatic colorectal cancer. Int J Colorectal
Dis. 34:13–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing MiRNA- LncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kumar MM and Goyal R: LncRNA as a
therapeutic target for angiogenesis. Curr Top Med Chem.
17:1750–1757. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Charles Richard JL and Eichhorn PJA:
Platforms for investigating LncRNA functions. SLAS Technol.
23:493–506. 2018.PubMed/NCBI
|
|
10
|
Qian X, Zhao J, Yeung PY, Zhang QC and
Kwok CK: Revealing lncRNA structures and interactions by
sequencing-based approaches. Trends Biochem Sci. 44:33–52. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma Y, Zhang J, Wen L and Lin A:
Membrane-lipid associated lncRNA: A new regulator in cancer
signaling. Cancer Lett. 419:27–29. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang HV and Chekanova JA: Long noncoding
RNAs in plants. Adv Exp Med Biol. 1008:133–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou H, Mangelsdorf M, Liu J, Zhu L and Wu
JY: RNA-binding proteins in neurological diseases. Sci China Life
Sci. 57:432–444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lujan DA, Ochoa JL and Hartley RS:
Cold-inducible RNA binding protein in cancer and inflammation.
Wiley Interdiscip Rev RNA. 9:10.1002/wrna.1462. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Köster T, Marondedze C, Meyer K and
Staiger D: RNA-binding proteins revisited-the emerging arabidopsis
mRNA interactome. Trends Plant Sci. 22:512–526. 2017. View Article : Google Scholar
|
|
16
|
Ferrè F, Colantoni A and Helmer-Citterich
M: Revealing protein-lncRNA interaction. Brief Bioinform.
17:106–116. 2016. View Article : Google Scholar
|
|
17
|
Panda AC, Martindale JL and Gorospe M:
Affinity pulldown of biotinylated RNA for detection of protein-RNA
complexes. Bio Protoc. 6:e20622016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao L, Zhang P, Li J and Wu M: LAST, a
c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote
CCND1 mRNA stability in human cells. Elife. 6:e304332017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gong C and Maquat LE: lncRNAs
transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs
via Alu elements. Nature. 470:284–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su W, Wang L, Niu F, Zou L, Guo C, Wang Z,
Yang X, Wu J, Lu Y, Zhang J, et al: LINC00857 knockdown inhibits
cell proliferation and induces apoptosis via involving STAT3 and
MET oncogenic proteins in esophageal adenocarcinoma. Aging (Albany
NY). 11:2812–2821. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia C, Zhang XY, Liu W, Ju M, Ju Y, Bu YZ,
Wang W and Shao H: LINC00857 contributes to hepatocellular
carcinoma malignancy via enhancing epithelial-mesenchymal
transition. J Cell Biochem. Dec 3–2018.(Epub ahead of print).
|
|
22
|
Dudek AM, van Kampen JG, Witjes JA,
Kiemeney LA and Verhaegh GW: LINC00857 expression predicts and
mediates the response to platinum-based chemotherapy in
muscle-invasive bladder cancer. Cancer Med. 7:3342–3350. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fléjou JF: WHO classification of digestive
tumors: The fourth edition. Ann Pathol. 31 (Suppl 5):S27–S31.
2011.(In French). View Article : Google Scholar
|
|
24
|
Gaspersz MP, Buettner S, van Vugt JL, de
Jonge J, Polak WG, Doukas M, Ijzermans JNM, Koerkamp BG and
Willemssen FE: Evaluation of the new American joint committee on
cancer staging manual 8th edition for perihilar cholangiocarcinoma.
J Gastrointest Surg. 24:1612–1618. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carlevaro-Fita J and Johnson R: Global
positioning system: Understanding long noncoding RNAs through
subcellular localization. Mol Cell. 73:869–883. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Z, Chao TC, Chang KY, Lin N, Patil VS,
Shimizu C, Head SR, Burns JC and Rana TM: The long noncoding RNA
THRIL regulates TNFα expression through its interaction with
hnRNPL. Proc Natl Acad Sci USA. 111:1002–1007. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Anders G, Mackowiak SD, Jens M, Maaskola
J, Kuntzagk A, Rajewsky N, Landthaler M and Dieterich C: doRiNA: A
database of RNA interactions in post-transcriptional regulation.
Nucleic Acids Res. 40:D180–D186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mura M, Hopkins TG, Michael T, Abd-Latip
N, Weir J, Aboagye E, Mauri F, Jameson C, Sturge J, Gabra H, et al:
LARP1 post-transcriptionally regulates mTOR and contributes to
cancer progression. Oncogene. 34:5025–5036. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Galgano A, Forrer M, Jaskiewicz L, Kanitz
A, Zavolan M and Gerber AP: Comparative analysis of mRNA targets
for human PUF-family proteins suggests extensive interaction with
the miRNA regulatory system. PLoS One. 3:e31642008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu S, Li G, Li Q, Zhang Q, Zhuo L, Chen
X, Zhai B, Sui X, Chen K and Xie T: The roles and mechanisms of YTH
domain-containing proteins in cancer development and progression.
Am J Cancer Res. 10:1068–1084. 2020.PubMed/NCBI
|
|
32
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tanabe A, Tanikawa K, Tsunetomi M, Takai
K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, et al:
RNA helicase YTHDC2 promotes cancer metastasis via the enhancement
of the efficiency by which HIF-1α mRNA is translated. Cancer Lett.
376:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tanabe A, Konno J, Tanikawa K and Sahara
H: Transcriptional machinery of TNF-α-inducible YTH domain
containing 2 (YTHDC2) gene. Gene. 535:24–32. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fanale D, Iovanna JL, Calvo EL, Berthezene
P, Belleau P, Dagorn JC, Bronte G, Cicero G, Bazan V, Rolfo C, et
al: Germline copy number variation in the YTHDC2 gene: Does it have
a role in finding a novel potential molecular target involved in
pancreatic adenocarcinoma susceptibility? Expert Opin Ther Targets.
18:841–850. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu
Y, Gregory BD, Schultz RM and Wang PJ: Nuclear m6A reader YTHDC1
regulates alternative polyadenylation and splicing during mouse
oocyte development. PLoS Genet. 14:e10074122018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X,
Ren F, Cui G and Sun R: Expression patterns and prognostic value of
m6A-related genes in colorectal cancer. Am J Transl Res.
11:3972–3991. 2019.PubMed/NCBI
|
|
38
|
Popova VV, Kurshakova MM and Kopytova DV:
Methods to study the RNA-protein interactions. Mol Biol (Mosk).
49:472–481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sakata T, Ferdous G, Tsuruta T, Satoh T,
Baba S, Muto T, Ueno A, Kanai Y, Endou H and Okayasu I: L-type
amino-acid transporter 1 as a novel biomarker for high-grade
malignancy in prostate cancer. Pathol Int. 59:7–18. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kaira K, Oriuchi N, Imai H, Shimizu K,
Yanagitani N, Sunaga N, Hisada T, Tanaka S, Ishizuka T, Kanai Y, et
al: Prognostic significance of L-type amino acid transporter 1
expression in resectable stage I–III nonsmall cell lung cancer. Br
J Cancer. 98:742–748. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang L, Cao L, Wen C, Li J, Yu G and Liu
C: LncRNA LINC00857 regulates lung adenocarcinoma progression,
apoptosis and glycolysis by targeting miR-1179/SPAG5 axis. Hum
Cell. 33:195–204. 2020. View Article : Google Scholar : PubMed/NCBI
|