Introduction
Breast cancer is a malignant tumor derived from
breast tissue (1). Breast cancer is
one of the leading causes of cancer-related death in women and the
second most common type of cancer in the world (2). There were ~266,120 newly diagnosed
cases and 40,920 deaths from breast cancer in the USA in 2018
(2). Although great progress has
been made in the treatment of breast cancer in the past decade, the
5-year survival rate of some patients is only 24% (3). The increase in tumor incidence and
tumor-specific death is almost always caused by recurrence and
metastasis (4). Hence, finding
viable breast cancer prognostic markers and potential treatment
targets is necessary for the treatment of patients with breast
cancer.
MicroRNAs (miRNAs) are small, highly conserved
non-coding RNAs consisting of 21–25 nucleotides that serve an
important role in RNA silencing and posttranscriptional regulation
of gene expression (5). Previous
studies have confirmed that dysregulated miRNAs are involved in the
progression of various cancers, including breast cancer (6,7). In
addition, studies in recent years have found that abnormally
expressed miRNAs in tissues and serum can be used as biomarkers for
tumor prognosis and diagnosis, for example, miR-519a was
downregulated in gastric cancer and was associated with poor
prognosis (8). The downregulation of
serum miR-218-5p is a biomarker for prostatic bone metastasis
(9).
miR-768-3p is a highly expressed miRNA whose role
has been described in a hepatitis B virus-related hepatocellular
carcinoma and hurthle cell carcinoma (10,11).
miR-768-3p is abnormally expressed in non-small cell lung cancer
tissues and cell lines A549 and HCC4006, and is involved in tumor
viability, migration and invasion (12). Compared with the expression level in
gastric cancer tissues, the expression level of miR-768-3p in
non-tumor tissues is higher (13).
Zheng et al (14) performed a
study with 77 breast cancer and 17 control tissues from frozen
mammoplasty samples in 2016 and found 34 differentially expressed
miRNAs, including miR-768-3p in the downregulated miRNAs. However,
the clinical and biological role of miR-768-3p in breast cancer
remains unclear.
The aim of the present study was to investigate the
expression and clinical association of miR-768-3p in breast cancer
and to explore the regulatory effect of miR-768-3p on breast cancer
cell function. The finding of the present study indicated that
miR-768-3p may be a new prognostic breast cancer marker and a
potential target for the molecular treatment of breast cancer.
Materials and methods
Patients and tissue specimens
Surgically resected tumor tissues and adjacent
tissues which were obtained 5 cm away from breast cancer tissues of
116 patients with breast cancer were collected from the Affiliated
Hospital of Weifang Medical University (Weifang, China) from June
2011 to June 2014. All patients' tissue specimens were confirmed to
be breast cancer and the patients did not receive any chemotherapy,
adjuvant therapy, or immunotherapy before surgery. Exclusion
criteria for the patients were: i) Multiple cancers or cancers of
other organs; ii) previous axillary surgery; or iii) history of
benign breast diseases. Patients were followed for 5 years and the
days from surgery to the last follow-up or death were recorded at
every 3 months for the first 2 years after surgery, then once every
6 months (between 2–4 years) and thereafter once yearly (after 4
years) by telephone to analyze the effects of miRNA changes on
overall survival. The present study was approved by the Research
Ethics Committee of Affiliated Hospital of Weifang Medical
University (Weifang, China; approval no. WYFY20110609001). All
participants in the study signed written informed consent. Patient
sample tissues were processed and anonymized according to ethical
and legal standards. The research methodology followed the
standards of the Helsinki Declaration. The clinicopathological
features of the patients (age range, 32–89 years; median age, 60
years) are listed in Table I. TNM
stage analysis of the patients was done according to the 2010
American Joint Committee on Cancer recommendation for Tumor-lymph
Node Metastasis Classification (AJCC 7th edition) (15).
 | Table I.Association between miR-768-3p
expression levels and clinicopathological features in patients with
breast cancer (n=116). |
Table I.
Association between miR-768-3p
expression levels and clinicopathological features in patients with
breast cancer (n=116).
|
|
| miR-768-3p
expression |
|
|---|
|
|
|
|
|
|---|
| Parameters | No of cases | Low (n=65) | High (n=51) | P-value |
|---|
| Age, years |
|
|
| 0.350 |
|
≤50 | 61 | 37 | 24 |
|
|
>50 | 55 | 28 | 27 |
|
| Tumor size, cm |
|
|
| 0.460 |
| ≤2 | 59 | 31 | 28 |
|
|
>2 | 57 | 34 | 23 |
|
|
Differentiation |
|
|
| 0.095 |
|
Well-defined + Moderate | 60 | 29 | 31 |
|
|
Poor | 56 | 36 | 20 |
|
| Lymph node
metastasis |
|
|
| 0.040 |
|
Negative | 62 | 29 | 33 |
|
|
Positive | 54 | 36 | 18 |
|
| TNM stage |
|
|
| 0.035 |
|
I–II | 71 | 34 | 37 |
|
|
III–IV | 45 | 31 | 14 |
|
| Subtypes |
|
|
| 0.008 |
| Luminal
A | 52 | 21 | 31 |
|
| Luminal
B | 22 | 13 | 9 |
|
| HER-2
upregulation | 14 | 9 | 5 |
|
|
Triple-negative breast
cancer | 28 | 22 | 6 |
|
Cell lines and transfection
MCF-10A, a normal breast cell line and breast cancer
cell lines MCF-7 and MDA-MB-231 were purchased from the Chinese
Academy of Science Cell Bank (Shanghai, China). The breast cancer
cell lines T-47D and SK-BR-3 were obtained from American Type
Culture Collection (ATCC). MCF-10A and all breast cancer cells were
cultured in DMEM (Thermo Fisher Scientific Inc.) containing 10%
fetal bovine serum (FBS, Invitrogen; Thermo Fisher Scientific Inc.)
and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific
Inc.) in a humidified incubator with 5% CO2 at 37°C. The
miR-768-3p mimic (50 nM), mimic negative control (NC, 50 nM),
miR-768-3p inhibitor (50 nM)and inhibitor NC (50 nM) were purchased
from Guangzhou RiboBio Co., Ltd. for in vitro regulation of
breast cancer cells. Untransfected cells were the control group.
The sequences of used were as follows: miR-768-3p mimic,
5′-UCACAAUGCUGACACUCAAACUGCUGAC-3′; miR-768-3p inhibitor,
5′-GUCAGCAGUUUGAGUGUCAGCAUUGUGA-3′ mimic NC,
5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′ and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′. The transfection reagent was
Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific Inc.). According to the requirements of the transfection
reagent, transfection was conducted in an incubator at 37°C for 6 h
and subsequently the medium was replaced with fresh DMEM medium.
Follow-up experiments were performed 48 h after transfection.
Reverse transcription-quantitative
(RT-q) PCR
TRIzol® (Invitrogen; Thermo Fisher
Scientific Inc.) reagent was added to patient tissues and cells
lines (MCF-10A, SK-BR-3, MCF-7, T-47D and MDA-MB-231) according to
the manufacturer's instructions. miRNA was purified from tissues
and cells using a miRNA Purification kit (CoWin Biosciences). The
extracted total RNA was then reverse transcribed into cDNA using
the miRNA cDNA Synthesis kit (CoWin Biosciences) according to the
manufacturer's protocol. qPCR was performed on a 7300 real-time PCR
system using a SYBR Green miRNA qPCR Assay kit (CoWin Biosciences).
The thermocycling condition used were as follows: 95°C for 10 min
followed by 40 cycles of 94°C for 15 sec, 55°C for 30 sec and 70°C
for 30 sec. The relative expression of miR-768-3p was calculated by
the 2−ΔΔCq method (16),
and U6 was used as the reference gene for mRNA quantification. The
following primer sequences were used: miR-768-3p forward
5′-GCCGAGUCACAAUGCUGACACUCA-3′ and reverse 5′-CTCGTTCGGCAGCACA-3′;
and U6 forward 5′-AACGCTTCACGAATTTGCGT-3′ and reverse
5′-CTCGTTCGGCAGCACA-3′. Each sample was tested in triplicate.
Cell viability assay
Breast cancer cells MCF-7 and MDA-MB-231 transfected
with the miR-768-3p mimic and miR-768-3p inhibitor and
corresponding NCs were seeded in 96-well plates at a density of
1×104 cells/well. The cells were dissociated into a
single cells suspensions with 0.05% trypsin (Invitrogen; Thermo
Fisher Scientific Inc.) at 24 h intervals, and then washed once
with PBS. Finally, the cells were counted manually using a
hemocytometer and observed under a light microscope (magnification,
×100). Experiments were repeated in triplicate.
Cell migration and invasion assay
Transwell assays were used to detect changes in cell
migration and invasion. Transfected MCF-7 and MDA-MB-231 cells were
prepared as a single cell suspension in serum-free medium and
8×104 cells were added to the upper chamber of the
Matrigel-coated Transwell chamber (37°C for 3 h to form the
Matrigel layer in the chamber) or the upper chamber of the
Matrigel-free chamber for determination of the invasion or
migration ability, respectively. DMEM medium (500 µl) containing
10% FBS was added to the lower chamber of the Transwell chamber.
After 24 h of culture in the incubator at 37°C, the non-migrated
and non-invaded cells in the upper chamber were wiped clean with
cotton swabs, fixed with 4% paraformaldehyde for 10 min at room
temperature and stained with 0.1% crystal violet for 10 min at room
temperature. Then the chamber was flushed with water until the
water was no longer purple. Five fields were randomly selected
under an light microscope (magnification, ×200) for counting.
Changes in cell migration and invasion were manually measured.
Luciferase reporter assay
Bioinformatics TargetScan v.7.2 was used to
(Whitehead Institute for Biomedical Research, Massachusetts
Institute of Technology) predict the target genes of miR-768-3p.
Then the dual-luciferase reporter assay (Promega Corporation) was
used to verify the results. The sequence Wild-type (WT) and mutant
(Mut) 3′UTR of eukaryotic translation initiation factor 4E (eIF4E)
was synthesized by Sangon Biotech, Co., Ltd, and then inserted into
the luciferase reporter pmiRGLO vector (Promega Corporation). MCF-7
cells were seeded in 24-well plates containing DMEM at a density of
5×104 and cultured overnight in an incubator at 37°C. WT
or Mut PmiRglo-3′-UTR-eIF4E, renin luciferase plasmid and
miR-768-3p mimic or inhibitor (Guangzhou RiboBio Co., Ltd.) were
transfected into cells. The transfection reagent was Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific Inc.). The
sequences used were as follows: miR-768-3p mimic,
5′-UCACAAUGCUGACACUCAAACUGCUGAC-3′; miR-768-3p inhibitor,
5′-GUCAGCAGUUUGAGUGUCAGCAUUGUGA-3′ mimic NC,
5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′ and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′. After 48 h, the MCF-7 cells were
collected and 200 µl reporter lysis buffer (Promega Corporation)
was added. Then, the luciferase activity was determined by the
Luciferase Assay System (Promega Corporation). Renilla
luciferase activity was used for normalization.
Statistical analysis
Statistical analysis of the present study was
performed using GraphPad Prism 7.0 software (GraphPad Software
Inc.) and SPSS 22.0 software (IBM Corp.) and all data are presented
as the mean ± SD. All experiments were repeated 3 times. The
difference between the breast cancer tissues and normal adjacent
tissues was detected by a paired Student's t-test. Comparisons
between multiple groups were performed using one-way ANOVA followed
by the post hoc Tukey's test. A χ2 test was used to
evaluate the association between miR-768-3p expression and the
clinicopathological features of patients with breast cancer.
Kaplan-Meier and log-rank tests were used for survival analyses.
Cox regression analysis was used to evaluate the effect of
miR-768-3p on the prognosis of patients with breast cancer.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of miR-768-3p in breast
cancer tissues and cell lines
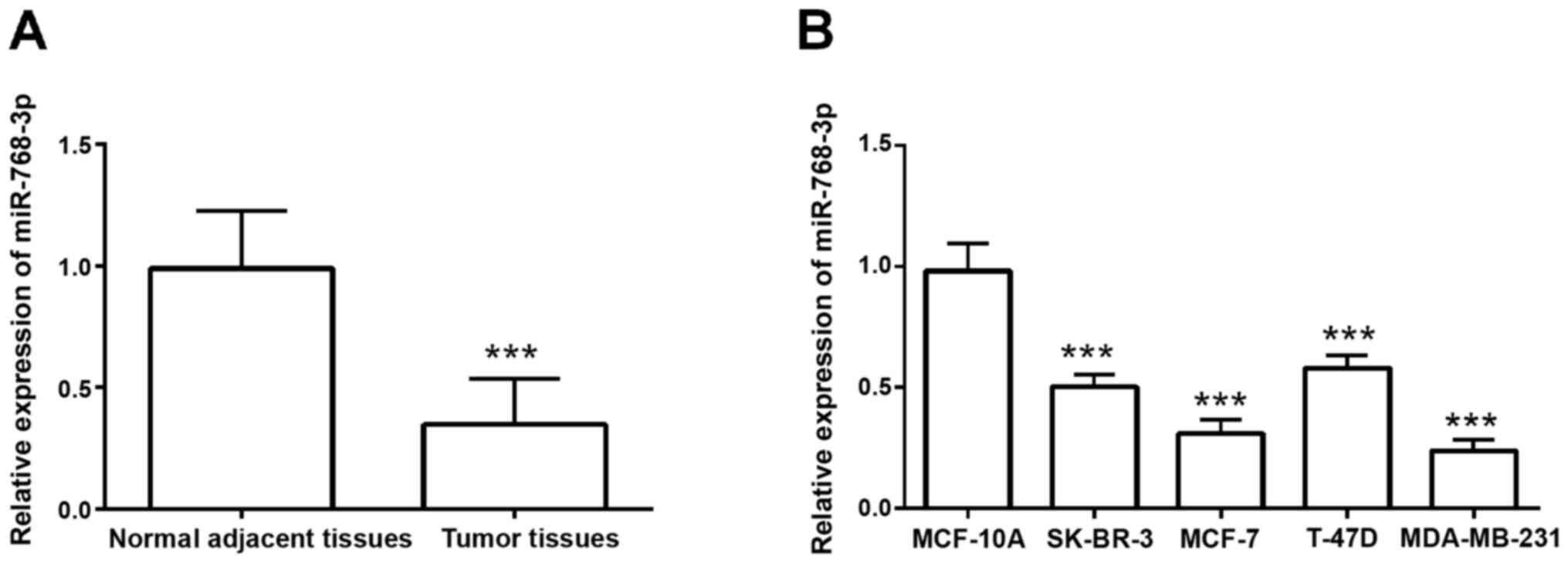
To detect the expression pattern of miR-768-3p in
breast cancer tissues, RT-qPCR was performed with 116 normal
adjacent and tumor tissues. Compared with normal adjacent tissues,
cancer tissues had significantly decreased miR-768-3p expression
(P<0.001; Fig. 1A). In addition,
compared with normal MCF-10A breast cells, breast cancer cells had
a significantly decreased expression level of miR-768-3p
(P<0.001; Fig. 1B), which was
consistent with the expression results in tissues. As the MCF-7 and
MDA-MB-231 had the lowest expression of miR-768-3p of the breast
cancer cells tested, they were used for subsequent experimentation.
The aforementioned results suggested that miR-768-3p is a tumor
suppressor miRNA in breast cancer.
miR-768-3p expression is associated
with the clinicopathological characteristics of patients with
breast cancer
Association between the expression level of
miR-768-3p and the clinicopathological characteristics of patients
with breast cancer was further explored. According to the average
expression level of miR-768-3p (0.346 ±0.189) in the tissue samples
of the patients as the cutoff value, the breast cancer patients
were divided into the high miR-768-3p expression group (n=51) and
the low miR-768-3p expression group (n=65). A χ2 test
was used to analyze the relationship between miR-768-3p and the
clinicopathological features of the patients and the results are
shown in Table I. Low expression of
miR-768-3p was significantly associated with lymph node metastasis,
tumor node metastasis (TNM) stage and cancer subtype (P<0.05;
Table I), but not with age, tumor
size or differentiation (P>0.05; Table I). The aforementioned findings
indicated that the reduction of miR-768-3p expression may serve a
crucial role in the breast cancer.
Low expression of miR-768-3p is
associated with poor prognosis in breast cancer
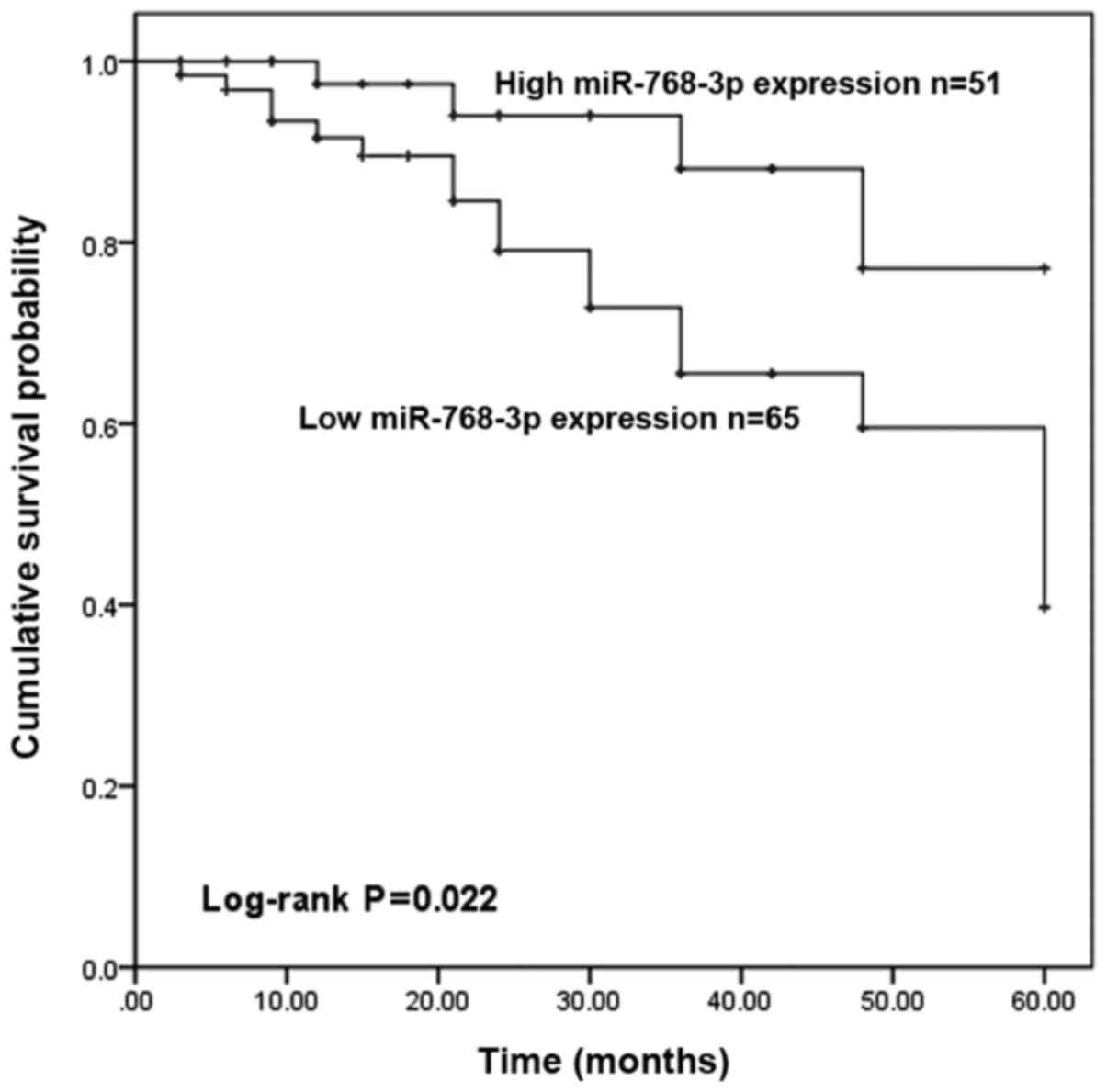
According to the 5-year follow-up information of
patients with breast cancer, the Kaplan-Meier method and log-rank
test were used to examine the relationship between miR-768-3p
expression and the survival time of patients with breast cancer and
to explore the prognostic value of miR-768-3p in patients with
breast cancer. The survival curve is shown in Fig. 2. Patients with low expression of
miR-768-3p had a shorter overall survival time (log-rank P=0.022;
Fig. 2) compared with patients with
high expression of miR-768-3p. In addition, multivariate Cox
regression was used to analyze the effect of miR-768-3p on the
prognosis of patients with breast cancer and the results are shown
in Table II. miR-768-3p [hazard
ratio (HR)=4.637; 95% confidence interval (CI)=1.296–16.597;
P=0.018], lymph node metastasis (HR=0.111; 95% CI=0.013–0.935;
P=0.043), TNM stage (HR=2.756; 95% CI=1.063–7.144; P=0.037),
subtypes (HR=2.789; 95% CI=1.055–7.376; P=0.039; Table II] can all be used as independent
prognostic factor for patients with breast cancer. In summary, the
results confirmed that low expression of miR-768-3p is associated
with poor prognosis of breast cancer.
 | Table II.Multivariate Cox analysis of
miR-768-3p and clinical parameters in relation to overall
survival. |
Table II.
Multivariate Cox analysis of
miR-768-3p and clinical parameters in relation to overall
survival.
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Features | HR | 95% CI | P-value |
|---|
| miR-768-3p
expression | 4.637 | 1.296–16.597 | 0.018 |
| Age | 0.398 | 0.133–1.196 | 0.101 |
| Tumor size | 0.531 | 0.188–1.500 | 0.232 |
|
Differentiation | 0.424 | 0.125–1.443 | 0.170 |
| Lymph node
metastasis | 0.111 | 0.013–0.935 | 0.043 |
| TNM stage | 2.756 | 1.063–7.144 | 0.037 |
| Subtypes | 2.789 | 1.055–7.376 | 0.039 |
miR-768-3p inhibits cell viability,
migration, and invasion in vitro
To explore the role of miR-768-3p in breast cancer,
this study also examined the effects of miR-768-3p on breast cancer
cell viability, migration, and invasion. miR-768-3p mimic and
miR-768-3p inhibitor and their corresponding NCs were transfected
into MCF-7 and MDA-MB-231 breast cancer cells. The changes in
miR-768-3p expression in the cells were detected by RT-qPCR. The
expression level of the miR-768-3p mimic group was significantly
higher compared with that in the control and mimic NC groups, while
that of the miR-768-3p inhibitor group was significantly lower
compared with the control and inhibitor NC groups (P<0.001;
Fig. 3A). The experimental results
confirmed the high transfection efficiency of the miR-768-3p mimic
and miR-768-3p inhibitor in breast cancer cells.
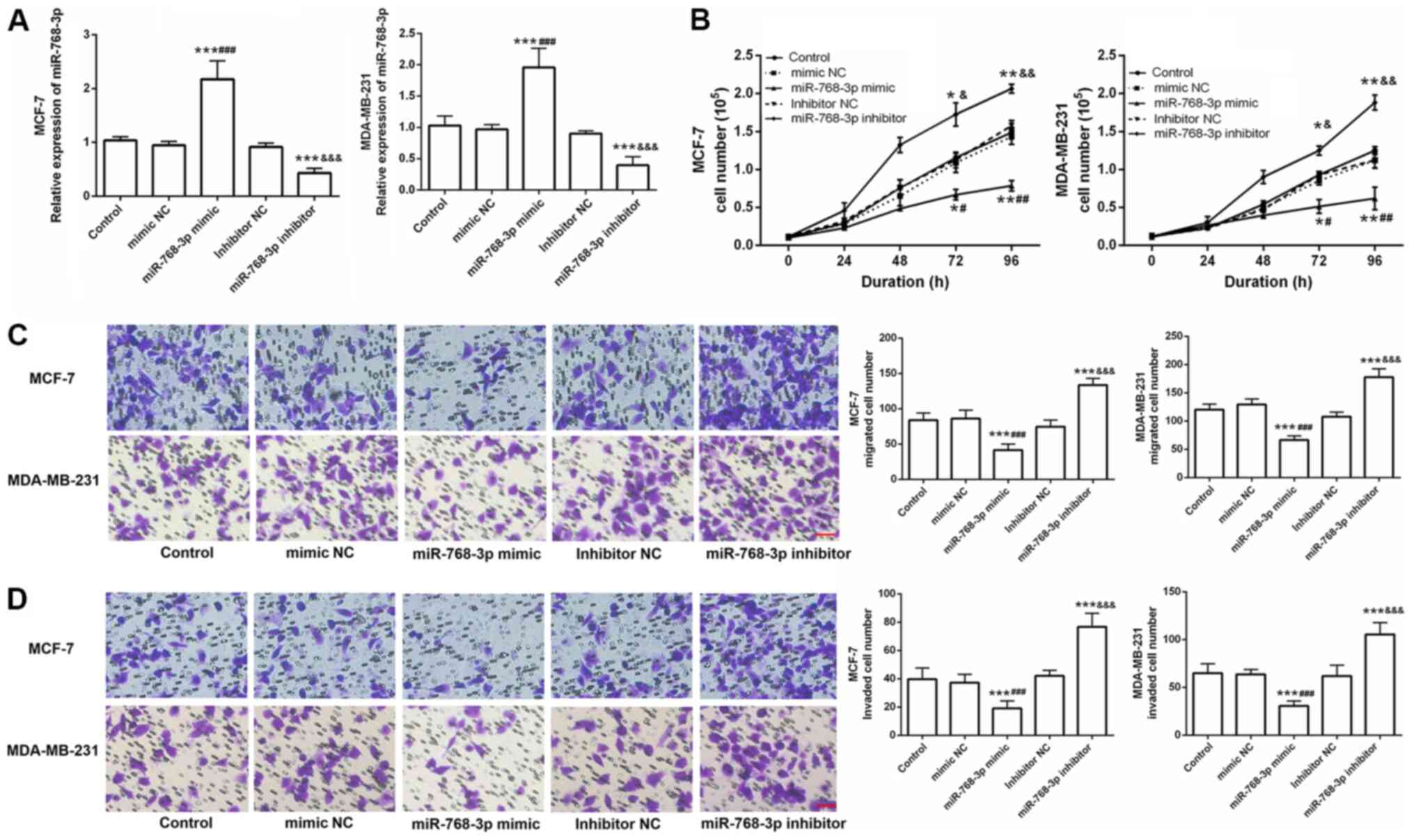 | Figure 3.Effects of miR-768-3p on the function
of breast cancer cells in vitro. (A) Expression level of
miR-768-3p in cells after transfection with the miR-768-3p mimic or
inhibitor and corresponding controls using RT-qPCR. The results
confirmed that the miR-768-3p mimic and inhibitor had a higher
transfection efficiency in cancer cells compared with the control,
mimic NC and inhibitor NC groups, respectively (***P<0.001,
compared with control; ###P<0.001, compared with
mimic NC; &&&P<0.001 compared with
inhibitor NC). (B) Viability ability was examined. Compared with
the control group, the miR-768-3p mimic group significantly
inhibited cell viability, while the miR-768-3p inhibitor group
significantly promoted cell viability (*P<0.05, **P<0.01,
compared with control; #P<0.05,
##P<0.001, compared with mimic NC;
&P<0.05, &&P<0.001,
compared with inhibitor NC). (C) Representative images and
quantitative analysis of cell migration by transwell assays. (D)
Representative images and quantitative analysis of cell invasion by
transwell assays. Compared with the control group, the miR-768-3p
mimic group significantly promoted cell migration and invasion,
while the miR-768-3p inhibitor group significantly promoted cell
migration and invasion. The randomly chosen fields were
photographed (magnification, ×200) (***P<0.001, compared with
control; ###P<0.001, compared with mimic NC;
&&&P<0.001, compared with inhibitor NC)
(scale bar=200 µm). miR, microRNA; RT-q, reverse
transcription-quantitative; NC, negative control; control,
untransfected cells. |
A hemocytometer was used to detect the effect of
miR-768-3p on cell viability. Compared with mimic NC or inhibitor
NC, overexpression of miR-768-3p significantly inhibited cell
viability in both MCF-7 and MDA-MB-231 cells, while knockdown of
miR-768-3p significantly promoted cell viability (P<0.05;
Fig. 3A). In addition, a Transwell
assay was used to analyze the effects of miR-768-3p expression on
cell migration and invasion abilities of breast cancer cells.
Compared with those of the control and mimic NC groups, the cell
migration and invasion abilities of the miR-768-3p mimic group were
inhibited, while cell migration and invasion abilities of the
miR-768-3p inhibitor group were enhanced in both cell lines
compared with the control and inhibitor NC groups (P<0.05;
Fig. 3C and D).
miR-768-3p targeted eIF4E in breast
cancer cells
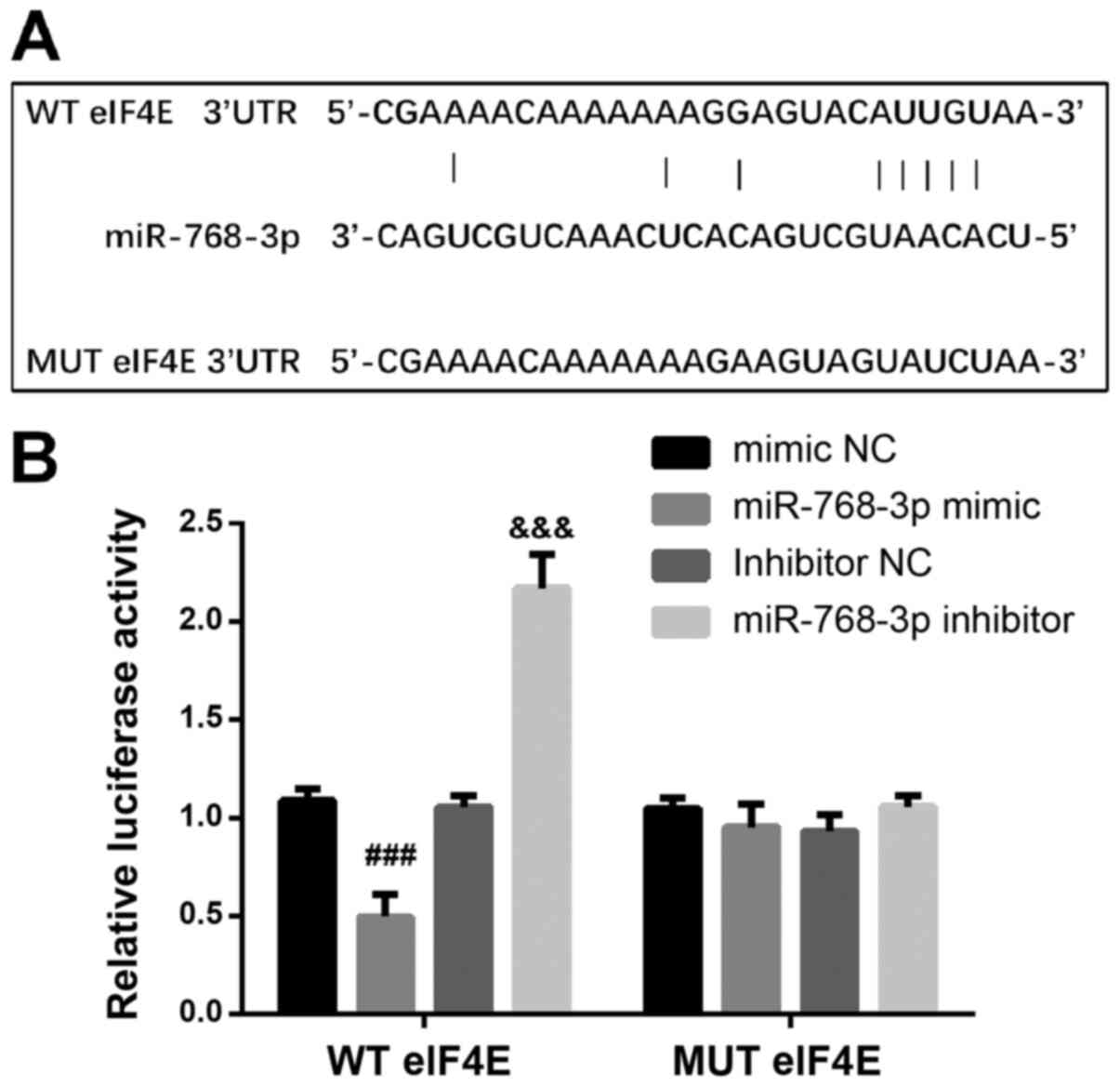
Finally, in order to understand the molecular
mechanism of miR-768-3p in breast cancer cells, bioinformatics
software was used to search for the target genes of miR-768-3p and
it was found that eukaryotic translation initiation factor 4E
(eIF4E) was the target gene of miR-768-3p (Fig. 4A). Luciferase reporter assay
demonstrated that luciferase activity was significantly inhibited
or increased when co-transfected with wild-type eIF4E and
miR-768-3p mimic or inhibitor compared with co-transfection of
those with mutant eIF4E (Fig. 4B).
The results indicated that miR-768-3p may affect the biological
function of breast cancer cells by targeting eIF4E.
Discussion
Cancer is one of the most common life-threatening
diseases in the world and breast cancer commonly occurs in women
(17). Although significant advances
have been made in the diagnosis and treatment of breast cancer in
recent years, there were 2.1 million new breast cancer cases
worldwide in 2018, so the prevention and treatment of breast cancer
remains a concern and challenge for oncologists worldwide (18,19). For
the early detection of breast cancer, surgical treatment,
chemotherapy and adjuvant therapy can significantly improve
survival, but for patients with metastasis and recurrence, the
identification of potentially valuable prognostic markers is
critical for the treatment of breast cancer (20).
Numerous previous studies have confirmed that miRNAs
are involved in a series of complex processes of tumor regulation,
including tumor viability, migration, invasion and apoptosis
(21–23). For example, miR-937 was upregulated
in breast cancer and regulated the viability and apoptosis of
breast cancer cells MCF-7 by targeting apoptotic protease
activating factor 1 (24). Recent
studies have found that abnormally expressed miRNAs in tumors can
be widely used as a biomarker for tumor diagnosis and prognosis
(25–27). For example, elevated miR-19b
expression can be used as a prognostic marker for breast cancer and
was involved in tumor progression through the PI3K/AKT pathway
(28). The upregulation of miR-206
and the downregulation of miR-145 were related to the poor
prognosis of patients with breast cancer and were important
indicators to predict the prognosis of patients with breast cancer
(29).
miR-768-3p has been shown to be abnormally expressed
in a variety of cancers. For example. Zheng et al (14) found 34 differentially expressed
miRNAs in 77 breast cancer and 17 control samples from frozen
mammoplasty samples in a 2016 study, and miR-768-3p was among the
downregulated miRNAs. The expression level of miR-768-3p in thyroid
cancer was downregulated and the difference in miR-768-3p
expression between cancerous and benign thyroid tissue was >5
fold (30). Similarly, miR-768-3p
was downregulated in melanoma and promoted the survival and
viability of melanoma cells by increasing the expression of target
gene elF4E (31).
In the present study, the expression level of
miR-768-3p in patients with breast cancer was assessed. The results
demonstrated that the expression of miR-768-3p was decreased in
cancer tissues compared with normal adjacent tissues of patients
with breast cancer, which was consistent with the results of a 2016
study by Zheng et al (14).
In addition, in the present study, miR-768-3p expression was also
decreased in 4 breast cancer cell lines compared with normal
MCF-10A cells. The findings of the present study indicated that
miR-768-3p acts as a tumor suppressor miRNA in breast cancer. To
test this hypothesis, the present study explored the relationship
between miR-768-3p expression and the clinicopathological
characteristics of patients with breast cancer and the results
demonstrated that low expression of miR-768-3p was significantly
associated with lymph node metastasis, TNM stage and breast cancer
subtype. Hence, miR-768-3p may be involved in the development of
this malignant tumor as a tumor suppressor miRNA in patients with
breast cancer. In addition, survival analysis in the present study
demonstrated that the overall survival time of patients with low
expression of miR-768-3p was shorter compared with that of patients
with high expression, suggesting that low expression of miR-768-3p
was associated with poor overall survival in patients with breast
cancer. Finally, the multivariate Cox regression model performed in
the present study confirmed that miR-768-3p was an independent
prognostic factor in patients with breast cancer. To the best of
our knowledge, the present study is the first study to examine the
clinical significance of miR-768-3p in patients with breast cancer
and to confirm that miR-768-3p can serve as a biomarker for breast
cancer prognosis.
In addition to the clinical application of
miR-768-3p in breast cancer, the effect of miR-768-3p on breast
cancer cell function in vitro was also assessed in the
present study. A study has demonstrated that inhibiting miR-768-3p
significantly reduced the viability, migration and invasion ability
in non-small cell lung cancer (12).
In the present study, it was first confirmed that the miR-768-3p
mimic and inhibitor were successfully transfected into breast
cancer cells. Viability and transwell assays performed in the
present study demonstrated that compared with their respective
control groups, overexpression of miR-768-3p could significantly
inhibit cell viability, migration and invasion, while
downregulation of miR-768-3p could significantly promote cell
viability, migration and invasion. The experimental results of the
present study confirmed that miR-768-3p, as a tumor suppressor
miRNA, was involved in the occurrence and development of breast
cancer. A previous study had confirmed that miR-768-3p serves an
important role in inhibiting eIF4E expression and mRNA response and
also regulates melanoma viability and survival in vitro
(31). eIF4E recognizes and bind to
the 5′cap structure of mRNA and delivers it to the eIF4E complex to
initiate translation (32). The
interaction between glycerol phosphate dehydrogenase and eIF4E
promotes the development of pancreatic cancer (33). Astrocyte-elevated gene-1 induced
gastric cancer metastasis by upregulation of eIF4E expression
(34). In addition, eIF4E can be
used as a therapeutic target for ovarian cancer, prostate cancer,
lung cancer and other cancers, and the eIF4E-directed therapies
LY2275796 (anti-sense oligonucleotides directed against eIF4E) and
ribavirin (which reduces eIF4E-dependent translation) are already
in clinical trials (35–39).
Hence, it was hypothesized that the regulatory
effect of miR-768-3p in breast cancer may be realized through
eIF4E. In the present study, it was confirmed by the luciferase
reporter assay that eIF4E was indeed the target gene of miR-768-3p.
The findings of the present study suggested that miR-768-3p may
affect the biological functions of breast cancer cells, such as
viability, migration and invasion by targeting eIF4E. It should be
noted that the present study had certain limitations. Firstly,
there was no significant association found between tumor
differentiation and prognosis in the present study, which may be
due to the small sample size. Therefore, a future study with a
large sample is needed for verification of the findings of the
present study. Secondly, the role of miR-768-3p in the regulation
of breast cancer was not verified in in vivo animal tumor
model experiments. Future studies should investigate the specific
mechanism of miR-768-3p in regulating breast cancer using in
vivo experiments.
In summary, the present study confirmed through a
series of experiments that miR-768-3p is a tumor suppressor in
breast cancer. In the present study, miR-768-3p was downregulated
in breast cancer and promoted cell viability, migration, and
invasion though eIF4E. Finally, in the present study the low
expression of miR-768-3p was significantly related to the poor
prognosis of patients with breast cancer and may be a potential
prognostic marker for breast cancer.
Acknowledgements
The authors would like to thank Dr Yunxia Liu
(Department of Internal Medicine, Fuyanshan Branch of Affiliated
Hospital of Weifang Medical University) for her contribution to the
data analysis of this study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ, YW, LW, JZ and XL contributed to the study
conception and design. YZ, LW and JZ were involved in material
preparation, performed the experiments and data collection and
analysis. XL supplied critical reagents. YW revised the manuscript
for important intellectual content. All the authors wrote the
manuscript. YZ and LW confirm the authenticity of all the raw data.
All the authors have read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Research Ethics
Committee of Affiliated Hospital of Weifang Medical University
(Weifang, China; approval no. WYFY20110609001). All participants in
the study signed written informed consent. The research methodology
meets the standards set out in the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Guo R, Chen Y, Borgard H, Jijiwa M, Nasu
M, He M and Deng Y: The function and mechanism of lipid molecules
and their roles in the diagnosis and prognosis of breast cancer.
Molecules. 25:48642020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gu Y, Wu G, Zou X, Huang P and Yi L:
Prognostic value of site-specific metastases and surgery in de novo
stage IV triple-negative breast cancer: A population-based
analysis. Med Sci Monit. 26:e9204322020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang M, Ji S, Shao G, Zhang J, Zhao K,
Wang Z and Wu A: Effect of exosome biomarkers for diagnosis and
prognosis of breast cancer patients. Clin Transl Oncol. 20:906–911.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li RH, Chen M, Liu J, Shao CC, Guo CP, Wei
XL, Li YC, Huang WH and Zhang GJ: Long noncoding RNA ATB promotes
the epithelial-mesenchymal transition by upregulating the
miR-200c/Twist1 axe and predicts poor prognosis in breast cancer.
Cell Death Dis. 9:11712018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su M, Niu Y, Dang Q, Qu J, Zhu D, Tang Z
and Gou D: Circulating microRNA profiles based on direct
S-Poly(T)Plus assay for detection of coronary heart disease. J Cell
Mol Med. 24:5984–5997. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiao Y, Humphries B, Yang C and Wang Z:
miR-205 dysregulations in breast cancer: The Complexity and
Opportunities. Noncoding RNA. 5:532019.PubMed/NCBI
|
|
7
|
Chong ZX, Yeap SK and Ho WY: Dysregulation
of miR-638 in the progression of cancers. Pathol Res Pract. Jan
29–2021.(Online ahead of print). View Article : Google Scholar
|
|
8
|
Cai H, Lin H, Cao W, Sun J, Huang Y and
Fang Y: Downregulation of miR-519a predicts poor prognosis and
contributes to tumor progression in gastric cancer. Oncol Res
Treat. 43:19–26. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peng P, Chen T, Wang Q, Zhang Y, Zheng F,
Huang S, Tang Y, Yang C, Ding W, Ren D, et al: Decreased miR-218-5p
levels as a serum biomarker in bone metastasis of prostate cancer.
Oncol Res Treat. 42:165–185. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z,
Wang JF, Zhang Z, Lu S, Huang X, et al: Plasma microRNA panel to
diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin
Oncol. 29:4781–4788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Petric R, Gazic B, Goricar K, Dolzan V,
Dzodic R and Besic N: Expression of miRNA and occurrence of distant
metastases in patients with hurthle cell carcinoma. Int J
Endocrinol. 2016:89452472016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie Z, Chen W, Chen Y, Wang X, Gao W and
Liu Y: miR-768-3p is involved in the viability, invasion and
migration of non-small cell lung carcinomas. Int J Oncol.
51:1574–1582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo J, Miao Y, Xiao B, Huan R, Jiang Z,
Meng D and Wang Y: Differential expression of microRNA species in
human gastric cancer versus non-tumorous tissues. J Gastroenterol
Hepatol. 24:652–657. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng T, Zhang X, Wang Y and Yu X:
Predicting associations between microRNAs and target genes in
breast cancer by bioinformatics analyses. Oncol Lett. 12:1067–1073.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang P, Fan C, Du J, Mo X and Zhao Q:
Association of miR-1247-5p expression with clinicopathological
parameters and prognosis in breast cancer. Int J Exp Pathol.
99:199–205. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fahad Ullah M: Breast cancer: Current
perspectives on the disease status. Adv Exp Med Biol. 1152:51–64.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berry DA, Cronin KA, Plevritis SK, Fryback
DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD and
Feuer EJ; Cancer Intervention and Surveillance Modeling Network
(CISNET) Collaborators, : Effect of screening and adjuvant therapy
on mortality from breast cancer. N Engl J Med. 353:1784–1792. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou W, Gong J, Chen Y, Chen J, Zhuang Q,
Cao J, Mei Z and Hu B: Long noncoding RNA LINC00899 suppresses
breast cancer progression by inhibiting miR-425. Aging (Albany NY).
11:10144–10153. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rahman MM, Brane AC and Tollefsbol TO:
MicroRNAs and epigenetics strategies to reverse breast cancer.
Cells. 8:12142019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weng YS, Tseng HY, Chen YA, Shen PC, Al
Haq AT, Chen LM, Tung YC and Hsu HL: MCT-1/miR-34a/IL-6/IL-6R
signaling axis promotes EMT progression, cancer stemness and M2
macrophage polarization in triple-negative breast cancer. Mol
Cancer. 18:422019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang H, Jiang W, Jing Z, Mu X and Xiong Z:
miR-937 regulates the viability and apoptosis via targeting APAF1
in breast cancer. Onco Targets Ther. 12:5687–5699. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li F: Expression of miR-221 and miR-489 in
breast cancer patients and their relationship with prognosis. Oncol
Lett. 19:1523–1529. 2020.PubMed/NCBI
|
|
26
|
Sun R, Muheremu A and Hu Y: miRNA-30c can
be used as a target in the diagnosis and treatment of osteosarcoma.
Onco Targets Ther. 11:9091–9099. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang F, Dai M, Chen H, Li Y, Zhang J, Zou
Z and Yang H: Prognostic value of hsa-mir-299 and hsa-mir-7706 in
hepatocellular carcinoma. Oncol Lett. 16:815–820. 2018.PubMed/NCBI
|
|
28
|
Li C, Zhang J, Ma Z, Zhang F and Yu W:
miR-19b serves as a prognostic biomarker of breast cancer and
promotes tumor progression through PI3K/AKT signaling pathway. Onco
Targets Ther. 11:4087–4095. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Quan Y, Huang X and Quan X: Expression of
miRNA-206 and miRNA-145 in breast cancer and correlation with
prognosis. Oncol Lett. 16:6638–6642. 2018.PubMed/NCBI
|
|
30
|
Vriens MR, Weng J, Suh I, Huynh N,
Guerrero MA, Shen WT, Duh QY, Clark OH and Kebebew E: MicroRNA
expression profiling is a potential diagnostic tool for thyroid
cancer. Cancer. 118:3426–3432. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang CC, Croft A, Tseng HY, Guo ST, Jin
L, Hersey P and Zhang XD: Repression of microRNA-768-3p by MEK/ERK
signalling contributes to enhanced mRNA translation in human
melanoma. Oncogene. 33:2577–2588. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pisera A, Campo A and Campo S: Structure
and functions of the translation initiation factor eIF4E and its
role in cancer development and treatment. J Genet Genomics.
45:13–24. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma X, Li B, Liu J, Fu Y and Luo Y:
Phosphoglycerate dehydrogenase promotes pancreatic cancer
development by interacting with eIF4A1 and eIF4E. J Exp Clin Cancer
Res. 38:662019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu S, Yang L, Wu D, Gao Z, Li P, Huang W
and Wang X: AEG-1 induces gastric cancer metastasis by upregulation
of eIF4E expression. J Cell Mol Med. 21:3481–3493. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jin J, Xiang W, Wu S, Wang M, Xiao M and
Deng A: Targeting eIF4E signaling with ribavirin as a sensitizing
strategy for ovarian cancer. Biochem Biophys Res Commun.
510:580–586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao Y, Yan M, Yun Y, Zhang J, Zhang R, Li
Y, Wu X, Liu Q, Miao W and Jiang H: MicroRNA-455-3p functions as a
tumor suppressor by targeting eIF4E in prostate cancer. Oncol Rep.
37:2449–2458. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu F, Wang X, Li J, Gu K, Lv L, Zhang S,
Che D, Cao J, Jin S and Yu Y: miR-34c-3p functions as a tumour
suppressor by inhibiting eIF4E expression in non-small cell lung
cancer. Cell Prolif. 48:582–592. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Karaki S, Andrieu C, Ziouziou H and Rocchi
P: The eukaryotic translation initiation Factor 4E (eIF4E) as a
therapeutic target for cancer. Adv Protein Chem Struct Biol.
101:1–26. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sobocan M, Smolle MA, Schatz C and
Haybaeck J: The interplay of tumor stroma and translational factors
in endometrial cancer. Cancers (Basel). 12:20742020. View Article : Google Scholar : PubMed/NCBI
|