Introduction
Colon cancer (colon and rectal) is the second most
frequent cause of cancer-related death in the world (1). The overall prognosis of colon cancer
has improved over the past decades owing better surgical techniques
as well as improved neo-adjuvant and adjuvant treatment. Despite
this, the prognosis is still poor when distant metastasis are
present (Stage IV colon cancer) with a 5 year relative survival
rate of 12% (2). Approximately 20%
of colon cancer patients have metastatic disease at diagnosis and
more than 30% of patients with colon cancer develop metastasis over
time (3). The process of metastasis
is complex and regulated by a wide range of cellular and molecular
changes including alterations of cytoskeleton protein and microRNA
(miRNA). Several studies have shown that stress conditions such as
hypoxia, lack of nutrients and pH alterations can induce
up-regulation of pro-metastatic genes (4–6).
Alterations of these genes can subsequently induce cancer cell
migration and invasion, further aggravating the disease.
Four and a half LIM domain 2 protein (FHL2) is a
multifunctional oncoprotein involved in cancer progression and
metastasis (7,8). FHL2 is known to modulate intracellular
signaling pathways by post-translational modifications of proteins
or by altering gene expression via interactions with transcription
factors (9). The expression patterns
of FHL2 are different in different cancer types and FHL2 has been
shown to be down-regulated in some cancer forms such as prostate
cancer (10) and acute myeloid
leukemia (11) whilst up-regulated
in others such as breast cancer (12), ovarian cancer (13), cervical cancer (14) and colon cancer (8). In one recent publication FHL2 was shown
to regulate ovarian cancer cell metastasis via Wnt/β-catenin
signaling (7). In addition, FHL2 has
been demonstrated to promote epithelial-mesenchymal transition
(EMT) in colon cancer cells (8)
which is known to be associated with cancer progression and
metastasis (15). Hence, FHL2 plays
an important role in cancer cell invasion and migration through
multiple different mechanisms.
E-cadherin (E-cad) is an important epithelial cell
adhesion molecule and loss of its function is associated with
epithelial cancer cell migration and invasion (16). E-cad suppresses cancer cell
metastasis by preventing the dislodging of cancer cells from the
primary tumor and thereby reduce cancer cell dissemination
(17). Thus, cancer cell migration
and E-cad levels are inversely related and invasion as well as
metastasis of cancer cells have been reported following loss of
E-cad (18,19). In contrast, E-cad has also been
reported to potentiate metastasis since it enhances cancer cell
survival (20). In addition,
down-regulation of E-cad by FHL2 is associated with EMT in colon
cancer (21). Thus, finding a way of
controlling both FHL2 and E-cad could potentially be a therapeutic
approach in antagonizing colon cancer metastasis.
miRNAs are 21–22 base pair long non-coding RNAs that
post-transcriptionally down-regulate mRNA by targeted degradation
or blocking translation initiation (22). It is estimated that more than 60% of
coding genes in humans have miRNA binding sites in their
3′-untranslated region (3′-UTR) (23). Accumulating data suggest that a
single miRNA may target multiple transcripts and a single
transcript can be regulated by multiple miRNAs within a cell type
(24,25). Some miRNAs are also known to regulate
cell adhesion process and thereby regulate cancer invasion and
metastasis (26). Interestingly,
some cancer-associated miRNAs can act as both oncomir and tumor
suppressive miRNA, dependent on the context and type of cancer
(27). For instance, up-regulation
of miR-340-5p has been shown to promote cancer cell proliferation
and progression in thyroid cancer (28) and gastric cancer (29). In contrast, several studies have
shown that miR-340-5p is down-regulated in glioblastoma multiforme
(30) and breast cancer (31). In addition, one study suggests that
lower expression of miR-340-5p is associated with higher levels of
FHL2 in ovarian cancer (7). However,
the interaction between miR-340-5p and FHL2 and their role in
regulating colon cancer cell migration and invasion has not yet
been investigated.
Based on the considerations above, we hypothesized
that miR-340-5p might regulate colon cancer cell migration and
invasion via targeting FHL2-E-cad axis. For this purpose, we used
metastatic human colon cancer cell lines to evaluate
miR-340-5p-mediated suppression of colon cancer cells migration and
invasion.
Materials and methods
Microarray database analysis
Four human colon cancer-related gene microarray
datasets (GDS4382, GSE115313, GDS4393 and GDS4516) were downloaded
from the Gene Expression Omnibus (GEO) database of NCBI (National
Center for Biotechnology Information). GDS4382 and GSE115313 were
used to compare FHL2 expression between normal colon mucosa and
colon cancer tissue. GDS4393 and GDS4516 were used to compare FHL2
expression between primary colon cancer, metastatic cancer and
metastatic recurrence samples. GDS4382 dataset contains 17 colon
cancer tumors and 17 adjacent non-cancerous tissues. GSE115313 data
set contains 42 paired tumor and normal colon mucosa samples from a
cohort study of 42 colon cancer patients. GDS4393 dataset contains
33 primary and 21 metastatic lesions from patients with
unresectable colon cancer. GDS4516 dataset contains 66 non
metastasized tumors, 20 metastasized tumors and 18 metastatic
recurrence tumors of colon cancer patients. GDS4382, GDS4393 and
GDS4516 datasets were from GPL570: [HG-U133_Plus_2] Affymetrix
Human Genome U133 Plus 2.0 Array platform. GSE115313 dataset was
from GPL16686: [HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array
[transcript (gene) version] platform. Qlucore Omics Explorer
version 3.6 (32) was used for gene
expression analysis and box plots generation. Tukey's post-hoc test
was used for statistical comparison between two groups.
Cells and reagents
The human epithelial colorectal cancer cell line
HT-29 was obtained from American Type Culture Collection (HTB-38,
ATCC). HT-29 cell line was characterized to have metastatic
properties (32,33). AZ-97 cell line was isolated from a
76-year-old female patient undergoing surgical resection and
established in our laboratory at Skåne University Hospital, Malmö,
Sweden (34). Cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich), in the
presence of 10% fetal bovine serum (FBS) and antibiotics (100 U/ml
penicillin, 100 µg/ml streptomycin) at 37°C and 5% CO2.
miR-340-5p mimic and mimic-Ctrl were purchased from Life
Technologies. TransIT-TKO transfection reagent (Mirus) was used to
evaluate the role of miR-340-5p. To study the biological function
of miR-340-5p, a target site blocker (TSB) was purchased from
Exiqon A/S (Vedbaek). The miRCURY LNA_TSB was designed to
specifically compete with the miR-340-5p.
Cell transfection
HT-29 and AZ-97 colon cancer cells were cultured to
70–80% confluency. After that cells were serum starved (0.1% serum)
overnight. On the next day 1×106 cells were seeded in a
6-well culture plate. Cells were then transfected with miR-340-5p
mimic (50 nM) or mimic-Ctrl (50 nM) for 24 h by using Mirus
transfection reagent in Opti-MEM reduced serum media according to
manufacturer's instructions. After 24 h, cells were harvested and
expression of miR-340-5p and FHL2 was analyzed by RT-qPCR. To
evaluate the effect of short serum exposure to serum starved cells,
transfected cells were cultured in Opti-MEM reduced serum media for
24 h and then exposed to 10% BSA for 30 min. RNA samples were
extracted by using TRIzol (Invitrogen, Thermo Fisher Scientific,
Inc.) and purified using Direct-zol RNA extraction kit (Zymo
Research) according to manufacturer's recommendations. cDNA was
synthesized using total RNA (0.4 µg) by Mir-XTM miRNA First-Strand
Synthesis Kit. Expression of miR-340-5p, FHL2 and E-cad mRNA were
quantified using miR-X™ miRNA RT-qPCR SYBR® kit
(Clontech). The PCR primers used were as follows; hsa-miR-340-5p
specific sense 5′-GGCTTATAAACGAATCACAGTCATTAAAA-3, FHL2 mRNA sense;
5′-GAAACTCACTGGTGGACAAGC-3′, antisense; 5′-GTGGCAGATGAAGCAGGTCT-3′,
E-cad mRNA sense; 5′-ACAGCCCCGCCTTATGATT-3′, antisense;
5′-TCGGAACCGCTTCCTTCA-3′, U6 sense; 5′-GCTTCGGCAGCACATATACTA-3′, U6
antisense; 5′-CGAATTTGCGTGTCATCCTTG-3′, β-actin sense;
5′-AGAGCCTCGCCTTTGCCGATCC-3′, antisense;
5′-CACATGCCGGAGCCGTTGTCG-3. Expression of miR-340-5p relative to U6
snRNA and expression of FHL2 as well as E-cad relative to β-actin
were determined using 2−ΔΔCq method.
Target site blocking of
miR-340-5p
Target Scan prediction tool was used to predict
binding site for miR-340-5p at the 3′-UTR of FHL2 mRNA (http://www.targetscan.org/). To evaluate the function
of the binding site, 20 nucleotides long target site blocker (TSB)
was designed to specifically compete with the mir-340-5p in the
3′-UTR of FHL2 mRNA. The miRCURY LNA_TSB The blocker was
synthesized as fully phosphorothiolated Locked Nucleic Acids (LNA)
in the DNA sequences to increase their affinity and selectivity for
the target. Under serum starved conditions, the target site blocker
TSB_FHL2_miR-340-5p; 5′-TTATAAAGTAGTTACAGCCT-3′ were co-transfected
with the miR-340-5p mimic (50 nM). FHL2 and E-cad mRNA and proteins
levels were quantified using RT-qPCR and confocal imaging,
respectively.
Chemotaxis and invasion assays
Migration and invasion assays were performed using
24-well cell migration chambers with 8 µm pore size inserts
(Corning Coster) as described previously (32). For invasion assay, each chamber was
coated with 30 µm of extracellular matrix (ECM) gel
(Sigma-Aldrich). HT-29 cancer cells were transfected with either
miR-340-5p-mimic (50 nM) alone or mimic-Ctrl (50 nM) alone or
miR-340-5p (50 nM) in the presence of TSB or TSB-Ctrl for 24 h in
Opti-MEM serum reduced media. Next day, transfected cells were
collected and 1×106 cells/ml were loaded into the
inserts and DMEM media containing 10% serum was added in the lower
chambers and incubated for 24 h at 37°C in 5% CO2.
Non-migrated cells were removed from the upper surface of the
insert using cotton swab and cells on the lower surface of the
insert membrane were fixed in ice-cold 100% methanol and stained
with 0.5% crystal violet. Cells were counted in five different high
power field (HPF). Data are expressed as the mean number of
migrated cells per high power field.
Confocal microscopy
For immunofluorescence imaging of E-cad and ki67,
cancer cells were grown to 60–70% confluency and then cells were
transfected with either miR-340-5p mimic (50 nM) alone or
mimic-Ctrl (50 nM) alone or miR-340-5p (50 nM) in the presence of
TSB or TSB-Ctrl for 24 h in Opti-MEM serum reduced media as on
glass coverslips as described above. Next day, cells were exposed
to 10% BSA for 30 min. Cells were then fixed with 2% formaldehyde
and permeabilized with 0.2% Triton X-100 for 20 min. After fixation
and permeabilization, cells are then washed two times with PBS
containing 2% fetal bovine serum. Samples were then incubated with
primary antibodies: Fluorescein isothiocyanate (FITC) conjugated
anti-ki67 antibody (ab206633; Abcam) and rabbit anti-human FHL2
antibody (ab12327; Abcam) in PBS containing 2% BSA overnight. In a
separate experiment for E-cad staining, samples were first
incubated with rabbit anti-human E-cad (ab40772; Abcam) primary
antibody in PBS containing 2% BSA overnight. After washing two
times, all samples were incubated with rat anti-rabbit
allophycocyanin (APC) conjugated secondary antibody (A-21038;
Thermo Fisher Scientific, Inc.) for 1 h. After immunostaining,
coverslips were rinsed with PBS twice and then stained with Hoechst
33258 (Thermo Fisher Scientific, Inc.) for 10 min. ProLong Diamond
Antifade Mountant (Thermo Fisher Scientific, Inc.) was added on all
coverslips before putting on the slides. Confocal z-stakes images
were taken using LSM 800 confocal microscope (Carl Zeiss) and
orthogonal projection images were created using all slices for a
total height of ~10 µm. Images were taken by using a ×63 oil
immersion objective (numeric aperture=1.25) and processed later
using ZEN2012 (Carl Zeiss) software.
Statistical analysis
Statistical analyses for in vitro experiments
were performed using GraphPad Prism 8 software. For multiple
comparisons, we used one-way analysis of variance (ANOVA) followed
by the Tukey's post hoc test. For comparison between two groups, we
used two-tailed t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
FHL2 expression in colon cancer
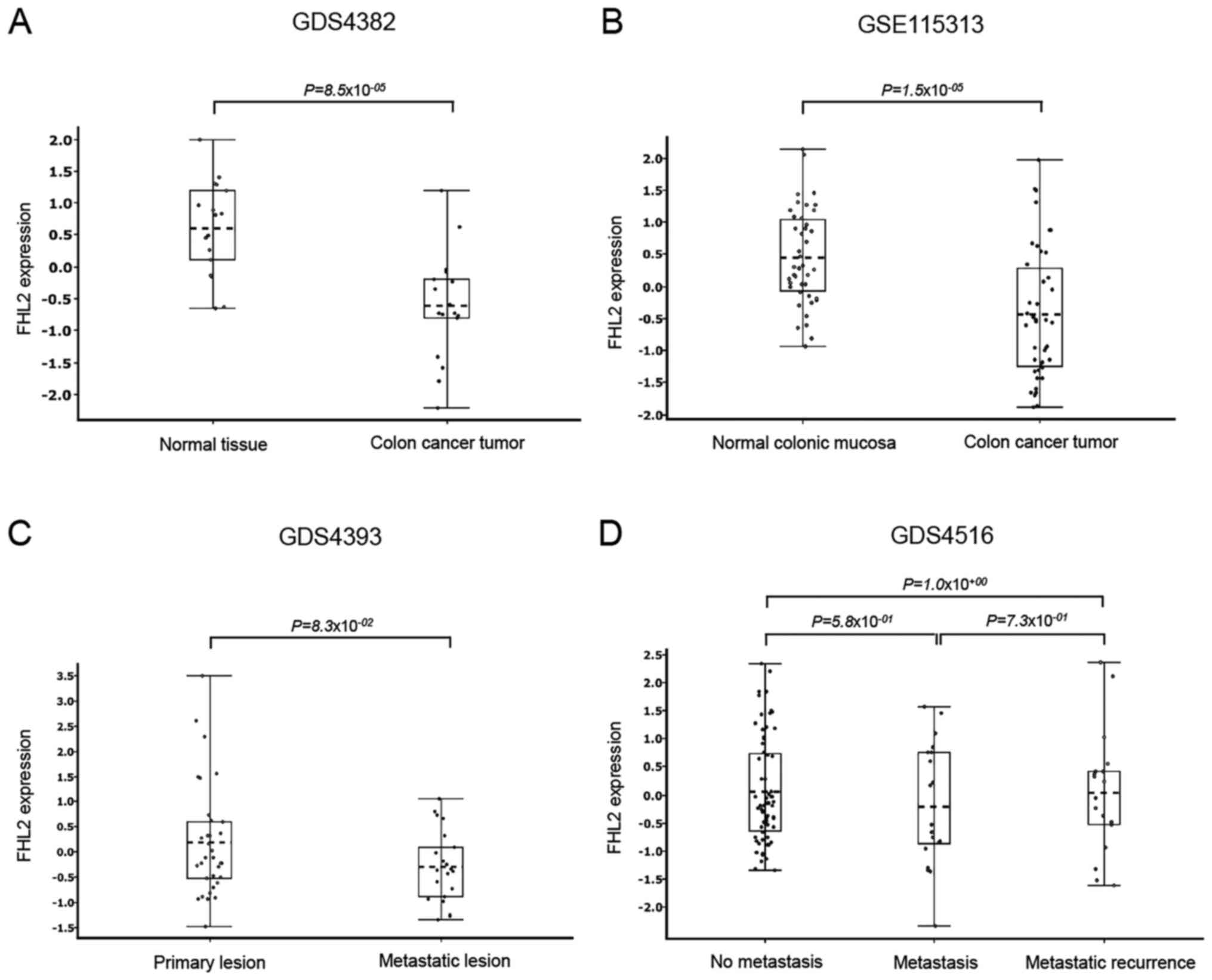
Knowing that FHL2 expression is different in
different tissues, we first compared FHL2 expression between normal
colon mucosa and colon cancer in different datasets. Datasets
GDS4382 and GSE115313 analysis revealed that colon cancer tissue
had significantly lower levels of FHL2 expression than matched
normal non-cancerous colonic tissue (Fig. 1A and B). We also compared primary
colon cancer samples with metastatic and recurrence metastatic
colonic tissues in two different datasets (GDS4393 and GDS4516), it
was found that there were no significant differences between
primary colon cancer and metastatic colon cancer tissues (Fig. 1C and D). In addition, we also
analyzed FHL2 expression among some common colon cancer cell lines
and found that FHL2 expression was relatively low in HT-29 and
HCC2998 cell lines compared to other cell lines such as CaCo2,
DLD-1, HCT116, HCT15, LoVo, SW480 and TC71 (Fig. S1A). We assessed FHL2 expression
between HT-29 and AZ-97 in low-serum condition and observed that
FHL2 expression is higher in AZ-97 cell line compared to HT-29 cell
line (Fig. S1B), suggesting that
AZ-97 cell line is similar to CaCo2, DLD-1, HCT116, HCT15, LoVo,
SW480 and TC71 cell lines in terms of FHL2 expression. Knowing that
FHL2 levels are different in different cancer types, we analyzed a
dataset (GSE103512) containing four different cancer samples, our
analysis revealed that colon cancer and prostate cancer had similar
levels of FHL2 expression while breast cancer and non-small cell
lung cancer had significantly lower levels of FHL2 expression than
colon cancer and prostate cancer (Fig.
S2).
miR-340-5p reduces FHL2 expression in
colon cancer
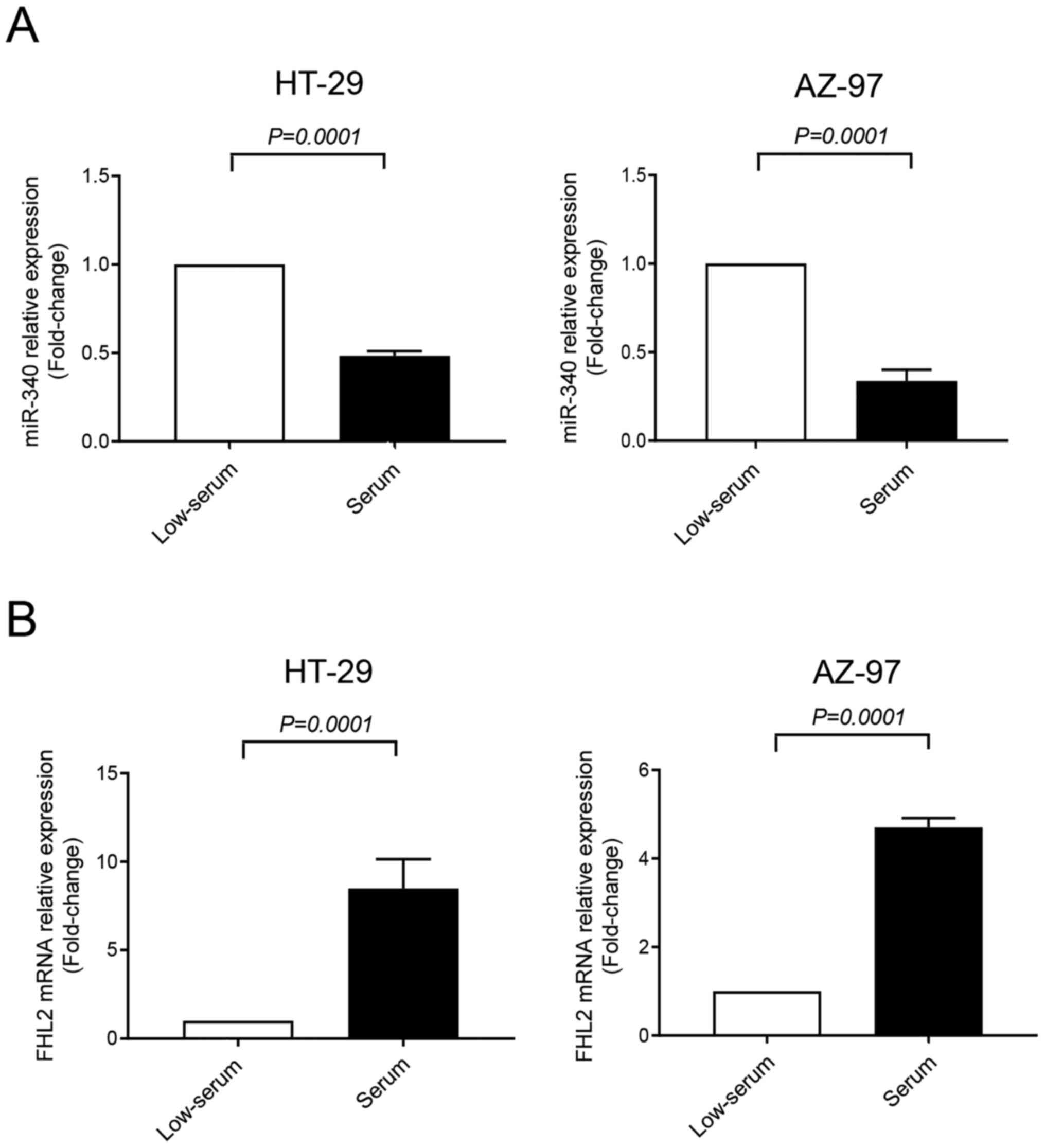
Expression of miR-340-5p and FHL2 were assessed in
HT-29 and AZ-97 colon cancer cell lines in low-serum (to mimic
stress condition of tumor microenvironment) and serum-grown culture
conditions by RT-qPCR. It was found that expression of FHL2 mRNA
was significantly higher in serum-grown HT-29 and AZ-79 cell lines
compared to low-serum condition (Fig.
2B). Furthermore, levels of miR-340-5p were significantly lower
in serum-grown cancer cells compared to low-serum cultured cells
(Fig. 2A), indicating an inverse
relationship between FHL2 mRNA and miR-340-5p in colon cancer
cells. Low-serum condition was used for the subsequent experiments
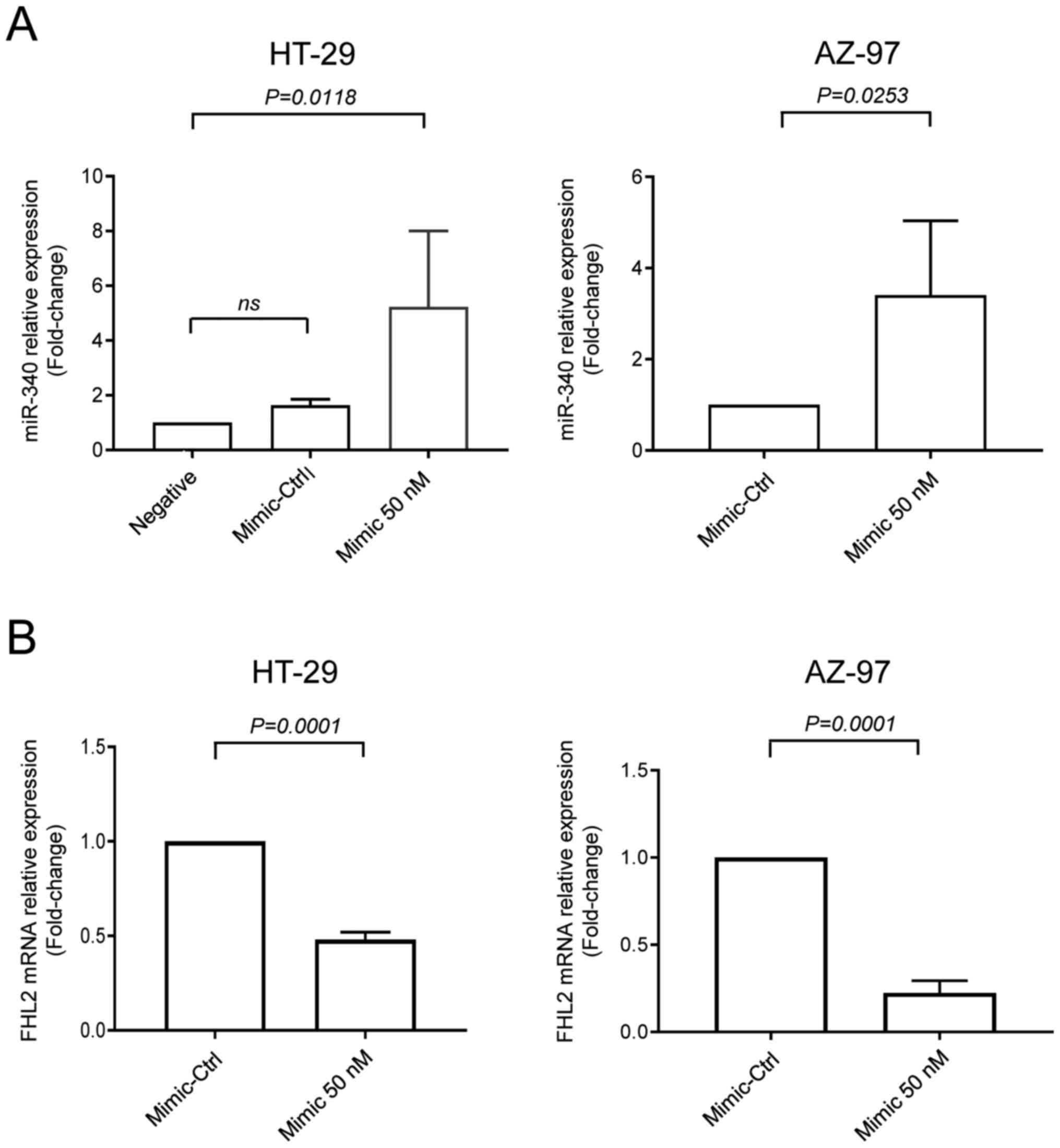
of the study. Next, cells were transfected with miR-340-5p mimic or
mimic-Ctrl. We found that levels of miR-340-5p were increased when
transfected with miR-340-5p mimic (Fig.
3A). Interestingly, transfection of both cell lines with
miR-340-5p mimic reduced levels of FHL2 mRNA (Fig. 3B).
FHL2 contains functional binding site
for miR-340-5p
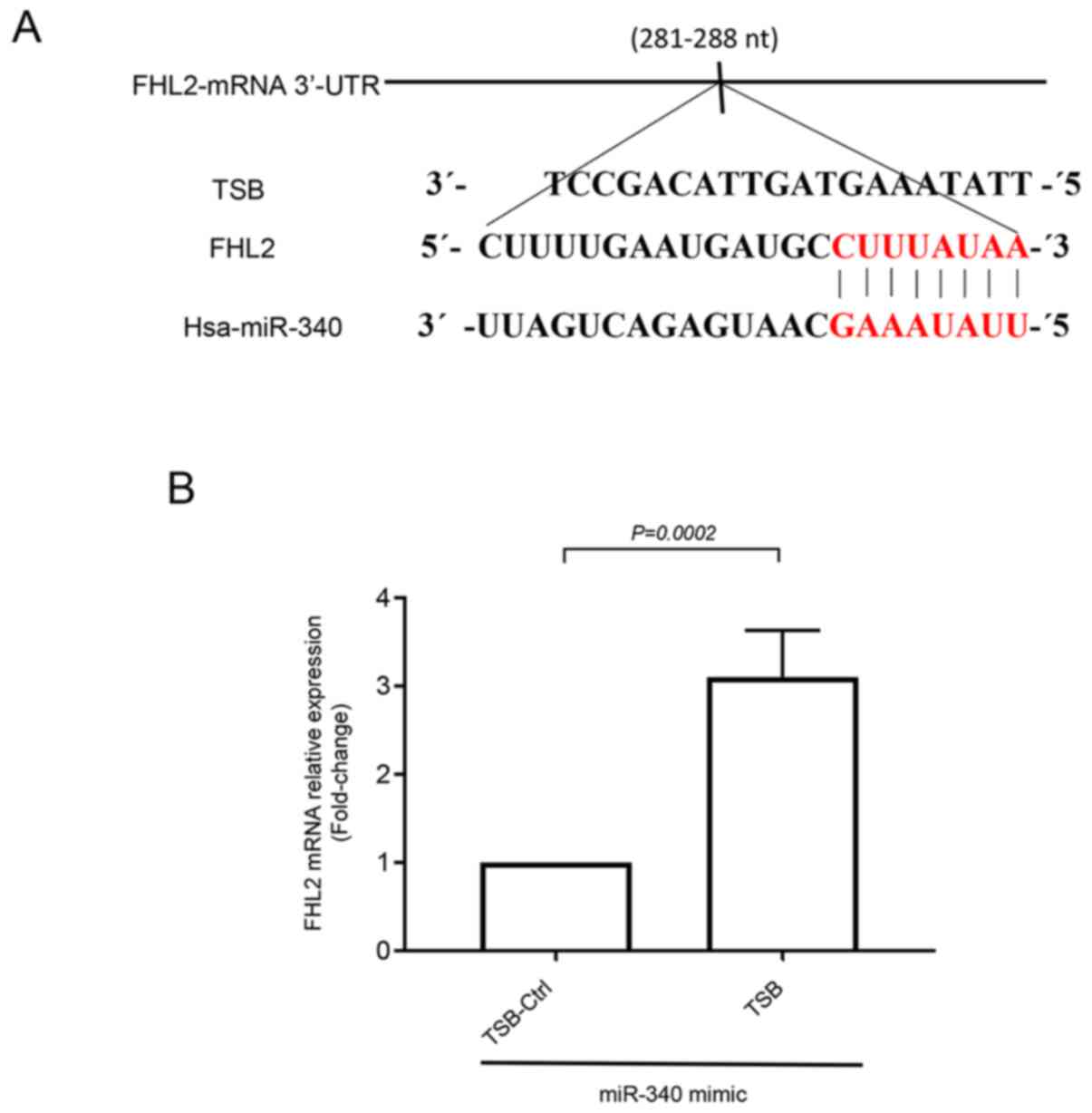
To investigate whether FHL2 oncogene had any binding
site for miR-340-5p, we utilized bioinformatics based target site
prediction tool (TargetScan). Our analysis revealed that there is
one potential binding site for miR-340-5p at 3′-UTR of FHL2 mRNA
where it contains complementary sequences of perfect 8′mer
base-pair match to the seeding region of miR-340-5p (Fig. 4A). We designed a specific target site
blocker (TSB) for FHL2 mRNA. We observed that co-transfection of
TSB with miR-340-5p mimic reversed the inhibitory effect of
miR-340-5p mimic on FHL2 mRNA expression. In contrast,
co-transection with a control TSB had no effect on the level of
FHL2 mRNA expression in colon cancer cells (Fig. 4B), confirming that miR-340-5p
specifically targets FHL2 mRNA in HT-29 colon cancer cell line.
miR-340-5p inhibits FHL2 expression
and proliferation of colon cancer cells
Short-time (30 min) serum stimulation increased FHL2
mRNA expression and transfection of miR-340-5p mimic was found to
reduce the expression of FHL2 mRNA (Fig.
5A). Moreover, co-transfection of miR-340-5p mimic with TSB
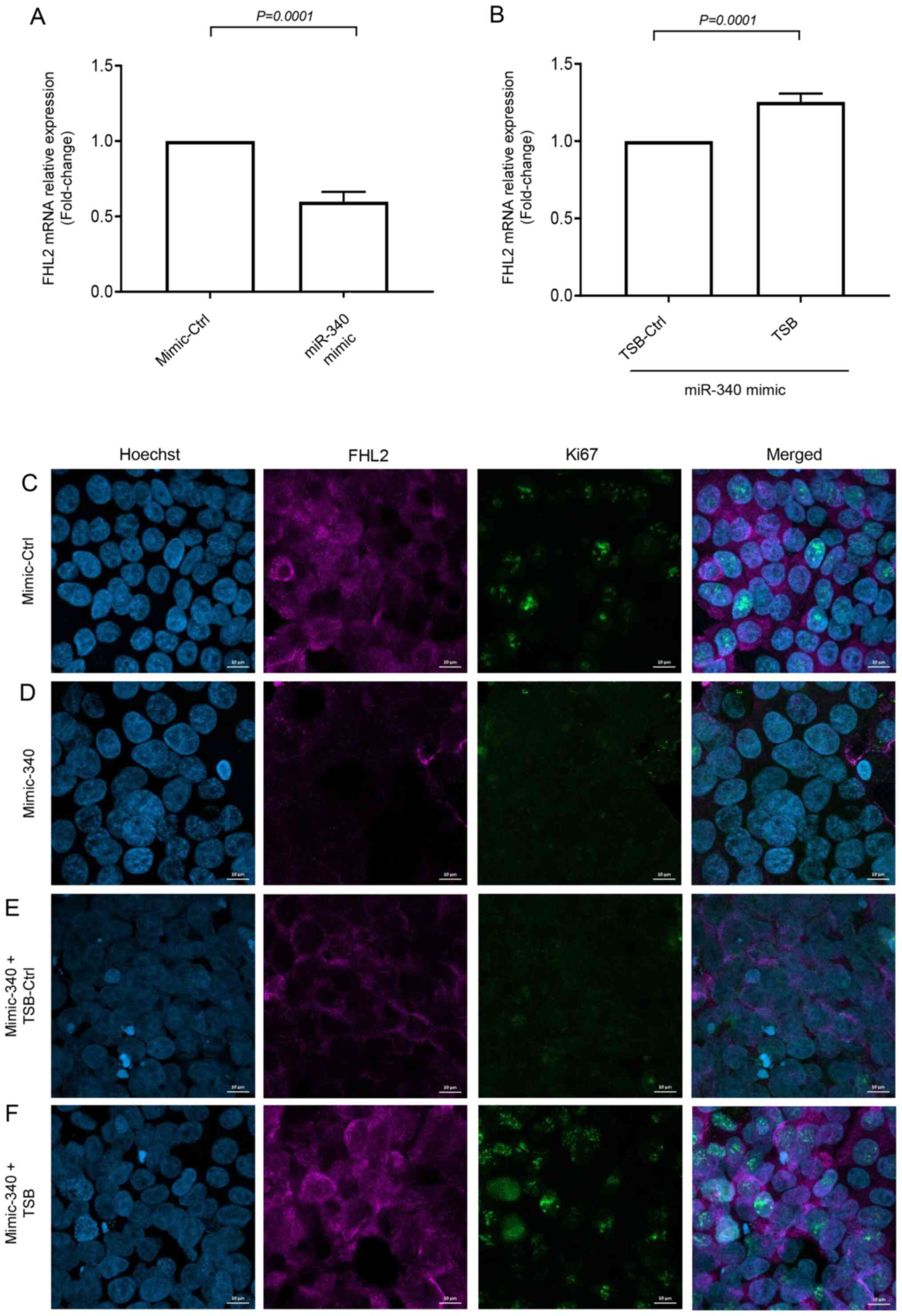
significantly increased levels of FHL2 mRNA expression (Fig. 5B). Confocal microscopy revealed that
serum stimulation increased FHL2 levels in the cytoplasm of the
cancer cells (Fig. 5C), and
miR-340-5p mimic treatment reduced FHL2 levels (Fig. 5D). Co-transfection of TSB with
miR-340-5p mimic reversed the effect of miR-340-5p mimic in
serum-stimulated cells but not by the TSB control (Fig. 5E and F). In addition, we found that
miR-340-5p mimic reduced cancer cell proliferation in terms of ki67
expression and co-transfection with TSB reversed miR-340-5p
mimic-induced inhibition of colon cancer cell proliferation but not
by TSB control (Fig. 5C-F).
miR-340-5p inhibits colon cancer cell
migration and invasion
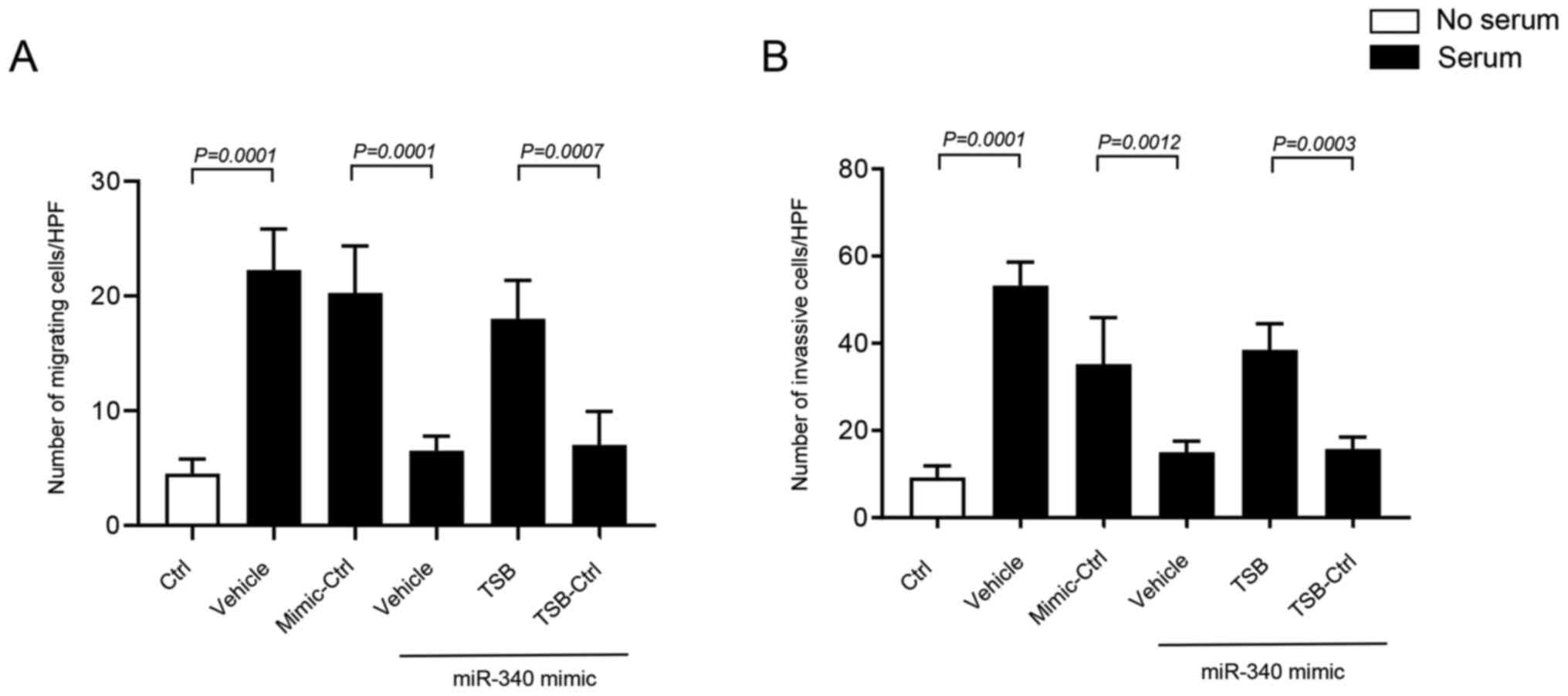
Transwell migration and invasion assays were
performed using 10% FBS as a chemoattractant in the lower chamber
of the wells. It was found that miR-340-5p regulates colon cancer
cell invasion and migration by targeting FHL2. Serum stimulant
significantly increased invasion and migration of HT-29 cells
(Fig. 6A, B). Interestingly,
transfection of HT-29 cells with miR-340-5p mimic significantly
reduced invasion and migration of cancer cells (Figs. 6A and B, and S3, S4).
Moreover, it was found that co-transfection of miR-340-5p mimic and
TSB, but not with control TSB, had significantly higher invasion
and migration of cancer cells (Figs. 6A
and B, and S3, S4), suggesting that miR-340-5p inhibits
cancer cell migration and invasion by targeting a specific binding
site at the 3′-UTR of FHL2 mRNA.
miR-340-5p up-regulates E-cadherin
expression in colon cancer cells
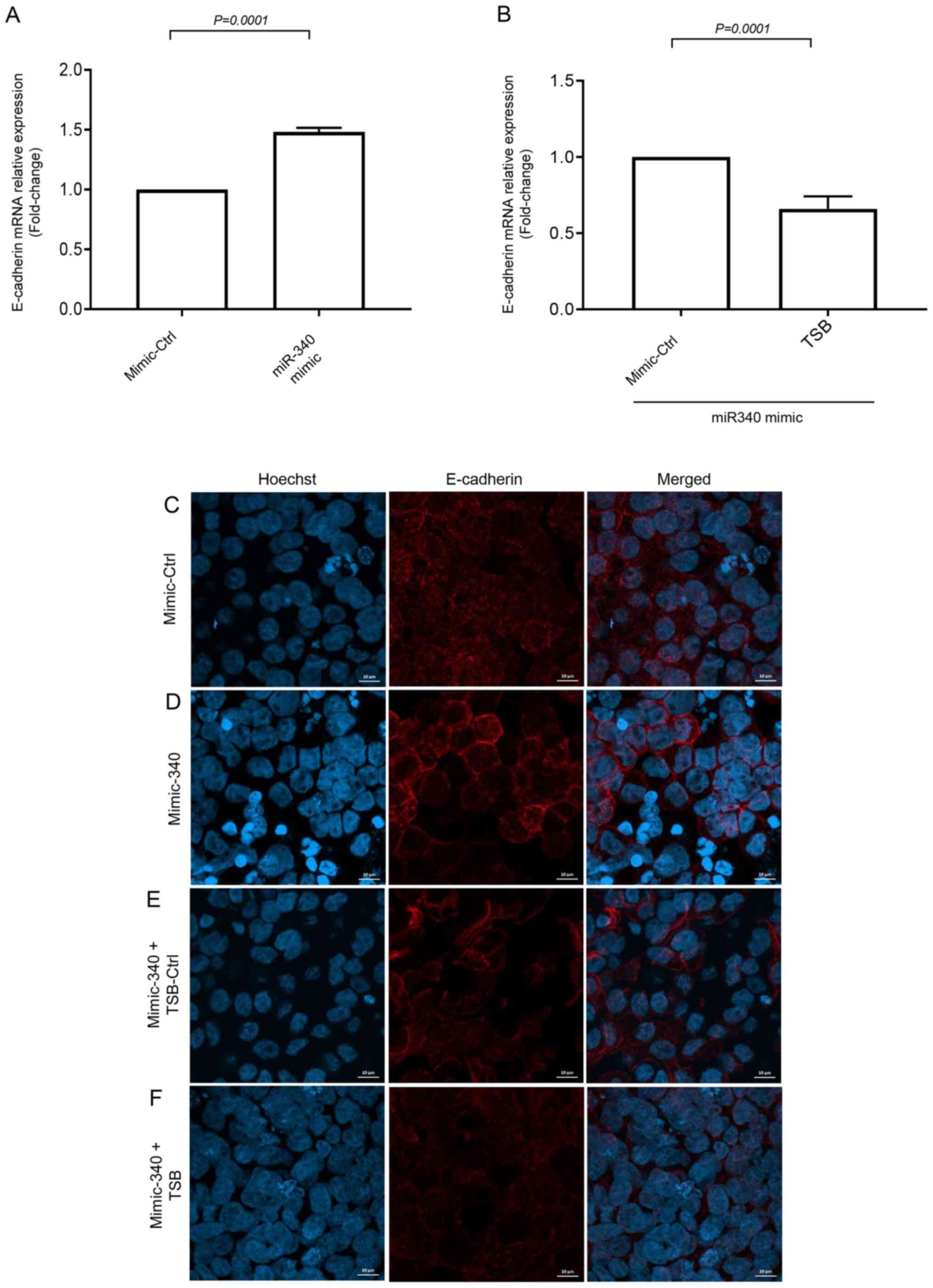
To further examine the downstream target protein of
FHL2 we examined E-cad expression, which is known to regulate
cancer cell metastasis. Short-term serum stimulation significantly
decreased E-cad mRNA expression in HT-29 cells (Fig. S5). The inhibition of serum-induced
FHL2 mRNA expression by miR-340-5p mimic was found to increase
expression of E-cad mRNA (Fig. 7A).
Moreover, co-transfection of miR-340-5p mimic with TSB
significantly decreased levels of E-cad mRNA expression (Fig. 7B), suggesting that miR-340-5p
inhibits FHL2-E-cad axis. Confocal microscopy revealed that
continuous distribution of E-cad in the cell-cell junction was
attenuated by short-serum stimulation in the stressed cancer cells
and miR-340-5p mimic treatment prevented the disruption of E-cad
distribution in cellular junction (Fig.
7C and D). As expected, co-transfection of TSB with miR-340-5p
mimic, reversed the effect of miR-340-5p mimic in serum-stimulated
cells (Fig. 7E and F).
Discussion
Metastasis of colon cancer is a major cause of colon
cancer-related deaths all over the world. Accumulating data suggest
that FHL2 oncogene plays an important role in cancer cells
migration and invasion. However, the role of FHL2 in colon cancer
migration and invasion has not been examined. Herein, we show for
the first time that FHL2 acts as an important regulator in colon
cancer cell migration and invasion under specific conditions.
Furthermore, we found that miR-340-5p has a functional binding site
on FHL2 mRNA and that targeting of FHL2 with miR-340-5p mimic
reduces cancer cell migration and invasion. Thus, targeting of FHL2
by miR-340-5p could be a useful strategy to inhibit colon cancer
metastasis.
FHL2 has been demonstrated to play important roles
in the cancer process including proliferation, differentiation and
migration (35). However, the
literature on FHL2 expression in different cancer types is somewhat
complicated and partly contradictory (8,10,12–14).
Herein, we first analyzed two colon cancer related microarray
datasets and found that FHL2 expression was down-regulated in
primary colon cancer compared to matched normal colonic mucosa.
This finding is in contrast to two previous studies reporting that
FHL2 expression is increased in colon cancer cells (8,36), which
could be related to differences in detection methodology. We used
two microarray based large datasets to compare FHL2 expression
between colon cancer and normal colon tissue while the other
studies, reporting higher expression of FHL2 in colon cancer, used
western blot and immunohistochemistry. Knowing that FHL2 expression
is different in different cancer types, we decided to analyze one
large dataset (GSE103512) containing four different cancer types.
Our analysis revealed that colon and prostate cancer had similar
and significantly higher levels of FHL2 expression compared to
breast and non-small cell lung cancer. In addition, we compared
primary colon cancer tissue with metastatic colon cancer tissue but
observed no significant differences in terms of FHL2 expression,
raising a question whether FHL2 plays any role in the pathogenesis
of colon cancer. Analysis of GSE97023 microarray dataset revealed
that commonly used colon cancer cell lines have two different
levels of FHL2 expression, indicating that colon cancer might have
two different levels of FHL2 expression depending on the colon cell
lines. This observation is in line with Amann et al
(37) reporting that HT-29 cell line
has lower levels of FHL2 expression compared to SW48, CaCo, Lovo,
SW480 and HCT116 colon cancer cell lines. We utilized two different
cells lines (HT-29 and AZ-97) representing low and relatively high
FHL2 expressing colon cancer cells for our subsequent stress
related study. To study stress-induced changes in HT-29 and AZ-97
colon cancer cell lines, we cultured both cell lines in low-serum
(stress) and serum-rich condition and evaluated FHL2 mRNA
expression. Surprisingly, we found that serum condition provoked
FHL2 expression in both colon cancer cell lines. Although there was
no significant difference in FHL2 expression between primary colon
cancer and metastatic colon cancer (microarray data analysis), our
observations herein suggest that sudden access to nutrients in
stressed cancer cells might increase FHL2 expression and mediate
colon cancer pathogenesis.
Accumulating data suggest that miRNAs have
characteristic expression patterns in different types of cancers
and can regulate cancer progression and metastasis (7,38–41). For
example, two recent studies reported that colon cancer tissue has
significantly lower levels of miR-340-5p compared to normal colon
tissue which is correlated to increased incidence of liver
(40) and lymph node (42) metastasis. In this context, we have
previously demonstrated that miR-340-5p can reduce colon cancer
cell migration by controlling RhoA activity (41), indicating that miR-340-5p can be used
to regulate colon cancer pathogenesis. In the present study, we
found that transfection of colon cancer cells with miR-340-5p mimic
reduced colon cancer cell migration and invasion via FHL2. FHL2 is
also shown to regulate cytoskeleton changes in cancer cells. We
anticipate that this finding is in line with other studies showing
that increased levels of miR-340-5p in breast (43), cervical (44), ovarian (7) and lung (45) cancer reduces metastasis.
To investigate the transient effect of serum on the
stressed cancer cells, we cultured transfected cells in low serum
conditions for 24 h and then exposed the cells to serum for 30 min.
We observed that this short serum exposure increased FHL2
expression in colon cancer cells and that transfection of cells
with miR-340-5p mimic significantly reduced serum-induced FHL2
expression both in mRNA and protein levels. Moreover, we found that
transfection of HT-29 cells with miR-340-5p reduced cancer cell
proliferation in terms of ki67 positive cells. Considering the fact
that FHL2 regulates cancer cell migration and proliferation
(7,46), these findings indicate that
miR-340-5p regulates colon cancer progression with an inverse
relationship to FHL2 expression and cell proliferation. Next, we
examined whether FHL2 had any binding site for miR-340-5p and
bioinformatics based analysis revealed that miR-340-5p had a
binding site at 3′-UTR for FHL2 mRNA. We next confirmed the
functionality of the binding site using a TSB that specifically
competes with miR-340-5p. We observed that exogenous delivery of
miR-340-5p into HT-29 cells reduced serum induced cancer cell
migration and invasion. The specific TSB also reversed the
miR-340-5p mimic-induced FHL2 expression in colon cancer cells,
suggesting that the binding site for miR-340-5p on FHL2 mRNA is a
functional binding site and it regulates the process of colon
cancer cells migration and invasion. It should be noted that FHL2
expression is documented to promote colon cancer cells invasiveness
by transforming epithelial cells to mesenchymal cells (8). Moreover, suppression of FHL2 has been
shown to down-regulate multiple oncogenes in gastrointestinal
cancer cells (36). Whether the
suppression of FHL2 by miR-340-5p in colon cancer also have the
ability to inhibit other oncogenes remains to be demonstrated.
Knowing that down-regulation of E-cad is an
important step in cancer cell progression and metastasis (20,47); we
next investigated E-cad expression in miR-340-5p mimic-transfected
HT-29 cells 30 min after exposure to serum. We observed that short
serum-induced down-regulation or disruption of E-cad in cancer
cells was reversed by miR-340-5p and TSB neutralized the effect of
miR-340-5p. This finding indicates that inhibition of colon cancer
cell migration and invasion by miR-340-5p might be via regulation
of E-cad expression. Our finding is in line with a study by Bai
et al reporting that irregular distribution of E-cad is
associated with EMT (48). In
addition, Zhang et al (21)
showed that FHL2 inversely regulates E-cad transcription via a
transcription repressor. In this context, it should be noted that
down-regulation of E-cad is associated with colon (49), breast (50) and prostate cancer metastasis
(51). Notably, our finding that
FHL2-E-cad axis plays an important role in colon cancer cell
migration and invasion, does not exclude the possibility that other
mechanisms might operate in parallel.
Taken together, we conclude that expression of FHL2
in stressed cancer cells play an important role in colon cancer
cell migration and invasion. This study shows that inhibition of
FHL2 expression by using miR-340-5p mimic reduces colon cancer
cells migration and invasion. Moreover, inhibition of FHL2
attenuates cancer cell proliferation and increases E-cad expression
in colon cancer cells, suggesting that targeting FHL2 by miR-340-5p
might be a useful approach to antagonize colon cancer cell
metastasis.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by funding from Swedish
Research Council (grant no. 537-2014-341) and the Maggie Stephens
foundation and Einar and Inga Nilsson foundation.
Availability of data and materials
The datasets analyzed during the current study are
available in the Gene Expression Omnibus database of the National
Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS4382,
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115313,
https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS4393
and http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS4516).
Authors' contributions
AA, RM, AH and CFR performed the experiments,
interpreted the results and contributed to manuscript writing. MR
conceived and designed the current study and contributed to the
writing of the manuscript. All authors read and approved the final
manuscript. AA and MR confirm the authenticity of all raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
FHL2
|
four and a half LIM domains protein
2
|
|
miRNA
|
microRNAs
|
|
E-cad
|
E-Cadherin
|
|
LNA
|
locked nucleic acids
|
|
TSB
|
target site blocker
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bockelman C, Engelmann BE, Kaprio T,
Hansen TF and Glimelius B: Risk of recurrence in patients with
colon cancer stage II and III: A systematic review and
meta-analysis of recent literature. Acta Oncol. 54:5–16. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Levin VA, Panchabhai SC, Shen L, Kornblau
SM, Qiu Y and Baggerly KA: Different changes in protein and
phosphoprotein levels result from serum starvation of high-grade
glioma and adenocarcinoma cell lines. J Proteome Res. 9:179–191.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ghosh T, Varshney A, Kumar P, Kaur M,
Kumar V, Shekhar R, Devi R, Priyanka P, Khan MM and Saxena S:
MicroRNA-874-mediated inhibition of the major G1/S phase cyclin,
CCNE1, is lost in osteosarcomas. J Biol Chem. 292:21264–21281.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rasool RU, Nayak D, Chakraborty S, Jamwal
VL, Mahajan V, Katoch A, Faheem MM, Iqra Z, Amin H, Gandhi SG and
Goswami A: Differential regulation of NM23-H1 under hypoxic and
serum starvation conditions in metastatic cancer cells and its
implication in EMT. Eur J Cell Biol. 96:164–171. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang Z, Li Q, Luo K, Zhang Q, Geng J,
Zhou X, Xu Y, Qian M, Zhang JA, Ji L and Wu J: miR-340-FHL2 axis
inhibits cell growth and metastasis in ovarian cancer. Cell Death
Dis. 10:3722019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang W, Jiang B, Guo Z, Sardet C, Zou B,
Lam CS, Li J, He M, Lan HY, Pang R, et al: Four-and-a-half LIM
protein 2 promotes invasive potential and epithelial-mesenchymal
transition in colon cancer. Carcinogenesis. 31:1220–1229. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tran MK, Kurakula K, Koenis DS and de
Vries CJ: Protein-protein interactions of the LIM-only protein FHL2
and functional implication of the interactions relevant in
cardiovascular disease. Biochim Biophys Acta. 1863:219–228. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kinoshita M, Nakagawa T, Shimizu A and
Katsuoka Y: Differently regulated androgen receptor transcriptional
complex in prostate cancer compared with normal prostate. Int J
Urol. 12:390–397. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hou Y, Wang X, Li L, Fan R, Chen J, Zhu T,
Li W, Jiang Y, Mittal N, Wu W, et al: FHL2 regulates hematopoietic
stem cell functions under stress conditions. Leukemia. 29:615–624.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gabriel B, Fischer DC, Orlowska-Volk M,
zur Hausen A, Schüle R, Müller JM and Hasenburg A: Expression of
the transcriptional coregulator FHL2 in human breast cancer: A
clinicopathologic study. J Soc Gynecol Investig. 13:69–75. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gabriel B, Mildenberger S, Weisser CW,
Metzger E, Gitsch G, Schüle R and Müller JM: Focal adhesion kinase
interacts with the transcriptional coactivator FHL2 and both are
overexpressed in epithelial ovarian cancer. Anticancer Res.
24:921–927. 2004.PubMed/NCBI
|
|
14
|
Jin H, Lee K, Kim YH, Oh HK, Maeng YI, Kim
TH, Suh DS and Bae J: Scaffold protein FHL2 facilitates
MDM2-mediated degradation of IER3 to regulate proliferation of
cervical cancer cells. Oncogene. 35:5106–5118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bates RC and Mercurio AM: The
epithelial-mesenchymal transition (EMT) and colorectal cancer
progression. Cancer Biol Ther. 4:365–370. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Beavon IR: The E-cadherin-catenin complex
in tumour metastasis: Structure, function and regulation. Eur J
Cancer. 36:1607–1620. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Christofori G and Semb H: The role of the
cell-adhesion molecule E-cadherin as a tumour-suppressor gene.
Trends Biochem Sci. 24:73–76. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frixen UH, Behrens J, Sachs M, Eberle G,
Voss B, Warda A, Löchner D and Birchmeier W: E-cadherin-mediated
cell-cell adhesion prevents invasiveness of human carcinoma cells.
J Cell Biol. 113:173–185. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berx G, Cleton-Jansen AM, Nollet F, de
Leeuw WJ, van de Vijver M, Cornelisse C and van Roy F: E-cadherin
is a tumour/invasion suppressor gene mutated in human lobular
breast cancers. EMBO J. 14:6107–6115. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Padmanaban V, Krol I, Suhail Y, Szczerba
BM, Aceto N, Bader JS and Ewald AJ: E-cadherin is required for
metastasis in multiple models of breast cancer. Nature.
573:439–444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang W, Wang J, Zou B, Sardet C, Li J,
Lam CS, Ng L, Pang R, Hung IF, Tan VP, et al: Four and a half LIM
protein 2 (FHL2) negatively regulates the transcription of
E-cadherin through interaction with Snail1. Eur J Cancer.
47:121–130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kreutziger KL and Kreutziger KL:
Comprehensive surgical management of mandibular fractures. South
Med J. 85:506–518. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valastyan S and Weinberg RA: Roles for
microRNAs in the regulation of cell adhesion molecules. J Cell Sci.
124((Pt 7)): 999–1006. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fridrichova I and Zmetakova I: MicroRNAs
contribute to breast cancer invasiveness. Cells. 8:13612019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao P, Ma W, Hu Z, Zhang Y, Zhang S and
Wang Y: Up-regulation of miR-340-5p promotes progression of thyroid
cancer by inhibiting BMP4. J Endocrinol Invest. 41:1165–1172. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi Z, Li Y, Qian X, Hu Y, Liu J, Zhang S
and Zhang J: miR-340 inhibits triple-negative breast cancer
progression by reversing EZH2 mediated miRNAs dysregulated
expressions. J Cancer. 8:3037–3048. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang D, Qiu S, Ge R, He L, Li M, Li Y and
Peng Y: miR-340 suppresses glioblastoma multiforme. Oncotarget.
6:9257–9270. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen CP, Sun ZL, Lu X, Wu WX, Guo WL, Lu
JJ, Han C, Huang JQ and Fang Y: miR-340 suppresses cell migration
and invasion by targeting MYO10 in breast cancer. Oncol Rep.
35:709–716. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Matsushita Y, Hoff SD, Nudelman ED, Otaka
M, Hakomori S, Ota DM, Cleary KR and Irimura T: Metastatic behavior
and cell surface properties of HT-29 human colon carcinoma variant
cells selected for their differential expression of sialyl-dimeric
Le(x)-antigen. Clin Exp Metastasis. 9:283–299. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bettenworth D, Mucke MM, Schwegmann K,
Faust A, Poremba C, Schäfers M, Domagk D and Lenz P:
Endoscopy-guided orthotopic implantation of colorectal cancer cells
results in metastatic colorectal cancer in mice. Clin Exp
Metastasis. 33:551–562. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zawadzki A, Liu Q, Wang Y, Melander A,
Jeppsson B and Thorlacius H: Verapamil inhibits L-type calcium
channel mediated apoptosis in human colon cancer cells. Dis Colon
Rectum. 51:1696–1702. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu Y, Guo Z, Zhang D, Zhang W, Yan Q, Shi
X, Zhang M, Zhao Y, Zhang Y, Jiang B, et al: A novel colon cancer
gene therapy using rAAVmediated expression of human shRNA-FHL2. Int
J Oncol. 43:1618–1626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang J, Yang Y, Xia HH, Gu Q, Lin MC,
Jiang B, Peng Y, Li G, An X, Zhang Y, et al: Suppression of FHL2
expression induces cell differentiation and inhibits gastric and
colon carcinogenesis. Gastroenterology. 132:1066–1076. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Amann T, Egle Y, Bosserhoff AK and
Hellerbrand C: FHL2 suppresses growth and differentiation of the
colon cancer cell line HT-29. Oncol Rep. 23:1669–1674.
2010.PubMed/NCBI
|
|
38
|
Al-Haidari A, Algaber A, Madhi R, Syk I
and Thorlacius H: miR-155-5p controls colon cancer cell migration
via post-transcriptional regulation of Human Antigen R (HuR).
Cancer Lett. 421:145–151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Takeyama H, Yamamoto H, Yamashita S, Wu X,
Takahashi H, Nishimura J, Haraguchi N, Miyake Y, Suzuki R, Murata
K, et al: Decreased miR-340 expression in bone marrow is associated
with liver metastasis of colorectal cancer. Mol Cancer Ther.
13:976–985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Algaber A, Al-Haidari A, Madhi R, Rahman
M, Syk I and Thorlacius H: MicroRNA-340-5p inhibits colon cancer
cell migration via targeting of RhoA. Sci Rep. 10:169342020.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang L, Men WL, Yan KM, Tie J, Nie YZ and
Xiao HJ: miR-340-5p is a potential prognostic indicator of
colorectal cancer and modulates ANXA3. Eur Rev Med Pharmacol Sci.
22:4837–4845. 2018.PubMed/NCBI
|
|
43
|
Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J,
Zhao JJ, Mao SS, Zhang GH, Xu XC and Zhang N: miR-340 inhibition of
breast cancer cell migration and invasion through targeting of
oncoprotein c-Met. Cancer. 117:2842–2852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xiao H, Yu L, Li F, Wang H, Li W and He X:
miR-340 suppresses the metastasis by targeting EphA3 in cervical
cancer. Cell Biol Int. 42:1115–1123. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fernandez S, Risolino M, Mandia N, Talotta
F, Soini Y, Incoronato M, Condorelli G, Banfi S and Verde P:
miR-340 inhibits tumor cell proliferation and induces apoptosis by
targeting multiple negative regulators of p27 in non-small cell
lung cancer. Oncogene. 34:3240–3250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu M, Wang J, Tang W, Zhan X, Li Y, Peng
Y, Huang X, Bai Y, Zhao J, Li A, et al: FOXK1 interaction with FHL2
promotes proliferation, invasion and metastasis in colorectal
cancer. Oncogenesis. 5:e2712016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Park SY, Lee SJ, Cho HJ, Kim TW, Kim JT,
Kim JW, Lee CH, Kim BY, Yeom YI, Lim JS, et al: Dehydropeptidase 1
promotes metastasis through regulation of E-cadherin expression in
colon cancer. Oncotarget. 7:9501–9512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bai S, Zeng R, Zhou Q, Liao W, Zhang Y, Xu
C, Han M, Pei G, Liu L, Liu X, et al: Cdc42-interacting protein-4
promotes TGF-B1-induced epithelial-mesenchymal transition and
extracellular matrix deposition in renal proximal tubular
epithelial cells. Int J Biol Sci. 8:859–869. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu
F, Peng Z and Yan D: LncRNA-ATB mediated E-cadherin repression
promotes the progression of colon cancer and predicts poor
prognosis. J Gastroenterol Hepatol. 31:595–603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wendt MK, Taylor MA, Schiemann BJ and
Schiemann WP: Down-regulation of epithelial cadherin is required to
initiate metastatic outgrowth of breast cancer. Mol Biol Cell.
22:2423–2435. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fan L, Wang H, Xia X, Rao Y, Ma X, Ma D,
Wu P and Chen G: Loss of E-cadherin promotes prostate cancer
metastasis via upregulation of metastasis-associated gene 1
expression. Oncol Lett. 4:1225–1233. 2012. View Article : Google Scholar : PubMed/NCBI
|