Introduction
Although mortality rates for estrogen
receptor-positive (ER+) breast cancer in the US have
been rapidly declining with the onset of successful endocrine
therapy [i.e., aromatase inhibitors (AI) and selective estrogen
receptor modulators], the development of resistance remains a
lingering challenge. Previously, it has been demonstrated that AI
resistance is associated with epithelial to mesenchymal transition
(EMT), increased growth factor signaling, and enhanced motility
(1–7). In cancer, EMT refers to several
phenotypic changes, with epithelial cells transitioning into a
mesenchymal phenotype (8). The
resultant phenotype reveals increased migration and invasiveness,
loss of polarity, and resistance to apoptosis (9). Furthermore, the emergence of a
subpopulation of radiation- and chemo-resistant breast CSCs within
AI-resistant breast tumors (1)
continues to complicate therapeutic interventions.
In tumors, CSCs dictate invasion, metastasis, and
drug resistance (10). Previous
reports have revealed that these highly tumorigenic CSCs are
involved in relapse, metastasis, and EMT (11). CSCs are characterized by their
preferential ability to initiate and propagate tumor growth and
their selective capacity for self-renewal and differentiation
(12). Reportedly, Al-Hajj et
al (13) were the first to
definitively identify and characterize human breast CSCs from
patients. They have demonstrated that human breast cancers contain
a subpopulation of CD44high/CD24low cells
that exhibit stem cell and malignant properties. Several reports
have revealed that CSCs are enriched among circulating tumors in
the peripheral blood of patients with breast cancer (14). Recent studies have shown that EMT, an
early step of tumor migration, can differentiate cancer cells into
a CSC-like state (15) thereby
establishing a functional link between CSCs and EMT. Currently,
effective targeted approaches to endocrine-resistant breast cancer
are lacking owing to an inability to inhibit breast CSCs and
completely unravel the rate-limiting proteins and pathways that
drive metastatic disease.
The mammosphere formation assay was established
based on the spheroid model (16).
Mammospheres represent a pre-cancerous state and also act as a
surrogate indicator for the presence of CSCs (17). This model is utilized based on the
rationale that only epithelial cells can survive in mammosphere
suspension cultures, whereas other cells undergo apoptosis owing to
the higher self-renewal capacity of stem cells when compared with
other cells (18–20). Considering the advantages and
appropriateness of this model, we evaluated the characteristics of
letrozole-resistant mammospheres and their implication toward a
more aggressive and migratory phenotype.
Materials and methods
Cell culture
Generation of the LTLT-Ca cell line (long-term
letrozole treated MCF-7 cells stably transfected with the human
aromatase gene) was previously described (4). Briefly, LTLT-Ca cells were isolated
from tumors of aromatase-transfected MCF-7 cells grown in
ovariectomized BALB/c athymic mice after 56 weeks of letrozole
treatment. The tumors start proliferating in the presence of the
drug after long-term treatment. Human LTLT-Ca cells were generously
provided by Dr Angela Brodie and were cultured in 75-cm2
flasks in phenol red-free IMEM (Improved Minimum Essential Medium;
Invitrogen; Thermo Fisher Scientific, Inc.), supplemented with 10%
charcoal-dextran-stripped fetal bovine serum (FBS), 100 U/ml
penicillin (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.), 25 µg/ml
amphotericin B (Gibco; Thermo Fisher Scientific, Inc.), 7.5 µg/ml
geneticin (Invitrogen; Thermo Fisher Scientific, Inc.) 1 µM
letrozole (Sigma-Aldrich; Merck KGaA). The culture flasks were
maintained in a humidified atmosphere of 5% CO2 at 37°C.
Letrozole-sensitive AC-1 cells were maintained in DMEM (Dulbecco's
modified Eagle's medium; Invitrogen; Thermo Fisher Scientific,
Inc.), supplemented with 5% FBS, 100 U/ml penicillin (Gibco; Thermo
Fisher Scientific, Inc.), 100 U/ml streptomycin (Gibco; Thermo
Fisher Scientific, Inc.), 25 µg/ml amphotericin B (Gibco; Thermo
Fisher Scientific, Inc.), 7.5 µg/ml geneticin (Invitrogen; Thermo
Fisher Scientific, Inc.) in a humidified atmosphere of 5%
CO2 at 37°C. T47D letrozole-sensitive (T47Darom) and
T47D letrozole-resistant (T47DaromlR) cells were cultured as
previously described by Gupta et al (21) and were a generous gift from IIT
Research Institute. Mycoplasma testing was performed for all cell
lines. All cells were authenticated by short tandem repeat (STR)
profiling by American Type Culture Collection (ATCC), confirming
that the AC-1 and LTLT-Ca cell lines shared more than 85% homology
with the MCF-7 cell lines, while the T47Darom and T47DaromLR cell
lines shared more than 85% homology with the T47D cell line. Cell
lines with 80% match are considered related (derived from a common
ancestry). In brief, 17 STR loci plus the sex-determining locus,
amelogenin, were amplified using the commercially available
PowerPlex® 18D Kit (Promega). The cell line samples were
processed using the ABI Prism® 3500 ×L Genetic Analyzer.
Data were analyzed using the GeneMapper® ID-X v1.2
software (Applied Biosystems). Appropriate positive controls (MCF-7
or T47D cell lines) were run and confirmed for each sample
submitted. Cell lines were authenticated using STR analysis as
described in 2012 in the ANSI Standard (ASN-0002) by the ATCC
Standards Development Organization (SDO), as well as Capes-Davis
et al (22).
Mammosphere culture and mammosphere
formation assay
AC-1 and LTLT-Ca cells were grown in regular media
to attain 80–90% confluency, and after media was removed, cells
were rinsed twice with Hank's Balanced Salt Solution (HBSS;
StemCell Technologies, Vancouver, Canada) to remove residual
culture media. Then, cells were gently scraped and resuspended in
10 ml of MammoCult™ media (StemCell Technologies). Next, cells were
centrifuged at 500 × g for 3 min at room temperature. The
supernatant was aspirated, and the pellet was resuspended into a
single cell suspension in 2 ml of MammoCult™ media. Cell viability
and concentration were determined by the Trypan Blue exclusion
assay. For mammosphere cultures, the seeding density for both cell
lines were 100,000 cells per 25-cm2 suspension flasks
(CellTreat Scientific Products). All flasks were incubated in a 5%
CO2 humidified incubator at 37°C for at least 7 days.
Once mammospheres were detected by light microscopy, the cells were
harvested as detailed below in western blot analysis.
Mammosphere self-renewal assay
For primary mammosphere formation, AC-1 and LTLT-Ca
cells were enumerated, and 20,000 cells/well were seeded in
ultra-low adhesion 6-well plates. The cultures were incubated in a
5% CO2 humidified incubator at 37°C for 7 days and
spheres ≥60 µm were counted and recorded. After primary spheres
were formed, secondary mammosphere formation was conducted for both
AC-1 and LTLT-Ca cells by dissociating the primary spheres with
Accutase (Sigma-Aldrich; Merck KGaA) according to the
manufacturer's instructions. Next, 20,000 cells/well were seeded in
ultra-low adhesion 6-well plates, with the remainder of the assay
conducted as described for primary mammosphere formation.
Cell proliferation assay
Proliferation assays were conducted as previously
described (23). Briefly, AC-1 or
LTLT-Ca cells were seeded in 96-well plates at a density of 1 ×
103 cells/well in a total volume of 100 µl and allowed
to attach overnight. Background levels were determined by preparing
blank samples, with media added to wells in the absence of cells.
On the following day, 10 µl of resazurin dye (Sigma-Aldrich; Merck
KGaA) was added to each well and incubated for 4 h at 5%
CO2 and 37°C. Samples were agitated for 1 min and the
fluorescence was measured at 24, 48, 72, 96 and 120 h using a
Biotek Synergy H1 microplate reader (BioTek Instruments, Inc.) to
measure fluorescence intensity at 550 nm excitation/590 nm emission
background wavelengths. All experiments were performed with n≥3 and
a total of 3 biological replicates were performed. The
proliferative activity was determined and calculated as described
below: Proliferative activity = [Fluorescence of viable
cells-Fluorescence of blank (media only)].
Wound healing assay
AC-1 and LTLT-Ca mammospheres were dissociated and
single cells were seeded at a density of 1.5×105
cells/well in 6-well tissue culture plates until 100% confluency
was attained. A scratch (wound) was made down the center of each
well using a 10 µl pipette tip, and new media was added. Each
scratch was immediately imaged (at 0 h) 3 times at different points
using the 5X objective on a Zeiss AX10 fluorescence microscope.
Images were captured using Olympus CellSens Standard 1.16 software.
Then, cells were grown at 37°C, 5% CO2, and images were
captured at 24 and 48 h. The wounds were measured and analyzed
using the WimScratch Wound healing Software.
Gel electrophoresis and western blot
analysis
Briefly, adherent cells were gently scraped and
homogenized in cold RIPA buffer supplemented with 2X protease and
phosphatase inhibitors (Thermo Fisher Scientific, Inc.).
Mammospheres were centrifuged at 400 × g for 7 min at 4°C and then
resuspended in cold RIPA buffer. All samples were incubated for 30
min on ice and then centrifuged for 20 min at 12,000 rpm. The
protein extract was quantified in each sample using the Bradford
assay. For gel electrophoresis, all samples were incubated with
Laemmli protein sample buffer (Bio-Rad) at 70°C for 10 min. Then,
75 µg of denatured protein was separated using 4–20%
Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-Rad) and
transferred to polyvinylidene difluoride (PVDF) membranes. All
blots were blocked for 1 h with 5% bovine serum albumin (BSA) in
phosphate-buffered saline (PBS) and 0.1% Tween-20 (PBS-T) buffer.
Following incubation with primary antibodies, anti-CD24 antibody at
1:1,000 dilution (catalog no. ab179821; Abcam) and anti-CD44
antibody 1:1,000 dilution (catalog no. ab157107; Abcam), the
membranes were incubated with the anti-rabbit secondary antibody
(catalog no. 7074S; Cell Signaling Technology). The protein bands
were detected using the Clarity Max Western ECL Substrate (Bio-Rad)
according to the manufacturer's instructions. Immunoreactive bands
were visualized using the ChemiDoc XRS imaging system (Bio-Rad).
The exposure time was automatically detected by the imaging system.
The protein bands were analyzed using Image Lab software (Bio-Rad).
Arbitrary densitometry units were quantified and expressed as mean
± standard deviation. The bands were normalized to the housekeeping
protein bands (GAPDH), whereby the density of the target protein in
each lane was multiplied by the ratio of the loading control
density from the control sample (lane 1) to the loading control
density of other lanes. The immunoblot images are representative of
more than three independent experiments with a minimum of 2
duplicates per sample.
Animal letrozole- sensitive and
letrozole-resistant cell tumors
SCID female ovariectomized mice (29–32 days old)
were obtained from Charles River Laboratories. All animals
underwent an adaptation period of 5–7 days in a pathogen-free and
sterile environment, with a phytoestrogen-free diet. AC-1 and
LTLT-Ca cells in the exponential phase of growth were harvested
using PBS supplemented with 2% EDTA solution and washed. Viable
cells (5×106) in a 50 µl sterile PBS suspension were
mixed with 100 µl Matrigel Reduced Factors (BD Biosciences). AC-1
and LTLT-Ca cells were injected into the mammary fat pad (n=5 for
each group). For AC-1 cells, estrogen pellets (0.72 mg, 60-day
release; Innovative Research of America) were subcutaneously
implanted in the lateral area of the neck, in the middle point
between the ear and shoulder, using a precision trocar (10 gauge).
All animal procedures were performed under anesthesia using a
ketamine/xylazine mixture consisting of 80 mg/kg of ketamine and 10
mg/kg of xylazine. Tumors were allowed to form over 10 days. Tumor
volume was measured weekly for 8 weeks using a digital caliper.
Tumor volume was calculated using the following formula:
4/3LM2, where L is the larger radius and M is the
smaller radius. At necropsy, animals were euthanized by exposure to
a CO2 chamber with a flow rate of 2L of
CO2/min at a displacement rate of 30–70% of the chamber
volume per minute. Death was confirmed by evidence of pale eyes and
the absence of a heartbeat and lack of respiration for at least 1
min. For further analysis, tumors were removed and fixed in 10%
formalin. All procedures involving animals were conducted in
compliance with state and federal laws, standards of the US
Department of Health and Human Services, and guidelines established
by the Xavier University of Louisiana University Animal Care and
Use Committee. The facilities and laboratory animal program of
Xavier University of Louisiana are accredited by the Association
for the Assessment and Accreditation of Laboratory Animal Care.
Immunohistochemistry of letrozole-
sensitive and letrozole-resistant tumors
For immunohistochemical analysis, AC-1 and LTLT-Ca
tumors were grown as described above, using groups of 5 animals,
each containing 2 tumors. Immunohistochemistry was performed as
previously described (24). Briefly,
a minimum of one tumor tissue per animal (n=5/group) was fixed,
deparaffinized, and rehydrated. Anti-CD24 (catalog no. MAB5248,
1:100 dilution; R&D Systems) and anti-CD44 (catalog number
MAB7045, 1:100 dilution; R&D Systems) antibodies were used as
potential markers. Staining was performed using EXPOSE mouse and
rabbit specific HRP/DAB detection IHC kit (catalog number: ab80436;
Abcam). Cells were counterstained with hematoxylin for 3 min,
dehydrated, and mounted. Once slides were prepared, at least 4
microscopic fields were randomly selected for each tumor and
visualized using ×20 magnification. Data are presented as a
semi-quantitative Histo-score, where the fractions were assigned as
negative (score 0), weakly positive (score 1), positive (score 2),
or strongly positive (score 3). All slides were scored blindly by
three individual investigators.
Immunofluorescence of letrozole-
sensitive and letrozole-resistant cells
Immunofluorescence was performed as previously
described (24). Briefly, three
replicates of LTLT-Ca cells were seeded in 8-well chamber slides
(Thermo Fisher Scientific, Inc.) pre-coated with 2% gelatin and
grown to 50% confluence. Cells were fixed with formaldehyde,
permeabilized with 0.5% NP-40 in PBS, and rinsed with PBS. The
slides were blocked with 10% goat serum (Invitrogen; Thermo Fisher
Scientific, Inc.) in PBS and incubated with anti-CD24 (1:200)
catalog no. sc-19585, anti-HCAM (1:200) catalog no. sc-7927, and
anti-Ki67 (1:200) catalog no. sc-23900, (Santa Cruz Biotechnology
Inc.). Then, samples were washed with 1% goat serum in PBS and
incubated with Alexa Fluor goat anti-rabbit-488 (1:1,000), catalog
number: A-11008, or Alexa Fluor goat anti-mouse-488 secondary
antibodies (1:1,000), catalog number: A-11001 (Invitrogen; Thermo
Fisher Scientific, Inc.) in 10% goat serum. The samples were washed
and stained with 300 nM DAPI (Invitrogen; Thermo Fisher Scientific,
Inc.). The slides were imaged using an Olympus Bx41 microscope
(Olympus) and captured using DP72 CCD driven by DP2 software
(Olympus); the color images were combined using ImageJ
software.
Reverse transcription-quantitative PCR
(RT-qPCR) for 2D vs. 3D letrozole-resistant cells
Briefly, LTLT-Ca cells were cultured adherently in
75-cm2 flasks in phenol red-free IMEM supplemented with
5% charcoal-stripped-FBS or as mammospheres as described above,
cultured until 70%–80% confluency. Total RNA was extracted from
cells using RNeasy (Qiagen) following the manufacturer's
recommendations. Each array profiles the expression of a panel of
84 genes including 7 internal controls and 5 housekeeping gene
controls. For each array, 2 µg of RNA was reverse-transcribed into
cDNA in the presence of gene-specific oligonucleotide primers using
the iScript cDNA Synthesis kit (Bio-Rad) as described in the
manufacturer's protocol. The cDNA template was mixed with the
appropriate ready-to-use PCR master mix (Bio-Rad). Equal volumes
were measured (in aliquots) into each well of the same plate, and
then the real-time PCR cycling program was run as described
previously (7). RT-qPCR was
performed using the manufacturer's protocols for Human Cell
Motility (PAHS-128ZD) and Human Epithelial to Mesenchymal
Transition (PAHS-090ZD) (EMT) RT2 Profiler PCR Array
(Qiagen). Relative gene expression was calculated using the
2−ΔΔCq method (25), in which Ct indicates the fractional
cycle number where the fluorescent signal reaches the detection
threshold. The ‘delta-delta’ method uses the normalized ΔCt value
of each sample, calculated using a total of five housekeeping gene
control genes (18S rRNA, HPRT1, RPL13A, GAPDH and ACTB) (26). Fold change values are presented as
average fold change = 2−(average ΔΔCt) for
genes in treated samples, relative to control samples. Differences
in gene expression between groups were calculated using Student's
t-test, in which fold changes ≤3 were considered significant. All
experiments were performed with a minimum of three biological
replicates.
Statistical analysis
Studies involving more than 2 groups were analyzed
by 1-way ANOVA with Tukey's posttest analysis; all others were
subjected to unpaired Student's t test and are summarized as the
mean ± standard error of the mean (SEM) using Graph Pad Prism V.6
(GraphPad Software, Inc.). Data are expressed as the mean unit ±
SEM (****P<0.0001, ***P<0.001, **P<0.01, *P<0.05).
Results
Increased presence of cancer stem cell
markers is associated with letrozole resistance
We have previously demonstrated that as breast
cancer cells transition from letrozole-sensitive to
letrozole-resistant, they are associated with estrogen
independence, enhanced cellular motility, and an EMT-like phenotype
(7,24). EMT is linked to the progression of
cancer, as well as increased stemness of tumors (27). To examine the expression of two
putative breast CSC markers in letrozole-sensitive (AC-1 cells) and
letrozole-resistant (LTLT-Ca cells) breast cancer cell lines, we
performed immunoblotting analysis. Considering that the mammosphere
formation assay is employed as a surrogate reporter of cancer stem
cell activity, both cell lines were cultured either adherently (2D)
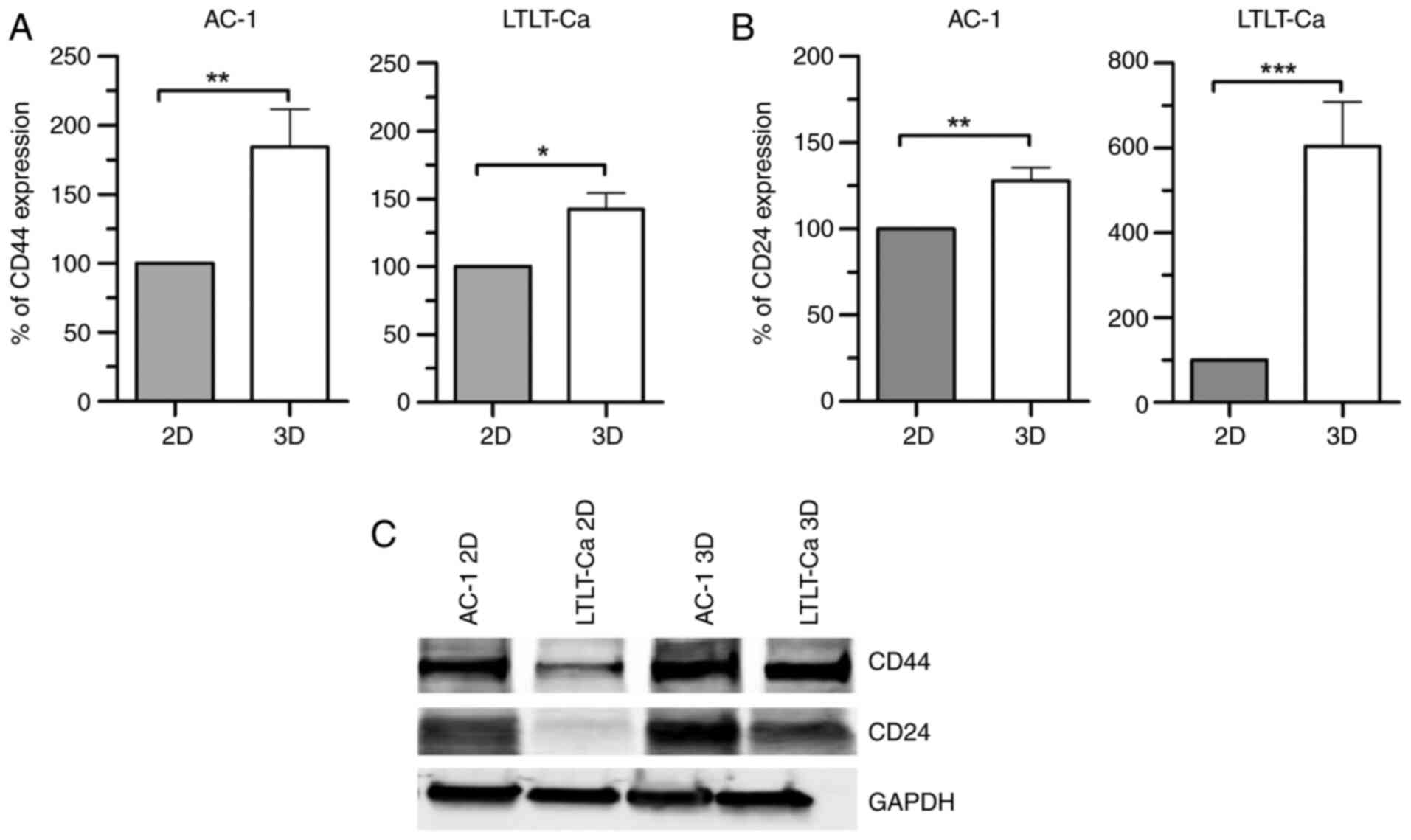
or in suspension (3D) as mammospheres. Our results demonstrated
that when both AC-1 and LTLT-Ca cells were cultured as
mammospheres, higher levels of CD24 and CD44 were expressed when
compared with their adherently cultured counterparts (Fig. 1). On comparing CD24 expression
between 2D and 3D cells, AC-1 3D cells exhibited an increase in
CD24 expression exceeding 25%, while LTLT-Ca 3D cells exhibited a
500% increase in CD24 expression when compared with their 2D
counterparts. When CD44 expression was examined, AC-1 3D and
LTLT-Ca 3D cells exhibited increased CD44 expression (75 and 50%,
respectively), suggesting that in both cell lines, mammosphere
cultures presented enriched CSC characteristics when compared with
cells cultured adherently, irrespective of their response to
letrozole. Furthermore, a similar result was observed in T47D
letrozole-sensitive and letrozole-resistant cell lines (Fig. S1).
Although in vitro human breast cancer cell
models are useful screening tools, they can be limited by the
absence of the breast tumor microenvironment. As in vitro
cultured cells exhibit less complexity when compared with those
in vivo, AC-1 and LTLT-Ca cells were inoculated into nude
mice and allowed to form tumors (Fig.
S2). The maximum diameter and volume of the AC-1 tumors were
6.36 × 6.78 mm and 365.66 mm3 respectively, while the
maximum diameter and volume of the LTLT-Ca tumors were 5.24 ×5.85
mm and 214.17 mm3 respectively. Then, tumors were
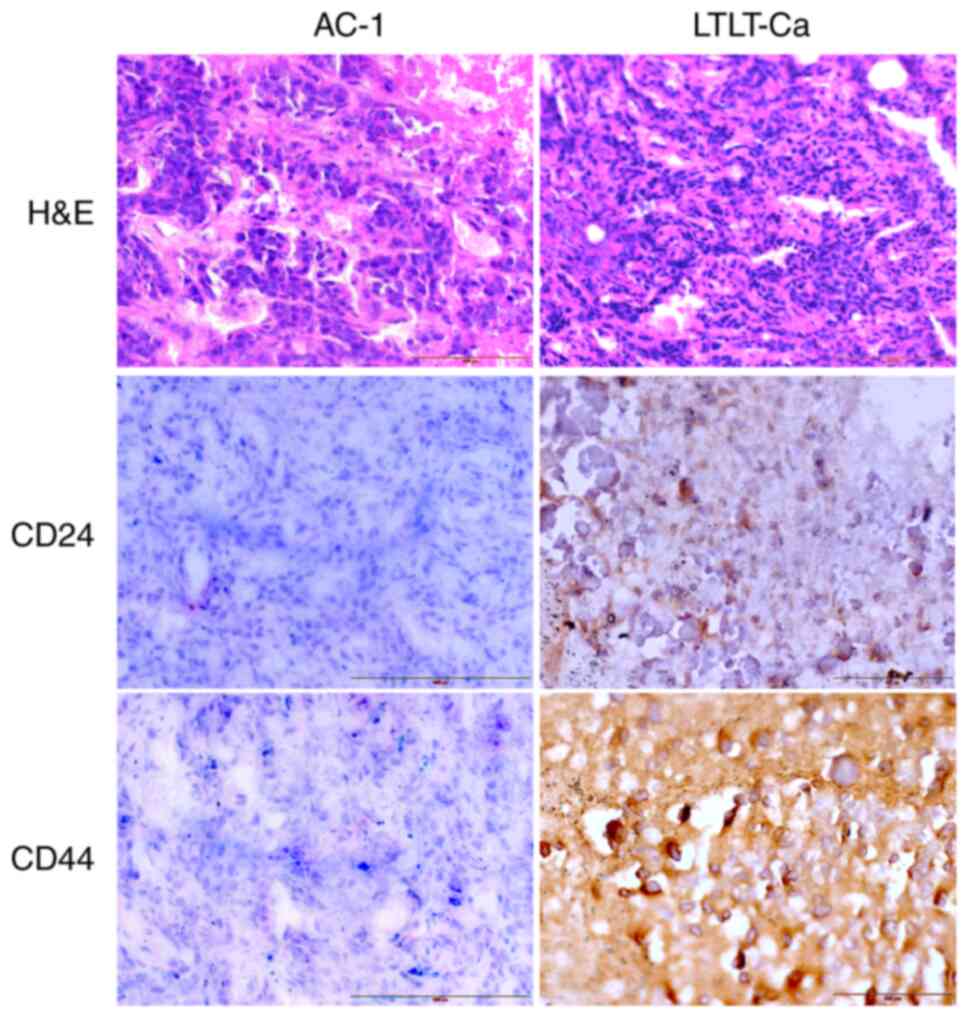
excised, and CD24 and CD44 protein expressions were examined by
immunohistochemistry. In addition, hematoxylin-eosin staining was
performed. Letrozole-sensitive tumors revealed markedly less CD44
expression when compared with letrozole-resistant tumors, scoring 1
and 3, respectively (Fig. 2),
whereas CD24 expression was higher in letrozole-resistant tumors
than in letrozole-sensitive tumors, scoring 2 and 1, respectively
(Fig. 2). In summary, the expression
of CSC markers in tumors was CD44+/CD24+ in
letrozole-resistant cells (LTLT-Ca) and low CD44/CD24 in
letrozole-sensitive cells (AC-1 cells); meaning that LTLT-Ca tumors
had higher expression of both CD44 and CD24 and AC-1 tumors had low
expression of CD44 and CD24.
Letrozole resistance is associated
with increased stemness
As marked differences were observed in the CD44/CD24
expression profile between AI-sensitive and AI-resistant tumors, we
examined whether these differences correlated with the self-renewal
capacity by using the mammosphere self-renewal assay. AC-1 and
LTLT-Ca cells were seeded at a low density in an environment that
prevented adherence, thus enabling proliferation in suspension as
spherical clusters (28).
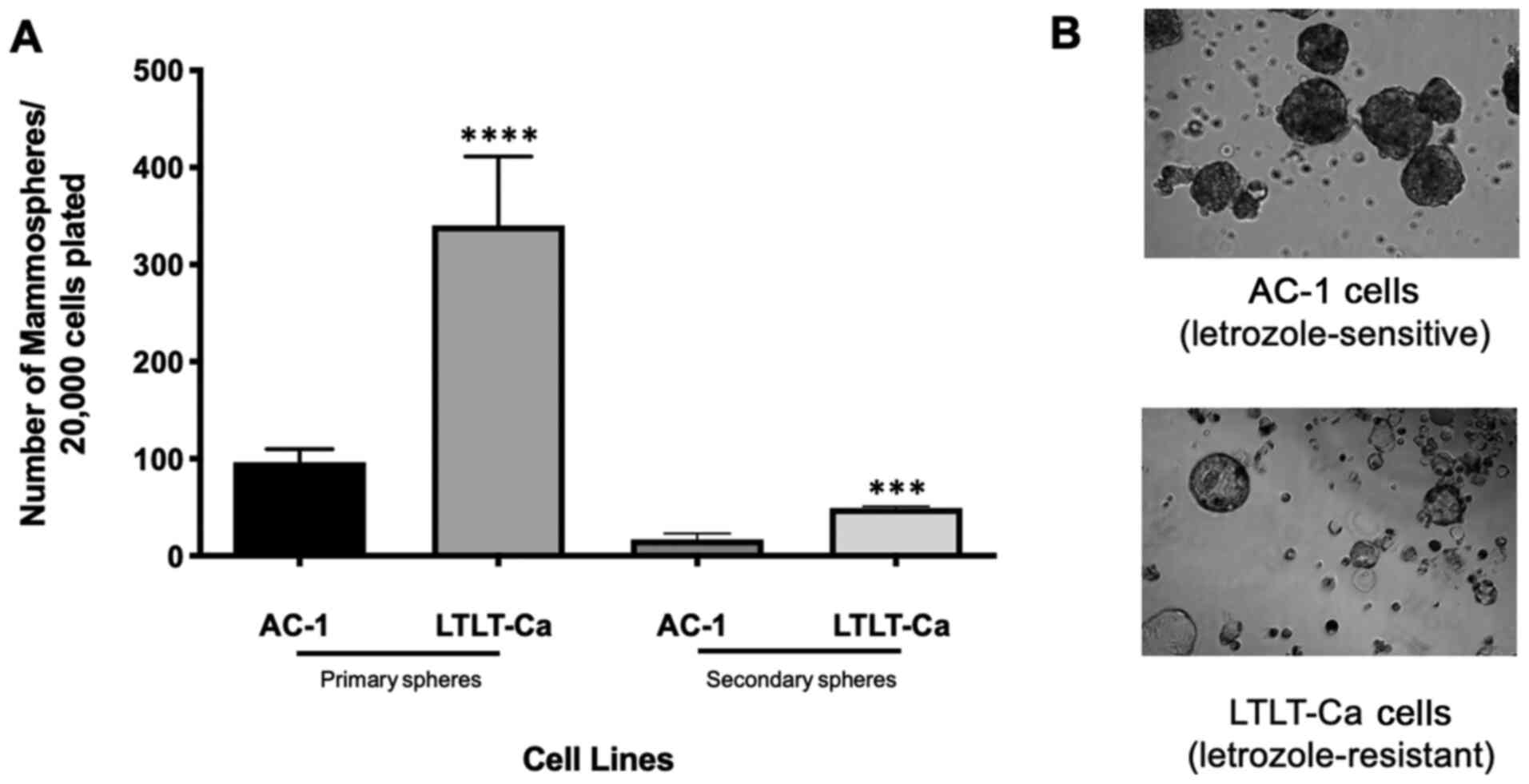
Interestingly, LTLT-Ca cells formed mammospheres at a 3.4-fold
higher index when compared with AC-1 cell mammospheres (Fig. 3A). Culture images were obtained to
examine their morphology, demonstrating that letrozole-resistant
cells formed hollow mammospheres, while letrozole sensitivity was
associated with solid mammospheres (Fig.
3B). Both cell lines formed symmetrical and tightly packed
mammospheres. The mammospheres underwent a second passage, and
LTLT-Ca mammospheres showed a 2.9-fold increase in mammosphere
formation when compared with AC-1 cells. Compared with primary
mammospheres, the total number of secondary mammospheres decreased;
however, the ratio of secondary mammosphere formation between AC-1
and the LTLT-Ca mammospheres was similar to the primary mammosphere
formation ratio. Based on this assay, letrozole-resistant cells
were more highly associated with increased self-renewal capacity
when compared with letrozole-sensitive cells. To determine whether
the increase in LTLT-Ca mammosphere formation could be attributed
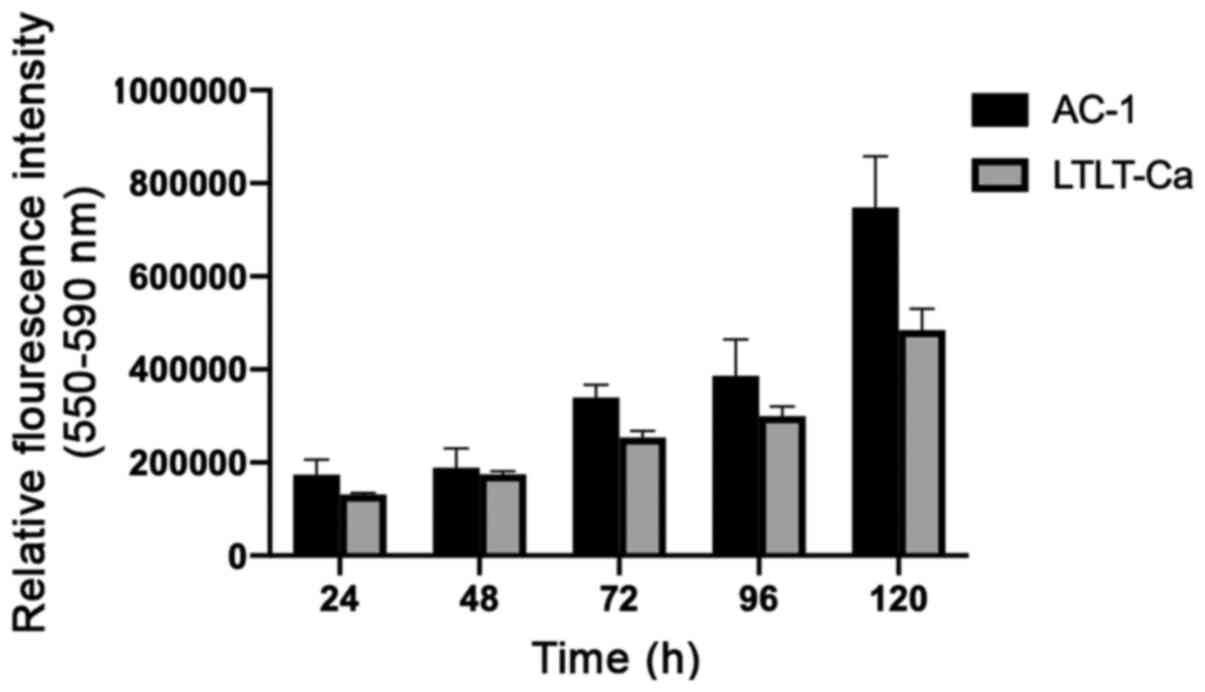
to enhanced cell growth, proliferation assays were performed to
compare both cell lines. AC-1 cells demonstrated a greater
proliferative capacity than LTLT-Ca cells, suggesting that the
increase in LTLT-Ca mammosphere formation was independent of cell
proliferation (Fig. 4). To
complement this finding, immunofluorescent analysis of LTLT-Ca
mammospheres was conducted, and the LTLT-Ca mammospheres were
CD44+/CD24 (Fig. S3),
similar to the tumors and adherent cultures. Furthermore, LTLT-Ca
mammospheres stained positive for Ki67, a common cell proliferation
marker, indicating their proliferative capacity.
Letrozole-resistant mammary cancer
stem cells promote an invasive phenotype
As previous reports have confirmed that LTLT-Ca
cells demonstrate greater migratory ability than AC-1 cells
(7), we measured the expression of
genes involved in motility and EMT, to assess whether the presence
of CSCs was associated with a migratory phenotype. Accordingly,
LTLT-Ca cells were cultured either adherently or as mammospheres,
with targeted gene expression arrays were conducted to measure the
expression of genes involved in cellular motility and EMT (genes
that were significantly altered where P<0.05 are shown in
Table I). When compared with LTLT-Ca
adherent cells, LTLT-Ca mammospheres displayed a −12.01-fold,
−6.44-fold, and −8.14-fold decrease in the expression of
caveolin-1, E-cadherin, and β-catenin, respectively. Interestingly,
LTLT-Ca mammospheres exhibited a −3.33-fold and −3.29-fold decrease
in epidermal growth factor receptor (EGFR) and integrin αV (CD51)
expression. In addition to genes involved in motility, we also
observed increased expression of TFPI-2 (4.42-fold) and STEAP1
(3.17-fold).
 | Table I.SuperArray analysis of gene
expression altered by letrozole-resistant mammospheres. |
Table I.
SuperArray analysis of gene
expression altered by letrozole-resistant mammospheres.
| Gene symbol | Gene
description | Fold change
(LTLT-Ca mammospheres/LTLT-Ca adherent cells) | Gene aliases |
|---|
| CAV1 | Caveolin 1 | −12.01 | BSCL3, CGL3,
MSTP085, VIP21 |
| CAV2 | Caveolin 2 | −3.33 | CAV, MGC12294 |
| CDH1 | E-cadherin | −6.44 | Arc-1, CD324, CDHE,
ECAD, LCAM, UVO |
| CTNNB1 | Catenin
(cadherin-associated protein), β1 | −8.14 | CTNNB,
DKFZp686D02253, FLJ25606, FLJ37923 |
| EGFR | Epidermal growth
factor receptor | −3.33 | ERBB, ERBB1, HER1,
PIG61, mENA |
| F11R | F 11 Receptor | −6.40 | CD321, JAM, JAM1,
JAMA, JCAM, KAT, PAM-1 |
| ITGAV | Integrin, αV | −3.29 | CD51,
DKFZp686A08142, MSK8, VNRA |
| KRT19 | Keratin 19 | −4.07 | CK19, K19, K1CS,
MGC15366 |
| MET | Met
proto-oncogene | −5.38 | AUTS9, HGFR, RCCP2,
c-Met |
| PTK2 | PTK2 protein
tyrosine kinase 2 | −3.05 | FADK, FAK, FAK1,
FRNK, pp125FAK |
| STEAP1 | Six transmembrane
epithelial antigen of the prostate 1 | 3.17 | MGC19484, PRSS24,
STEAP |
| TFPI2 | Tissue factor
pathway inhibitor 2 | 4.42 | FLJ21164, PP5,
REF1, TFPI-2 |
| TGFB2 | Transforming growth
factor, β2 | −4.23 | MGC116892,
TGF-β2 |
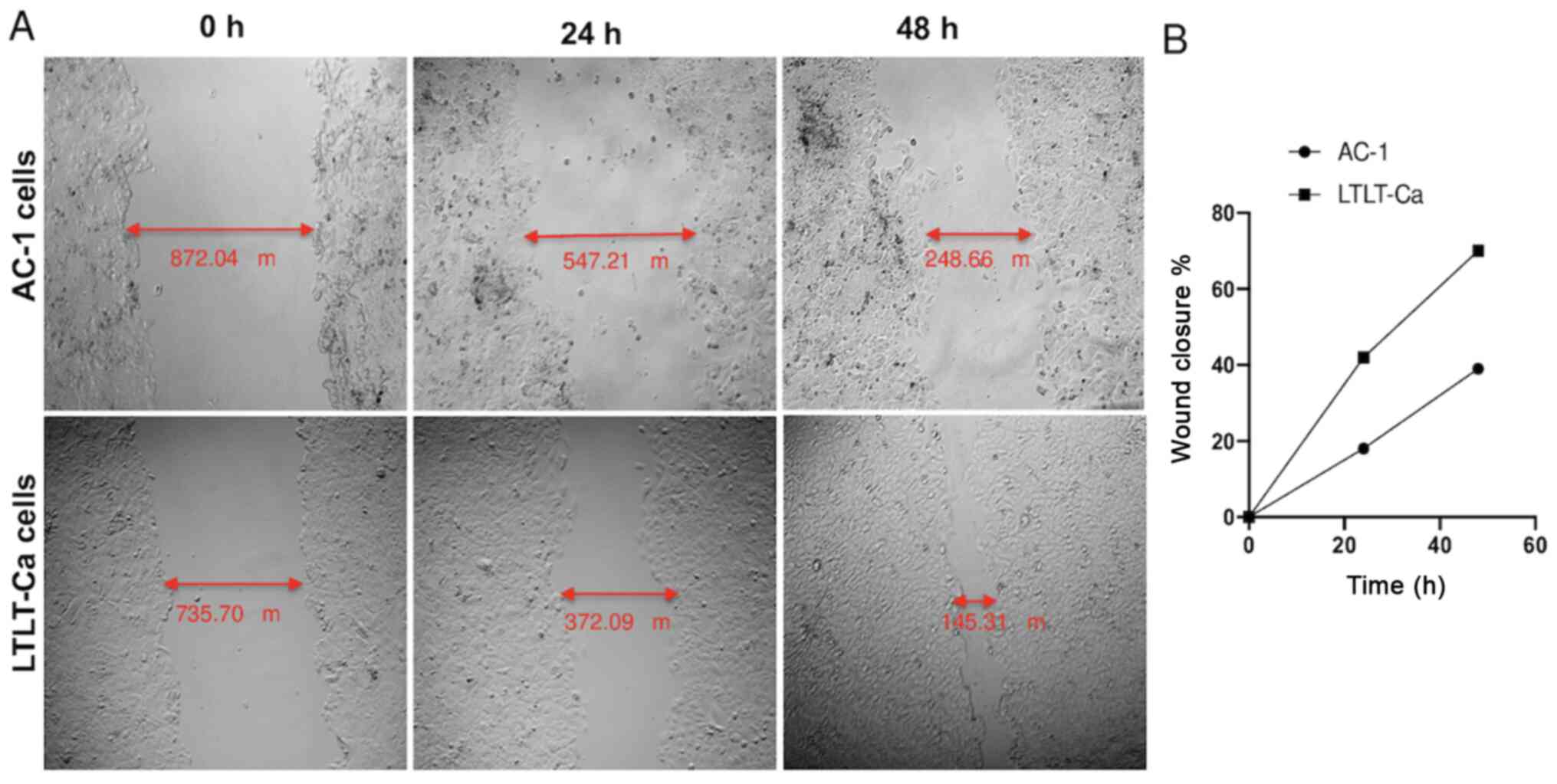
As proof of concept, a scratch assay (i.e., wound
healing assay) was performed to assess whether the
letrozole-resistant mammospheres impacted migratory behavior
(Fig. 5). The results demonstrated
that as early as 24 h, the letrozole-resistant mammospheres began
migrating faster than the letrozole-sensitive mammospheres
(Fig. 5). After 48 h, this effect
was even more pronounced, and the LTLT-Ca cell wound closure was
70%, whereas the AC-1 wound closure was 39%, suggesting that as
cells acquire resistance and express putative breast CSC markers,
they become more aggressive through increased motility. Taken
together, as letrozole-resistant cells acquire CSC characteristics,
they are less associated with epithelial-like features, progressing
toward a more mesenchymal phenotype.
Discussion
Letrozole resistance remains a major clinical
obstacle. Although newer targeted therapeutic approaches that
combine letrozole and palbociclib (a CDK4/6 inhibitor) are
available, this therapeutic strategy has been reserved for
ER-positive metastatic breast cancer patients. Unfortunately, there
are no effective targeted therapies currently available for
ER-negative, letrozole-resistant metastatic breast cancer patients.
Therefore, identifying mechanisms of resistance among this
population is highly significant. Previously, Al-Hajj et al
(13) have demonstrated that
CD44+/CD24−/low cells within a breast tumor
possess self-renewal properties and are capable of tumor formation.
Therefore, it is crucial to understand this cell subpopulation, as
they are associated with cancer recurrence and treatment
resistance. Thus, these cells must be targeted for eradication to
prevent tumor relapse. To comprehensively understand the role of
mammospheres in breast cancer resistance, we performed a series of
studies to characterize CSC markers in letrozole-resistant breast
cancer cells. Although CD44 is considered the most established CSC
marker in a majority of cancers (29), CD24 remains controversial owing to
its prognostic value and significance (30).
Immunoblots were performed and revealed a marked
difference in the CD44 and CD24 expression profiles between
adherent cells and mammospheres of cells, as well as between
letrozole-resistant vs. letrozole-sensitive tumors. Letrozole-
resistant mammospheres demonstrated a higher expression of both
CD44 and CD24 than adherent cells. Our in vitro findings are
in accordance with previous studies reporting high CD44 expression
in half of the breast cancer cell lines studied and that most of
the cell lines expressed increased amounts of CD24 (30). Some results obtained by Ricardo et
al (30) were also corroborated
by Li et al (31,32), where it was suggested that the
CD44/CD24 ratio would serve as a more effective marker to identify
stemness of cancer cells. Our letrozole-resistant tumors were
CD44+/CD24+, while the letrozole-sensitive
tumors had low levels of CD44/CD24. To further analyze the LTLT-Ca
mammospheres, immunofluorescent staining was performed, and results
were consistent with the immunoblots. Although we expected that
LTLT-Ca tumors would exhibit reduced CD24 expression, our findings
were substantiated by previous reports demonstrating that the
MDA-MB 468 triple-negative breast cancer cell line exhibits a
similar CD44+/CD24+ expression profile
(30). This phenotype is indicative
of a highly differentiated basal/epithelial cell type. In this
study, our results regarding the CD44+/CD24+
phenotype may indicate interconversion between phenotypes.
Furthermore, our findings suggest that the epithelial-like
CD44+/CD24+ phenotype can readily give rise
to CD44+/CD24− cells during tumor initiation
(33). The
CD44+/CD24+ phenotype may represent a
transient state as cells progress to a more mesenchymal
CD44+/CD24− state.
An additional consideration is that as tumors were
formed, the mice may be affected by other factors, including the
tumor microenvironment and the potential contribution of mouse stem
cells. The former plays a major role in cellular signaling,
cell-cell communication, and cell surface markers such as CD24 and
CD44, which exhibit variable expression levels at different stages
of tumorigenesis. Since mouse specific stem cell markers were not
explicitly examined, this may represent another avenue contributing
to tumor formation and a limitation to this present study.
One of the most striking morphological features
observed was that LTLT-Ca cells formed hollow mammospheres, whereas
AC-1 cells formed solid mammospheres. Although still unclear, this
change in morphology is likely associated with changes that occur
as cells transition from a letrozole-sensitive to a
letrozole-resistant phenotype. Previous reports from our research
group have revealed that compared with letrozole-sensitive AC-1
cells, LTLT-Ca cells exhibit a change in cell morphology from a
rounded, uniform cell body to a less organized cell body with
protrusions indicative of EMT (7).
The mammosphere formation assay revealed that LTLT-Ca cells were
able to form more mammospheres than AC-1 cells in both primary and
secondary passages independent of proliferation.
Considering these morphological alterations along
with the increased mammospheres formation potential of LTLT-Ca
cells, we aimed to clarify whether mammosphere culture conditions
revealed novel changes in motility and gene expression that are
absent in adherent cultures and results demonstrated LTLT-Ca cells
exhibited increased migratory potential. Gene expression studies
between LTLT-Ca mammospheres and LTLT-Ca adherent cells were
performed, and our findings undoubtedly demonstrated that
caveolin-associated genes were significantly downregulated.
Reportedly, loss of caveolin 1 (CAV1) is found to be associated
with poor patient outcomes. More specifically, the absence of CAV1
in breast cancer stroma is associated with poor clinical outcomes
(34–36), including early tumor recurrence,
lymph node metastasis, and tamoxifen resistance. Additionally, when
CAV1 was silenced in stromal cells, it promoted tumor growth in
breast cancer xenograft mouse models (37), suggesting that CAV1 functions as a
tumor suppressor. This is a significant finding as CAV1
downregulation leads to the loss of E-cadherin and increased
transcriptional activity of β-catenin, as well as enhanced tumor
cell invasion (38). The loss of
E-cadherin in LTLT-Ca mammospheres was expected as previous in
vivo studies by our group have demonstrated that
letrozole-resistant tumors express low levels of E-cadherin and
high levels of N-cadherin (24).
Herein, further loss of cell-cell adhesion, as indicated by the
−6.44-fold decrease in E-cadherin expression, is associated with
tumor progression, invasion, and metastasis, all of which are
clinically relevant features of letrozole-resistant breast cancer.
Consequently, decreased expression of genes that collectively
promote cell adhesion (CAV1, CAV2, CDH1 and CTNNB1) enables cells
to dissociate from the primary tumor and transition to a more
mesenchymal phenotype. Ultimately, the cadherin/catenin complex is
critical for epithelial integrity, while the consequences of
β-catenin deletion have not been experimentally investigated, and
the loss of E-cadherin-bound β-catenin correlates significantly
with poor outcomes in breast cancer (39). While additional motility assays, like
the invasion assay, were not performed, based on the LTLT-Ca
mammosphere gene expression profile and migration assay results, it
is likely the invasive behavior of the LTLT-Ca cells will follow a
similar trend as the migratory behavior.
Furthermore, gene expression studies revealed the
upregulation of two genes: six transmembrane epithelial antigen of
prostate 1 (STEAP1) and tissue factor pathway inhibitor 2 (TFPI2).
STEAP1 has roles in intercellular communication, serves as a
channel or transporter, and is involved in cell adhesion (40). Although it has been previously
reported that low STEAP1 expression is associated with a malignant
phenotype and poor prognosis (41),
the relationship between STEAP1 and breast cancer remains unclear.
Moreover, previous reports have indicated conflicting roles for
STEAP1 in breast cancer; however, our finding revealing increased
STEAP1 expression in LTLT-Ca mammospheres supports the findings of
Maia et al (42), which
demonstrated that STEAP1 is overexpressed in the MCF-7 breast
cancer cell line, human breast cancer epithelial cells, and rat
mammary glands. Further studies are needed to confirm the
contribution of STEAP1 in various subtypes of breast cancer. TFPI-2
was increased by 4.42-fold and is a serine protease inhibitor
involved in preventing the release of matrix metalloproteinases.
Additional reports have revealed that TFPI-2 suppresses breast
cancer cell proliferation (43). As
mammospheres are a surrogate reporter of CSCs, the increased TFPI-2
expression was not surprising as this cell subpopulation is
relatively dormant and not highly proliferative.
In summary, we characterized letrozole-resistant
mammospheres using the mammospheres formation assay, immunoblotting
of cells and tumors, and gene expression arrays.
Letrozole-resistant mammospheres were associated with the
expression of CD44+/CD24+, increased
stemness, invasive markers, and increased migration. Collectively,
as letrozole resistance is more highly associated with CSCs, this
may provide mechanistic insight into a new strategy to target the
drug-resistant nature of AI-resistant breast cancer. Future studies
may require analysis of letrozole sensitive mammospheres to
pin-point the molecular pathways contributing to drug sensitivity
within the cancer stem cell population which ultimately may reveal
new targets to exploit for breast cancer patients.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Mrs. Tonya Bryant
and Dr Boyada Tep from the College of Pharmacy and Pharmaceutical
Sciences, Institute of Public Health, Florida A&M University,
Tallahassee, FL, USA for assisting with the immunohistochemistry
slides.
Funding
This work was supported in part by the National
Institutes of Health (NIH; grant no. 1SC1GM126617). This
publication was supported by funding from the Louisiana Cancer
Research Consortium and the National Institutes of Health Research
Centers at Minority Institutes (grant nos. 8G12MD007595 and
U54MD007582) from the National Institute on Minority Health and
Health Disparities. The contents are solely the responsibility of
the authors and do not necessarily represent the official views of
the Louisiana Cancer Research Consortium or the NIH.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SLT directed the project and conceived and designed
the experiments. KMG, JRP, RRW and ID performed the experiments.
KMG, JRP, RRW, AMD and ID contributed to data acquisition. AMD
provided technical editing. JRP and KMG confirmed the authenticity
of all the raw data. SLT, JRP, and KMG analyzed and interpreted the
data. SLT, JRP and KMG wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were approved by the Xavier
University of Louisiana Institutional Animal Care and Use Committee
(New Orleans, USA).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gilani RA, Kazi AA, Shah P, Schech AJ,
Chumsri S, Sabnis G, Jaiswal AK and Brodie AH: The importance of
her2 signaling in the tumor-initiating cell population in aromatase
inhibitor-resistant breast cancer. Breast Cancer Res Treat.
135:681–692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brodie A, Jelovac D, Macedo L, Sabnis G,
Tilghman S and Goloubeva O: Therapeutic observations in MCF-7
aromatase xenografts. Clin Cancer Res. 11:884s–888s.
2005.PubMed/NCBI
|
|
3
|
Brodie A, Jelovac D, Sabnis G, Long B,
Macedo L and Goloubeva O: Model systems: Mechanisms involved in the
loss of sensitivity to letrozole. J Steroid Biochem Mol Biol.
95:41–48. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jelovac D, Sabnis G, Long BJ, Macedo L,
Goloubeva OG and Brodie AM: Activation of mitogen-activated protein
kinase in xenografts and cells during prolonged treatment with
aromatase inhibitor letrozole. Cancer Res. 65:5380–5389. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sabnis G and Brodie A: Adaptive changes
results in activation of alternate signaling pathways and
resistance to aromatase inhibitor resistance. Mol Cell Endocrinol.
340:142–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sabnis G, Schayowitz A, Goloubeva O,
Macedo L and Brodie A: Trastuzumab reverses letrozole resistance
and amplifies the sensitivity of breast cancer cells to estrogen.
Cancer Res. 69:1416–1428. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tilghman SL, Townley I, Zhong Q, Carriere
PP, Zou J, Llopis SD, Preyan LC, Williams CC, Skripnikova E,
Bratton MR, et al: Proteomic signatures of acquired letrozole
resistance in breast cancer: Suppressed estrogen signaling and
increased cell motility and invasiveness. Mol Cell Proteomics.
12:2440–2455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsai JH and Yang J: Epithelial-mesenchymal
plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gupta PB, Chaffer CL and Weinberg RA:
Cancer stem cells: Mirage or reality? Nat Med. 15:1010–1012. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell
LL, Polyak K, Brisken C, Yang J and Weinberg RA: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Al-Hajj M and Clarke MF: Self-renewal and
solid tumor stem cells. Oncogene. 23:7274–7282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Armstrong AJ, Marengo MS, Oltean S, Kemeny
G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ and
Garcia-Blanco MA: Circulating tumor cells from patients with
advanced prostate and breast cancer display both epithelial and
mesenchymal markers. Mol Cancer Res. 9:997–1007. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van der Horst G, Bos L and van der Pluijm
G: Epithelial plasticity, cancer stem cells, and the
tumor-supportive stroma in bladder carcinoma. Mol Cancer Res.
10:995–1009. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weiswald LB, Bellet D and Dangles-Marie V:
Spherical cancer models in tumor biology. Neoplasia. 17:1–15. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Serrano M: The ink4a/arf locus in murine
tumorigenesis. Carcinogenesis. 21:865–869. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cicalese A, Bonizzi G, Pasi CE, Faretta M,
Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP and Pelicci
PG: The tumor suppressor p53 regulates polarity of self-renewing
divisions in mammary stem cells. Cell. 138:1083–1095. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dey D, Saxena M, Paranjape AN, Krishnan V,
Giraddi R, Kumar MV, Mukherjee G and Rangarajan A: Phenotypic and
functional characterization of human mammary stem/progenitor cells
in long term culture. PLoS One. 4:e53292009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Manuel Iglesias J, Beloqui I,
Garcia-Garcia F, Leis O, Vazquez-Martin A, Eguiara A, Cufi S, Pavon
A, Menendez JA, Dopazo J and Martin AG: Mammosphere formation in
breast carcinoma cell lines depends upon expression of e-cadherin.
PLoS One. 8:e772812013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gupta A, Mehta R, Alimirah F, Peng X,
Murillo G, Wiehle R and Mehta RG: Efficacy and mechanism of action
of proellex, an antiprogestin in aromatase overexpressing and
letrozole resistant t47d breast cancer cells. J Steroid Biochem Mol
Biol. 133:30–42. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Capes-Davis A, Reid YA, Kline MC, Storts
DR, Strauss E, Dirks WG, Drexler HG, MacLeod RA, Sykes G, Kohara A,
et al: Match criteria for human cell line authentication: Where do
we draw the line? Int J Cancer. 132:2510–2519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Johnson KP, Yearby LA, Stoute D, Burow ME,
Rhodes LV, Gray M, Carriere P, Tilghman SL, McLachlan JA, Ochieng
J, et al: In vitro and in vivo evaluation of novel anticancer
agents in triple negative breast cancer models. J Health Care Poor
Underserved. 24 (Suppl 1):104–111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carriere PP, Llopis SD, Naiki AC, Nguyen
G, Phan T, Nguyen MM, Preyan LC, Yearby L, Pratt J, Burks H, et al:
Glyceollin i reverses epithelial to mesenchymal transition in
letrozole resistant breast cancer through zeb1. Int J Environ Res
Public Health. 13:ijerph130100102016.PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative pcr and
the 2(−delta delta c(t)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pfaffl MW, Lange IG, Daxenberger A and
Meyer HH: Tissue-specific expression pattern of estrogen receptors
(ER): Quantification of ER alpha and ER beta mRNA with real-time
RT-PCR. Acta Pathol Microbiol Scand Suppl. 109:345–355. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Creighton CJ, Li X, Landis M, Dixon JM,
Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A,
Herschkowitz JI, et al: Residual breast cancers after conventional
therapy display mesenchymal as well as tumor-initiating features.
Proc Natl Acad Sci USA. 106:13820–13825. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dontu G, Abdallah WM, Foley JM, Jackson
KW, Clarke MF, Kawamura MJ and Wicha MS: In vitro propagation and
transcriptional profiling of human mammary stem/progenitor cells.
Genes Dev. 17:1253–1270. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du L, Wang H, He L, Zhang J, Ni B, Wang X,
Jin H, Cahuzac N, Mehrpour M, Lu Y and Chen Q: Cd44 is of
functional importance for colorectal cancer stem cells. Clin Cancer
Res. 14:6751–6760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ricardo S, Vieira AF, Gerhard R, Leitao D,
Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F and Paredes
J: Breast cancer stem cell markers cd44, cd24 and aldh1: Expression
distribution within intrinsic molecular subtype. J Clin Pathol.
64:937–946. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li W, Ma H, Zhang J, Zhu L, Wang C and
Yang Y: Unraveling the roles of cd44/cd24 and aldh1 as cancer stem
cell markers in tumorigenesis and metastasis. Sci Rep. 7:138562017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li W, Ma H, Zhang J, Zhu L, Wang C and
Yang Y: Author correction: Unraveling the roles of cd44/cd24 and
aldh1 as cancer stem cell markers in tumorigenesis and metastasis.
Sci Rep. 8:42762018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oliveras-Ferraros C, Vazquez-Martin A,
Martin-Castillo B, Cufí S, Del Barco S, Lopez-Bonet E, Brunet J and
Menendez JA: Dynamic emergence of the mesenchymal
cd44(pos)cd24(neg/low) phenotype in her2-gene amplified breast
cancer cells with de novo resistance to trastuzumab (herceptin).
Biochem Biophys Res Commun. 397:27–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Simpkins SA, Hanby AM, Holliday DL and
Speirs V: Clinical and functional significance of loss of
caveolin-1 expression in breast cancer-associated fibroblasts. J
Pathol. 227:490–498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sloan EK, Ciocca DR, Pouliot N, Natoli A,
Restall C, Henderson MA, Fanelli MA, Cuello-Carrión FD, Gago FE and
Anderson RL: Stromal cell expression of caveolin-1 predicts outcome
in breast cancer. Am J Pathol. 174:2035–2043. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Witkiewicz AK, Dasgupta A, Sotgia F,
Mercier I, Pestell RG, Sabel M, Kleer CG, Brody JR and Lisanti MP:
An absence of stromal caveolin-1 expression predicts early tumor
recurrence and poor clinical outcome in human breast cancers. Am J
Pathol. 174:2023–2034. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Trimmer C, Sotgia F, Whitaker-Menezes D,
Balliet RM, Eaton G, Martinez-Outschoorn UE, Pavlides S, Howell A,
Iozzo RV, Pestell RG, et al: Caveolin-1 and mitochondrial sod2
(mnsod) function as tumor suppressors in the stromal
microenvironment: A new genetically tractable model for human
cancer associated fibroblasts. Cancer Biol Ther. 11:383–394. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu Z, Ghosh S, Wang Z and Hunter T:
Downregulation of caveolin-1 function by egf leads to the loss of
e-cadherin, increased transcriptional activity of beta-catenin, and
enhanced tumor cell invasion. Cancer Cell. 4:499–515. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dolled-Filhart M, McCabe A, Giltnane J,
Cregger M, Camp RL and Rimm DL: Quantitative in situ analysis of
beta-catenin expression in breast cancer shows decreased expression
is associated with poor outcome. Cancer Res. 66:5487–5494. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gomes IM, Maia CJ and Santos CR: Steap
proteins: From structure to applications in cancer therapy. Mol
Cancer Res. 10:573–587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie J, Yang Y, Sun J, Jiao Z, Zhang H and
Chen J: Steap1 inhibits breast cancer metastasis and is associated
with epithelial-mesenchymal transition procession. Clin Breast
Cancer. 19:e195–e207. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Maia CJ, Socorro S, Schmitt F and Santos
CR: Steap1 is over-expressed in breast cancer and down-regulated by
17beta-estradiol in mcf-7 cells and in the rat mammary gland.
Endocrine. 34:108–116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang G, Huang W, Li W, Chen S, Chen W,
Zhou Y, Peng P and Gu W: Tfpi-2 suppresses breast cancer cell
proliferation and invasion through regulation of erk signaling and
interaction with actinin-4 and myosin-9. Sci Rep. 8:144022018.
View Article : Google Scholar : PubMed/NCBI
|