Introduction
Skin cancer arises from the uncontrolled growth of
keratinocytes and primarily develops upon exposure to ultraviolet
radiation from tanning beds or sunlight (1). It can be classified as basal cell
carcinoma, melanoma or cutaneous squamous cell carcinoma (CSCC)
(2). Epidemiological studies
indicate that CSCC accounts for 20% of all skin cancers and is
associated with a continuously increasing incidence worldwide,
posing a threat to public health (3). CSCC is a common type of malignant skin
tumor derived from epidermal malpighian cells (4). Furthermore, the metastasis, invasion
and local recurrence of CSCC are still the major reasons for
treatment failure in most patients (5).
Transmembrane protein 40 (TMEM40) is a multi-pass
membrane protein consisting of 233 amino acids, with two isoforms
(6–9). Its coding gene is located on chromosome
3p25.2 in humans. TMEM40 plays a role in collagen-induced arthritis
(10–12). Notably, in bladder cancer and tongue
squamous cell carcinoma, TMEM40 expression levels are significantly
associated with clinical stage, histological grade, pathological
grade and primary tumor (pT) status (13,14).
However, the specific function of TMEM40 in CSCC, especially the
underlying molecular mechanisms by which TMEM40 performs its
functions and modulates the malignant behavior of CSCC cells,
requires further study. Several recent studies have focused on
CSCC, with the aim of reducing mortality rates; however, the
overall pathophysiological and molecular mechanisms underlying the
role of TMEM40 in CSCC remain largely unknown (15,16).
In the present study, to evaluate the role of TMEM40
in CSCC development, the expression levels of TMEM40 were examined
in CSCC tissue compared with matched adjacent normal tissue
samples. The potential associations between TMEM40 levels and
clinicopathological features were also determined. Furthermore, the
potential role of TMEM40 in the proliferation, migration and
invasion of CSCC cells was also evaluated in vitro. The
results suggested that TMEM40 serves an important role in CSCC cell
proliferation and migration and may represent a potential
therapeutic target in patients with CSCC.
Materials and methods
Tissue sample collection
A total of 40 participants (17 females and 23 males)
diagnosed with CSCC were included in the present study. The mean
(±SD) age was 61.9 (±11.2) years, ranging between 33 and 87 years.
The inclusion criteria were: i) Diagnosis as CSCC (17); ii) initial diagnosis and no previous
treatment; and iii) availability of detailed medical records and
follow-up visit. Exclusion criteria were non-cutaneous SCC, CSCC
that had been previously treated, cutaneous carcinoma in
situ, unavailability of detailed medical records and follow-up
visit. Paraffin-embedded tissues were obtained from patients with
CSCC. A total of 40 CSCC tissue samples and 40 matched adjacent
normal cutaneous tissue samples (10 mm from cancer) were collected
from preoperative treatment-naïve patients with CSCC who had
undergone surgical intervention between May 2018 and June 2019 at
Zhujiang Hospital of Southern Medical University (Guangzhou,
China). All tissue samples were obtained with written informed
consent from the patients involved in this research project. The
present study was approved by the Medical Ethics Committee of
Zhujiang Hospital of Southern Medical University (approval no.
2020-KY-059-01).
Tissue microarray (TMA) construction
and immunohistochemistry
According to standard methods (10,14,18), a
TMA was constructed using 40 CSCC paraffin-embedded tissues and 40
normal cutaneous paraffin-embedded tissue samples. The tissues were
fixed with 4% paraformaldehyde for 12 h at 4°C. The slice thickness
was 6 µm and sections were stained by immunohistochemistry. The
sections were deparaffinized in xylene and hydrated using gradient
ethanol solutions. The sections were then blocked in PBS containing
5% normal goat serum (cat. no. 16210072; Gibco; Thermo Fisher
Scientific, Inc.) for 10 min at room temperature and subsequently
incubated with TMEM40-specific primary antibody (1:100; cat. no.
sc-393601; Santa Cruz Biotechnology, Inc.) overnight at 4°C. After
being washed three times in 1X PBS, the sections were incubated
with horseradish peroxidase-conjugated secondary antibody (1:1,000;
cat. no. sc-2357; Santa Cruz Biotechnology, Inc.) for 1 h at room
temperature. After additional 1X PBS washing, the sections were
visualized using a DAB Horseradish Peroxidase Color Development kit
(cat. no. P0202; Beyotime Institute of Biotechnology) and
counterstained with hematoxylin for 2 min at room temperature.
TMEM40 expression was assessed semi-quantitatively
by scoring the percentage positive cells and staining intensity by
two observers under a light microscope (magnifications, ×40 and
×100; Nikon Eclipse TiS; Nikon Corporation). The extent of TMEM40
immunoreactivity was scored as: i) 0, for <5% positive cells;
ii) 1, for 5–25% positive cells; iii) 2, for 26–50% positive cells;
iv) 3, 51–75% positive cells; and v) 4, for 76–100% positive cells.
The intensity of TMEM40 was scored as: i) 0, no staining; ii) 1,
light yellow; iii) 2, brown; and iv) 3, dark brown. The overall
staining score was obtained by multiplying the immunoreactivity
score by the intensity score to obtain four grades: i) Negative,
for an overall score of 0; ii) weakly positive, for an overall
score of 1–4; iii) moderately positive, for an overall score of
5–8; and iv) strongly positive for an overall score of 9–12
(10).
Cell culture
The CSCC cell lines (A431 and SCL-1) were purchased
from the China Center for Type Culture Collection. A431 and SCL1
cells were cultured in high-glucose DMEM (Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin/streptomycin (Biowest) in an incubator at
37°C with a humidified atmosphere of 5% CO2 and 95%
air.
TMEM40-small interfering RNA (siRNA)
transfection
The siRNA targeting TMEM40 and negative control
(NC)-siRNA were supplied by Invitrogen (Thermo Fisher Scientific,
Inc.), and the sequence was verified by sequencing. A total of 5
nmol siRNA and NC were transfected into CSCC cells with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 20 min at room temperature according to the
manufacturer's instructions. The control group was treated with
PBS. The transfection efficiency was assessed by flow cytometry
(MoFlo XDP; Beckman Coulter, Inc.). Briefly, after 48 h of
transfection with fluorescently labeled with 6-carboxy-fluorescein
(FAM) TMEM40-siRNA, 1×105 cells were collected and the
percentage of FAM-positive cells was obtained using a flow
cytometer, representing the transfection efficiency, and was
analyzed with FlowJo software (version 7.6; FlowJo LLC). The
sequence of TMEM40-siRNA was 5′-GUGGACGCCUCUCAGUUAA-3′, and the
non-targeting NC-siRNA sequence was 5′-TTCTCCGAACGTGTCACGT-3′.
Western blotting
To detect TMEM40 protein expression, tissue samples
(8 paired normal and tumor tissues) were lysed, and proteins were
extracted using radioimmunoprecipitation assay buffer containing 1
mM PMSF (Beyotime Institute of Biotechnology) according to standard
protocols. The protein concentrations in the supernatants were
determined using a BCA Protein Assay kit (cat. no. P0006; Beyotime
Institute of Biotechnology). Subsequently, ~40 µg of each total
protein sample were separated by SDS-PAGE on 10% gels and
transferred to nitrocellulose membranes, which were then blocked
with 5% fat-free milk for 2 h at room temperature. The membranes
were then washed with TBST and incubated overnight at 4°C with a
mouse anti-TMEM40 primary antibody (1:500; cat. no. sc-393601;
Santa Cruz Biotechnology, Inc.) and mouse anti-GAPDH primary
antibody (1:1,000; cat. no. 60004-1-Ig; ProteinTech Group, Inc.).
After washing, membranes were incubated with appropriate
HRP-conjugated secondary antibodies (1:5,000; cat. no. sc-2357;
Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. The
signals on the membrane were detected using an enhanced
chemiluminescent reagent kit (cat. no. 36222ES60; Shanghai Yeasen
Biotechnology Co., Ltd.).
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
To determine TMEM40 mRNA expression levels, total
RNA was extracted from clinical samples or from A431 and SCL-1
cells 48 h after transfection using a PureLink RNA Mini kit (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Total RNA was quantified using a NanoDrop™ 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.). A TaqMan MicroRNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) was used to reverse transcribe cDNA from total RNA according
to the manufacturer's instructions. SYBR-Green PCR kit (Takara
Biotechnology Co., Ltd.) and an ABI 7500 realtime PCR amplifier
(Applied Biosystems; Thermo Fisher Scientific, Inc.) were used for
qPCR. The thermocycling conditions were as follows: 95°C for 10
min, followed by 40 cycles at 95°C for 15 sec and 60°C for 34 sec.
GAPDH was used for normalization. The relative gene expression data
were detected by qPCR and analysed with the 2−ΔΔCq
method (19). The RT-qPCR primers
and their sequences are listed in Table
I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer name | Primer sequence
(5′-3′) |
|---|
| TMEM40-F |
GCGGTAGGGGTGTACGGT |
| TMEM40-R |
CCGGACACGCTGAACTTGT |
| GAPDH-F |
CAGCCTCAAGATCATCAGCA |
| GAPDH-R |
TGTGGTCATGAGTCCTTCCA |
Cell proliferation assay
Cell proliferation was measured using Cell Counting
Kit-8 (Nanjing KeyGen Biotech Co., Ltd.). Briefly, a total of
1.5×103 A431 and SCL-1 cells were plated into 96-well
microplates (Corning, Inc.). After transfection, cells were
incubated with CCK-8 reagent (Beyotime Institute of Biotechnology)
for 2 h at 37°C, and the absorbance was measured at 450 nm using a
microplate reader (Infinite® M200; Tecan Group, Ltd.)
according to the manufacturer's instructions.
Wound healing assay
A wound healing assay was used to assess cell
migration. A total of 1×104 A431 and SCL-1 cells were
seeded in 6-well plates, transfected for 48 h and cultured with
serum-free medium to 85–90% confluence. The cell layers were
scratched using a 10 µl RNase-free pipette tip to create wound
gaps. The wound gaps were imaged at two different time points (0
and 48 h) using an light microscope (magnification, ×40) and
analyzed by measuring the distance that the cells migrated in three
different areas of each wound.
Transwell migration and invasion
assays
The migration and invasion (inserts were pre-coated
with Matrigel for 4 h at 37°C) of A431 and SCL-1 cells were
examined using Transwell assays with modified 24-well Boyden
chambers (8-µm pore size; Corning, Inc.). A total of
3×104 cells in 300 µl of DMEM were seeded into the upper
chamber of the Transwell inserts 24 h after TMEM40-siRNA
transfection, and 700 µl of DMEM supplemented with 20% FBS was
added to the lower chambers. Cells were incubated at 37°C in 5%
CO2 for 36 h after plating. The cells were then fixed
with 100% methanol for 5 min at room temperature and stained with
0.5% crystal violet at room temperature for 15 min. The numbers of
migrating or invading cells that passed through the membrane were
counted in three randomly selected fields under a light microscope
(magnification, ×40), and the average value was calculated with
ImageJ software (version 1.8; National Institutes of Health).
Cell cycle analysis
A431 and SCL-1 cells were fixed overnight in 70%
ethanol at −20°C after digestion with 0.05% trypsin for 3 min at
room temperature. (Nanjing KeyGen Biotech Co., Ltd.). The fixed
cells were stained with 450 µl propidium iodide (PI; Nanjing KeyGen
Biotech Co., Ltd.) and 50 µl RNase A (KeyGen Biotech, Nanjing) at
room temperature in the dark for 30 min. Cell cycle progression was
examined using a flow cytometer (MoFlo XDP; Beckman Coulter, Inc.)
and analyzed using FlowJo software (version 7.6; FlowJo LLC).
Apoptosis assessment
Flow cytometry (MoFlo XDP; Beckman Coulter, Inc.)
was used to evaluate the apoptosis rate in transfected cells. SCL1
and A431 cells (6×105) were plated in 60-mm dishes and
incubated overnight. The cells were then transiently transfected
with TMEM40-siRNA or control vector for 48 h. The cells were then
collected by trypsinization, washed with cold PBS and stained using
a FITC Annexin V/Dead Cell Apoptosis kit (Nanjing KeyGen Biotech
Co., Ltd.) at room temperature for 30 min. The apoptosis rates of
the CSCC cells were analyzed using FlowJo software (version 7.6;
FlowJo LLC).
Statistical analysis
Statistical analysis was performed using the SPSS
(version 20.0; IBM Corp.) or GraphPad Prism software (version 5.0;
GraphPad Software, Inc.). The differences between the CSCC tissue
and paired normal tissue groups were evaluated using a paired
Student's t-test. One-way ANOVA followed by Tukey's post hoc test
were used for multiple comparisons. Differences between groups were
tested using the χ2 or Fisher's exact test (n<5) test
for categorical variables. The results are presented as the mean ±
SD. P<0.05 was considered to indicate a statistically
significant difference.
Results
TMEM40 expression is upregulated in
CSCC tissue samples
To investigate the role of TMEM40 in CSCC, the
expression of TMEM40 was evaluated in patient samples using
immunohistochemistry. TMEM40 expression was significantly
upregulated in CSCC tissue compared with adjacent normal skin
tissue samples (Fig. 1A-H; Table II). As shown in Table III, TMEM40 protein expression was
significantly associated with tumor size (P=0.007). However, there
was no significant association between TMEM40 expression and age,
pathological grade, lymph node metastasis or pT status (all
P>0.05). To determine the expression of TMEM40 in CSCC at the
mRNA and protein levels, western blot and RT-qPCR analysis were
performed in CSCC tissue and paired adjacent normal skin tissue
samples (n=8; Fig. 1I and J). All of
these results suggested that TMEM40 expression was upregulated in
CSCC tissue compared with normal adjacent tissue.
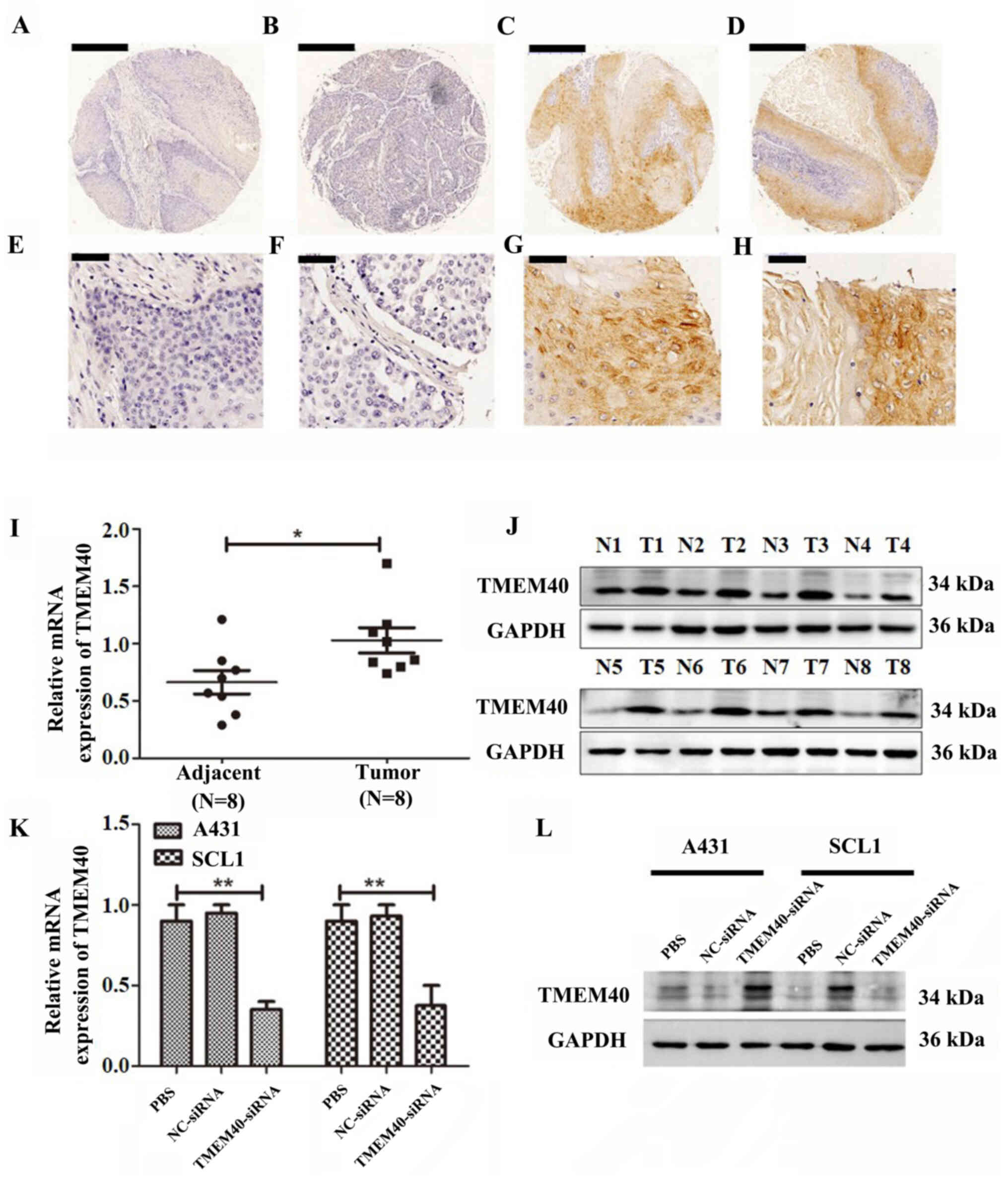 | Figure 1.TMEM40 was expressed in CSCC tissues
and cell lines. (A-D) Immunohistochemical of TMEM40 protein
expression was significantly increased in CSCC compared with normal
cutaneous squamous cells. Magnification, ×5. Scale bar, 500 µM.
(E-H) High-magnification immunohistochemical staining of TMEM40.
Magnification, ×40. Scale bar, 50 µm. TMEM40 (I) mRNA and (J)
protein expression was significantly increased in CSCC compared
with normal cervical tissue. n=8 in each group. TMEM40-siRNA
significantly downregulates the expression of TMEM40 in A431 and
SCL1 cells at the (K) mRNA and (L) protein levels. *P<0.05;
**P<0.01. CSCC, cutaneous squamous cell carcinoma; NC, negative
control; N, normal tissue; T, tumor tissue; TMEM40, transmembrane
protein 40; siRNA, small interfering RNA. |
 | Table II.TMEM40 expression in paired CSCC and
normal skin tissue samples (n=40). |
Table II.
TMEM40 expression in paired CSCC and
normal skin tissue samples (n=40).
| Tissue type | Negative TMEM40
expression, n (%) | Positive TMEM40
expression, n (%) |
P-valuea |
|---|
| CSCC | 29 (72.5) | 11 (27.5) | <0.001 |
| Normal skin | 9 (22.5) | 31 (77.5) |
|
 | Table III.Association between TMEM40 expression
and clinicopathological features in squamous cell carcinoma
tissues. |
Table III.
Association between TMEM40 expression
and clinicopathological features in squamous cell carcinoma
tissues.
|
|
| TMEM40 protein
expression |
|
|---|
| Clinicopathological
features | Total patients,
n | Negative, n
(%) | Positive, n
(%) |
P-valuea |
|---|
| All patients | 40 | 9 (22.5) | 31 (77.5) | Not applicable |
| Age, years |
|
|
| 0.456 |
|
≤48 | 23 | 4 (17.4) | 19 (82.6) |
|
|
>48 | 17 | 5 (29.4) | 12 (70.6) |
|
| Lymph node
metastasis |
|
|
| 0.061 |
| No | 24 | 8 (33.3) | 16 (66.7) |
|
|
Yes | 16 | 1 (6.3) | 15 (93.7) |
|
| Tumor size, cm |
|
|
| 0.007 |
|
<3 | 11 | 6 (54.5) | 5 (45.5) |
|
| ≥3 | 29 | 3 (10.3) | 26 (89.7) |
|
| Pathological
grade |
|
|
| 0.265 |
| I | 19 | 6 (31.6) | 13 (68.4) |
|
|
II–III | 21 | 3 (14.2) | 18 (85.8) |
|
| pT status |
|
|
| 0.120 |
| T1 | 13 | 5 (38.5) | 8 (61.5) |
|
|
T2-T3 | 27 | 4 (14.8) | 23 (85.2) |
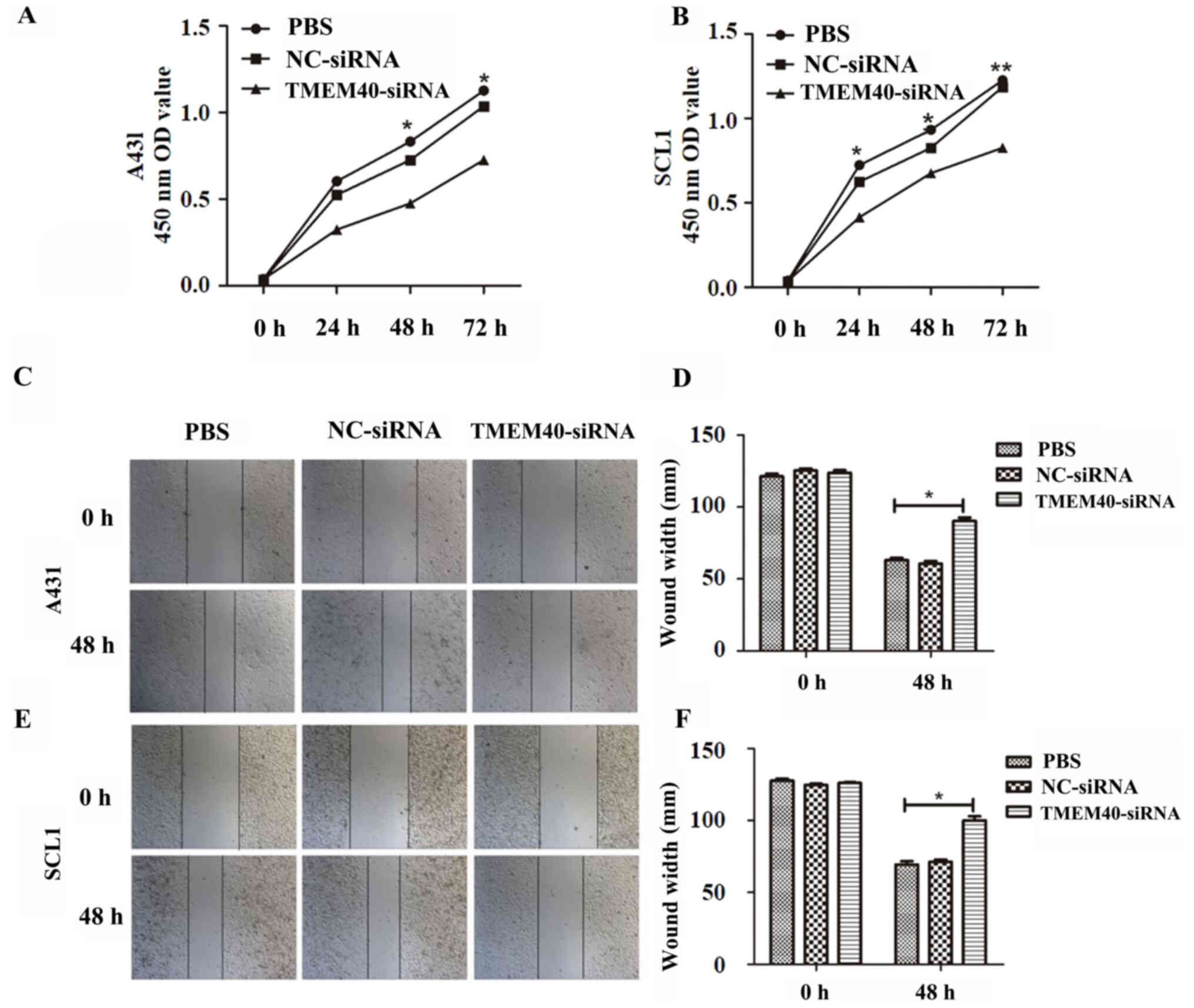
Downregulation of TMEM40 inhibits CSCC
cell proliferation
TMEM40-siRNA transfection was used to silence the
expression of the TMEM40 gene in A431 and SCL1 cells. TMEM40-siRNA
group exhibited significantly decreased TMEM40 expression compared
with the other groups (Fig. 1K and
L). TMEM40-siRNA and NC transfection efficiency were ~77.9 and
73.8% in A431 cells, respectively, and 74.1 and 75.7% in SCL1
cells, respectively (Fig. S1).
These transfected cells were used in the following experiments. A
CCK-8 assay was performed to study the effect of TMEM40 expression
on CSCC cell growth. The growth curves demonstrated that cell
proliferation was significantly decreased in the TMEM40-siRNA group
compared with in the NC-siRNA and PBS groups in both A431 (at 48
and 72 h) and SCL1 cells (at 24, 48 and 72 h) (Fig. 2A and B).
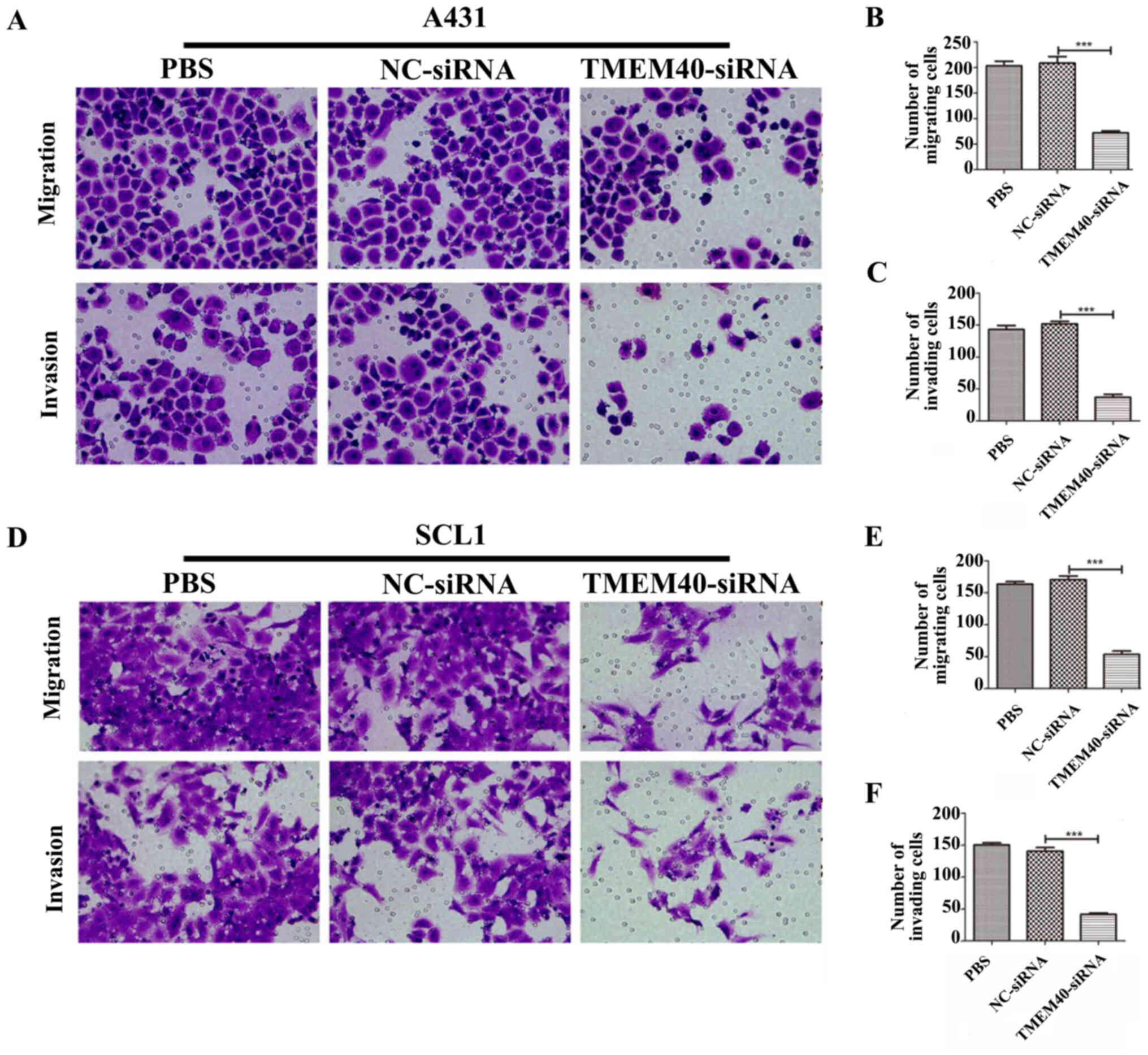
TMEM40 silencing inhibits the
migration and invasion of CSCC cells
Wound healing and Transwell assays were performed to
determine whether the TMEM40 gene could affect the migration and
invasion of CSCC cells. Wound healing assay shown in Fig. 2C and E suggested that
TMEM40-knockdown significantly slowed wound closure. The width of
the wound of A431 (Fig. 2D) and SCL1
(Fig. 2F) cells was significantly
higher in the TMEM40-silenced groups than in the other groups.
Subsequently, cell migration and invasion was also detected with
Transwell assays. As shown in Fig.
3, significantly fewer migrating and invading cells were
observed in the TMEM40-siRNA group compared with in the other
groups.
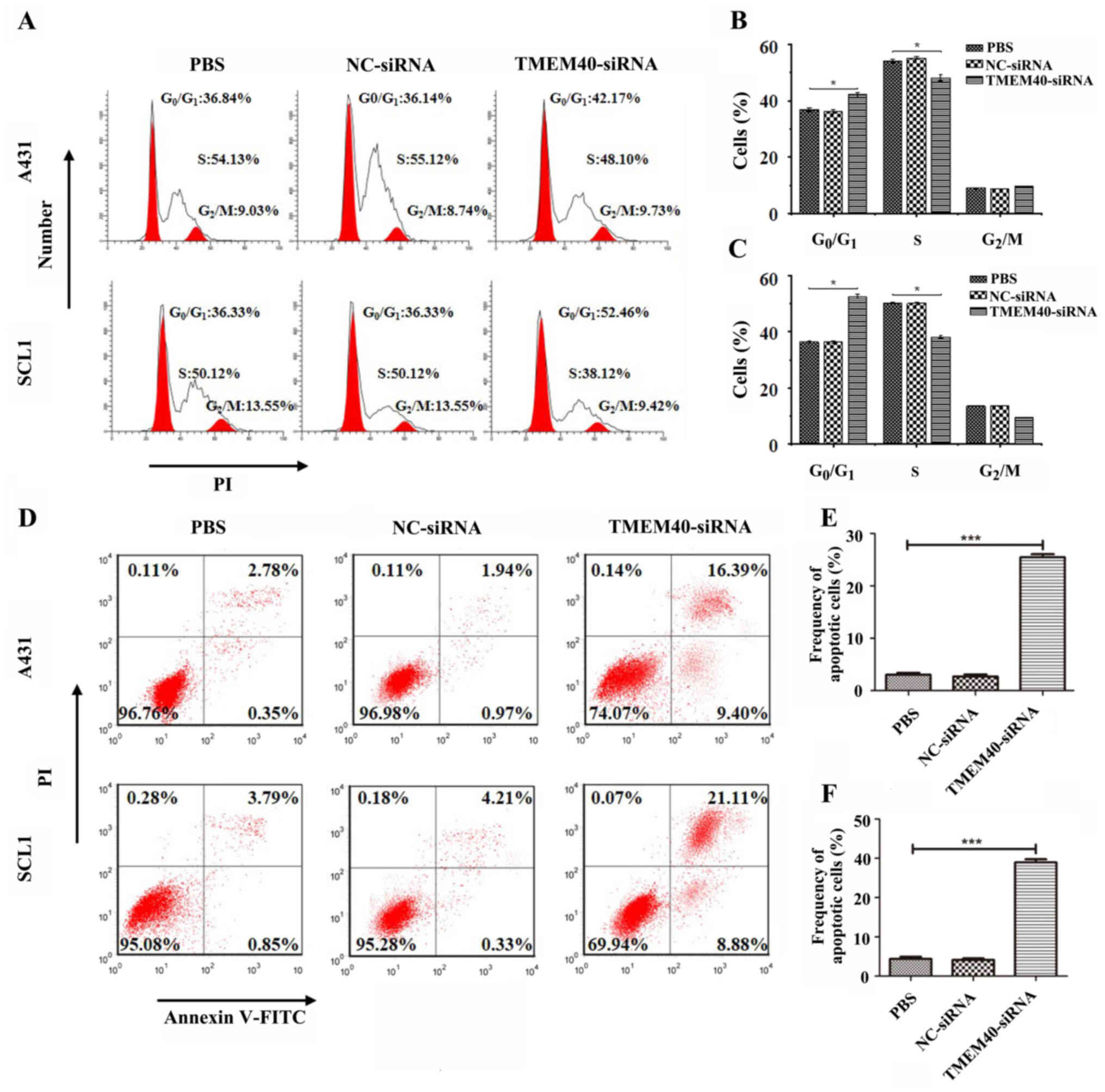
Silencing TMEM40 is associated with
cell cycle regulation and apoptosis
To determine the role of TMEM40 in cell growth,
apoptosis and cell cycle distribution were assessed in A431 and
SCL1 following TMEM40 knockdown. The results demonstrated that
TMEM40 silencing induced G0/G1-phase arrest
in A431 and SCL1 cells (Fig. 4A),
and the percentage of cells in G0/G1 phase in
the TMEM40-silenced group was higher than that in the other groups
(Fig. 4B and C). Moreover, there was
a significantly higher percentage of AnnexinV+
PI+ (late apoptotic) cells and AnnexinV+
PI− (early apoptotic) cells among TMEM40-silenced CSCC
cells than in the other groups (Fig.
4D-F), indicating that TMEM40- silencing significantly
increased apoptosis.
Discussion
CSCC is a major public health concern due to its
associated medical costs and high incidence (20). The known predisposing factors of CSCC
include ultraviolet radiation exposure, chronic immunosuppressed
state, inherited genetic conditions, ionizing radiation exposure,
human papillomavirus infection and chronic arsenic exposure
(21–25). Moreover, it is necessary to
differentiate benign lesions and reactive squamo-proliferative
lesions from CSCC (26,27) and to identify the high-risk features
associated with invasive tumor progression (28). Therefore, a clear understanding of
the molecular mechanism that induces the development and
progression of CSCC is essential for the development of diagnostic
and prognostic tools, as well as targeted therapies (29,30).
Previous studies have indicated that TMEM40 is
upregulated in bladder cancer and tongue squamous cell carcinoma,
compared with normal tissue, and that it is involved in cell
invasion and migration (6,10,13,21).
Therefore, TMEM40 may represent a potential biomarker for new
therapeutic strategies and diagnostic development. However, to the
best of our knowledge, the molecular mechanism underlying the
biological function of TMEM40 in CSCC has not been characterized
yet. In the current study, the expression of TMEM40 was evaluated
in two CSCC cell lines, in addition to tissue samples, both at the
protein and the mRNA levels. TMEM40 gene expression was
significantly upregulated in CSCC tissues, compared with adjacent
normal skin, and was associated with tumor size, but not with age
or sex. Immunohistochemical detection of TMEM40 expression in
tissue samples from patients with CSCC further supported these
results at the protein level. In order to assess the role of TMEM40
on CSCC cell phenotype and to evaluate possible biological
functions, TMEM40-siRNA was transfected into A431 and SCL1
cells.
Furthermore, TMEM40 knockdown inhibited the
proliferation, migration and invasion of A431 and SCL1 cells.
Invasion and migration are the most critical characteristics of
malignant tumors. It has been demonstrated that knockdown of TMEM40
expression in clear cell renal cell carcinoma, bladder cancer and
tongue squamous cell carcinoma results in significant inhibition of
cell proliferation, migration and invasion in vitro
(10,13). In the present study, a similar result
was also observed, and TMEM40 knockdown resulted in marked
reduction in migration and invasion of CSCC cells. These findings
provide evidence for the role of TMEM40 in CSCC cell proliferation
and motility. Matrix metalloproteinases (MMPs) can promote tumor
cell relocation by degrading certain components of the
extracellular matrix (31,32). It has been reported that four
collagenolytic MMPs, namely MMP-1 and MMP-2, MMP-9 and MMP-13, are
associated with CSCC invasion (32,33). It
is therefore possible that reduction in MMP production following
TMEM40 knockdown impairs implantation of CSCC cells, thus delaying
tumor cell growth. Moreover, TMEM40 knockdown may favor the
invasion and migration of CSCC cells via multiple downstream
MMP-mediated pathways. However, the mechanism underlying this
regulatory effect remains to be elucidated in future studies.
TMEM40 silencing inhibited the G1/S cell
cycle transition and induced apoptosis. Cell cycle analysis
following TMEM40 knockdown revealed significant
G0/G1 phase arrest. In addition,
downregulation of several genes coding for proteins regulating cell
cycle, including cyclin D1, cyclin E, CDK2, CDK4 and CDK6, is
associated with tumor progression (34). Cyclin D1 is a key protein in cell
cycle regulation that has been shown to be sufficient to drive cell
cycle progression (35–37). The present study demonstrated that
TMEM40 was involved in the regulation of the cell cycle in CSCC
cells and may therefore influence the expression levels of cell
cycle proteins.
Furthermore, the role of TMEM40 in the proliferation
and apoptosis A431 and SCL1 cells was also evaluated. Functional
experiments using CSCC cells revealed that TMEM40 silencing
resulted in decreased cell proliferation and increased apoptosis.
Apoptosis is a complex process that involves several signaling
pathways. Accordingly, the findings of the current study warrant
further investigation of TMEM40-related signaling pathways for the
clinical treatment of CSCC.
In summary, the present study demonstrated that
TMEM40 was significantly upregulated in CSCC cells and tissue, and
associated with the pathogenic mechanism of CSCC. Thus, TMEM40 may
serve an important role in the development and progression of CSCC.
Taken together, the present findings provide insight into the
function of TMEM40 in CSCC, which may be used as a potential
therapeutic option for CSCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LY and LLG conceived and designed the study. LY and
JL performed the experiments, wrote the manuscript and revised it
critically for important intellectual content. XFZ coordinated the
research and analyzed the data. WLZ and DLL performed western blot
and immunohistochemistry analysis, and were involved in drafting
the manuscript. XWQ and TDZ performed the statistical analysis. LLG
and TDZ confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
All tissue samples were obtained with written
informed consent from the patients involved in this research
project. This study was approved by the Medical Ethics Committee of
Zhujiang Hospital of Southern Medical University (approval no.
2020-KY-059-01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CSCC
|
cutaneous squamous cell carcinoma
|
|
IHC
|
immunohistochemistry
|
|
TMEM40
|
transmembrane protein 40
|
|
CCK-8
|
Cell Counting Kit-8
|
|
NC
|
negative control
|
|
siRNA
|
small interfering RNA
|
References
|
1
|
Kivisaari A and Kähäri VM: Squamous cell
carcinoma of the skin: Emerging need for novel biomarkers. World J
Clin Oncol. 4:85–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Y, Huang C and Yang X: Characterization
of TCF4-mediated oncogenic role in cutaneous squamous cell
carcinoma. Int J Clin Exp Pathol. 12:3583–3594. 2019.PubMed/NCBI
|
|
3
|
Zhang L, Qin H, Wu Z, Chen W and Zhang G:
Pathogenic genes related to the progression of actinic keratoses to
cutaneous squamous cell carcinoma. Int J Dermatol. 57:1208–1217.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Niu T, Tian Y, Wang G, Guo G, Tong Y and
Shi Y: Inhibition of ROS-NF-κB-dependent autophagy enhances
Hypocrellin A united LED red light-induced apoptosis in squamous
carcinoma A431 cells. Cell Signal. 69:1095502020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burton KA, Ashack KA and Khachemoune A:
Cutaneous squamous cell carcinoma: A review of high-risk and
metastatic disease. Am J Clin Dermatol. 17:491–508. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Q, Huang D, Zhang Z, Feng Y, Fu M,
Wei M, Zhou J, Huang Y, Liu S and Shi R: High expression of TMEM40
contributes to progressive features of tongue squamous cell
carcinoma. Oncol Rep. 41:154–164. 2019.PubMed/NCBI
|
|
7
|
Yue Y, Grossmann B, Ferguson-Smith M, Yang
F and Haaf T: Comparative cytogenetics of human chromosome 3q21.3
reveals a hot spot for ectopic recombination in hominoid evolution.
Genomics. 85:36–47. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yue Y, Grossmann B, Tsend-Ayush E,
Grutzner F, Ferguson-Smith MA, Yang F and Haaf T: Genomic structure
and paralogous regions of the inversion breakpoint occurring
between human chromosome 3p12.3 and orangutan chromosome 2.
Cytogenet Genome Res. 108:98–105. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Müller S, Stanyon R, Finelli P,
Archidiacono N and Wienberg J: Molecular cytogenetic dissection of
human chromosomes 3 and 21 evolution. Proc Natl Acad Sci USA.
97:206–211. 2000. View Article : Google Scholar
|
|
10
|
Zhang QY, Fu MT, Zhang ZF, Feng YZ, Wei M,
Zhou JY and Shi R: Expression of TMEM40 in bladder cancer and its
correlation with clinicopathological parameters. Int J Clin Exp
Pathol. 10:8050–8057. 2017.PubMed/NCBI
|
|
11
|
Darai E, Kost-Alimova M, Kiss H, Kansoul
H, Klein G and Imreh S: Evolutionarily plastic regions at human
3p21.3 coincide with tumor breakpoints identified by the
‘elimination test’. Genomics. 86:1–12. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu X, Teng H, Marques A, Ashgari F and
Ibrahim SM: High resolution mapping of Cia3: A common arthritis
quantitative trait loci in different species. J Immunol.
182:3016–3023. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang ZF, Zhang HR, Zhang QY, Lai SY, Feng
YZ, Zhou Y, Zheng SR, Shi R and Zhou JY: High expression of TMEM40
is associated with the malignant behavior and tumorigenesis in
bladder cancer. J Transl Med. 16:92018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Farshchian M, Nissinen L, Siljamäki E,
Riihilä P, Piipponen M, Kivisaari A, Kallajoki M, Grénman R,
Peltonen J, Peltonen S, et al: Tumor cell-specific AIM2 regulates
growth and invasion of cutaneous squamous cell carcinoma.
Oncotarget. 8:45825–45836. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bottomley MJ, Thomson J, Harwood C and
Leigh I: The role of the immune system in cutaneous squamous cell
carcinoma. Int J Mol Sci. 20:20092019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin N, Zhou Y, Lian X and Tu Y:
MicroRNA-31 functions as an oncogenic microRNA in cutaneous
squamous cell carcinoma cells by targeting RhoTBT1. Oncol Lett.
13:1078–1082. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stratigos A, Garbe C, Lebbe C, Malvehy J,
del Marmol V, Pehamberger H, Peris K, Becker JC, Zalaudek I, Saiag
P, et al: Diagnosis and treatment of invasive squamous cell
carcinoma of the skin: European consensus-based interdisciplinary
guideline. Eur J Cancer. 51:1989–2007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Farshchian M, Nissinen L, Siljamäki E,
Riihilä P, Toriseva M, Kivisaari A, Ala-Aho R, Kallajoki M,
Veräjänkorva E, Honkanen HK, et al: EphB2 promotes progression of
cutaneous squamous cell carcinoma. J Invest Dermatol.
135:1882–1892. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
Relative Gene Expression Data Using Real-Time Quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou M, Liu W, Ma S, Cao H, Peng X, Guo L,
Zhou X, Zheng L, Guo L, Wan M, et al: A novel onco-miR-365 induces
cutaneous squamous cell carcinoma. Carcinogenesis. 34:1653–1659.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McGuire JF, Ge NN and Dyson S: Nonmelanoma
skin cancer of the head and neck I: Histopathology and clinical
behavior. Am J Otolaryngol. 30:121–133. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Martinez JC and Cook JL: High-risk
cutaneous squamous cell carcinoma without palpable lymphadenopathy:
Is there a therapeutic role for elective neck dissection? Dermatol
Surg. 33:410–420. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weinberg AS, Ogle CA and Shim EK:
Metastatic cutaneous squamous cell carcinoma: An update. Dermatol
Surg. 33:885–899. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Zhou C, Zhang C, Xie X, Zhou Z, Zhou
M, Chen L and Ding Z: MicroRNA-664 functions as an oncogene in
cutaneous squamous cell carcinomas (cSCC) via suppressing
interferon regulatory factor 2. J Dermatol Sci. 94:330–338. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trakatelli M, Ulrich C, Del MV, Euvrard S,
Stockfleth E and Abeni D: Epidemiology of nonmelanoma skin cancer
(NMSC) in Europe: Accurate and comparable data are needed for
effective public health monitoring and interventions. Br J
Dermatol. 156 (Suppl 3):S1–S7. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alam M and Ratner D: Cutaneous
squamous-cell carcinoma. N Engl J Med. 344:975–983. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng C, Zhang HL, Zeng A, Bai M and Wang
XJ: Tumor-suppressive microRNA-216b binds to TPX2, activating the
p53 signaling in human cutaneous squamous cell carcinoma. Mol Ther
Nucleic Acids. 20:186–195. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tian K, Liu W, Zhang J, Fan X, Liu J, Zhao
N, Yao C and Miao G: MicroRNA-125b exerts antitumor functions in
cutaneous squamous cell carcinoma by targeting the STAT3 pathway.
Cell Mol Biol Lett. 25:122020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dlugosz A, Merlino G and Yuspa SH:
Progress in cutaneous cancer research. J Investig Dermatol Symp
Proc. 7:17–26. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mavropoulos JC, Aldabagh B and Arron ST:
Prospects for personalized targeted therapies for cutaneous
squamous cell carcinoma. Semin Cutan Med Surg. 33:72–75. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu D, Zhou G, Shi H, Chen B, Sun X and
Zhang X: Downregulation of transmembrane protein 40 by miR-138-5p
suppresses cell proliferation and mobility in clear cell renal cell
carcinoma. Iran J Biotechnol. 18:e22702020.PubMed/NCBI
|
|
32
|
Ala-aho R, Ahonen M, George SJ, Heikkilä
J, Grénman R, Kallajoki M and Kähäri VM: Targeted inhibition of
human collagenase-3 (MMP-13) expression inhibits squamous cell
carcinoma growth in vivo. Oncogene. 23:5111–5123. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Johansson N, Airola K, Grénman R,
Kariniemi AL, Saarialho-Kere U and Kähäri VM: Expression of
collagenase-3 (matrix metalloproteinase-13) in squamous cell
carcinomas of the head and neck. Am J Pathol. 151:499–508.
1997.PubMed/NCBI
|
|
34
|
Le XF, Bedrosian I, Mao W, Murray M, Lu Z,
Keyomarsi K, Lee MH, Zhao J and Bast RC Jr: Anti-HER2 antibody
trastuzumab inhibits CDK2-mediated NPAT and histone H4 expression
via the PI3K pathway. Cell Cycle. 5:1654–1661. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Ding R, Han Z, Ma Z and Wang Y:
Targeting of cell cycle and let-7a/STAT3 pathway by niclosamide
inhibits proliferation, migration and invasion in oral squamous
cell carcinoma cells. Biomed Pharmacother. 96:434–442. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang XQ, Feng H, Li ZY, Guo J and Li M:
Aspirin is involved in the cell cycle arrest, apoptosis, cell
migration, and invasion of oral squamous cell carcinoma. Int J Mol
Sci. 19:20292018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Santamaría D, Barrière C, Cerqueira A,
Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M and
Barbacid M: Cdk1 is sufficient to drive the mammalian cell cycle.
Nature. 448:811–815. 2007. View Article : Google Scholar
|