Introduction
Gastric cancer was the third most common cause of
cancer-associated mortality in Asia in 2018 (1). Asia has a high incidence of gastric
cancer globally, and approximately half of the world's gastric
cancer cases and deaths occur in China (1). In China, gastric cancer is the second
most common malignancy and the third leading cause of death from
malignant neoplasms (1). In 2012,
Gansu province in China had a gastric cancer age-standardized
incidence rate by Chinese standard population and a mortality rate
of 62.34/100,000 and 36.94/100,000 (2), respectively, which is much higher than
the average levels in China of 22.06/100,000 and 15.16/100,000
(3). The prevalence of gastric
cancer in Wuwei, Gansu, China is almost five times higher than that
nationwide (4). Our previous study
established a large-scale natural population cohort involving
24,115 individuals from Wuwei, Gansu, China, and has conducted
research regarding various aspects to explore the causes of the
high incidence of gastric cancer in this region and to provide a
theoretical basis for the formulation of control policies (5).
Great efforts, such as the development of more
effective biomarkers for diagnosis, prognosis, monitoring and
prediction, have been made regarding the clinical management of
patients with gastric cancer (6).
Gastric cancer has atypical symptoms in the early stage and lacks
effective early screening markers; therefore, most patients have
entered the advanced stage when they are detected (7). A retrospective study revealed that the
5-year rate of relative survival of patients with early gastric
cancer (EGC) with treatment is 105.0% compared with the expected
survival of individuals from the general population matched for age
and sex (8). By contrast, few EGCs
are discovered in China and the West, leading to 5-year relative
survival rates of 10–40% (9–11). In Japan, the male mortality rate of
gastric cancer has fallen by more than half since a mass screening
program was introduced in the early 1970s (9). Therefore, early diagnosis and treatment
of gastric cancer, and screening are important. Gastroscopy is a
valuable tool for reducing the mortality associated with gastric
cancer (12). However, due to its
acceptability and cost, large-scale screening and early detection
with gastroscopy might not be easy. At present, available tumor
markers for gastric cancer, including carcinoembryonic antigen and
cancer antigen 19-9, are useful for detecting recurrence and
distant metastasis or predicting patient survival; however, these
are inadequate for gastric cancer screening due to their low
sensitivity, particularly for EGC (13). Therefore, there is an urgent
requirement for novel non-invasive methods for the screening of
patients with gastric cancer, and microRNAs (miRNAs/miRs) have been
increasingly studied for this.
miRNAs are endogenous 18–24 nucleotide RNAs, which
can serve critical regulatory roles in animals and plants (14). miRNAs can combine with other
associated proteins to form an active RNA-induced silencing complex
(RISC), and RISC combines with the 5′ untranslated region, open
reading frames or 3′ untranslated region of a target gene mRNA to
suppress its translation or to induce its degradation (15,16). An
increasing number of studies have reported that miRNAs can be used
as biomarkers for gastric cancer diagnosis, as well as targets for
disease treatment (17–19). For example, a study of 682
participants examined the expression levels of 578 miRNAs in serum
and demonstrated that the combination of 12 miRNAs in serum has
excellent diagnostic value for gastric cancer (13). Microarray technology has been widely
used to investigate miRNA expression in multiple tumor types, such
as gastric cancer and lung cancer (20,21).
The present study, as a part of the aforementioned
Wuwei cohort research project, utilized samples collected from
patients diagnosed with EGC during the screening of this disease in
Wuwei. miRNA profiles in five pairs of EGC tissues and adjacent
non-cancerous tissues were explored using a miRNA microarray.
Bioinformatics methods were used to analyze the functions and
mechanisms of the dysregulated miRNAs, as well as their potential
as prognostic factors. The present study aimed to provide a basis
for the identification of EGC screening biomarkers and to discuss
the particularity of gastric cancer in Gansu province from the
perspective of miRNAs.
Materials and methods
Patients and samples
EGC tissues and adjacent non-cancerous mucosa
tissues (5 cm from the cancer tissues) were obtained from 5
patients with EGC who were treated with endoscopic submucosal
dissection at the Department of Gastroenterology, The Gansu Wuwei
Tumor Hospital (Wuwei, China) and Department of Gastroenterology,
The First Hospital of Lanzhou University (Lanzhou, China) between
November 2014 and April 2015. The inclusion criteria were: i)
Patients who agreed to participate in the study and signed the
informed consent were included; ii) all patients were
pathologically diagnosed as EGC. The tumor stage was confirmed
according to the Eighth Edition of the The American Joint Committee
on Cancer (AJCC) TNM staging system guidelines (22); and iii) the patients did not receive
radiotherapy or chemotherapy before surgery. The exclusion criteria
were: i) Patients who did not agree to participate in the study;
ii) patients who had other malignancies; and iii) patients who had
other diseases, such as diabetes mellitus and hypertension. The
patients included 3 men and 2 women. Clinicopathological features
of the patients are listed in Table
I. The present study was approved by the Ethics Committee of
The First Hospital of Lanzhou University (approval no.
LDYYLL2012001; Lanzhou, China). All patients and their family
members signed an informed consent form. The tissue samples were
stored and transported at −80°C.
 | Table I.Clinicopathological features of
patients with early gastric cancer whose tissues underwent microRNA
microarray testing. |
Table I.
Clinicopathological features of
patients with early gastric cancer whose tissues underwent microRNA
microarray testing.
| Patient | Sex | Age, years | Location | Lesion
location | Histology | T stage | N stage | M stage | AJCC pathologic
stage (8th edition) |
|---|
| Patient 1 | Female | 65 | Wuwei, Gansu,
China | Cardia | Adenocarcinoma | T1a | 0 | 0 | IA |
| Patient 2 | Male | 72 | Wuwei, Gansu,
China | Gastric antrum | Adenocarcinoma | Tis | 0 | 0 | 0 |
| Patient 3 | Male | 46 | Wuwei, Gansu,
China | Gastric body | Adenocarcinoma | T1b | 0 | 0 | IA |
| Patient 4 | Male | 45 | Wuwei, Gansu,
China | Cardia | Adenocarcinoma | T1a | 0 | 0 | IA |
| Patient 5 | Female | 67 | Lanzhou, Gansu,
China | Gastric antrum | Adenocarcinoma | Tis | 0 | 0 | 0 |
RNA extraction
Total RNA from 10 tissue samples was isolated using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and
purified using a RNeasy mini kit (Qiagen, Inc.) according to the
manufacturer's protocols. RNA quality and quantity were measured
using a NanoDrop spectrophotometer (ND-1000; NanoDrop Technologies;
Thermo Fisher Scientific, Inc.). RNA integrity was determined by
formaldehyde agarose gel electrophoresis in
3-(N-morpholino)propanesulfonic acid buffer with a 1.2% gel.
Ethidium bromide was used as a fluorescent dye, and the GelDoc Go
Gel Imaging System (Bio-Rad Laboratories, Inc.) was used for
imaging.
miRNA labeling and array
hybridization
miRNA microarray assays were performed by Aksomics,
Inc. The miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon; Qiagen,
Inc.) was used with total RNA samples according to the
manufacturer's guidelines for miRNA labeling. After termination of
the labeling procedure, the Hy3™-labeled samples were hybridized on
the array in miRCURY LNA™ microRNA Array Kit, 7th generation-hsa,
mmu and rno (Exiqon; Qiagen, Inc.). Following hybridization, the
slides were washed several times using Wash buffer kit (Exiqon;
Qiagen, Inc.). The slides were scanned using an Axon GenePix 4000 B
microarray scanner (Molecular Devices, LLC).
Microarray data processing
Scanned images were subsequently imported into
GenePix Pro 6.0 (Molecular Devices, LLC) for grid alignment and
data extraction. Data analyses were performed using R software
(v3.6.3; http://cran.r-project.org/src/base/R-3/). The ‘limma’
package (v3.42.2) (23) in R was
used for background correction and normalization between arrays.
The robust multi-array average algorithm was selected when
performing background correction, and the ‘offset’ parameter was
set to 50. Expression data were normalized using median
normalization. The landing lights (probe ID 13138; annotated as
Hy3™) were only included for gal-file orientation, and their
corresponding data points were removed prior to the normalization
of the dataset, according to the manual of the miRCURY LNA™
microRNA Array Kit, 7th generation-hsa, mmu and rno (Exiqon;
Qiagen, Inc.). For the same probes, the expression data of all
samples corresponding to each probe were averaged. The probe with
the maximum average value was retained among the same probes. The
‘limma’ package was used to give a microarray linear model fit,
compute moderated t-statistics, moderated F-statistic, and log-odds
of differential expression by empirical Bayes moderation of the
standard errors towards a common value. The cutoff criteria for
differentially expressed miRNAs (DEMs) were a |log2 fold
change (log2FC)| ≥1 and an adjusted P-value (adj. P.
val) <0.05. Data were submitted to the Gene Expression Omnibus
database (24) (dataset accession
no. GSE158315).
Functional annotation of DEMs
The functions of DEMs were annotated using TAM
(v2.0), a web-based program for annotations of human miRNAs
(25).
mRNA expression profiles of gastric
cancer at stage I/II based on data from The Cancer Genome Atlas
(TCGA)
The mRNA expression profiles of three pairs of
gastric cancer tissues and their matched adjacent non-cancerous
mucosa tissues were obtained from TCGA [project ID, TCGA-stomach
adenocarcinoma (STAD)] using the Genomic Data Commons Data Transfer
Tool (v1.5.0; http://gdc.cancer.gov/access-data/gdc-data-transfer-tool).
The samples were obtained from 3 Asian patients with stage I/II
gastric cancer (AJCC, Eighth Edition; Table II). The ‘DESeq2’ package (v1.26.0;
http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html)
(26) in R was used to perform the
differential analysis. Differentially expressed genes (DEGs) with
|log2FC| ≥2 and adj. P. val <0.05 in cancer tissues
compared with non-cancerous tissues were filtered.
 | Table II.Clinicopathological features of
patients with gastric cancer at stage I/II whose gene expression
profiles were obtained from TCGA. |
Table II.
Clinicopathological features of
patients with gastric cancer at stage I/II whose gene expression
profiles were obtained from TCGA.
| Patient | Sex | Age, years | Ethnicity | Lesion
location | Histology | T stage | N stage | M stage | AJCC pathologic
stage (8th Edition) |
|---|
| TCGA-HU-A4GH | Male | 75 | Asian | Gastric body | Adenocarcinoma | T1b | N0 | M0 | IA |
| TCGA-HU-A4GP | Female | 62 | Asian | Gastric antrum | Adenocarcinoma | T2 | N1 | M0 | IIA |
| TCGA-HU-A4HB | Male | 68 | Asian | Gastric antrum | Adenocarcinoma | T2 | N2 | M0 | IIB |
Functional enrichment and pathway
analysis of DEGs
Gene Ontology (GO) analysis (27), Kyoto Encyclopedia of Genes and
Genomes (KEGG) analysis (28) and
Gene Set Enrichment Analysis (GSEA) (29) were performed on the DEGs in the
cancer tissues and adjacent non-cancerous tissues of 3 Asian
patients with stage I/II gastric cancer. First, the ‘org.Hs.eg.db’
package (v3.10.0; http://www.bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html)
was used to convert the gene symbols into entrezIDs. Subsequently,
‘clusterProfiler’ (v3.14.3; http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
(30), ‘ggplot2’ (v3.3.2) (31) and ‘enrichplot’ (v1.6.1; http://bioconductor.org/packages/devel/bioc/html/enrichplot.html)
packages were utilized to find the enriched GO terms and KEGG
pathways, where a false discovery rate <0.05 was considered to
be statistically significant. A ‘gmt’ format file called ‘Hallmark
gene sets, NCBI (Entrez) gene IDs’, which includes the information
of the gene sets and their associated pathways, was downloaded from
the GSEA website (https://www.gsea-msigdb.org/gsea/index.jsp) and was
used to perform GSEA in R software. GO and KEGG analyses of the
predicted DEM target genes were performed in the same manner.
Prediction of DEM target genes
The miRDB database (http://mirdb.org/), an online database for miRNA
target prediction and functional annotation, was used to predict
the target relationships between DEMs and DEGs (32). DEMs and DEGs were controlled to have
opposite trends in expression level variations between cancer
tissues and non-cancerous tissues. In other words, for the
upregulated DEMs, target genes were predicted among the
downregulated DEGs and vice versa. Subsequently, a miRNA-gene
regulatory network was constructed and visualized using Cytoscape
software (v3.7.2) (33).
Construction of the target gene
protein-protein interaction (PPI) network and hub genes association
network
The target genes were input into the Search Tool for
the Retrieval of Interacting Genes/Proteins database (v11.0;
http://string-db.org) (34), an online protein interaction search
tool for the retrieval of interacting genes/proteins, to construct
a PPI network. ‘Experiments’, ‘Databases’, ‘Co-experiment’ and
‘Co-occurrence’ were set as the parameter of ‘active interaction
sources’, and the interactions with a combined score >0.7 were
selected for the PPI network. The PPI network was visualized using
Cytoscape software, and the tight link hub mRNAs within the PPI
network were calculated using Molecular Complex Detection in
Cytoscape (v2.0.0; http://apps.cytoscape.org/apps/mcode) (35) with default parameters.
Survival analysis validation
To verify the results obtained, clinical data and
miRNA expression data of 80 Asian patients with gastric cancer were
downloaded from TCGA (project ID, TCGA-STAD). For each miRNA, which
was in the miRNA-gene network, the ‘survminer’ package (v0.4.8;
http://cran.r-project.org/web/packages/survminer/index.html)
was used to find a best separation cutoff value, and then patients
were divided into two groups according to the cutoff value. The
prognostic values of partial DEMs were identified by Kaplan-Meier
analysis with the ‘survival’ package (v3.2.3) (36) in R software, and P<0.05 (log-rank
test) was considered to indicate a statistically significant
difference.
Statistical analysis
The statistical analyses were performed using R
software. A moderated t-test was performed to screen the DEMs with
the ‘limma’ package, and a paired samples t-test was performed to
screen the DEGs with the ‘DESeq2’ package. The expression levels of
miRNAs and genes are presented as log2 FC. The log-rank
test was performed in the survival analysis with the ‘survival’
package. P<0.05 was considered to indicate a statistically
significant difference.
Results
Microarray data processing and
identification of DEMs
A flowchart of the research design is shown in
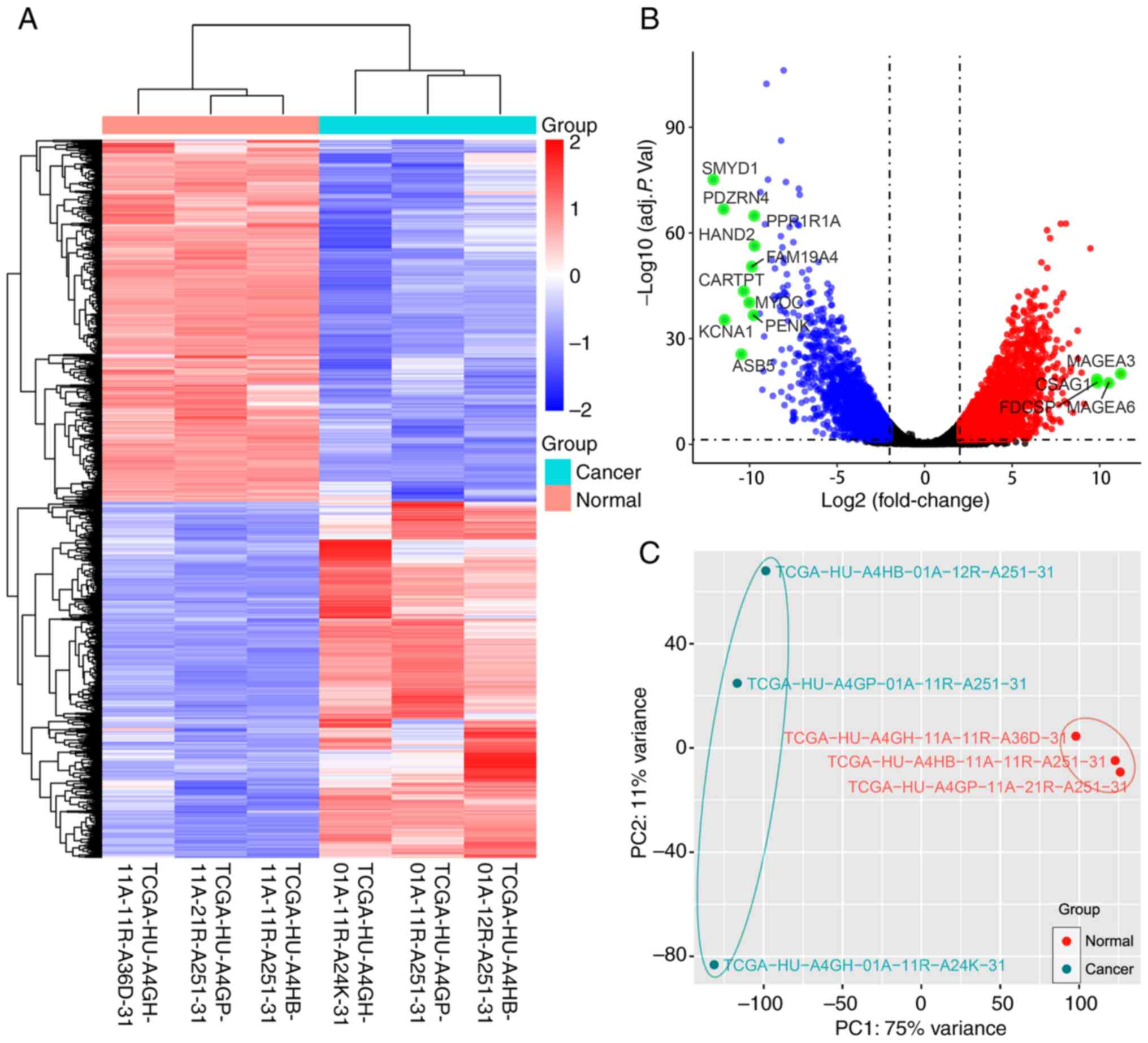
Fig. 1. The box and density plots
indicated the normalization results of miRNA microarray data for 10
tissue samples (Fig. 2A and B). A
total of seven upregulated and 40 downregulated human miRNAs were
identified using the criteria of log2FC ≥1 and adj. P.
val <0.05 [seven downregulated miRNAs of human viruses and one
downregulated small nuclear RNA (snRNA) were also detected]. The
heatmap demonstrated that the samples could be separated into two
groups based on the 55 dysregulated RNA molecules (Fig. 2C). The volcano plot revealed the
expression distribution of miRNAs (Fig.
2D), and the principal component analysis plot is shown in
Fig. 2E.
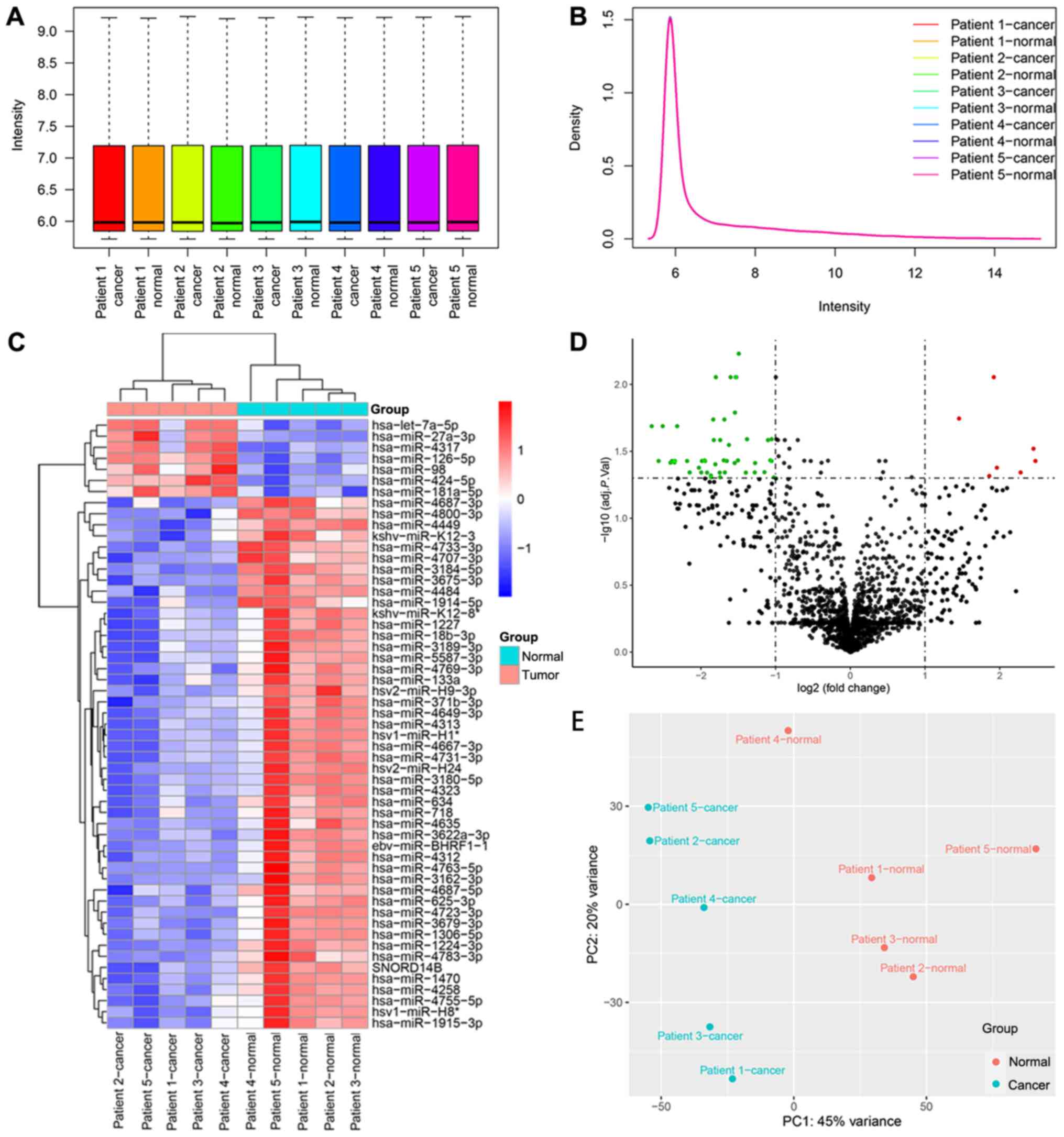 | Figure 2.Data analysis of the miRNA microarray
using five early gastric cancer tissues and five adjacent
non-cancerous tissues, and the identification of DEMs. (A) Box plot
after normalization, which shows the maximum, upper quartile,
median, lower quartile and minimum of different datasets. (B)
Density plot after normalization, which shows a probability
distribution at different miRNA expression levels. (C) Heat map
generated by unsupervised hierarchical clustering for DEMs between
two groups. The color varies from red to blue, indicating
upregulated or downregulated expression of DEMs, respectively. (D)
Volcano plot of the DEMs in the miRNA microarray. Red and green
dots represent upregulated and downregulated molecules (54 miRNAs
and one small nuclear RNA), respectively. (E) Principal component
analysis result of the tissue samples. adj. P. Val., adjusted
P-value; DEMs, differentially expressed miRNAs; miRNA/miR,
microRNA; PC, principal component. |
Functional annotations of DEMs
The functions of the DEMs were annotated using TAM.
‘Hormones regulation’, ‘Human embryonic stem cell (hESC)
regulation’, ‘Immune response’, ‘Inflammation’, ‘Cell cycle
related’, ‘Epithelial-mesenchymal transition’, ‘Cell death’,
‘Hematopoiesis’ and ‘MiRNA tumor suppressors’ were the most
enriched function terms (Table
III).
 | Table III.Functional annotation of
differentially expressed miRNAs using the tool for annotations of
human miRNAs. |
Table III.
Functional annotation of
differentially expressed miRNAs using the tool for annotations of
human miRNAs.
| Function term | Count | Percent | Fold | P-value |
|---|
| Hormones
regulation | 9 | 0.14516129 | 3.27217742 | 0.00062950 |
| Human embryonic
stem cell (hESC) regulation | 8 | 0.09411765 | 2.12156863 | 0.02291040 |
| Immune
response | 8 | 0.16666667 | 3.75694444 | 0.00052898 |
| Inflammation | 8 | 0.19512195 | 4.39837398 | 0.00016338 |
| Cell cycle
related | 7 | 0.10606061 | 2.39078283 | 0.01856736 |
|
Epithelial-mesenchymal transition | 5 | 0.12195122 | 2.74898374 | 0.02820963 |
| Cell death | 5 | 0.09090909 | 2.04924242 | 0.08525479 |
| Hematopoiesis | 5 | 0.16129032 | 3.63575269 | 0.00867962 |
| MiRNA tumor
suppressors | 5 | 0.13513514 | 3.04617117 | 0.01854272 |
| Lipid
metabolism | 4 | 0.20000000 | 4.50833333 | 0.00898287 |
| Angiogenesis | 3 | 0.12500000 | 2.81770833 | 0.08400821 |
| Cell
proliferation | 3 | 0.10714286 | 2.41517857 | 0.12100311 |
| HIV latency | 3 | 0.14285714 | 3.22023810 | 0.06027038 |
| Adipocyte
differentiation | 3 | 0.11111111 | 2.50462963 | 0.11122976 |
| Onco-miRNAs | 3 | 0.09677419 | 2.18145161 | 0.15217690 |
| Apoptosis | 2 | 0.04545455 | 1.02462121 | 0.59774380 |
| Bone
regeneration | 2 | 0.05882353 | 1.32598039 | 0.45394525 |
|
Folliculogenesis | 2 | 0.28571429 | 6.44047619 | 0.03458783 |
| Anti-cell
proliferation (Hwang etal BJC2006) | 2 | 0.18181818 | 4.09848485 | 0.08125429 |
| Cell division | 2 | 0.11764706 | 2.65196078 | 0.17103576 |
| Immune system
(Xiao's Cell2009) | 2 | 0.11111111 | 2.50462963 | 0.18736736 |
| Cell
differentiation | 1 | 0.05882353 | 1.32598039 | 0.54311212 |
| Chemosensitivity of
tumor cells | 1 | 0.25000000 | 5.63541667 | 0.16641762 |
| DNA repair | 1 | 0.10000000 | 2.25416667 | 0.36724517 |
| Granulopoiesis | 1 | 0.10000000 | 2.25416667 | 0.36724517 |
| Carbohydrate
metabolism | 1 | 0.14285714 | 3.22023810 | 0.27345270 |
| Cell fate
determination | 1 | 0.03846154 | 0.86698718 | 0.70138621 |
| Heart
development | 1 | 0.14285714 | 3.22023810 | 0.27345270 |
Identification of DEGs using samples
from TCGA
To further investigate the functions of DEMs via
their target genes, the present study aimed to obtain DEGs between
EGC and non-cancerous tissues. Since there was only 1 Asian patient
with EGC, the scope was expanded to stage I/II. A total of 3 Asian
patients with stage I/II gastric cancer were included in the
present study. The clinicopathological features of the selected
patients are shown in Table II. A
total of 2,097 upregulated genes and 2,131 downregulated genes were
identified, and are shown in a heat map and a volcano plot
(Fig. 3A and B). Principal component
analysis indicated that the gastric cancer tissue samples at stage
I/II and their matched non-cancerous tissue samples could be
divided into two groups based on the gene profile (Fig. 3C).
GO analysis, KEGG analysis and GSEA of
DEGs
To explore the functions and pathways of DEGs in the
cancer tissues and non-cancerous tissues of 3 Asian patients with
stage I/II gastric cancer, GO analysis, KEGG analysis and GSEA were
performed. Functional enrichment results revealed that DEGs were
mainly associated with ‘Regulation of membrane potential’ and
‘Muscle system process’ in the biological process category. In the
cellular component category, the DEGs were mainly enriched in
‘Neuronal cell body’, ‘Collagen-containing extracellular matrix’
and ‘Cell-cell junction’. ‘Channel activity’ and ‘Passive
transmembrane transporter activity’ were enriched terms in the
molecular function (MF) category (Fig.
4A). In addition, KEGG pathway analysis indicated that the
enrichment of DEGs was associated with ‘Neuroactive ligand-receptor
interaction’ and ‘Cytokine-cytokine receptor interaction’. ‘Cell
adhesion molecules (CAMs)’ and ‘Calcium signaling pathway’ were
also enriched (Fig. 4B). Finally,
GSEA results suggested that pathways of the ‘cell cycle’, ‘DNA
replication’, ‘P53 signaling pathway’ and ‘natural killer
cell-mediated cytotoxicity’ tended to be activated in the cancer
group (Fig. 4C-F).
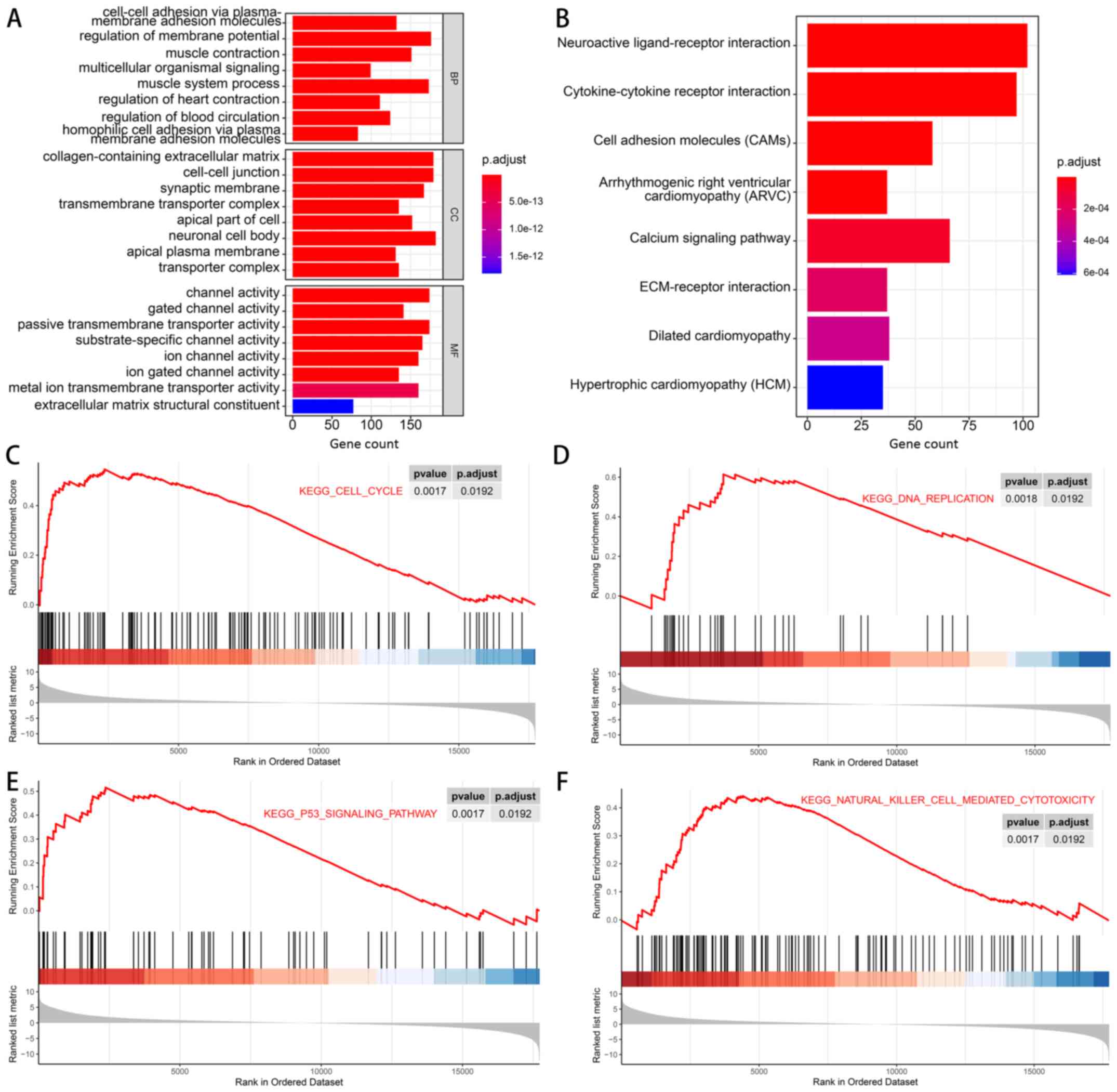 | Figure 4.GO analysis, KEGG analysis and GSEA
of the differentially expressed genes between the cancer tissues
and non-cancerous tissues of 3 Asian patients with stage I/II
gastric cancer. (A) Results of GO analysis, including BP, CC and MF
aspects. (B) Results of KEGG analysis. (C-F) Results of GSEA
revealing that the KEGG pathways of (C) ‘cell cycle’, (D) ‘DNA
replication’, (E) ‘P53 signaling pathway’ and (F) ‘natural killer
cell-mediated cytotoxicity’ were activated in the gastric cancer
tissues at stage I/II. BP, biological process; CC, cellular
component; GO, Gene Ontology; GSEA, Gene Set Enrichment Analysis;
KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular
function; p.adjust, adjusted P-value. |
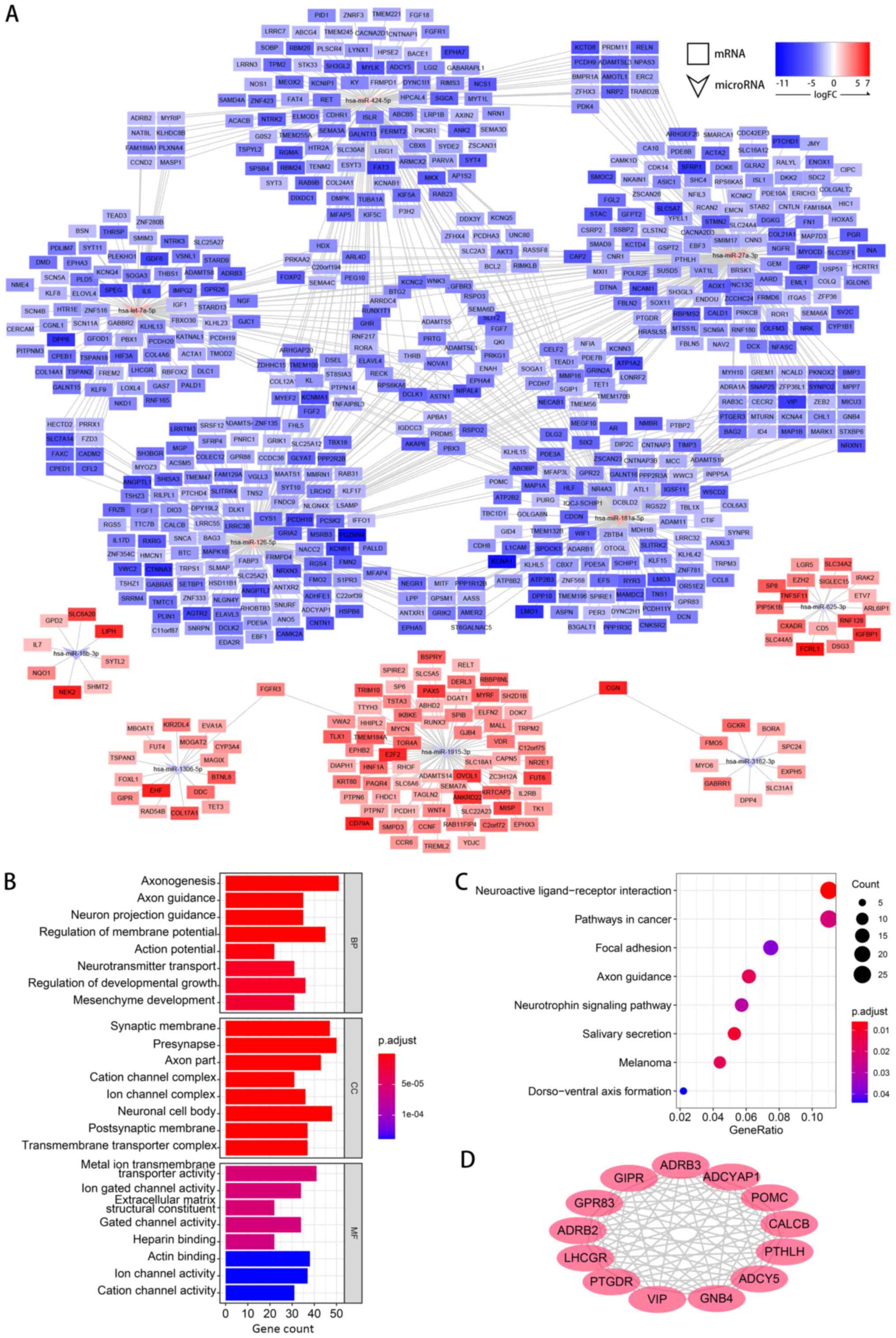
miRNA-gene network construction and
functions of target genes
After the DEMs and DEGs were identified, miRDB was
used to predict the possible target relationships between them. For
the upregulated miRNAs, target genes were predicted among the
downregulated DEGs and vice versa. Subsequently, a miRNA-gene
network was constructed (Fig.
5A).
GO and KEGG analyses of the predicted target genes
were performed to learn more about the functions of the DEMs. In
the MF category of GO analysis, the terms with the highest gene
counts were ‘Metal ion transmembrane transporter activity’, ‘Actin
binding’ and ‘Ion channel activity’ (Fig. 5B). KEGG terms, such as ‘Neuroactive
ligand-receptor interaction’, ‘Pathways in cancer’ and ‘Focal
adhesion’, were significantly enriched (Fig. 5C).
The results of PPI analysis are shown in Fig. 5D. A total of 13 genes, including
ADRB3, ADCYAP1, POMC, CALCB, PTHLH, ADCY5, GNB4, VIP, PTGDR, LHCGR,
ADRB2, GPR83 and GIPR were filtered as hub genes in the network,
which indicated that the DEMs may mainly work by regulating these
hub genes.
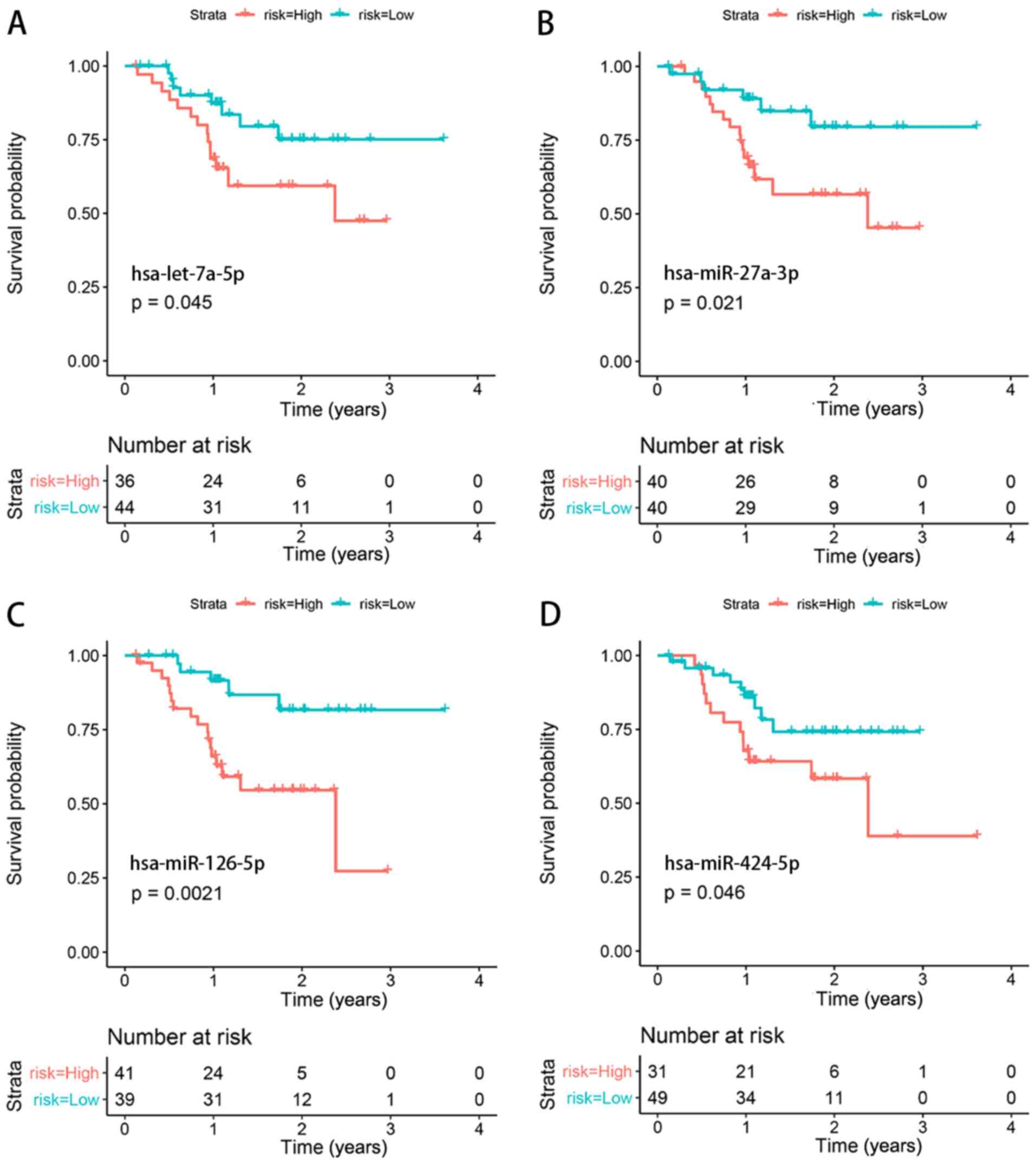
Survival analysis validation
Clinical data and RNA expression data of Asian
patients with gastric cancer (irrespective of the stage) from TCGA
were used for survival analysis. A total of four miRNAs
(hsa-let-7a-5p, hsa-miR-27a-3p, hsa-miR-126-5p and hsa-miR-424-5p)
were identified to be significantly associated with the prognosis
of patients with gastric cancer (Fig.
6).
Discussion
A total of seven upregulated and 40 downregulated
human miRNAs were identified in EGC tissues compared with adjacent
non-cancerous tissues. The functions of the DEMs were annotated
using the TAM webtool. ‘Hormones regulation’ was the most enriched
function term, which indicated that hormones may be regulated in
EGC by miRNAs. A population-based matched cohort study in Sweden
suggested that menopausal hormone therapy users are at a decreased
risk of gastric adenocarcinoma (37). In addition, gonadotropin-releasing
hormone (38),
corticotropin-releasing hormone (39), steroid hormones (40) and growth hormone (41) have been reported to be associated
with gastric cancer. However, more evidence is required to verify
the role of hormone regulation in EGC.
Expression profiles of three pairs of cancer tissues
and adjacent non-cancerous tissues from patients with stage I/II
gastric cancer were downloaded. Gastric cancer exhibits biological
and epidemiological differences between Asian and non-Asian
populations (42,43). To improve the understanding of the
roles of DEMs, the present study aimed to identify target mRNAs
among DEGs in Asian patients. Since only 1 Asian patient with EGC
has been included in the TCGA database to date, the scope was
expanded to patients at stage I/II (AJCC, Eighth Edition). Notably,
‘Neuroactive ligand-receptor interaction’ and ‘Calcium signaling
pathway’ were enriched KEGG terms for DEGs. ‘Neuroactive
ligand-receptor interaction’, ‘Axon guidance’ and ‘Neurotrophin
signaling pathway’ were enriched KEGG terms for the target genes of
DEMs. Furthermore, ‘muscle contraction’ was an enriched biological
process term in GO analysis of DEGs. These results suggest that
pathways associated with nerves, muscle contraction and calcium
signaling may serve a role in gastric cancer. There is much
evidence regarding the association between the calcium signaling
pathway and gastric cancer (44,45).
Regarding nerve and muscle contraction, certain proteins involved
in neurodegenerative events are considered to be associated with
gastric cancer, according to previous reports (46–48).
Based on the target gene results, ‘pathways in cancer’ and other
pathways associated with cancer, such as ‘Focal adhesion’ (49) and ‘Melanoma’, were also affected.
Since the results obtained by this method are closely associated
with the gene set, the two methods of detecting functional
enrichment complement each other.
The miRNA-gene network was constructed. However,
>90% of the target genes were identified to be potentially
modulated by hsa-let-7a-5p, hsa-miR-27a-3p, hsa-miR-126-5p,
hsa-miR-424-5p, hsa-miR-181a-5p and hsa-miR-1915-3p in the network.
Survival analysis of Asian patients demonstrated that four of the
miRNAs (hsa-let-7a-5p, hsa-miR-27a-3p, hsa-miR-126-5p and
hsa-miR-424-5p) were significantly associated with the prognosis of
patients with gastric cancer. Multiple studies have demonstrated
that hsa-miR-27a-3p (50–53) and hsa-miR-424-5p (54–56) are
upregulated in gastric cancer and act as tumor activators. The
present results demonstrated that hsa-miR-27a-3p and has-miR-424-5p
could be upregulated in EGC and may affect the prognosis of
patients with gastric cancer. Liang et al (57) reported that the hsa-let-7 family
inhibits tumor invasion and metastasis by targeting myosin heavy
chain 9 in human gastric cancer, which is different from the
results of the present study. Since the sample size included in the
present study was small, the results should be verified with larger
sample sizes. To the best of our knowledge, there have been no
studies on hsa-miR-126-5p in gastric cancer. Although a study using
expression profile analysis of miRNAs in prostate cancer by
next-generation sequencing demonstrated that hsa-miR-126-5p is
highly expressed in tumor tissues (58), other studies came to different
conclusions and have suggested that it may be a tumor suppressor
(59,60). The functions of hsa-miR-126-5p in
gastric cancer and EGC need to be verified. Experiments have
demonstrated that hsa-miR-181a-5p can promote the progression of
gastric cancer via Ras association domain family member 6-mediated
mitogen activated kinase-like protein signaling activation
(61) or by regulating protein
tyrosine phosphatase non-receptor type 9 (62). Furthermore, hsa-miR-1915-3p inhibits
Bcl-2 expression in the development of gastric cancer (63). hsa-miR-1915-3p serves a role in
breast cancer inhibition (64) and
increases drug sensitivity in colorectal cancer (65). The present results support the role
of hsa-miR-181a-5p as a tumor activator and the role of
hsa-miR-1915-3p as a tumor suppressor. Overall, the studies of
miRNAs in EGC are still limited, and the present study provided
some evidence for this. There are also differentially expressed
viral miRNAs and snRNAs. Further research is required to clarify
their functions.
The present study intended to perform preliminary
screening of miRNAs and the results need to be verified in a larger
cohort. Additionally, the present study was based on individuals in
Gansu province and Asia, which is helpful to explore the
particularity of gastric cancer in these regions. However, the
population is also a limitation of the present study when the
results need to be extended to other regions or other high
incidence areas of gastric cancer. Comparing the results with other
population cohorts should be a direction of future research.
In conclusion, in the present study, the miRNA
profiles in five pairs of EGC tissues and adjacent non-cancerous
tissues were investigated using a miRNA microarray. The possible
mechanisms and abilities as prognostic factors of DEMs were
assessed using bioinformatics methods. The present study revealed
that certain miRNAs, including hsa-let-7a-5p, hsa-miR-27a-3p,
hsa-miR-126-5p and hsa-miR-424-5p, were upregulated in EGC, and
were associated with the prognosis of gastric cancer based on
analysis of the miRNA expression profiles of patients from Gansu
province, China. The present study could provide a basis for the
identification of EGC screening biomarkers and for exploring the
cause of the high incidence of gastric cancer in Gansu province,
China from the perspective of miRNAs.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key
R&D Program of China (grant no. 2016YFC1302201), the Natural
Science Foundation of Gansu Province, China (grant nos. 18JR3RA336
and 17JR5RA272) and the Fundamental Research Funds for the Central
Universities (grant no. lzujbky-2020-kb16).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in the Gene Expression Omnibus
repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE158315).
Authors' contributions
YL, WS and YW conceived the study, and YZ and LL
designed the study. YZ, HW and HY were involved in the sample and
data collection. YL, LL and TH performed the RNA extraction
experiments and the quality control. YL, LL and HW performed the
bioinformatics analysis and prepared the figures. YZ, HZ, HY and XC
performed the statistical analysis. YL, YZ and TH drafted the
manuscript. HZ, XC and WS contributed substantially to the revision
of the manuscript. The authenticity of all the raw data has been
assessed by YL, WS and YW. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of The First Hospital of Lanzhou University (approval no.
LDYYLL2012001; Lanzhou, China). Written informed consent forms were
signed by the patients and their family members.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu Y, Zhang X, Chen L, Zhao Q and Xia X:
Cancer incidence and mortality in Gansu province, 2012. Chin J
Cancer Res. 28:301–310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Zhang S, Zeng H, Zuo T,
Jia M, Xia C, Zou X and He J: Report of cancer incidence and
mortality in China, 2012. China Cancer. 25:1–8. 2016.
|
|
4
|
Guo Q, Liu X, Lu L, Yuan H, Wang Y, Chen
Z, Ji R and Zhou Y: Comprehensive evaluation of clinical efficacy
and safety of celecoxib combined with chemotherapy in management of
gastric cancer. Medicine (Baltimore). 96:e88572017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ji R: Gastric cancer screening and related
risk factors analysis in the Wuwei Natrual population cohort.
Lanzhou University; 2018
|
|
6
|
Pasechnikov V, Chukov S, Fedorov E,
Kikuste I and Leja M: Gastric cancer: Prevention, screening and
early diagnosis. World J Gastroenterol. 20:13842–13862. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Everett SM and Axon AT: Early gastric
cancer in Europe. Gut. 41:142–150. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki H, Oda I, Abe S, Sekiguchi M, Mori
G, Nonaka S, Yoshinaga S and Saito Y: High rate of 5-year survival
among patients with early gastric cancer undergoing curative
endoscopic submucosal dissection. Gastric Cancer. 19:198–205. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gong W, Luo S, Hu R, Wang H, Pan J, Fei F,
Wu H and Yu M: Survival analysis of gastric cancer patients during
2005–2010 in Zhejiang Province, China. Zhonghua Zhong Liu Za Zhi.
36:636–639. 2014.(In Chinese). PubMed/NCBI
|
|
11
|
Zheng L, Wu C, Xi P, Zhu M, Zhang L, Chen
S, Li X, Gu J and Zheng Y: The survival and the long-term trends of
patients with gastric cancer in Shanghai, China. BMC Cancer.
14:3002014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hamashima C, Shabana M, Okada K, Okamoto M
and Osaki Y: Mortality reduction from gastric cancer by endoscopic
and radiographic screening. Cancer Sci. 106:1744–1749. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
So JBY, Kapoor R, Zhu F, Koh C, Zhou L,
Zou R, Tang YC, Goo PCK, Rha SY, Chung HC, et al: Development and
validation of a serum microRNA biomarker panel for detecting
gastric cancer in a high-risk population. Gut. 70:829–837. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han J, Lee Y, Yeom KH, Kim YK, Jin H and
Kim VN: The Drosha-DGCR8 complex in primary microRNA processing.
Genes Dev. 18:3016–3027. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng GD, Xu ZY, Hu C, Lv H, Xie XH, Huang
T, Zhang YQ, Chen GP, Fu YF and Cheng XD: Exosomal miR-590-5p in
serum as a biomarker for the diagnosis and prognosis of gastric
cancer front. Mol Biosci. 8:6365662021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang
J, Yang X and Liu Z: Exosomal miR-1246 in serum as a potential
biomarker for early diagnosis of gastric cancer. Int J Clin Oncol.
25:89–99. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: miR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shin VY and Chu KM: MiRNA as potential
biomarkers and therapeutic targets for gastric cancer. World J
Gastroenterol. 20:10432–10439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu M, Hu C, Wu F, Shu L, Pan Y, Liu X, Liu
P, Ma F, Deng C and Huang M: MiR-320a is associated with cisplatin
resistance in lung adenocarcinoma and its clinical value in
non-small cell lung cancer: A comprehensive analysis based on
microarray data. Lung Cancer. 147:193–197. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Amin MB, Edge SB, Greene FI, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC cancer staging manual. Springer-Verlag New
York, the United States: 2016
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Han X, Wan Y, Zhang S, Zhao Y, Fan
R, Cui Q and Zhou Y: TAM 2.0: Tool for MicroRNA set analysis.
Nucleic Acids Res. 46:W180–W185. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gaudet P and Dessimoz C: Gene ontology:
Pitfalls, biases, and remedies. Methods Mol Biol. 1446:189–205.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wickham H: ggplot2: Elegant Graphics for
Data Analysis. Springer-Verlag; New York, NY: 2016
|
|
32
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48:D127–D131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Therneau TM and Grambsch PM: Modeling
Survival Data: Extending the Cox Model. Springer-Verlag; New York,
NY: 2000, View Article : Google Scholar
|
|
37
|
Brusselaers N, Maret-Ouda J, Konings P,
El-Serag HB and Lagergren J: Menopausal hormone therapy and the
risk of esophageal and gastric cancer. Int J Cancer. 140:1693–1699.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu M, Zhu J, Ling Y, Shi W, Zhang C and Wu
H: The lower expression of gonadotropin-releasing hormone receptor
associated with poor prognosis in gastric cancer. Int J Clin Exp
Med. 8:13365–13370. 2015.PubMed/NCBI
|
|
39
|
Yang S, Liu W, Wen J, Zhu M and Xu S:
Corticotropin releasing hormone is correlated with tumorigenesis of
gastric cancer. Cancer Invest. 31:167–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Frycz BA, Murawa D, Borejsza-Wysocki M,
Wichtowski M, Spychała A, Marciniak R, Murawa P, Drews M and
Jagodziński PP: mRNA expression of steroidogenic enzymes, steroid
hormone receptors and their coregulators in gastric cancer. Oncol
Lett. 13:3369–3378. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gan J, Ke X, Jiang J, Dong H, Yao Z, Lin
Y, Lin W, Wu X, Yan S, Zhuang Y, et al: Growth hormone-releasing
hormone receptor antagonists inhibit human gastric cancer through
downregulation of PAK1-STAT3/NF-κB signaling. Proc Natl Acad Sci
USA. 113:14745–14750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yamada T, Yoshikawa T, Taguri M, Hayashi
T, Aoyama T, Sue-Ling HM, Bonam K, Hayden JD and Grabsch HI: The
survival difference between gastric cancer patients from the UK and
Japan remains after weighted propensity score analysis considering
all background factors. Gastric Cancer. 19:479–489. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kerckhoffs KGP, Liu DHW, Saragoni L, van
der Post RS, Langer R, Bencivenga M, Iglesias M, Gallo G, Hewitt
LC, Fazzi GE, et al: Mucin expression in gastric- and
gastro-oesophageal signet-ring cell cancer: Results from a
comprehensive literature review and a large cohort study of
Caucasian and Asian gastric cancer. Gastric Cancer. 23:765–779.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang ZL, Li ZR, Li JS and Wang SR:
Calcium-sensing receptor antagonist NPS-2143 suppresses
proliferation and invasion of gastric cancer cells. Cancer Gene
Ther. 27:548–557. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang B, Wu J, Zhu MX, Sun X, Liu J, Xie R,
Dong TX, Xiao Y, Carethers JM, Yang S and Dong H: VPAC1 couples
with TRPV4 channel to promote calcium-dependent gastric cancer
progression via a novel autocrine mechanism. Oncogene.
38:3946–3961. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ge C, Li Q, Wang L and Xu X: The role of
axon guidance factor semaphorin 6B in the invasion and metastasis
of gastric cancer. J Int Med Res. 41:284–292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pan G, Zhang X, Ren J, Lu J, Li W, Fu H,
Zhang S and Li J: Semaphorin 5A, an axon guidance molecule,
enhances the invasion and metastasis of human gastric cancer
through activation of MMP9. Pathol Oncol Res. 19:11–18. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Park T, Lee YJ, Jeong SH, Choi SK, Jung
EJ, Ju YT, Jeong CY, Park M, Hah YS, Yoo J, et al: Overexpression
of neuron-specific enolase as a prognostic factor in patients with
gastric cancer. J Gastric Cancer. 17:228–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Eke I and Cordes N: Focal adhesion
signaling and therapy resistance in cancer. Semin Cancer Biol.
31:65–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Du M, Zheng R, Ma G, Chu H, Lu J, Li S,
Xin J, Tong N, Zhang G, Wang W, et al: Remote modulation of lncRNA
GCLET by risk variant at 16p13 underlying genetic susceptibility to
gastric cancer. Sci Adv. 6:eaay55252020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang F, Chen X, Li X, Chen J, Tang Y, Cai
Y, Wang Y, Chen Z, Li L, Li R and Deng Z: Long intergenic
non-protein coding RNA 1089 suppresses cell proliferation and
metastasis in gastric cancer by regulating
miRNA-27a-3p/epithelial-mesenchymal transition (EMT) axis. Cancer
Manag Res. 12:5587–5596. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Geng G, Liu X, Xu A, Lu Z, Chen K, He J,
Qi D and Yuan X: Low abundance of TFPI-2 by both promoter
methylation and miR-27a-3p regulation is linked with poor clinical
outcome in gastric cancer. J Gene Med. 22:e31662020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou L, Liang X, Zhang L, Yang L, Nagao N,
Wu H, Liu C, Lin S, Cai G and Liu J: MiR-27a-3p functions as an
oncogene in gastric cancer by targeting BTG2. Oncotarget.
7:51943–51954. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wei S, Li Q, Li Z, Wang L, Zhang L and Xu
Z: Correction: miR-424-5p promotes proliferation of gastric cancer
by targeting Smad3 through TGF-β signaling pathway. Oncotarget.
8:340182017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wei S, Li Q, Li Z, Wang L, Zhang L and Xu
Z: miR-424-5p promotes proliferation of gastric cancer by targeting
Smad3 through TGF-β signaling pathway. Oncotarget. 7:75185–75196.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liang S, He L, Zhao X, Miao Y, Gu Y, Guo
C, Xue Z, Dou W, Hu F, Wu K, et al: MicroRNA let-7f inhibits tumor
invasion and metastasis by targeting MYH9 in human gastric cancer.
PLoS One. 6:e184092011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Song C, Chen H, Wang T, Zhang W, Ru G and
Lang J: Expression profile analysis of microRNAs in prostate cancer
by next-generation sequencing. Prostate. 75:500–516. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Queiroz AL, Zhang B, Comstock DE, Hao Y,
Eriksson M, Hydbring P, Vakifahmetoglu-Norberg H and Norberg E:
miR-126-5p targets Malate Dehydrogenase 1 in non-small cell lung
carcinomas. Biochem Biophys Res Commun. 499:314–320. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang C, Zhou B, Liu M, Liu Y and Gao R:
miR-126-5p restoration promotes cell apoptosis in cervical cancer
by targeting Bcl2l2. Oncol Res. 25:463–470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Mi Y, Zhang D, Jiang W, Weng J, Zhou C,
Huang K, Tang H, Yu Y, Liu X, Cui W, et al: miR-181a-5p promotes
the progression of gastric cancer via RASSF6-mediated MAPK
signalling activation. Cancer Lett. 389:11–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu Z, Sun F, Hong Y, Liu Y, Fen M, Yin K,
Ge X, Wang F, Chen X and Guan W: MEG2 is regulated by miR-181a-5p
and functions as a tumour suppressor gene to suppress the
proliferation and migration of gastric cancer cells. Mol Cancer.
16:1332017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cui HW, Han WY, Hou LN, Yang L, Li X and
Su XL: miR-1915-3p inhibits Bcl-2 expression in the development of
gastric cancer. Biosci Rep. 39:BSR201823212019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Jin ML, Kim YW, Jin HL, Kang H, Lee EK,
Stallcup MR and Jeong KW: Aberrant expression of SETD1A promotes
survival and migration of estrogen receptor α-positive breast
cancer cells. Int J Cancer. 143:2871–2883. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xu K, Liang X, Cui D, Wu Y, Shi W and Liu
J: miR-1915 inhibits Bcl-2 to modulate multidrug resistance by
increasing drug-sensitivity in human colorectal carcinoma cells.
Mol Carcinog. 52:70–78. 2013. View Article : Google Scholar : PubMed/NCBI
|