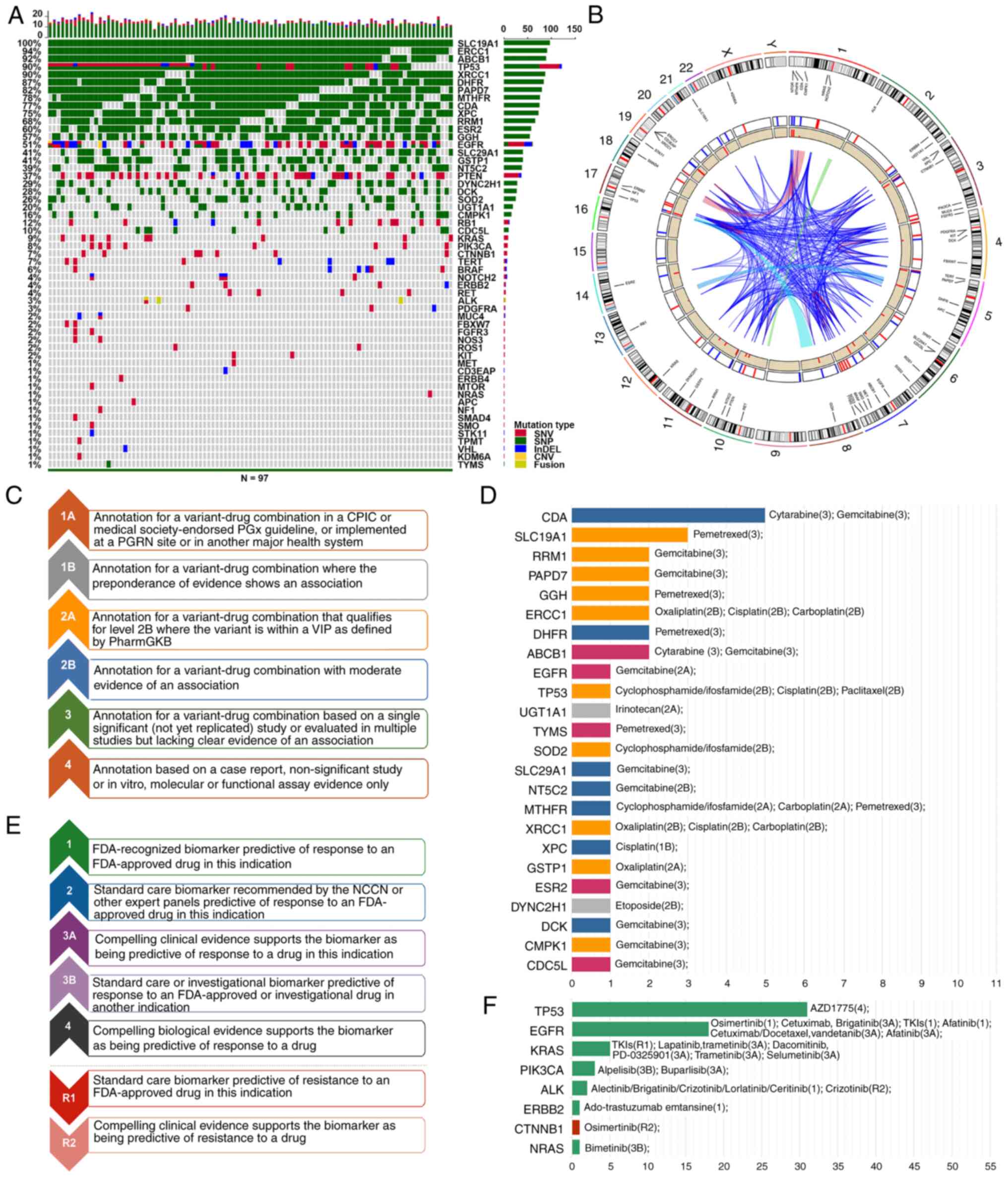
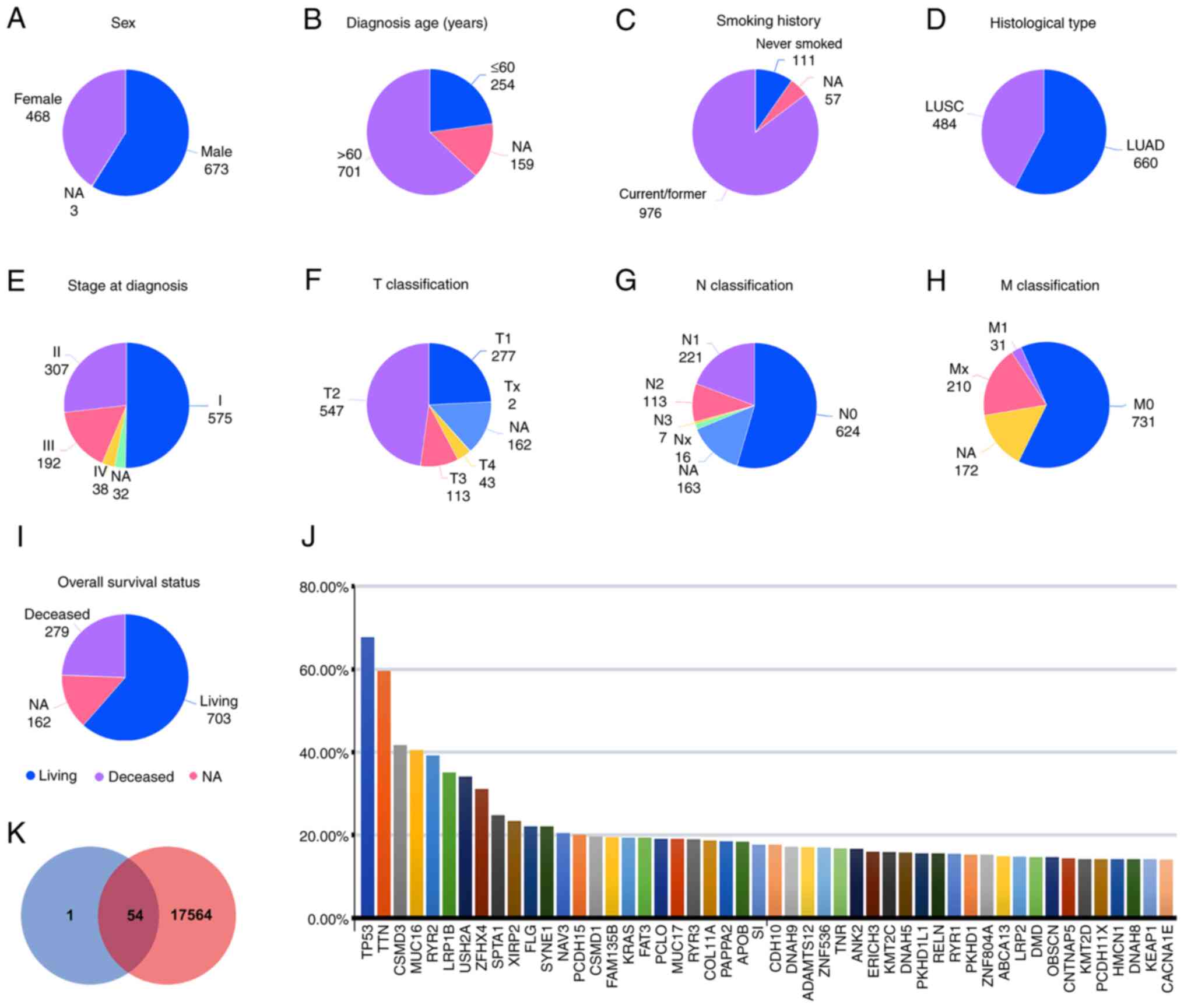
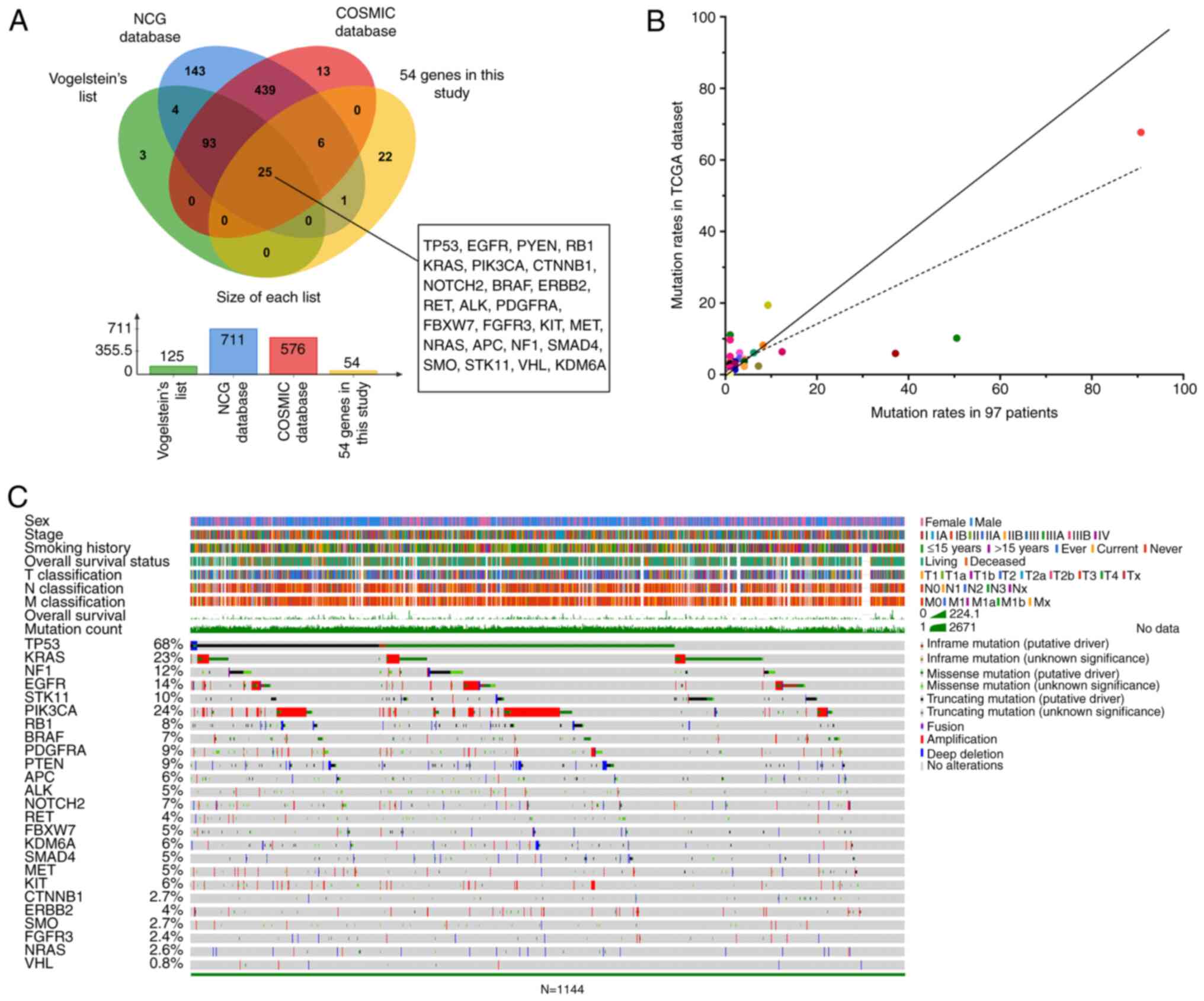
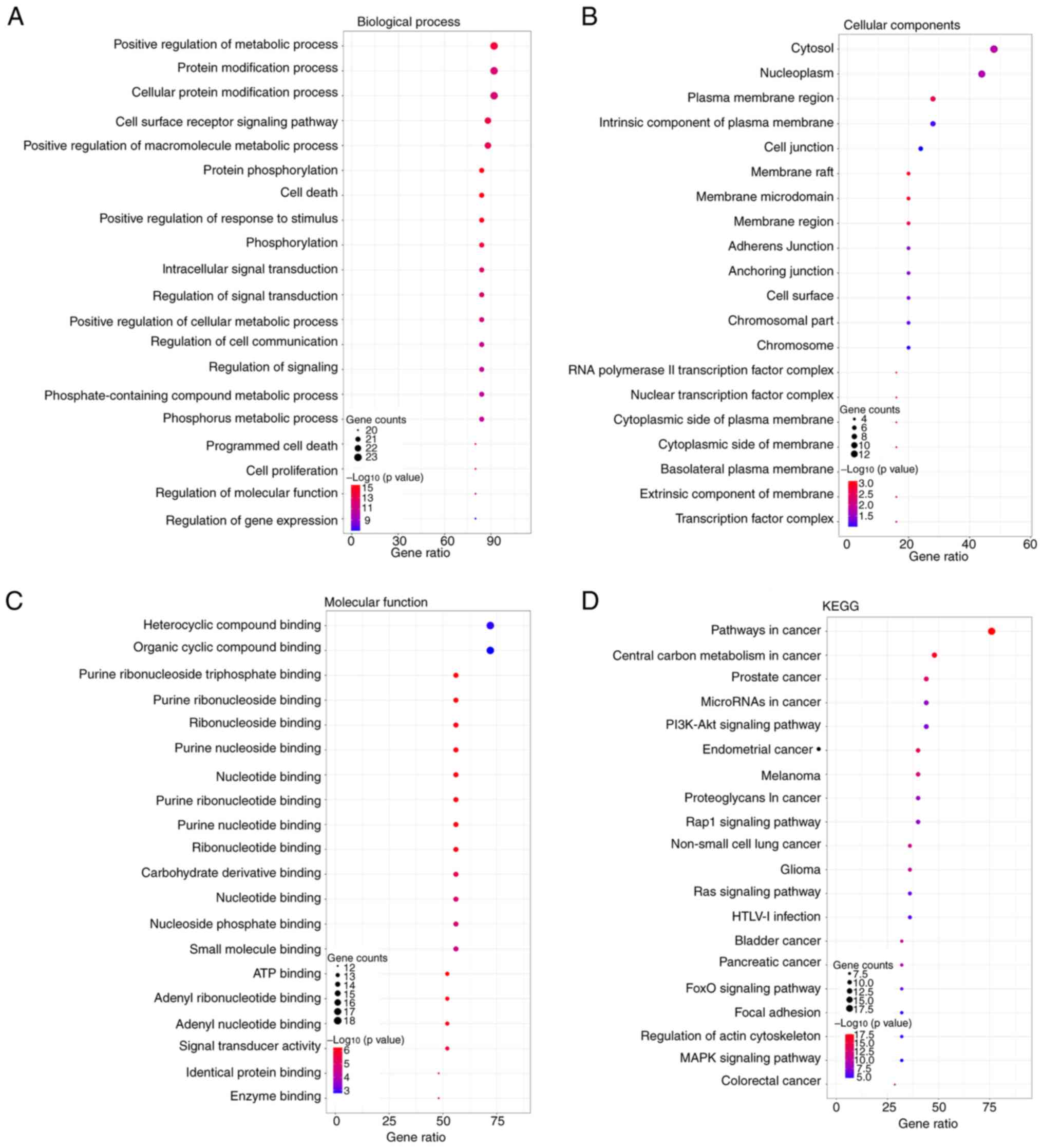
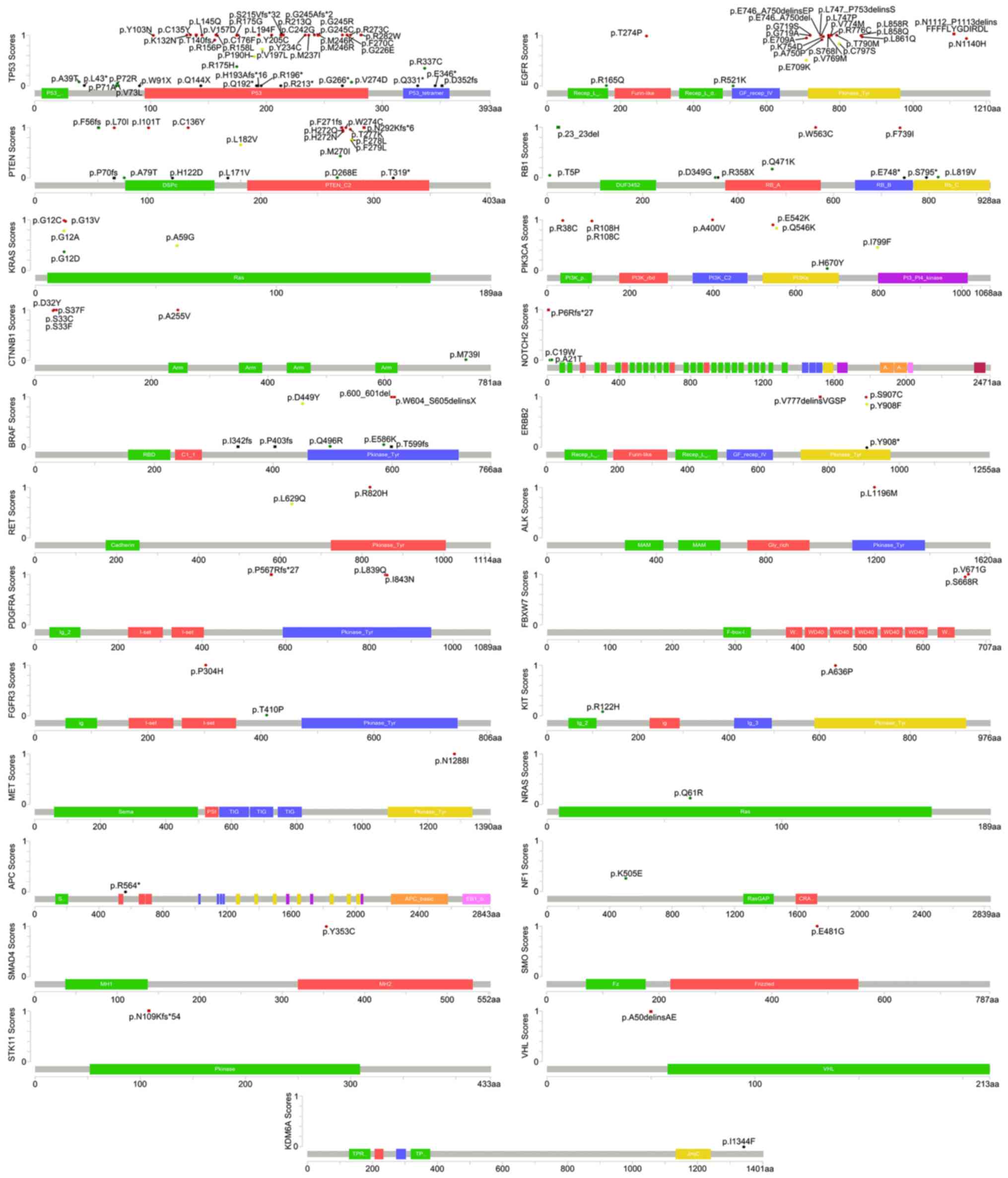
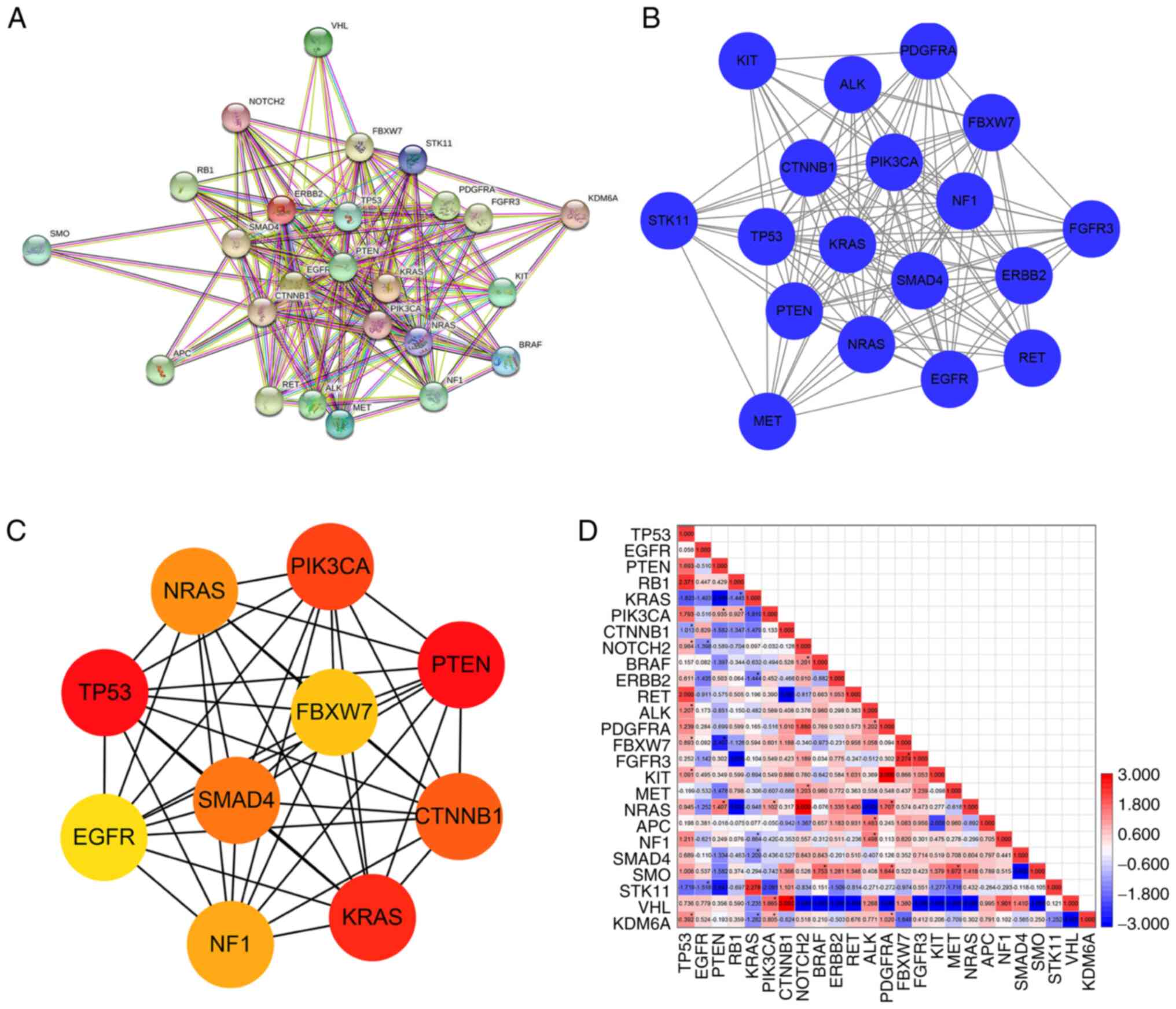
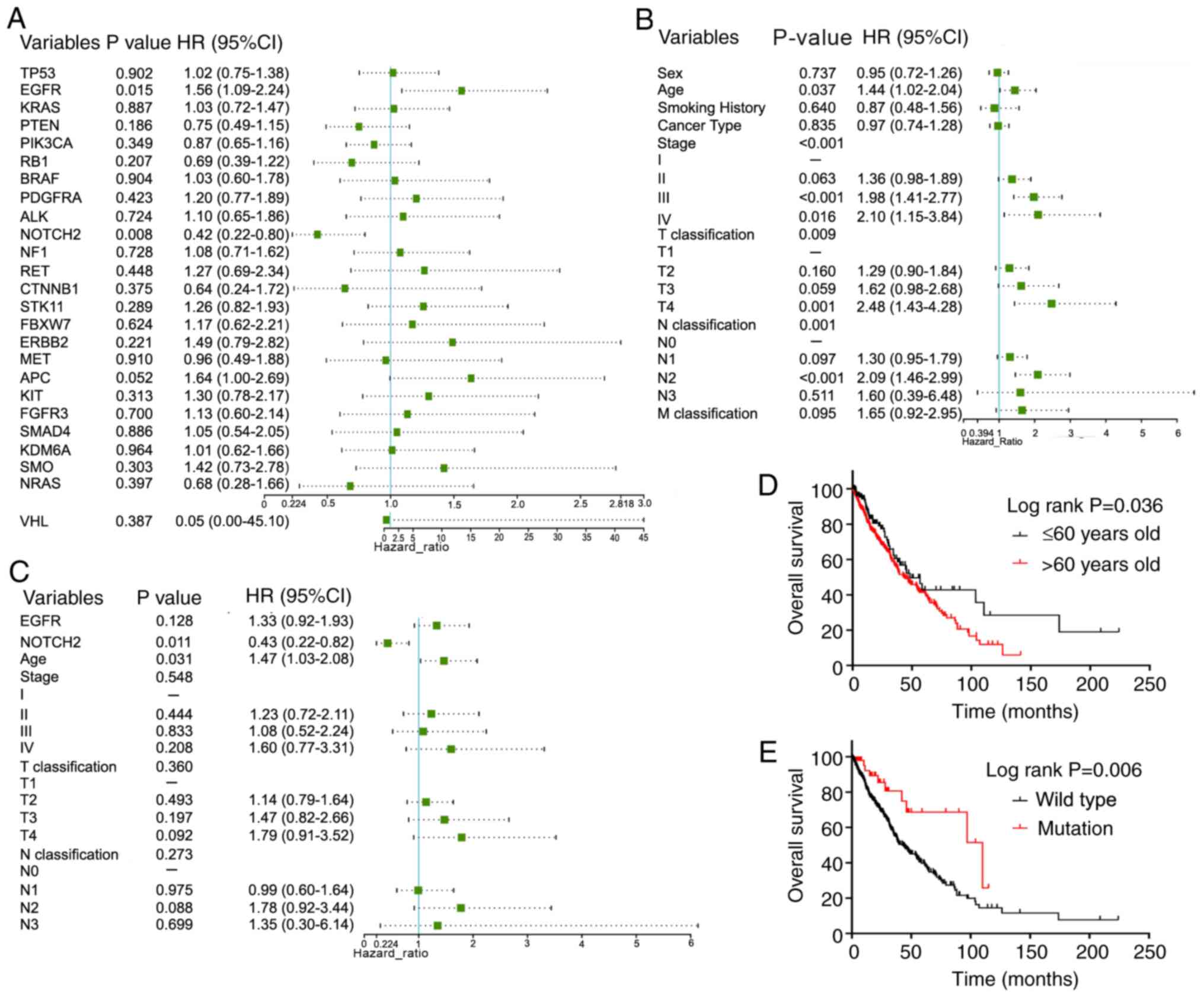
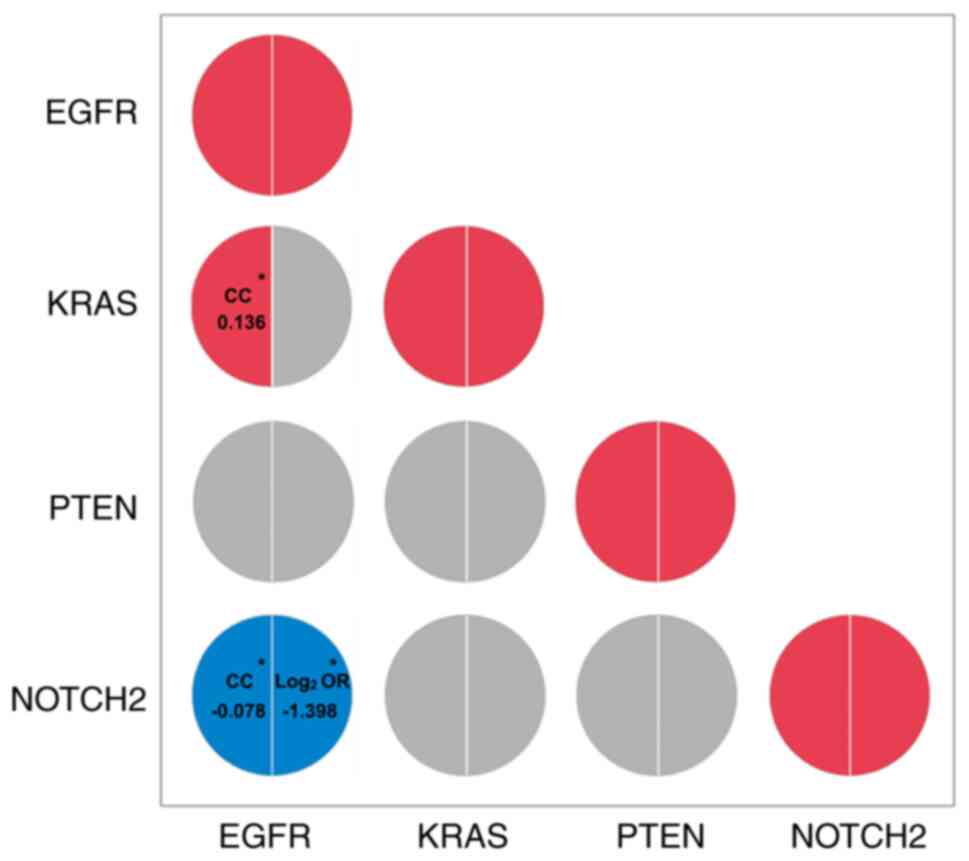
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feng RM, Zong YN, Cao SM and Xu RH:
Current cancer situation in China: Good or bad news from the 2018
Global Cancer Statistics? Cancer Commun. 39:222019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang TJ, Saad S, Qureshi YH, Jani A, Nanda
T, Yaeh AM, Rozenblat T, Sisti MB, Bruce JN, McKhann GM, et al:
Does lung cancer mutation status and targeted therapy predict for
outcomes and local control in the setting of brain metastases
treated with radiation? Neuro Oncol. 17:1022–1028. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsang YH, Dogruluk T, Tedeschi PM,
Wardwell-Ozgo J, Lu H, Espitia M, Nair N, Minelli R, Chong Z, Chen
F, et al: Functional annotation of rare gene aberration drivers of
pancreatic cancer. Nat Commun. 7:105002016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu J, Lee W, Jiang Z, Chen Z,
Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J,
et al: Genome and transcriptome sequencing of lung cancers reveal
diverse mutational and splicing events. Genome Res. 22:2315–2327.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Metzker ML: Sequencing technologies-the
next generation. Nat Rev Genet. 11:31–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhai H, Zhong W, Yang X and Wu YL:
Neoadjuvant and adjuvant epidermal growth factor receptor tyrosine
kinase inhibitor (EGFR-TKI) therapy for lung cancer. Transl Lung
Cancer Res. 4:82–93. 2015.PubMed/NCBI
|
|
10
|
Yuan J, Zhang N, Yin L, Zhu H, Zhang L,
Zhou L and Yang M: Clinical implications of the autophagy core gene
variations in advanced lung adenocarcinoma treated with gefitinib.
Sci Rep. 7:178142017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shaw AT, Solomon BJ, Besse B, Bauer TM,
Lin CC, Soo RA, Riely GJ, Ou SI, Clancy JS, Li S, et al: ALK
resistance mutations and efficacy of lorlatinib in advanced
anaplastic lymphoma kinase-positive non-small-cell lung cancer. J
Clin Oncol. 37:1370–1379. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kalemkerian GP, Narula N, Kennedy EB,
Biermann WA, Donington J, Leighl NB, Lew M, Pantelas J, Ramalingam
SS, Reck M, et al: Molecular testing guideline for the selection of
patients with lung cancer for treatment with targeted tyrosine
kinase inhibitors: American society of clinical oncology
endorsement of the college of American pathologists/international
association for the study of lung cancer/association for molecular
pathology clinical practice guideline update. J Clin Oncol.
36:911–919. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu YL, Planchard D, Lu S, Sun H, Yamamoto
N, Kim DW, Tan DSW, Yang JC, Azrif M, Mitsudomi T, et al: Pan-Asian
adapted Clinical Practice Guidelines for the management of patients
with metastatic non-small-cell lung cancer: A CSCO-ESMO initiative
endorsed by JSMO, KSMO, MOS, SSO and TOS. Ann Oncol. 30:171–210.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Savas P, Hughes B and Solomon B: Targeted
therapy in lung cancer: IPASS and beyond, keeping abreast of the
explosion of targeted therapies for lung cancer. J Thorac Dis. 5
(Suppl 5):S579–S92. 2013.PubMed/NCBI
|
|
15
|
Tian L, Chen Q, Yi X, Wang G, Chen J, Ning
P, Yang K and Liu Z: Radionuclide I-131 labeled albumin-paclitaxel
nanoparticles for synergistic combined chemo-radioisotope therapy
of cancer. Theranostics. 7:614–623. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LJ and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Campbell JD, Alexandrov A, Kim J, Wala J,
Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et
al: Distinct patterns of somatic genome alterations in lung
adenocarcinomas and squamous cell carcinomas. Nat Genet.
48:607–616. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Repana D, Nulsen J, Dressler L,
Bortolomeazzi M, Venkata SK, Tourna A, Yakovleva A, Palmieri T and
Ciccarelli FD: The Network of Cancer Genes (NCG): A comprehensive
catalogue of known and candidate cancer genes from cancer
sequencing screens. Genome Biol. 20:12019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guibert N, Hu Y, Feeney N, Kuang Y,
Plagnol V, Jones G, Howarth K, Beeler JF, Paweletz CP and Oxnard
GR: Amplicon-based next-generation sequencing of plasma cell-free
DNA for detection of driver and resistance mutations in advanced
non-small cell lung cancer. Ann Oncol. 29:1049–1055. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Craig DJ, Morrison T, Khuder SA, Crawford
EL, Wu L, Xu J, Blomquist TM and Willey JC: Technical advance in
targeted NGS analysis enables identification of lung cancer
risk-associated low frequency TP53, PIK3CA, and BRAF mutations in
airway epithelial cells. BMC Cancer. 19:10812019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Liu L, Guo Z, Liang W, He J, Huang
L, Deng Q, Tang H, Pan H, Guo M, et al: UGT1A1 polymorphisms with
irinotecan-induced toxicities and treatment outcome in Asians with
Lung Cancer: A meta-analysis. Cancer Chemother Pharmacol.
79:1109–1117. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bai Y, Wu HW, Ma X, Liu Y and Zhang YH:
Relationship between UGT1A1*6/*28 gene polymorphisms and the
efficacy and toxicity of irinotecan-based chemotherapy. Onco
Targets Ther. 10:3071–3081. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Choi YJ and Lee SH, Kim MS, Jung SH, Hur
SY, Chung YJ and Lee SH: Whole-exome sequencing identified the
genetic origin of a mucinous neoplasm in a mature cystic teratoma.
Pathology. 48:372–376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan AC, Lai GGY, Tan GS, Poon SY, Doble B,
Lim TH, Aung ZW, Takano A, Tan WL, Ang MK, et al: Utility of
incorporating next-generation sequencing (NGS) in an Asian
non-small cell lung cancer (NSCLC) population: Incremental yield of
actionable alterations and cost-effectiveness analysis. Lung
Cancer. 139:207–215. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
DiBardino DM, Rawson DW, Saqi A, Heymann
JJ, Pagan CA and Bulman WA: Next-generation sequencing of non-small
cell lung cancer using a customized, targeted sequencing panel:
Emphasis on small biopsy and cytology. Cytojournal. 14:72017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Preusser M, Berghoff AS, Koller R,
Zielinski CC, Hainfellner JA, Liebmann-Reindl S, Popitsch N, Geier
CB, Streubel B and Birner P: Spectrum of gene mutations detected by
next generation exome sequencing in brain metastases of lung
adenocarcinoma. Eur J Cancer. 51:1803–1811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sonmezler O, Boga I and Bisgin A:
Integration of liquid biopsies into clinical laboratory
applications via NGS in cancer diagnostics. Clin Lab. doi:
10.7754/Clin.Lab.2019.190836.
|
|
31
|
Wang Y, Lai H, Fan X, Luo L, Duan F, Jiang
Z, Wang Q, Leung ELH, Liu L and Yao X: Gossypol inhibits non-small
cell lung cancer cells proliferation by targeting
EGFRL858R/T790M. Front Pharmacol. 9:7282018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Centeno I, Blay P, Santamaría I, Astudillo
A, Pitiot AS, Osorio FG, González-Arriaga P, Iglesias F, Menéndez
P, Tardón A, et al: Germ-line mutations in epidermal growth factor
receptor (EGFR) are rare but may contribute to oncogenesis: A novel
germ-line mutation in EGFR detected in a patient with lung
adenocarcinoma. BMC Cancer. 11:1722011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song P, Zhang F, Li Y, Yang G, Li W, Ying
J and Gao S: Concomitant TP53 mutations with response to crizotinib
treatment in patients with ALK-rearranged non-small-cell lung
cancer. Cancer Med. 8:1551–1557. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Halvorsen AR, Silwal-Pandit L, Meza-Zepeda
LA, Vodak D, Vu P, Sagerup C, Hovig E, Myklebost O, Børresen-Dale
AL, Brustugun OT and Helland Å: TP53 mutation spectrum in smokers
and never smoking lung cancer patients. Front Genet. 7:852016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Leijen S, van Geel RM, Pavlick AC, Tibes
R, Rosen L, Razak AR, Lam R, Demuth T, Rose S, Lee MA, et al: Phase
I study evaluating WEE1 inhibitor AZD1775 as monotherapy and in
combination with gemcitabine, cisplatin, or carboplatin in patients
with advanced solid tumors. J Clin Oncol. 34:4371–4380. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Minamimoto R, Jamali M, Gevaert O,
Echegaray S, Khuong A, Hoang CD, Shrager JB, Plevritis SK, Rubin
DL, Leung AN, et al: Prediction of EGFR and KRAS mutation in
non-small cell lung cancer using quantitative 18F
FDG-PET/CT metrics. Oncotarget. 8:52792–52801. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Raso MG, Behrens C, Herynk MH, Liu S,
Prudkin L, Ozburn NC, Woods DM, Tang X, Mehran RJ, Moran C, et al:
Immunohistochemical expression of estrogen and progesterone
receptors identifies a subset of NSCLCs and correlates with EGFR
mutation. Clin Cancer Res. 15:5359–5368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao J, Gao J, Guo L, Hu X, Liu Q, Zhao J,
Liu L, Jiang J, Wang M, Liang Z, et al: EGFR and KRAS gene
mutations in 754 patients with resectable stage I–IIIa non-small
cell lung cancer and its clinical significance. Zhongguo Fei Ai Za
Zhi. 20:617–622. 2017.(In Chinese). PubMed/NCBI
|
|
39
|
Carey AM, Pramanik R, Nicholson LJ, Dew
TK, Martin FL, Muir GH and Morris JD: Ras-MEK-ERK signaling cascade
regulates androgen receptor element-inducible gene transcription
and DNA synthesis in prostate cancer cells. Int J Cancer.
121:520–527. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hsu F, De Caluwe A, Anderson D, Nichol A,
Toriumi T and Ho C: EGFR mutation status on brain metastases from
non-small cell lung cancer. Lung Cancer. 96:101–107. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Zhu T, Liu J, Liu J, Gao D, Su T
and Zhao R: FLNa negatively regulated proliferation and metastasis
in lung adenocarcinoma A549 cells via suppression of EGFR. Acta
Biochim Biophys Sin. 50:164–170. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang ZM, Ding XP, Pen L, Mei L and Liu T:
Analysis of CEA expression and EGFR mutation status in non-small
cell lung cancers. Asian Pac J Cancer Prev. 15:3451–3455. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Barak V, Goike H, Panaretakis KW and
Einarsson R: Clinical utility of cytokeratins as tumor markers.
Clin Biochem. 37:529–540. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cho A, Hur J, Moon YW, Hong SR, Suh YJ,
Kim YJ, Im DJ, Hong YJ, Lee HJ, Kim YJ, et al: Correlation between
EGFR gene mutation, cytologic tumor markers, 18F-FDG uptake in
non-small cell lung cancer. BMC Cancer. 16:2242016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Molinier O, Goupil F, Debieuvre D, Auliac
JB, Jeandeau S, Lacroix S, Martin F and Grivaux M: Five-year
survival and prognostic factors according to histology in 6101
non-small-cell lung cancer patients. Respir Med Res. 77:46–54.
2020.PubMed/NCBI
|
|
46
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Oshita F, Kasajima R and Miyagi Y:
Multiplex genomic test of mutation and fusion genes in small biopsy
specimen of lung cancer. J Exp Ther Oncol. 11:189–194.
2016.PubMed/NCBI
|
|
48
|
Liao L, Ji X, Ge M, Zhan Q, Huang R, Liang
X and Zhou X: Characterization of genetic alterations in brain
metastases from non-small cell lung cancer. FEBS Open Bio.
8:1544–1552. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen CY, Chen YY, Hsieh MS, Ho CC, Chen
KY, Shih JY and Yu CJ: Expression of notch gene and its impact on
survival of patients with resectable Non-small cell lung cancer. J
Cancer. 8:1292–1300. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Motooka Y, Fujino K, Sato Y, Kudoh S,
Suzuki M and Ito T: Pathobiology of Notch2 in lung cancer.
Pathology. 49:486–493. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mimae T, Okada M, Hagiyama M, Miyata Y,
Tsutani Y, Inoue T, Murakami Y and Ito A: Upregulation of notch2
and six1 is associated with progression of early-stage lung
adenocarcinoma and a more aggressive phenotype at advanced stages.
Clin Cancer Res. 18:945–955. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Baumgart A, Mazur PK, Anton M, Rudelius M,
Schwamborn K, Feuchtinger A, Behnke K, Walch A, Braren R, Peschel
C, et al: Opposing role of Notch1 and Notch2 in a Kras(G12D)-driven
murine non-small cell lung cancer model. Oncogene. 34:578–588.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ding XY, Ding J, Wu K, Wen W, Liu C, Yan
HX, Chen C, Wang S, Tang H, Gao CK, et al: Cross-talk between
endothelial cells and tumor via delta-like ligand 4/Notch/PTEN
signaling inhibits lung cancer growth. Oncogene. 31:2899–2906.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sriuranpong V, Borges MW, Ravi RK, Arnold
DR, Nelkin BD, Baylin SB and Ball DW: Notch signaling induces cell
cycle arrest in small cell lung cancer cells. Cancer Res.
61:3200–3205. 2001.PubMed/NCBI
|
|
55
|
Baumgartner U, Berger F, Hashemi Gheinani
A, Burgener SS, Monastyrskaya K and Vassella E: MiR-19b enhances
proliferation and apoptosis resistance via the EGFR signaling
pathway by targeting PP2A and BIM in non-small cell lung cancer.
Mol Cancer. 17:442018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Karachaliou N, Cardona AF, Bracht JWP,
Aldeguer E, Drozdowskyj A, Fernandez-Bruno M, Chaib I, Berenguer J,
Santarpia M, Ito M, et al: Integrin-linked kinase (ILK) and src
homology 2 domain-containing phosphatase 2 (SHP2): Novel targets in
EGFR-mutation positive non-small cell lung cancer (NSCLC).
EBioMedicine. 39:207–214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yu S, Liu D, Shen B, Shi M and Feng J:
Immunotherapy strategy of EGFR mutant lung cancer. Am J Cancer Res.
8:2106–2115. 2018.PubMed/NCBI
|
|
58
|
Katoh M and Katoh M: Precision medicine
for human cancers with Notch signaling dysregulation (Review). Int
J Mol Med. 45:279–297. 2020.PubMed/NCBI
|