Introduction
Lung cancer is one of the most frequently diagnosed
types of cancer in both men and women worldwide; ~1.8 million new
people were diagnosed in 2012, with 1.6 million fatalities
(1). It is estimated that the United
States alone had >230,000 new cases in 2018 (1). Lung cancer is a very heterogeneous
disease at a cellular and histological level. It is well known that
80–85% of lung cancer cases are classified as non-small cell lung
cancer (NSCLC), which includes several subtypes, including
undifferentiated carcinoma or large cell carcinoma, squamous cell
carcinoma, adenocarcinoma and other subtypes (2). The average 5-year overall survival rate
for patients with lung cancer is 18% due to the advanced stages of
disease diagnosed and its known heterogeneity, including tumor
microenvironmental factors, complex molecular characteristics and
the limited therapy options (3). New
therapeutic strategies are emerging with the introduction of
several lines of tyrosine kinase inhibitors in patients with ALK,
EGFR, NTRK and ROS1 mutations (4).
In addition, immune checkpoint inhibitors have markedly improved
the NSCLC treatment (5). However,
there are increasing problems, such as optimal patient selection,
optimal duration of treatment and clinical implications of combined
treatments, that limit its discriminatory potency. Therefore, it is
important to identify the molecular mechanisms responsible for the
pathogenesis of NSCLC.
Exosomal RNA is involved in biological processes in
tumor cells, immune cells and stromal cells (6). A diverse collection of the exosomal RNA
species has been identified by RNA sequencing analysis, and
microRNAs (miRNAs/miRs) are the most abundant in extracellular
vesicles (7). miRNAs, a group of
small non-coding RNAs (18–22 bp), negatively regulate the
expression of target genes by reducing the stability of the target
mRNA or post-transcriptionally inhibiting the translation process
(8). Exosomal miRNAs serve important
roles in proliferation, invasion, metastasis and drug resistance by
regulating gene expression in target cells (9,10). In
addition, numerous studies have revealed that downregulation or
upregulation of miRNAs is associated with the initiation and
development of cancer, and that miRNAs can act as either an
oncogene or a tumor suppressor gene (11,12). For
example, Han et al (13) has
reported that exosomal miR-26b-5p decreases activating
transcription factor 2 expression to enhance the radiosensitivity
of X-radiation in lung adenocarcinoma cells. Wang et al
(14) has reported that miR-23b
inhibits cell proliferation, invasion and migration by
downregulating RUNX family transcription factor 2 through the
Wnt/β-catenin signaling pathway in NSCLC. miR-433 expression has
been shown to be downregulated in glioma (15), cervical cancer (16), colorectal cancer (17), breast cancer (18), esophageal squamous cell carcinoma
(ESCC) (19) and oral squamous cell
carcinoma (20), among others.
Previously, Liu et al (21)
and Li et al (22) have
reported that miR-433 decreases cell proliferation and invasion, as
well as tumor progression in NSCLC (21,22).
However, the characteristic molecular mechanism and the underlying
role of miR-433 require further investigation, especially in the
tumor microenvironment of NSCLC.
In the present study, exosomes were isolated from
the plasma of patients with NSCLC after the combined chemotherapy
of gemcitabine and cisplatin treatment, and miR-433 expression was
analyzed in the plasma of patients with resistant NSCLC, patients
with sensitive NSCLC and in normal serum. However, the exact
function and underlying mechanism of miR-433 in NSCLC remains
unclear. Therefore, the present study was performed to investigate
the effects of miR-433 on NSCLC cell proliferation, apoptosis, DNA
damage and tumor growth in vivo.
Materials and methods
Collection of NSCLC tissues
A total of 50 NSCLC tissues and adjacent normal lung
tissues (2 cm away from the tumor edge) from patients (n=50; median
age, 59 years; age range, 29–83 years; 16 females and 34 males)
were collected between January 2018 and January 2020. All patients
were pathologically diagnosed and underwent therapeutic surgery at
the Tianjin Medical University Cancer Institute and Hospital
(Tianjin, China). Serum samples (4 ml) were collected from patients
diagnosed with NSCLC (n=33; 18 chemotherapy-resistant patients and
15 chemotherapy-sensitive patients) and healthy individuals (n=19;
median age, 46 years; age range, 25–19 years; 9 females and 10
males). The current study was conducted under the International
Ethical Guidelines for Biomedical Research Involving Human Subjects
with the approval of the Ethics Committee of Tianjin Medical
University Cancer Institute and Hospital and carried out in
accordance with the Declaration of Helsinki (approval no.
EK2019034), and all participants gave their written informed
consent.
Cell culture and transfection
A549, H1299 and LLC cells were purchased from iCell
Bioscience, Inc., and were cultured in DMEM (Invitrogen; Thermo
Fisher Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 µg/ml streptomycin and 100 U/ml penicillin
(both Beyotime Institute of Biotechnology) at 37°C in 5%
CO2. Briefly, once cells reached 70–80% confluence,
antisense oligonucleotide (ASO)-miR-433 (100 nM;
5′-ACACCGAGGAGCCCAUCAUGAU-3′; Shanghai GenePharma Co., Ltd.),
pri-miR-433 (2 µg; Shanghai GenePharma Co., Ltd.) or pTMED5 (2 µg;
Shanghai GenePharma Co., Ltd.) were transfected into A549 and H1299
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. As non-targeting negative controls (NCs), ASO-NC (100
nM; 5′-CAGUACUUUUGUGUAGUACAA-3′; Shanghai GenePharma Co., Ltd.) for
the ASO-miR-433 or the empty vector pcDNA3 for pri-miR-433 and
pTMED5 (2 µg; Shanghai GenePharma Co., Ltd.) were also transfected
into A549 and H1299 cells. The cells were incubated for 6 h at
37°C. Subsequently, the transfection medium was replaced with
complete medium containing 10% FBS. After 48 h at 37°C, cells were
collected for subsequent analyses. Recombinant lentiviruses
expressing lenti-miR-433 or the empty lentiviral vector
pLENT-Puro-MIR-GFP were produced by Shanghai GeneChem Co., Ltd. A
total of 6×105 LLC cells were infected with concentrated
lentivirus (MOI=40) in the presence of 8 µg/ml polybrene (Sigma
Aldrich; Merck KGaA). After 60 h of incubation at 37°C, cells were
selected using puromycin (3 µg/ml; Sigma Aldrich; Merck KGaA) for 5
days before subsequent experiments.
Isolation of exosomes from serum
Exosome fractions were prepared using the exoEasy
Maxi kit (Qiagen China Co., Ltd.) according to the manufacturer's
instructions.
Reverse transcription-quantitative
(RT-qPCR)
Total RNA was extracted from tissues and A549 and
H1299 cells using TRIzol® Reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). RT was performed using the RT-PCR Quick
Master Mix kit (Toyobo Life Science) according to the
manufacturer's protocol. The resultant cDNA was amplified by qPCR
using the following primers: miR-433 forward,
5′-TCGGCAATCATGATGGGCTCCTC-3′ and reverse,
5′-CTCAACTGGTGTCGTGGAGTC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; TMED5 forward,
5′-CCTTTCTACCCTTGATTT-3′ and reverse, 5′-TATAGCCACATTCTCCTT-3′;
β-actin forward, 5′-CGTGACATTAAGGAGAAGCTG-3′ and reverse,
5′-CTAGAAGCATTTGCGGTGGAC-3′. Amplification and detection were
performed with a SYBR-Green PCR kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.) for TMED5 and β-actin, and
SYBR® Green Realtime PCR Master Mix (Toyobo Life
Science) for miR-433 and U6 on the ABI PRISM 7700 Sequence
Detection System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were as follows: 95°C for 4
min, followed by 40 cycles of 95°C for 30 sec, 59°C for 30 sec and
72°C for 1 min, with a final extension at 72°C for 5 min. The
relative expression levels were normalized to the endogenous
controls U6 for miR-433 and β-actin for TMED5, and were expressed
using the 2−ΔΔCq method (23).
Western blotting
The exosomes were mixed with 1X SDS loading buffer
and A549 and H1299 cells were washed in cold 1X PBS and lysed using
RIPA lysis buffer (Beyotime Institute of Biotechnology)
supplemented with a protease phosphatase inhibitor, boiled at 95°C
for 5 min and centrifuged in 12,000 × g for 5 min at 4°C. Protein
concentration was determined using the BCA Protein Assay kit
(Beyotime Institute of Biotechnology). Proteins in supernatants (30
µg/lane) were separated via 10% SDS-PAGE and transferred onto PVDF
membranes (EMD Millipore), which were blocked with 5% skimmed milk
(Solarbio Science & Technology Co., Ltd.) for 1 h at room
temperature. Subsequently, the membranes were incubated overnight
at 4°C with primary antibodies against CD63 (cat. no. ab213090;
1:1,000), Alix (cat. no. ab225555; 1:2,000), tumor susceptibility
101 (TSG101; cat. no. ab125011; 1:2000), cleaved caspase3 (cat. no.
ab49822; 1:600), cleaved poly(ADP-ribose)polymerase (PARP; cat. no.
ab32064; 1:5,000), tumor protein p53 binding protein 1 (53BP1; cat.
no. ab175188; 1:5,000), H2AX (cat. no. ab124781; 1:3,000),
phosphorylated (p)-β-catenin (cat. no. ab75777; 1:1,000), β-catenin
(cat. no. ab68183; 1:3,000), c-myc (cat. no. ab32072; 1:2,000),
cyclin D1 (cat. no. ab16663; 1:1,000), TMED5 (cat. no. ab228920;
1:2,000) and GAPDH (cat. no. ab8245; 1:5,000) (all from Abcam).
After washing, protein samples were incubated with HRP-conjugated
secondary antibodies (cat. no. ab6789; 1:3,000; Abcam) for 1 h at
room temperature. Finally, an ECL kit (Beyotime Institute of
Biotechnology) was used to assess protein bands. The data were
analyzed via densitometry using ImageJ software (version 1.8.0;
National Institutes of Health) and normalized to the expression of
the internal control GAPDH.
Flow cytometry assay for cell cycle
and apoptosis analysis
For the analysis of cell cycle, the cells were
digested using trypsin (Invitrogen; Thermo Fisher Scientific, Inc.)
and stained with PI (Beyotime Institute of Biotechnology) using a
Cycletest™ Plus DNA Reagent kit (BD Biosciences) according to the
manufacturer's manual, and cells were detected by flow cytometry
assay. Briefly, a total of 2×106 cells/well were seeded
onto 6-well plates. On the next day, A549 and H1299 cells were
harvested and fixed in 70% ethanol at 4°C overnight. The fixed
cells were then incubated with PBS containing 0.1% Triton X-100 for
30 min at room temperature, and then stained with 20 µg/ml PI for
30 min in the dark at room temperature. The stained cells were
analyzed using the NovoCyte flow cytometer with the NovoCyte 1.4.1
software (both ACEA Biosciences, Inc.; Agilent Technologies, Inc.).
The proliferation index (PI) was calculated according to the
following formula: PI=(S +
G2/M)/(G0/G1 + S+
G2/M).
Apoptosis was detected using an Annexin V-FITC kit
(Solarbio Science & Technology Co., Ltd.) according to the
manufacturer's protocol. Briefly, A549 and H1299 cells were washed
three times with PBS and then fixed with 100 µl binding buffer
(Solarbio Science & Technology Co., Ltd.). Subsequently, 5 µl
PI and 5 µl Annexin V-FITC were used to stain the cells for 15 min
in the dark at room temperature. Finally, cells were analyzed with
a flow cytometer (Epics XL; Beckman Coulter, Inc.) and FlowJo
software (v.10.4.2; FlowJo LLC).
Cell Counting Kit-8 (CCK-8) assay
Briefly, A549 and H1299 cells were plated into the
96-well culture plates at a density of 3×103 cells/well
in 100 µl culture medium one day prior to transfection. At 0 and 48
h of culture, 10 µl CCK-8 reagent (Solarbio Science &
Technology Co., Ltd.) was added into each well and incubated at
37°C for 2 h. In addition, A549 and H1299 cells were plated into
the 96-well culture plates at a density of 3×103
cells/well in 100 µl culture medium one day prior to transfection
with pcDNA and miR-433, and 1, 2, 3 or 4 µg/ml cisplatin (Solarbio
Science & Technology Co., Ltd.) was added at 37°C for 24 h.
Relative cell proliferating rate was measured using a microplate
reader (Thermo Fisher Scientific, Inc.) at an absorbance of 450
nm.
Colony formation assay
Following transfection with the pri-miR-433,
ASO-miR-433, pTMED5 and the control group, NSCLC cells were seeded
into 12-well culture dishes at a density of 500 cells/dish and
cultured in a 5% CO2 incubator at 37°C. The medium was
replaced every 3 days. After 14 days, cells were stained using 1%
crystal violet at room temperature for 15 min (Beyotime Institute
of Biotechnology).
In vivo experiments
A total of 10 6-week-old male C57BL/6J mice (20±2 g;
6–8 weeks old) were purchased from Beijing Vital River Laboratory
Animal Technologies Co., Ltd. All mice were housed under a fixed
12-h light-dark cycle in a laminar flow room at constant
temperature (23°C) and humidity (40–70%), with ad libitum access to
sterilized food and water. The mice were randomly divided into 2
groups (5 mice/group): Lenti-Con and Lenti-miR-433.
Lentiviral-transduced LLC cells were resuspended in PBS and
subcutaneously injected into the ventral forearm of mice. The
behavior of mice and changes in body weight were recorded daily.
During 1–4 week of in vivo explant assay, the subcutaneous
lengths and widths of tumors were measured weekly. Mice were
sacrificed 30 days post-injection, and the tumors were extracted
and visually compared. The mice were euthanized using carbon
dioxide (at a range rate of 30–70% of the chamber volume per
minute), and their deaths were verified by checking eye color and
reflex action. Tumor volume was calculated using the equation: V =
(LxW2)/2, where V represents the tumor volume, L
represents the length and W represents the width. The study was
approved by the Institutional Animal Care and Use Committee of
Tianjin Medical University Cancer Institute and Hospital (approval
no. LLSP2019017).
Immunohistochemistry (IHC)
For IHC assays, tumor tissues were fixed with 4%
formalin for 24 h at room temperature, paraffin-embedded and cut
into 4-µm-thick sections. Subsequently, the slides were
deparaffinized in xylene and rehydrated through a descending
ethanol series. Antigen retrieval was performed with citrate buffer
in a microwave oven. Subsequently, slides were blocked with 3%
hydrogen peroxidase in methanol and 10% goat serum (Solarbio
Science & Technology Co., Ltd.) for 1 h at room temperature.
Slides were incubated with CD4 (cat. no. ab183685; 1:1,000; Abcam),
CD8 (cat. no. ab217344; 1:1,000; Abcam), TMED5 (cat. no. SRP08852;
1:1,1000; Tianjin Saier Biotechnology Co., Ltd.) and β-catenin
antibodies (cat. no. ab223075; 1:1,000; Abcam) overnight at 4°C and
then with HRP-labeled goat anti-rabbit IgG (cat. no. ab6721;
1:2,000; Abcam) for 30 min at room temperature. The slides were
stained with DAB (1:50) as the chromogen and counterstained with
hematoxylin for 1 min at room temperature. Slides were evaluated
using an optic Olympus BX51 microscope at light field (Olympus
Corporation; magnification, ×200) with a Micro-Publisher 3.3 RTV
camera (Q Imaging Scientific & Industrial Camera; Teledyne
Photometrics).
Transcriptome analysis
Global characterization of T cells in NSCLC by
single-cell sequencing was used to analyze the immune cell
infiltration for TMED5 (Global characterization of T cells in
non-small cell lung cancer by single-cell sequencing; http://lung.cancer-pku.cn/index.php).
Bioinformatics analysis
The Cancer Genome Atlas (TCGA) portal (https://tcgadata.nci.nih.gov/tcga/) was used for
the gene expression data of 233 patients with
chemotherapy-sensitive NSCLC, 279 patients with
chemotherapy-resistant NSCLC and 59 normal samples. Gene Expression
Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/) and StarBase V3.0
(http://starbase.sysu.edu.cn/) were used
to analyze TMED5 expression in NSCLC. Kaplan-Meier Plotter
(http://kmplot.com/analysis/) was used to
analyze the overall survival of patients with NSCLC with high or
low miR-433 expression (504 patients with lung adenocarcinoma and
472 patients with lung squamous cell carcinoma). The Human Protein
Atlas (https://www.proteinatlas.org/) was
used to analyze the overall survival of patients with NSCLC with
high (n=557) or low (n=437) TMED5 expression.
EGFP fluorescent reporter assay
This assay was performed as previously described
(24). A549 and H1299 cells were
co-transfected with pri-miR-433 (0.5 µg) or ASO-miR-433 (100 nM)
together with pcDNA3/EGFP-TMED5 3′-untranslated region (UTR)
wild-type or mutated reporter plasmids (0.5 µg) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
pDsRed2-N1 (0.05 µg; Clontech Laboratories, Inc.) was used for
normalization. After 48 h of transfection, cells were lysed and the
intensity of EGFP and RFP fluorescence was determined using an
F-4500 fluorescence spectrophotometer (Hitachi, Ltd.).
Comet assay
The evaluation of DNA damage was analyzed with the
alkaline comet assay using the Reagent kit for Single Cell Gel
Electrophoresis Assay (Trevigen, Inc.) according to the
manufacturer's instructions. A549 and H1299 cells were seeded at a
density of 105 cells/well in 12-well plates. Cells were
treated with bersaldegenin-1,3,5-orthoacetate (Sigma-Aldrich; Merck
KGaA) at a concentration of 1.0 µg/ml for 6 h and 10 µg/ml taxol
(Solarbio Science & Technology Co., Ltd.), used to induce DNA
damage, for 4 h at 37°C. Subsequently, A549 and H1299 cells were
collected by centrifugation at 500 × g for 5 min at 4°C and
combined with low melting agarose at a ratio of 1:10, and 50 µl
cell suspension was spread onto the CometSlides (Invitrogen; Thermo
Fisher Scientific, Inc.). Slides were immersed in the lysis
solution for 1 h at 4°C, after which they were placed in the
alkaline unwinding solution (Trevigen, Inc.) for 1 h at 4°C. Slides
were placed in the electrophoresis slide tray and immersed in the
alkaline electrophoresis solution in the CometAssay ES unit
(Trevigen, Inc.). Slides were submitted to electrophoresis at 1
V/cm for 30 min. Following electrophoresis, slides were washed and
then stained with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.)
for 30 min at room temperature. Slides were analyzed under a
fluorescence microscope (magnification, ×200, Nikon PCM-2000; Nikon
Corporation).
Top/fop luciferase assay
To analyze β-catenin transcriptional activity,
TOP-flash luciferase (plasmid 16558; Addgene, Inc.) and FOP
luciferase (plasmid 16559; Addgene, Inc.) and the indicated
plasmids (pcDNA3 + pcDNA3, pcDNA3 + miR-433, miR-433 + pTMED5) were
transfected into A549 and H1299 cells using Lipofectamine 2000
according to the manufacturer's protocol. Luciferase activity was
measured after 48 h using a Dual-Luciferase Reporter Assay kit
(Promega Corporation) according to the manufacturer's protocol, and
Renilla luciferase activity was used as an internal
control.
Transmission electron microscopy (TEM)
analysis and exosome particle size detection
For TEM analysis, isolated exosomes were fixed with
4% paraformaldehyde at room temperature for 30 min and 4%
glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) and then placed
on a carbon-coated copper grid and immersed in a 2% phosphotungstic
acid solution for examination (JEM-1200EX; JEOL, Ltd.). For exosome
particle size detection, a total of 1 ml filtered PBS was used to
mix the exosome and was then stored on ice. A disposable clean
sample cell was selected and wiped with dust-free paper to ensure
that no particles adhered to the outer tube wall. The exosome
solution was slowly injected to avoid air bubbles, slightly tilting
the sample cell. The sample cell was sealed with a lid. The
instrument operating procedure was followed using the Nanosight NS
300 system (NanoSight; Malvern Panalytical).
Statistical analysis
SPSS 19.0 (IBM Corp.) statistical software was used
for data analysis and GraphPad Prism V (GraphPad Software, Inc.)
was used for image editing. Measurement data were expressed as the
mean ± SD and compared using the unpaired t-test. Differences among
multiple groups were determined using one-way ANOVA with Tukey's
post-hoc test. The survival data was analyzed using Kaplan-Meier
curves with the log-rank test. The χ2 test was used to
analyze whether miR-433 expression was associated with
clinicopathological characteristics. P<0.05 was considered to
indicate a statistically significant difference.
Results
Exosome-derived miR-433 expression is
downregulated in NSCLC, especially in chemotherapy-resistant
NSCLC
First, exosomes were isolated using the exoEasy Maxi
kit from the serum of patients with NSCLC. Subsequently, TEM was
used to verify the exosomal cup-shaped membranes (Fig. 1A). Nanosight particle tracking
revealed that the sizes of these exosomes were ~80-150 nm (Fig. 1B). Western blot analysis confirmed
the expression of specific exosomal markers such as CD63, Alix and
TSG101 (Fig. 1C). miR-433 expression
was lower in the serum of patients with chemotherapy-resistant
NSCLC compared with that of patients with chemotherapy-sensitive
NSCLC and with normal serum (Fig.
1D). miR-433 expression was negatively associated with a large
tumor size, severe TNM stage and distant metastasis in patients
with NSCLC (Table I). In addition,
TCGA database showed lower miR-433 expression in patients with
chemotherapy-resistant NSCLC (Fig.
1E). Furthermore, patients with NSCLC with low miR-433
expression exhibited a poor prognosis using Kaplan-Meier Plotter
(Fig. 1F).
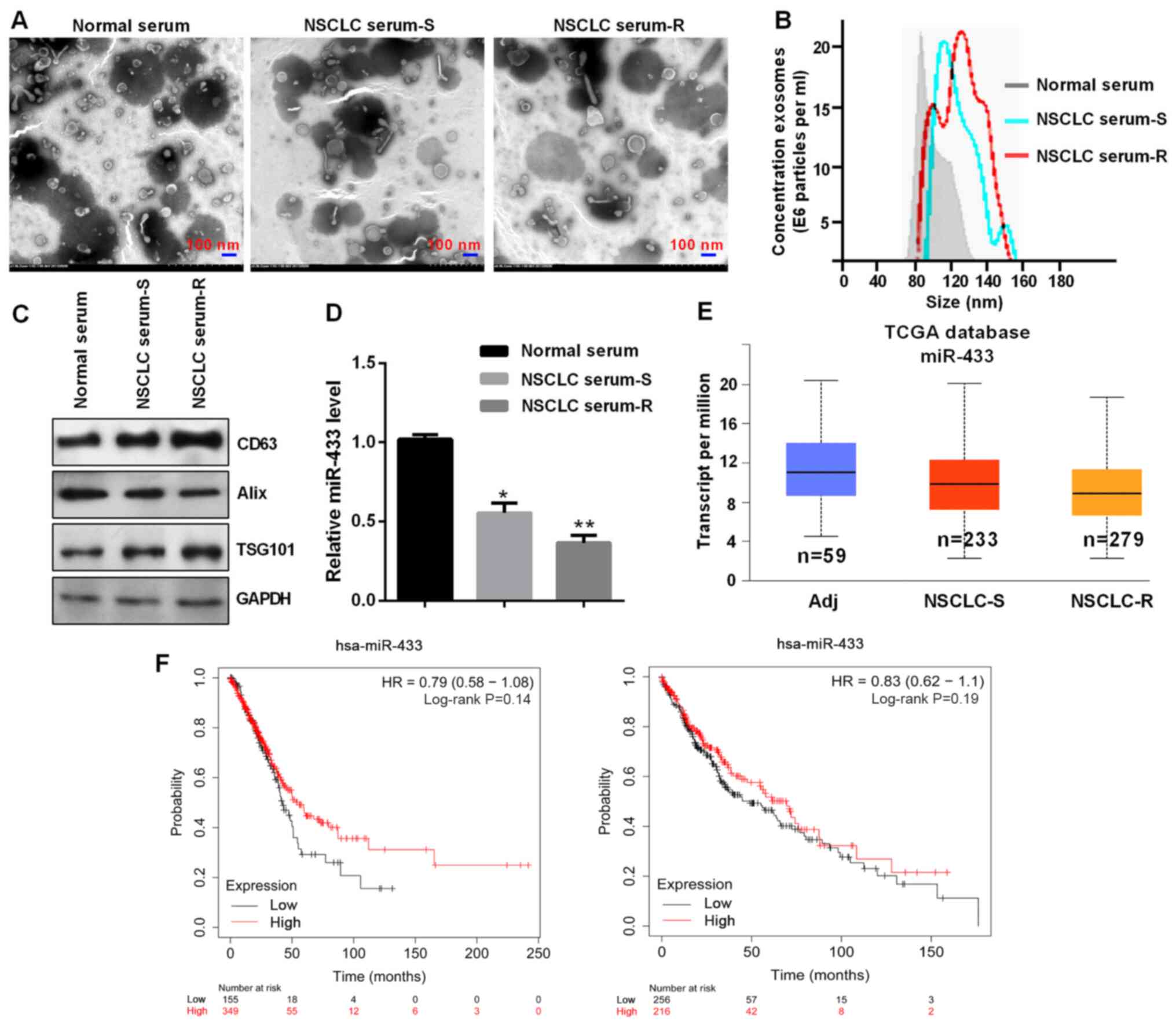 | Figure 1.Exosome-derived miR-433 was
downregulated. (A) Exosomes were assessed using transmission
electron microscopy. Scale bar, 100 nm. (B) Western blot analysis
of the exosomal markers CD63, Alix and TSG101. (C) Exosomes were
detected by Nanosight particle tracking. (D) Reverse
transcription-quantitative PCR of miR-433 expression in the
indicated groups. *P<0.05 and **P<0.01 vs. normal serum. (E)
TCGA database showed the miR-433 expression in the indicated
groups. (F) Low expression levels of exosomal miR-433 predicted a
poor prognosis in patients with NSCLC. Left panel, 504 patients
with lung adenocarcinoma; right panel, 472 patients with lung
squamous cell carcinoma. NSCLC, non-small cell lung cancer; S,
sensitive; R, resistant; Adj, adjacent; miR, microRNA; TCGA, The
Cancer Genome Atlas; HR, hazard ratio; TSG101, tumor susceptibility
101. |
 | Table I.Association of miR-433 expression
with the clinicopathological features of 50 patients with non-small
cell lung cancer. |
Table I.
Association of miR-433 expression
with the clinicopathological features of 50 patients with non-small
cell lung cancer.
|
|
| miR-433
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | No. | Low (n=31) | High (n=19) | P-value |
|---|
| Age, years |
|
|
|
|
|
<60 | 21 | 12 | 9 | 0.547 |
|
≥60 | 29 | 19 | 10 |
|
| Sex |
|
|
|
|
|
Male | 34 | 22 | 12 | 0.566 |
|
Female | 16 | 9 | 7 |
|
| Tumor size |
|
|
|
|
| ≥5
cm | 30 | 22 | 8 | 0.043a |
| <5
cm | 20 | 9 | 11 |
|
| TNM stage |
|
|
|
|
|
I–II | 19 | 8 | 11 | 0.023a |
|
III–IV | 31 | 23 | 8 |
|
| Distant
metastasis |
|
|
|
|
| No | 24 | 11 | 13 | 0.026a |
|
Yes | 26 | 20 | 6 |
|
| Histological
type |
|
|
|
|
|
Squamous | 28 | 16 | 12 | 0.425 |
|
Adenocarcinoma | 22 | 15 | 7 |
|
Suppressive role of miR-433 in
NSCLC
To assess the role of miR-433 in NSCLC cells,
gain-of-function and knockdown approaches were performed. RT-qPCR
was used to confirm the transfection efficiency (Fig. 2A). As shown in Fig. 2B, CCK-8 assay indicated that miR-433
overexpression inhibited cell viability compared with the control
group, while its inhibitor increased cell viability in H1299 and
A549 cells (Fig. 2B). Similarly,
pri-miR-433 and ASO-miR-433 significantly decreased or enhanced,
respectively, the colony formation ability of A549 and H1299 cells
compared with their respective control groups (Fig. 2C). Flow cytometry demonstrated that
overexpression of miR-433 blocked cell cycle progression, while
miR-433-knockdown promoted cell cycle progression of A549 and H1299
cells by regulating the PI index (Figs.
2D and E, and S1A and B). Flow
cytometry demonstrated that overexpression of miR-433 accelerated
apoptosis in NSCLC cells (Figs. 2F
and S1C and D), also demonstrated
by the increased levels of cleaved caspase 3 and cleaved PARP
(Fig. 2G). Knockdown of miR-433
inhibited apoptosis in NSCLC cells (Figs. 2F and S1C
and D), also demonstrated by the decreased expression levels of
cleaved caspase 3 and cleaved PARP (Fig.
2G). Subsequently, the effect of miR-433 on the regulation of
chemoresistance to cisplatin was assessed. Cisplatin
chemoresistance was decreased when cells were treated with
pri-miR-433 in A549 and H1299 cells compared with that in cells
transduced with pcDNA3 in a dose-dependent manner with 0–4 µg/ml
cisplatin, as assessed by CCK-8 assay (Fig. 2H). In addition,
miR-433-overexpressing A549 and H1299 cells repaired DNA breaks
more slowly than control vector-treated cells (Fig. 2I). Furthermore, miR-433
overexpression promoted the protein expression of 53BP1 or H2AX in
A549 and H1299 cells (Fig. 2J).
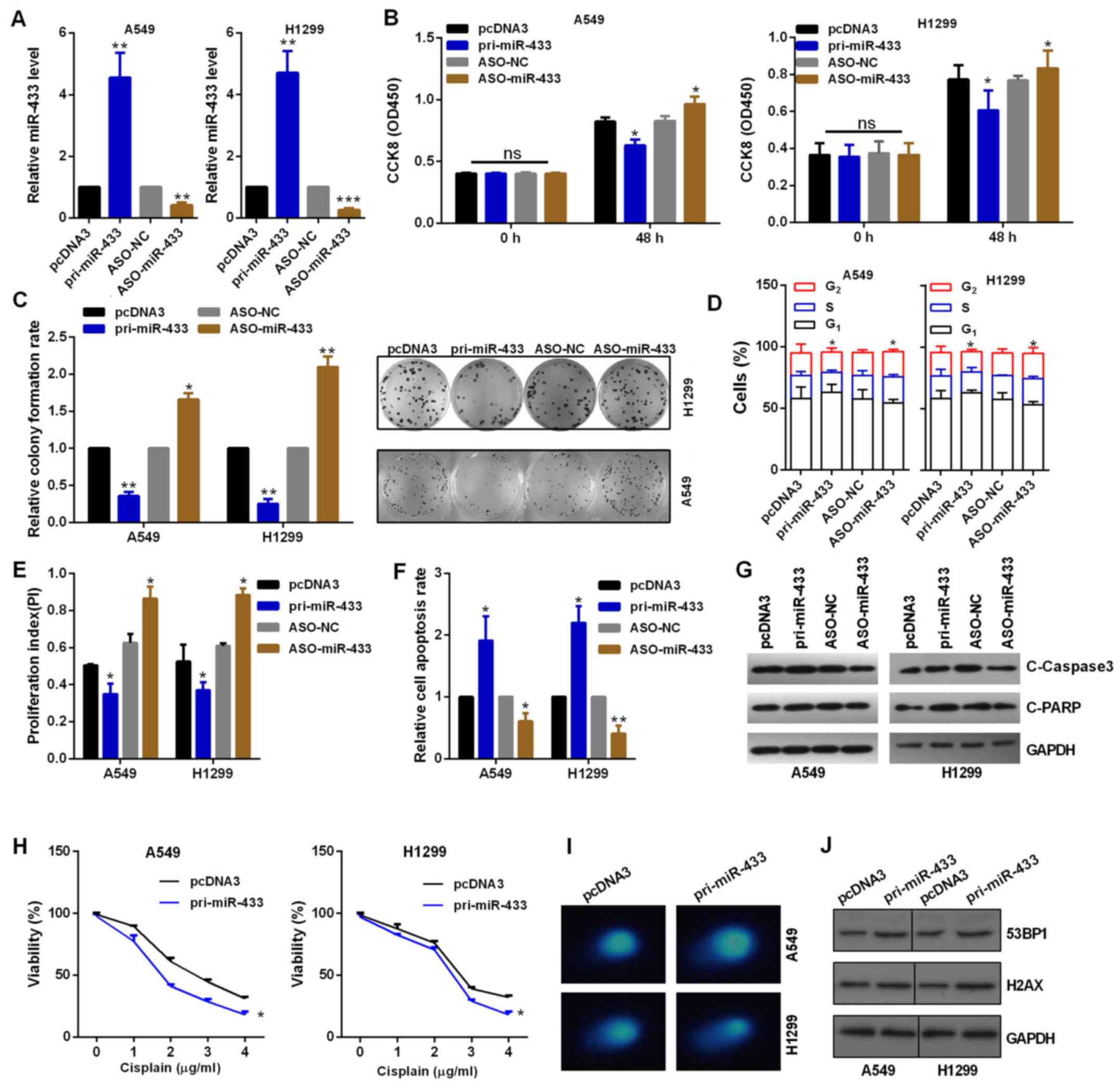 | Figure 2.Suppressive role of miR-433 in
vitro. (A) Efficiency of pri-miR-433 or ASO-miR-433 was
detected by reverse transcription-quantitative PCR assay in A549
and H1299 cells. (B) Role of altered miR-433 expression on the
viability of A549 and H1299 cells was detected by CCK-8 assay. (C)
Colony formation assay showed the proliferation ability of cells
transfected with pri-miR-433 or ASO-miR-433. (D) Flow cytometric
analysis showed that miR-433 overexpression in A549 and H1299 cells
resulted in an increase of cells in the G0/G1
phase and a decrease of cells in the S and G2 phases.
(E) miR-433 overexpression inhibited the PI, while
miR-433-knockdown promoted the PI. (F) Flow cytometric assay showed
miR-433 overexpression promoted apoptosis and ASO-miR-433 inhibited
apoptosis in A549 and H1299 cells. (G) Western blot analysis showed
the protein expression levels of C-caspase 3 and C-PARP in A549 and
H1299 cells. (H) Transfected A549 and H1299 cells were treated with
0–4 µg/ml cisplatin in 96-well plates for 24 h. CCK-8 assay was
then used to investigate cell viability. (I) Comet assays showed
the degree of DNA breaks in A549 and H1299 cells transfected with
the indicated plasmids treated with 10 µg/ml taxol for 4 h. (J)
Western blot analysis showed the protein expression levels of 53BP1
and H2AX in A549 and H1299 cells transfected with the indicated
plasmids treated with 10 µg/ml taxol for 4 h. *P<0.05,
**P<0.01 and ***P<0.001 vs. pcDNA3 or ASO-NC. ns, not
significant; CCK-8, Cell Counting Kit-8; OD, optical density; miR,
microRNA; NC, negative control; ASO, antisense oligonucleotide; C,
cleaved; PARP, poly(ADP-ribose)polymerase; 53BP1, tumor protein p53
binding protein 1; PI, proliferation index. |
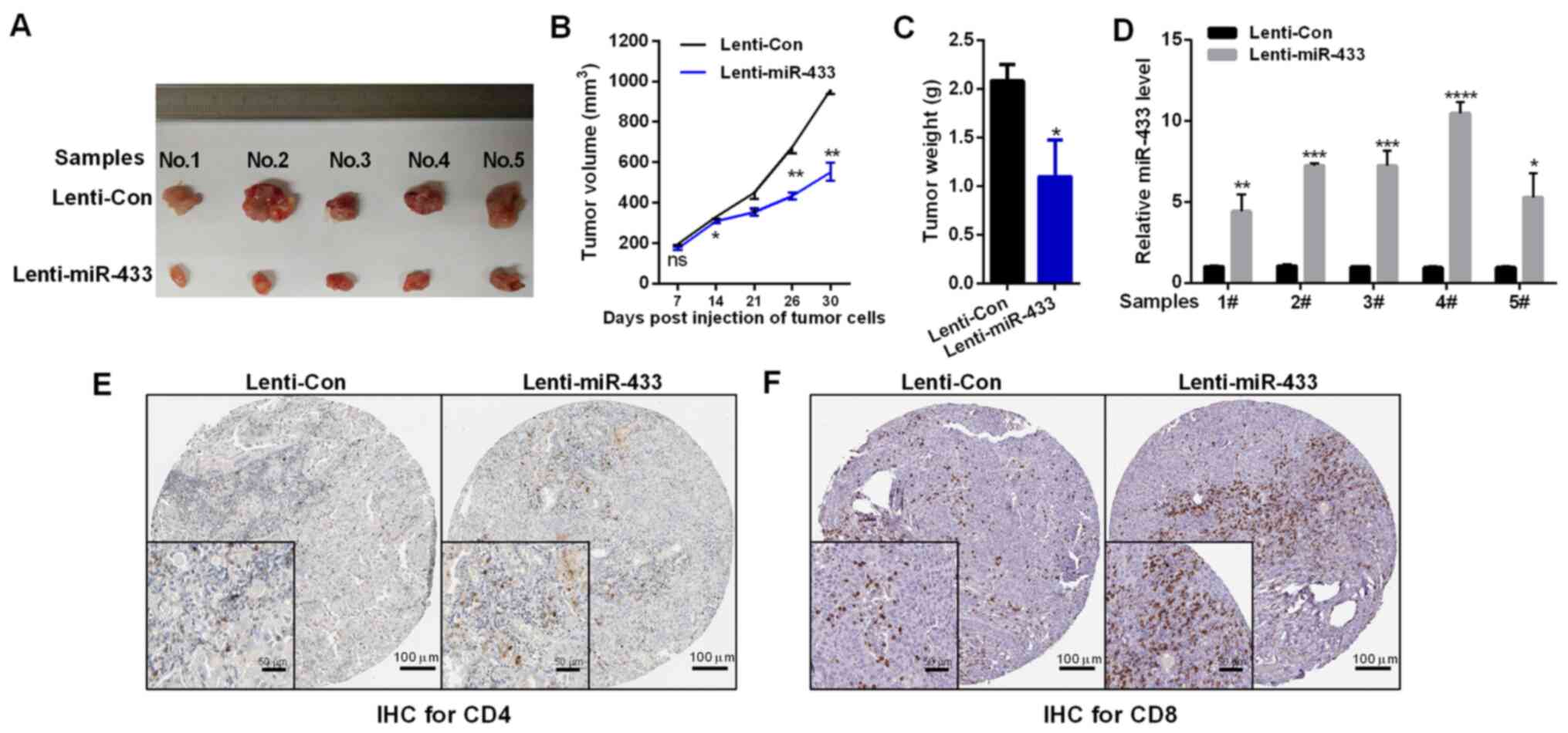
miR-433 inhibits tumor growth by promoting CD4 and
CD8 T-cell infiltration in vivo. To test whether miR-433 could
enhance the antitumor effects of lung cancer, LLC cells were stably
transfected with lenti-miR-433 or empty vector (lenti-Con). Mice
injected with lenti-miR-433 exhibited decreased tumor volume and
tumor weight compared with those injected with lenti-Con (Fig. 3A-C). The expression levels of miR-433
in tumors were confirmed by RT-qPCR assay (Fig. 3D). The effects of miR-433 on T-cell
infiltration were evaluated, and it was found that miR-433
overexpression increased the CD4 and CD8 T-cell infiltration
intratumorally, as assessed using IHC assay (Fig. 3E).
TMED5 is a direct target gene of
miR-433
TMED5 was identified to have a binding site with
miR-433 using StarBase V3.0. Highly conserved predicted binding
sites and the mutant sites are shown in Fig. 4A in the 3′-UTR of TMED5. EGFP
reporter assay indicated that the EGFP activity of TMED5 was
decreased by miR-433 overexpression and increased by miR-433
knockdown in A549 and H1299 cells, indicating that miR-433 directly
targeted TMED5 (Fig. 4B). In
addition, altered miR-433 failed to affect the EGFP intensity of
the plasmid carrying TMED5 3′-UTR MUT in A549 and H1299 cells
(Fig. 4C). Moreover, TMED5 mRNA and
protein expression was decreased by miR-433 overexpression and
enhanced by miR-433-knockdown in A549 and H1299 cells (Fig. 4D and E). The protein expression
levels of TMED5 were markedly increased in NSCLC tissues compared
with in adjacent normal tissues using the GEPIA database (Fig. 4F). The mRNA expression levels of
TMED5 were also markedly increased in NSCLC tissues according to
TCGA database, especially in chemotherapy-resistant NSCLC tissues
(Fig. 4G). In addition, patients
with NSCLC with high TMED5 expression exhibited a poor prognosis
according to The Human Protein Atlas (Fig. 4H).
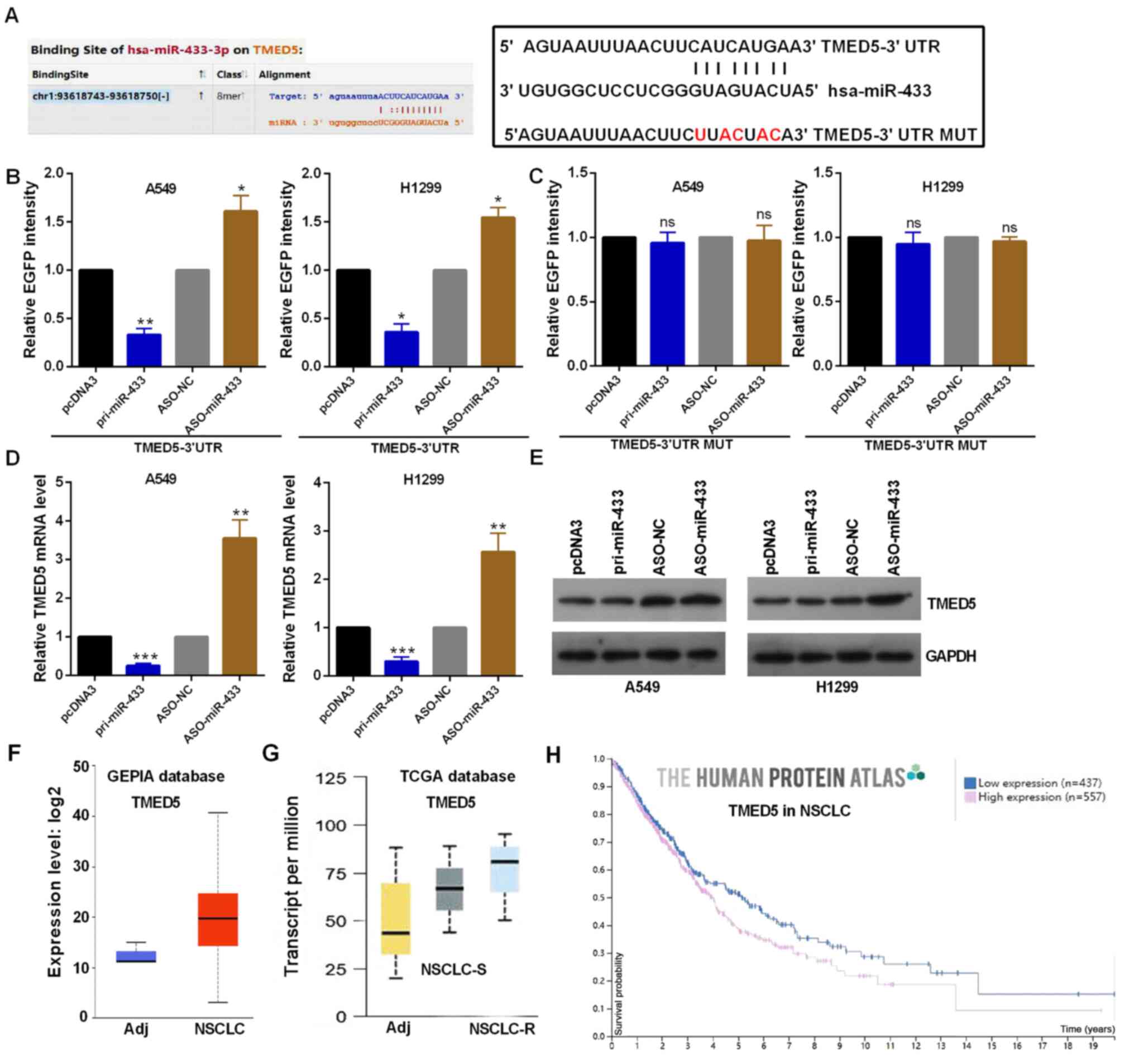 | Figure 4.miR-433 directly targets TMED5. (A)
Predicted miR-433 binding sites in TMED5 mRNA WT and MUT using
StarBase V3.0. (B and C) EGFP intensity in A549 and H1299 cells
co-transfected with pri-miR-433 or ASO-miR-433 and the WT or MUT
3′-UTR of TMED5. (D) TMED5 mRNA expression with the indicated
transfections was measured by reverse transcription-quantitative
PCR assay in A549 and H1299 cells. (E) TMED5 protein expression in
A549 and H1299 cells transfected with pri-miR-433 or ASO-miR-433
and respective controls was determined by western blotting. (F)
GEPIA database showing the TMED5 protein expression. (G)TCGA
database showing the TMED5 mRNA expression. (H) Human Protein Atlas
showing that high protein expression levels of TMED5 predicted a
poor prognosis in patients with NSCLC. *P<0.05, **P<0.01 and
***P<0.001 vs. pcDNA3 or ASO-NC. ns, not significant; WT,
wild-type; MUT, mutated; 3′-UTR, 3′-untranslated region; miR,
microRNA; TMED5, transmembrane p24 trafficking protein 5; NC,
negative control; ASO, antisense oligonucleotide; TCGA, The Cancer
Genome Atlas; NSCLC, non-small cell lung cancer; R, resistant; Adj,
adjacent. |
TMED5 functions as an oncogene in
NSCLC
The plasmid of TMED5 overexpression (pTMED5) was
transfected into NSCLC cells. pTMED5 transfection enhanced TMED5
mRNA and protein expression in A549 and H1299 cells (Fig. 5A and B). CCK-8 assay revealed that
pTMED5 promoted cell viability in A549 and H1299 cells (Fig. 5C). Colony formation assay indicated
that pTMED5 induced a significant increase in colony formation in
A549 and H1299 cells (Fig. 5D).
Additionally, pTMED5 accelerated G1 to S and
G2 transition in NSCLC cells (Figs. 5E and F, and S2A and B). Transfected pTMED5
significantly decreased the apoptotic rate (Figs. 5G and S2C), as well as decreased the levels of
cleaved caspase 3 and cleaved PARP in NSCLC cells (Fig. 5H). Cisplatin chemoresistance was
increased in A549 and H1299 cells treated with pTMED5 than that in
cells transduced with pcDNA3 in a dose-dependent manner with 0–4
µg/ml cisplatin, as assessed by CCK-8 assay (Fig. 5I). In addition, TMED5-overexpressing
A549 and H1299 cells repaired DNA breaks more quickly than control
vector-treated cells (Fig. 5J).
TMED5 overexpression inhibited the protein expression levels of
53BP1 and H2AX in A549 and H1299 cells (Fig. 5K). Furthermore, deep single cell
transcriptome data in NSCLC tissues revealed upregulated TMED5
expression in the subset of CD4-CD69 (Non-traditional regulatory T
cells), CD4-CXCL13 (exhausted CD4 T cells), CD4-CTLA4 (naïve CD4 T
cells and Treg cells), CD8-ZNF683 (pre-exhaustion CD8 T cells) and
CD8-LAYN (exhausted CD8+ T cells) compared with the
normal tissues, indicating that CD69, CXCL13, CTLA4, ZNF683 and
LAYN may be negative regulatory genes in cellular immune response
(Fig. 5L and M).
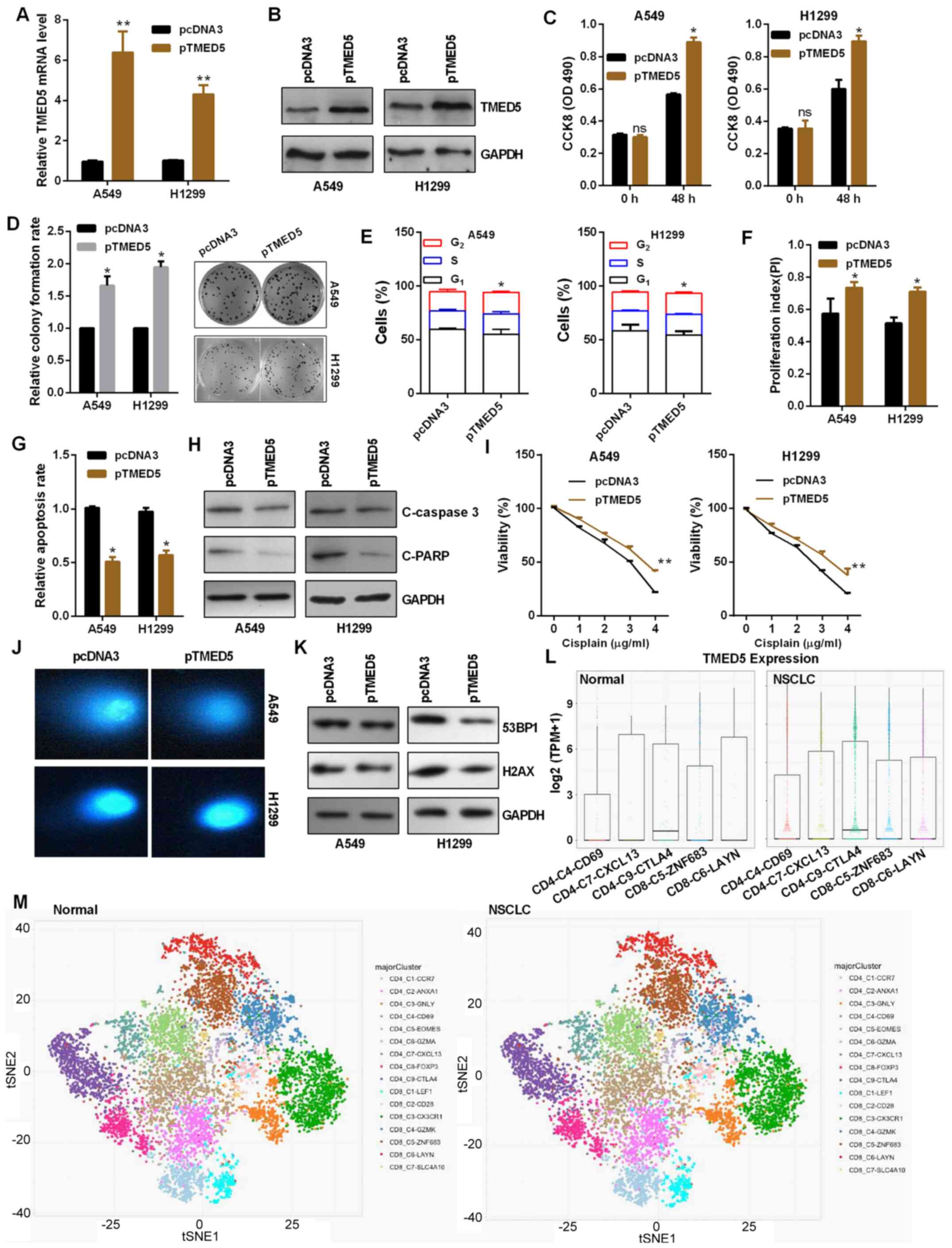 | Figure 5.TMED5 functions as an oncogene in
NSCLC. Efficiency of pTMED5 was detected by (A) reverse
transcription-quantitative PCR and (B) western blot assay in A549
and H1299 cells. (C) Role of altered TMED5 expression on the
viability of A549 and H1299 cells was detected by CCK-8 assay. (D)
Colony formation assay showed the proliferation ability of A549 and
H1299 cells transfected with pTMED5. (E) Flow cytometric analysis
showed that TMED5 overexpression in A549 and H1299 cells resulted
in a decrease of cells in the G0/G1 phase and
an increase of cells in the S and G2 phases. (F) TMED5
overexpression promoted the proliferation index. (G) Flow
cytometric assay showed that TMED5 overexpression decreased
apoptosis in A549 and H1299 cells. (H) Western blotting showed the
protein expression levels of C-caspase 3 and C-PARP in A549 and
H1299 cells. (I) Transfected A549 and H1299 cells were treated with
0–4 µg/ml cisplatin in 96-well plates for 24 h. CCK-8 assay was
then used to investigate cell viability. (J) Comet assays showed
the degree of DNA breaks in A549 and H1299 cells transfected with
the indicated plasmids treated with 10 µg/ml taxol for 4 h. (K)
Western blotting showed the protein expression levels of 53BP1 and
H2AX in A549 and H1299 cells transfected with the indicated
plasmids treated with 10 µg/ml taxol for 4 h. (L and M) Deep single
cell transcriptome data showed the immune cell infiltration in
NSCLC and the control group. These figures were taken form ‘Global
characterization of T cells in non-small cell lung cancer by
single-cell sequencing’ (http://lung.cancer-pku.cn/index.php). *P<0.05 and
**P<0.01 vs. pcDNA3. ns, not significant; p, plasmid; CCK-8,
Cell Counting Kit-8; OD, optical density; C, cleaved; PARP,
poly(ADP-ribose)polymerase; 53BP1, tumor protein p53 binding
protein 1; NSCLC, non-small cell lung cancer; TMED5, transmembrane
p24 trafficking protein 5; TPM, transcript per million. |
miR-433 regulates the WNT/β-catenin
signaling pathway through TMED5
As shown in Fig. 6A,
TMED5 overexpression markedly promoted the phosphorylation of
β-catenin (p-β-catenin), but not total β-catenin expression, as
well as significantly increased c-myc and cyclin D1 expression.
miR-433 overexpression in A549 and H1299 cells co-transfected with
pTMED5 blocked the TMED5-induced increases of protein expression
levels, including those of p-β-catenin, c-myc and cyclin D1
(Fig. 6A). Additionally, TOP/FOP
luciferase reporter assays identified that miR-433 overexpression
significantly inhibited the Top/Fop radio, indicating the
inactivation of the WNT signaling pathway by miR-433 overexpression
in A549 and H1299 cells (Fig. 6B).
Finally, IHC was performed to detect the expression levels of TMED5
and β-catenin in miR-433-mediated tumors in vivo. The
results revealed that Lenti-miR-433 inhibited the protein
expression levels of TMED5 and β-catenin (Fig. 6C).
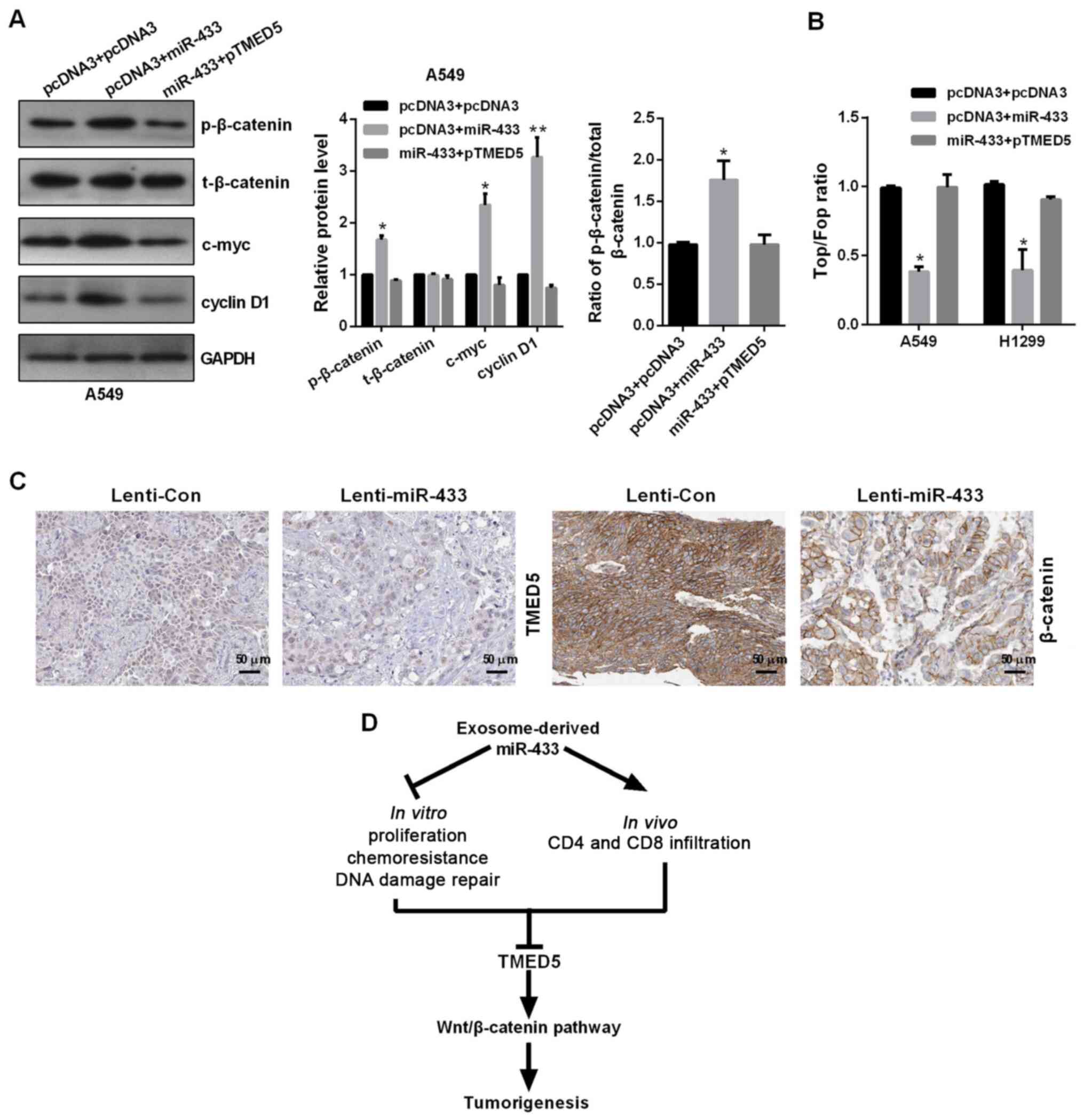 | Figure 6.miR-433 inhibits the WNT signaling
pathway through TMED5. (A) Western blotting showed the protein
levels of p-β-catenin, β-catenin, c-myc, cyclin D1 and GAPDH in
A549 and H1299 cells transfected with the indicated plasmids. (B)
Top/Fop luciferase reporter assay was performed to detect WNT
activity. (C) Immunohistochemistry showed the protein expression
levels of TMED5 and β-catenin. Scale bar, 50 µm. (D) Model of
potential association of miR-433 and its target gene TMED5.
*P<0.05 and **P<0.01 vs. pcDNA3 + pcDNA3. p, plasmid; TMED5,
transmembrane p24 trafficking protein 5; miR, microRNA; p,
phosphorylated; t, total; Lenti, lentivirusl Con, control. |
Discussion
Emerging evidence has identified that miRNAs serve
crucial roles in various biological functions, such as
differentiation, proliferation, chemotherapy resistance,
metastasis, autophagy and DNA damage, in a number of diseases,
including intervertebral disc degeneration, Alzheimer's disease and
Huntington's disease (25).
Differentially expressed miRNAs are closely associated with tumors
and the tumor microenvironment (25,26).
Furthermore, exosomal miRNAs released by cancer cells can directly
induce a series of phenotypes. For example, exosomal miR-766-3p
promotes cell migration and invasion through homeobox A13 in ESCC
(27). Exosomal miR-500a-3p promotes
cisplatin resistance and stemness via negatively regulating FBXW7
in gastric cancer (28). In ovarian
cancer, high miR-433-expressing cells release miR-433 into the
growth media via exosomes, which in turn can induce a senescence
bystander effect (29). The present
study reported that exosomal miR-433 expression was lower in plasma
of patients with chemotherapy-resistant NSCLC compared with in
plasma of patients with chemotherapy-sensitive NSCLC and in normal
serum, and miR-433 expression was significantly negatively
associated large tumor size, advanced TNM stage, distant metastasis
and a poor prognosis in patients with NSCLC. Notably, Weng et
al (30) also reported that
PCGEM1 prostate-specific transcript (PCGEM1) was highly expressed
in NSCLC cells, while miR-433-3p was lowly expressed in NSCLC
cells. PCGEM1 silencing or miR-433-3p overexpression inhibit cell
proliferation, migration and invasion, but accelerate apoptosis in
NSCLC cells (30).
The tumor microenvironment (TME) represents a key
factor for tumor heterogeneity maintenance, tumor progression and
drug resistance, and is composed of different cellular populations,
such as stromal, immune, endothelial and cancer stem cells
(31). Exosomal miRNAs from tumor
cells disturb T-cell metabolism and immune response, and increase
the production of cytokines in the TME (32). miRNAs in exosomes promote Treg
expansion accompanying ERK, STAT1 and STAT3 expression, but inhibit
the differentiation of T helper (Th)1 and Th17 cells, as well as
leading to an amplification of cytokine production in the TME
(33). The present study reported
that exosomal miR-433 inhibited tumor growth in vitro and
in vivo by blocking the cell cycle and promoting apoptosis
and CD4 and CD8 T-cell infiltration. However, the underlying
mechanism requires further investigation. Exosomal miR-433 also
inhibited chemoresistance to cisplatin by regulating DNA
damage.
The Wnt signaling pathway is one of the most
important signaling pathways during development of a number of
tumors and other diseases, including embryonic development and
maturation of the central nervous system (34,35).
Increasing evidence has indicated miRNA dysregulation and the
Wnt/β-catenin signaling pathway jointly drive carcinogenesis,
cancer metastasis and drug-resistance (36–40). The
present study reported that miR-433 inactivated the Wnt/β-catenin
signaling pathway by targeting TMED5 in NSCLC.
In conclusion, the findings of the present study
provide novel insight into the role of tumor-derived exosomal
miR-433 in NSCLC progression (Fig.
6D). NSCLC-derived exosomal miR-433 may mediate cell
proliferation, tumor growth, cell cycle, apoptosis, chemoresistance
and CD4 and CD8 T-cell infiltration, which may be associated with
the enrichment of exosomal miR-433 targeting the downregulation of
TMED5 and the WNT/β-catenin signaling pathway. Therefore, based on
the current findings, targeting exosomes and exosomal miRNAs may be
used as an effective anticancer strategy.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Shujuan Yuan
(Tianjin Medical University, Tianjin, China) for her effort with
the revision of the language of the manuscript.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RH conceived and designed the current study, and
contributed to writing the manuscript. RH and BL performed the
experiments and confirm the authenticity of all the raw data. RZ
and YZ analyzed and interpreted the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The current study was conducted under the
International Ethical Guidelines for Biomedical Research Involving
Human Subjects with the approval of the Ethics Committee of Tianjin
Medical University Cancer Institute and Hospital and performed in
accordance with the Declaration of Helsinki (approval no.
EK2019034), and all participants provided their written informed
consent. The animal experiments were approved by the Institutional
Animal Care and Use Committee of Tianjin Medical University Cancer
Institute and Hospital (approval no. LLSP2019017).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nasim F, Sabath BF and Eapen GA: Lung
cancer. Med Clin North Am. 103:463–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duma N, Santana-Davila R and Molina JR:
Non-small cell lung cancer: Epidemiology, screening, diagnosis, and
treatment. Mayo Clin Proc. 94:1623–1640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hoy H, Lynch T and Beck M: Surgical
treatment of lung cancer. Crit Care Nurs Clin North Am. 31:303–313.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Skoulidis F and Heymach JV: Co-occurring
genomic alterations in non-small-cell lung cancer biology and
therapy. Nat Rev Cancer. 19:495–509. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yoneda K, Imanishi N, Ichiki Y and Tanaka
F: Immune checkpoint inhibitors (ICIs) in non-small cell lung
cancer (NSCLC). J UOEH. 40:173–189. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xie Y, Dang W, Zhang S, Yue W, Yang L,
Zhai X, Yan Q and Lu J: The role of exosomal noncoding RNAs in
cancer. Mol Cancer. 18:372019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu C, Meiners S, Lukas C, Stathopoulos GT
and Chen J: Role of exosomal microRNAs in lung cancer biology and
clinical applications. Cell Prolif. 53:e128282020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Williams M, Cheng YY, Blenkiron C and Reid
G: Exploring mechanisms of microRNA downregulation in cancer.
Microrna. 6:2–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deng S, Calin GA, Croce CM, Coukos G and
Zhang L: Mechanisms of microRNA deregulation in human cancer. Cell
Cycle. 7:2643–2646. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of microRNAs in cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han F, Huang D, Huang X, Wang W, Yang S
and Chen S: Exosomal microRNA-26b-5p down-regulates ATF2 to enhance
radiosensitivity of lung adenocarcinoma cells. J Cell Mol Med.
24:7730–7742. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang HX, Wang XY, Fei JW, Li FH, Han J and
Qin X: MicroRNA-23B inhibits non-small cell lung cancer
proliferation, invasion and migration via downregulation of RUNX2
and inhibition of Wnt/Β-catenin signaling pathway. J Biol Regul
Homeost Agents. 34:825–835. 2020.PubMed/NCBI
|
|
15
|
Qu Y, Zhu J, Liu J and Qi L: Circular RNA
circ_0079593 indicates a poor prognosis and facilitates cell growth
and invasion by sponging miR-182 and miR-433 in glioma. J Cell
Biochem. 120:18005–18013. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding L and Zhang H: Circ-ATP8A2 promotes
cell proliferation and invasion as a ceRNA to target EGFR by
sponging miR-433 in cervical cancer. Gene. 705:103–108. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan L, You WQ, Sheng NQ, Gong JF, Hu LD,
Tan GW, Chen HQ and Wang ZG: A CREB1/miR-433 reciprocal feedback
loop modulates proliferation and metastasis in colorectal cancer.
Aging (Albany NY). 10:3774–3793. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang T, Jiang K, Zhu X, Zhao G, Wu H,
Deng G and Qiu C: miR-433 inhibits breast cancer cell growth via
the MAPK signaling pathway by targeting Rap1a. Int J Biol Sci.
14:622–632. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Q, Wang Y, Mu Y, Wang X and Fan Q:
miR-433-3p inhibits proliferation and invasion of esophageal
squamous cell carcinoma by targeting GRB2. Cell Physiol Biochem.
46:2187–2196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang YJ, Zhang ZF, Fan SH, Zhuang J, Shan
Q, Han XR, Wen X, Li MQ, Hu B, Sun CH, et al: MicroRNA-433 inhibits
oral squamous cell carcinoma cells by targeting FAK. Oncotarget.
8:100227–100241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu N, Liu Z, Zhang W, Li Y, Cao J, Yang H
and Li X: MicroRNA-433 reduces cell proliferation and invasion in
non-small cell lung cancer via directly targeting E2F transcription
factor 3. Mol Med Rep. 18:1155–1164. 2018.PubMed/NCBI
|
|
22
|
Li J, Chen M and Yu B: miR-433 suppresses
tumor progression via Smad2 in non-small cell lung cancer. Pathol
Res Pract. 215:1525912019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang Z, Sun Q, Guo J, Wang S, Song G, Liu
W, Liu M and Tang H: GRSF1-mediated MIR-G-1 promotes malignant
behavior and nuclear autophagy by directly upregulating TMED5 and
LMNB1 in cervical cancer cells. Autophagy. 4:668–685. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vishnoi A and Rani S: miRNA biogenesis and
regulation of diseases: An overview. Methods Mol Biol. 1509:1–10.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sandiford OA, Moore CA, Du J, Boulad M,
Gergues M, Eltouky H and Rameshwar P: Human aging and cancer: Role
of miRNA in tumor microenvironment. Adv Exp Med Biol. 1056:137–152.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu S, Lin Z, Zheng Z, Rao W, Lin Y, Chen
H, Xie Q, Chen Y and Hu Z: Serum exsomal miR-766-3p expression is
associated with poor prognosis of esophageal squamous cell
carcinoma. Cancer Sci. 110:3881–3892. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin H, Zhang L, Zhang C and Liu P:
Exosomal miR-500a-3p promotes cisplatin resistance and stemness via
negatively regulating FBXW7 in gastric cancer. J Cell Mol Med.
24:8930–8941. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Weiner-Gorzel K, Dempsey E, Milewska M,
McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A,
Sharpe D, et al: Overexpression of the microRNA miR-433 promotes
resistance to paclitaxel through the induction of cellular
senescence in ovarian cancer cells. Cancer Med. 4:745–758. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weng L, Qiu K, Gao W, Shi C and Shu F:
LncRNA PCGEM1 accelerates non-small cell lung cancer progression
via sponging miR-433-3p to upregulate WTAP. BMC Pulm Med.
20:2132020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Santos P and Almeida F: Role of exosomal
miRNAs and the tumor microenvironment in drug resistance. Cells.
9:14502020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bland CL, Byrne-Hoffman CN, Fernandez A,
Rellick SL, Deng W and Klinke DJ 2nd: Exosomes derived from B16F0
melanoma cells alter the transcriptome of cytotoxic T cells that
impacts mitochondrial respiration. FEBS J. 285:1033–1050. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS,
Zhang XS, Cui J, Zeng YX and Li J: Tumor-derived exosomes promote
tumor progression and T-cell dysfunction through the regulation of
enriched exosomal microRNAs in human nasopharyngeal carcinoma.
Oncotarget. 5:5439–5452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Taciak B, Pruszynska I, Kiraga L, Bialasek
M and Krol M: Wnt signaling pathway in development and cancer. J
Physiol Pharmacol. 69:185–196. 2018.PubMed/NCBI
|
|
35
|
Kassumeh S, Weber GR, Nobl M, Priglinger
SG and Ohlmann A: The neuroprotective role of Wnt signaling in the
retina. Neural Regen Res. 16:1524–1528. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stewart DJ: Wnt signaling pathway in
non-small cell lung cancer. J Natl Cancer Inst. 106:djt3562014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang J, Gao W, Liu G, Sheng W, Zhou J,
Dong Q and Dong M: miR-944 suppresses EGF-induced EMT in colorectal
cancer cells by directly targeting GATA6. Onco Targets Ther.
14:2311–2325. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu Z, Zhang Y, Yang Z, Zhu Y, Xie Y, Zhou
F and Cai L: Elevation of miR-302b prevents multiple myeloma cell
growth and bone destruction by blocking DKK1 secretion. Cancer Cell
Int. 21:1872021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xie Y, Xue C, Guo S and Yang L:
MicroRNA-520a suppresses pathogenesis and progression of
non-small-cell lung cancer through targeting the RRM2/Wnt axis.
Anal Cell Pathol (Amst). 2021:96524202021.PubMed/NCBI
|
|
40
|
Wang H, Yang Q, Li J, Chen W, Jin X and
Wang Y: MicroRNA-15a-5p inhibits endometrial carcinoma
proliferation, invasion and migration via downregulation of VEGFA
and inhibition of the Wnt/β-catenin signaling pathway. Oncol Lett.
21:3102021. View Article : Google Scholar : PubMed/NCBI
|