Introduction
In 1998, claudins were identified as the major
integral membrane proteins essential for tight junction (TJ)
assembly (1). Since then, 27 members
of this gene family have been found in mammals. Our contemporary
understanding of claudins is based on their canonical barrier and
fence functions. Recently, this view has expanded considerably
following increasing evidence suggesting that claudins may be
involved in signal transduction and that they may be causally
important in tumorigenesis (2).
Obvious dysregulation of claudin expression has been found in a
number of cancer tissues. Owing to the specific expression profile
and differences between normal and tumor cells, claudins are
attractive targets that can theoretically enable selective drug
delivery with minimal adverse events (3). To effectively target claudins for
clinical applications, it is essential to understand their roles
and fully elucidate the complicated mechanisms by which the
expression of claudins is dysregulated in cancer. The present study
reviews the current knowledge describing the dysregulation of
claudin expression in cancer and discusses the intrinsic and
extrinsic determinants of the context-specific expression patterns
of claudins.
TJs and claudins
Epithelial and endothelial cells in most living
systems form a sheet-like structural interface between the external
environment and internal compartments that not only protects the
internal organs, but also acts as a selective barrier between the
body and the external environment that restricts free exchange
across the paracellular space (4).
The paracellular space between neighboring cells in the epithelial
sheet is connected by several types of cell-cell junctions, which
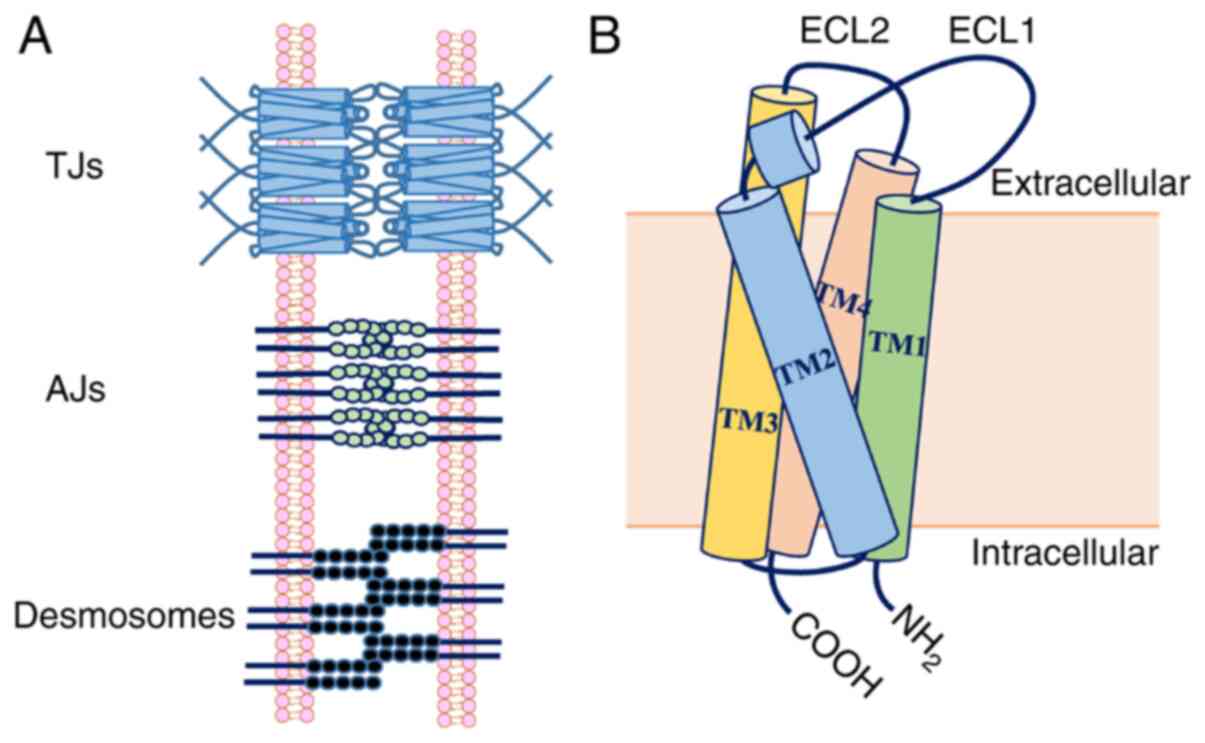
can be categorized as TJs, adherens junctions (AJs) and desmosomes
(Fig. 1) (5). AJs bind with the cytoskeleton to impart
support and serve as signaling hubs, playing important roles in
regulating gene expression (6).
Desmosomes create a strong structural network that binds cells
together, but is not continuous and cannot prevent solute transport
(7). Therefore, TJs are the chief
intercellular junctions that act as permeability barriers and
confer polarity to epithelial cells in close proximity to adjacent
plasma membranes containing TJ strands formed by TJ proteins
(8). TJ proteins are diverse and can
be divided into three groups: Transmembrane proteins, adaptor
proteins and signaling proteins associated with TJs (9). These proteins interact with each other
and other proteins to form a multimeric protein architecture. As
key proteins, claudins are a family of integral membrane proteins
that are essential for TJ strand assembly.
Claudin (which means ‘to close’, named after the
Latin word ‘claudere’) was identified as the major integral
membrane protein forming TJ strands by Tsukita in 1998 (1). A total of 27 homologous claudin genes
have been identified in mammals, and numerous more claudin proteins
are selectively expressed by alternative splicing in various
tissues. According to homology, claudins 1–10, 14, 15, 17 and 19
are homologous classic claudins, while claudins with less homology
(claudins 11–13, 16, 18 and 20–27) are non-classical claudins
(10). Based on their functions,
claudins can also be classified as paracellular barrier-forming
claudins function in intracellular space sealing, and paracellular
channel-forming claudins function in selective permeability. The
paracellular barrier-forming group includes claudins 1, 3–5, 7, 11,
14 and 18, which are responsible for restricting transport and
enhancing the barrier function of the cell. The claudins involved
in paracellular channel-forming ensure proper availability of water
and ions for cellular functions; for example, claudins 10a and 17
allow transport of anions, claudins 10b and 15 allow passage of
cations, and claudin-2 allows transport of both anions and water
(11). The structure of claudins
includes four transmembrane domains, the intracellular N and C
termini, and two extracellular loops (ECLs) (ECL1 and ECL2)
(Fig. 1). ECLs are involved in the
formation of interactions between claudin strands and determine the
characteristics of claudin-based TJs, as their homology is low
among claudins (12). ECLs are also
specific binding sites for Clostridium perfringens
enterotoxin and monoclonal antibodies, which have been utilized for
imaging and therapy in pathological conditions (9).
Physiological functions of claudins
Our contemporary understanding of claudins is based
on their canonical barrier and fence functions, which work on the
basis of forming TJ complexes. Based on the crystallographic
structure of claudin-15 and cysteine cross-linking experiments with
mutated claudin-15 and claudin-2, as well as freeze-fracture
electron microscopy images, Suzuki et al (13) proposed a model of paracellular pores
in which β-barrel-like channels are formed by the apposition of two
anti-parallel double rows of claudins in the membrane of two
adjacent cells. Given the results of the crystallization of some
other claudins (claudins 3, 4 and 19), this structure seems to be
common to other claudins, with differences in residues of their
extracellular regions accounting for their different permeabilities
and selectivities (14–16). According to physiological studies,
two mechanisms have been proposed for paracellular permeability:
The ‘pore’ mechanism, in which solutes or ions pass through a
paracellular channel formed by the TJ strands (enabling transport
of molecules with an estimated diameter of ~4 Angstrom), and the
‘leak’ mechanism, in which solutes supposedly pass through breaks
in the TJ strands (permitting permeation of molecules up to a size
limit of ~60 Angstrom) (17).
Claudins are also essential for the asymmetric localization of
membrane proteins and lipids in the exoplasmic leaflet, acting as a
membrane fence in epithelial cells (18).
Our understanding of claudins has expanded
considerably over decades of research, following increasing
evidence suggesting that claudins may be involved in signal
transduction. As the PDZ domain-binding motif located in the
cytoplasmic C-terminal tail of claudins can directly interact with
zonula occluden (ZO) family proteins, which play important roles in
a number of cellular processes, it is not unexpected that claudins
participate in signal transduction (9). Claudins mainly function as inhibitory
factors through interaction with numerous other signaling pathway
elements (such as yes-associated protein/transcriptional
co-activator with PDZ-binding motif and β-catenin), retaining them
in the submembrane compartment (19,20). On
the other hand, a number of signaling pathways can modify claudins
to regulate their protein localization, protein interactions and
overall turnover (2). Moreover,
mounting evidence suggests that claudins are localized to sites
outside the TJ complex, such as the basolateral membrane and
nucleus (21). Little is known about
the functions of these claudins, and available data support a
cell-extracellular matrix interaction and signaling function that
integrates, codifies and transports information into the cell
(21). These functions are mainly
achieved through the formation of a complex between claudins and
other molecules, such as integrins, epithelial cell adhesion
molecules and matrix metalloproteinases, which leads to the
activation of signal transduction (Fig.
2) (22–24).
 | Figure 2.Physiological functions of claudins.
a, TJ assembly: Close apposition of adjacent plasma membranes where
TJ strands are formed by claudins. b, The ‘pore’ pathway: Solutes
or ions pass through a paracellular channel determined by the
charged residue status in the first ECL (ECL1) of claudins. c, The
‘leak’ pathway, in which solutes supposedly pass through the
paracellular space, dependent on the breaking and reorganization of
claudin strands. d, Fence function: Proteins and lipids in the
exoplasmic leaflet cannot freely diffuse across claudin strands. e,
Signaling transduction: Claudins mainly function as inhibitory
factors and retain signaling molecules in the submembrane
compartment. f, Claudins outside TJs always associate with other
molecules to engage in cell-ECM interactions and signal
transduction. TJ, tight junction; ECM, extracellular matrix; ECL,
extraceullar loop. |
In line with their aforementioned important
functions, dysregulation of claudin-mediated barrier function and
signaling is a precursor for the pathogenesis of numerous human
diseases, including a number of cancer types (9). For example, mutations of CLDN1,
CLDN5, CLDN14, CLDN16, and CLDN19 have been reported to
cause human neonatal ichthyosis, sclerosing cholangitis,
non-syndromic deafness, familial hypomagnesaemia and other symptoms
(9). Knockout of CLDN18 leads
to increased abundance and proliferation of known distal lung
progenitors, the alveolar epithelial type II (AT2) cells,
activation of Yes-associated protein (YAP), and tumorigenesis in
mice (20). The dysregulated
expression of claudins in cancer has attracted attention, given the
increased burden of cancer worldwide.
Expression patterns of claudins
The claudin expression profile of numerous different
tissues and cells, including normal and tumor tissues, has been
assessed at the mRNA and protein levels. Studies have revealed a
range of outcomes that reflect the complexity of claudins in terms
of spatial localization, tumor type and stage of disease. Through a
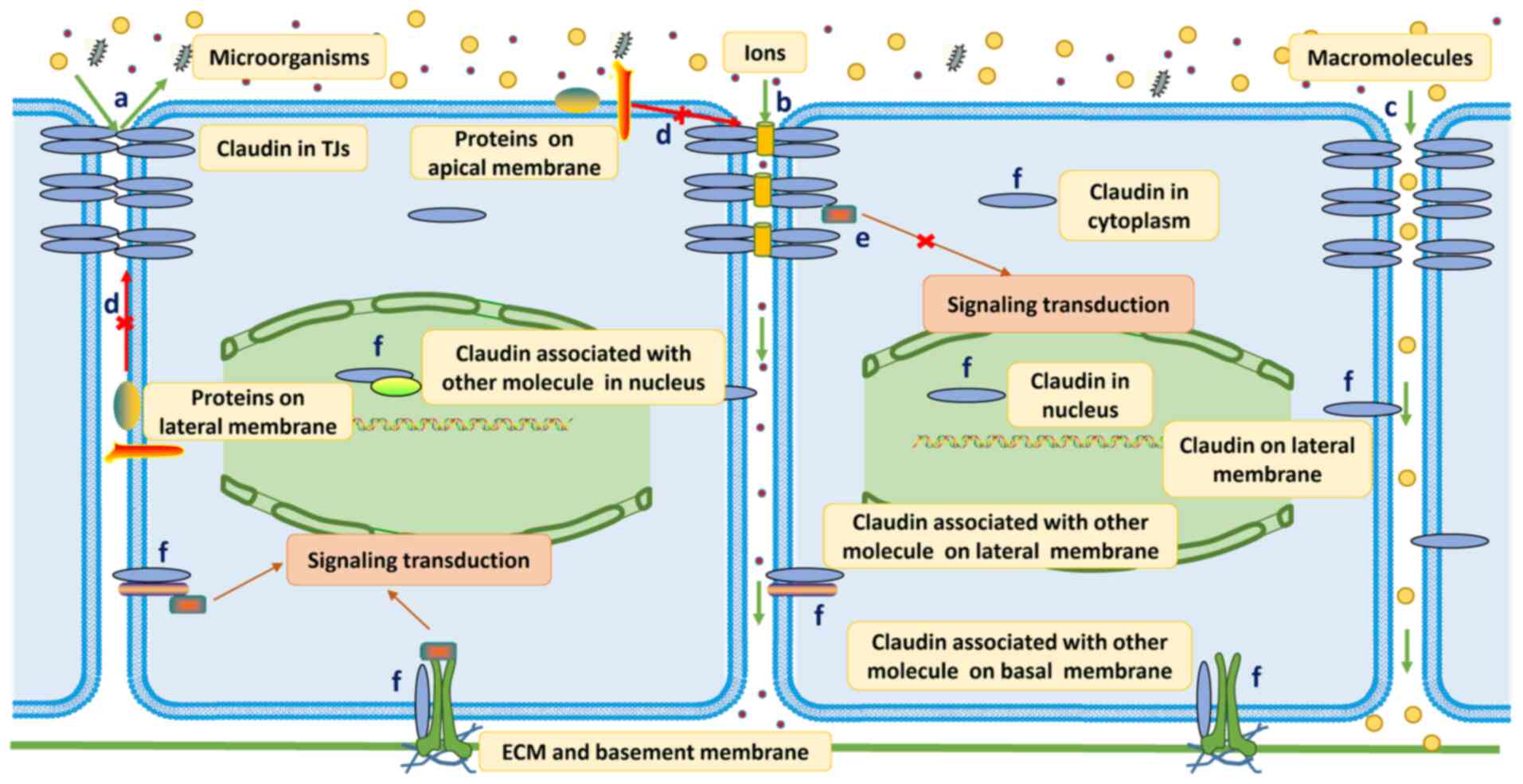
comprehensive review of the transcription data in The Cancer Genome
Atlas (TCGA), using the online tool GEPIA (http://gepia.cancer-pku.cn) (Fig. 3), the protein expression data in the
Human Protein Atlas (https://www.proteinatlas.org) and individual studies,
most of the claudins, if not all, have been reported to show the
following expression features.
 | Figure 3.Expression of claudin genes in human
cancer. The figure shows the expression of claudin genes in various
common cancer types and subtypes. The green color corresponds to
normal tissues, and the red color corresponds to tumor tissues. The
depth of color represents the strength of expression. The numbers
in each box are the median transcripts per million of certain tumor
and normal tissues. This figure was compiled using data generated
by The Cancer Genome Atlas and Genotype-Tissue Expression projects,
accessible through GEPIA. BRCA, invasive breast carcinoma; CESC,
cervical squamous carcinoma; COAD, colon adenocarcinoma; ESCA,
esophageal carcinoma; HNSC, head and neck squamous cell carcinoma;
KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal
papillary cell carcinoma; LUAD, lung adenocarcinoma; LUSC, lung
squamous carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD,
pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; READ,
rectum adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach
adenocarcinoma; TGCT, testicular germ cell tumor; UCEC, uterine
corpus endometrial carcinoma; N, normal tissues; T, tumor
tissues. |
First, the protein level of claudins is not
necessarily correlated with the mRNA level. Although 26 claudin
genes have been found in humans (CLDN13 is absent), not each
one has been validated at the protein level. Currently, only
claudins 1–7, 10–12, 15, 16, 18, 19 and 23 have been identified at
the mass spectrometry level, although claudins 8 and 9 have been
identified at the protein level by immunoblotting, and some
claudins may be expressed in tissues that have not been probed
(25). Of them, claudins 1, 3, 4, 6,
7 and 18 are the most extensively studied in tumors.
Second, claudin transcripts can be alternatively
spliced to produce isoforms, and some of these isoforms are only
found in cancer. Alternatively spliced transcript variants have
been found for the majority of CLDN family members, although
the protein forms of some of these isoforms have not been detected.
Currently, credible immunological evidence exists for the presence
of the claudin 10 and 18 protein isoforms claudins 10a and 10b and
claudins 18.1 and 18.2, respectively (26–28). In
addition to splice variants that result from differing
physiological conditions, many more transcript variants that do not
exist in normal tissues may result from alterations that affect RNA
splicing, which dysregulate the expression of CLDN genes,
contributing to cancer progression (29).
Third, the expression of claudins changes
dynamically during development and differentiation. Claudin-6
expression was reported to be 90-fold higher in undifferentiated
human pluripotent stem cells than in differentiated cells, while in
adult tissues, it was nearly completely absent (30,31). The
expression of claudin-1 declines with differentiation, whereas the
expression of claudin-4 increases and is highest in the late stage,
in which human embryonic stem cells transition into hepatocyte-like
cells (32). In the colon, claudin-7
expression gradually increases with the differentiation of
epithelial cells into luminal cells, which results in an expression
gradient from the basal side to the surface of the intestinal lumen
(33). Age-related changes in
expression have also been reported, including changes in claudin-3
expression, which is downregulated with aging (34).
Fourth, although various claudins are expressed in
the same tissue simultaneously, distinct claudin members are
expressed in a tissue-specific manner. Claudins 18 and 19 are two
typical examples. Claudin-18 expression is restricted to the lung,
where it is present as the isoform claudin-18.1, and to the
stomach, where it is present as the isoform claudin-18.2 (27). Claudin-19 is detected mainly in the
thick ascending limb of the loop of Henle, the retinal epithelium
and peripheral neurons in normal tissues (35). The expression of claudins 1, 4 and 7
is relatively widespread and closely related to cancer (36,37).
Single-site specificity within one organ was also discovered for
claudin-1 expression, as claudin-1 mRNA expression was found to be
higher in the distal site than in the proximal site of the colon
(38).
Fifth, the expression of claudins appears to be
reversed with carcinogenesis; that is, a claudin that is
downregulated in a specific tissue will be upregulated in tumors
arising from that tissue, while a claudin that is upregulated in a
specific tissue will be expressed at low levels in tumors derived
from that tissue. This is especially true for claudin-18 and −6. In
one study, the expression of claudin-18 nearly disappeared in lung
cancer tissues and was significantly downregulated in gastric
cancer tissues, while it was ectopically upregulated in pancreatic
cancer tissues (27). Claudin-6 is a
strictly oncofetal cell surface antigen whose expression is tightly
regulated and completely absent in normal human tissues, but
reactivated in germline tumors such as testicular, ovarian and
uterine cancer (31).
Sixth, the expression levels of some claudins are
heterogeneous within tumors and may differ in different cancer
subtypes. For example, in estrogen receptor (ER)-positive luminal A
and luminal B breast cancer, the expression of claudin-1 was found
to be downregulated, while in the ER-negative basal-like breast
cancer subtype, increased expression and cytoplasmic delocalization
were observed (39). Consensus
molecular subtype 2 and the transit-amplifying and C5 subtypes of
metastatic colorectal cancer exhibit higher expression of claudin-1
than other subtypes (40). The
expression levels of claudins are also not homogeneous
intratumorally. Claudin-1 expression was shown to be retained in
the central regions, but lost in the invasive edges in colorectal
cancer (CRC) tissues (41). Although
the majority of studies have agreed with these findings, some
studies have reported the opposite results, especially for claudins
that are widely expressed in a number of cancer types (claudins 1,
4 and 7) (36,37).
Seventh, changes in the expression of claudins are a
very early event, although these changes are always exacerbated
with the progression of the cancer. Claudin-1 was shown to be
upregulated in the course of colitis, while claudin-7 mRNA levels
were decreased in adenomas, suggesting that changes in claudin-7
mRNA levels are an early event in the development of CRC (42,43).
Claudin-18 mRNA levels were also found to be decreased in
intestinal metaplasia and intraepithelial neoplasia tissues before
gastric cancer formation, but were commonly increased in precursor
lesions of pancreatic cancer (27,44).
Regulation of claudin expression in
cancer
Since the expression patterns of claudins are
diverse and dynamic, the transcription and function of claudins
must be tightly controlled via a wide range of regulatory
mechanisms, including genetic background, epigenetic alterations,
transcriptional changes and post-translational modifications
(PTMs), which are commonly modulated by oncogenic signaling
pathways. Extrinsic factors, including the cancer cell of origin,
differentiation status, pre-existing or acquired tumor
microenvironment and environmental factors, also determine the
expression profile of claudins (Table
I; Fig. 4).
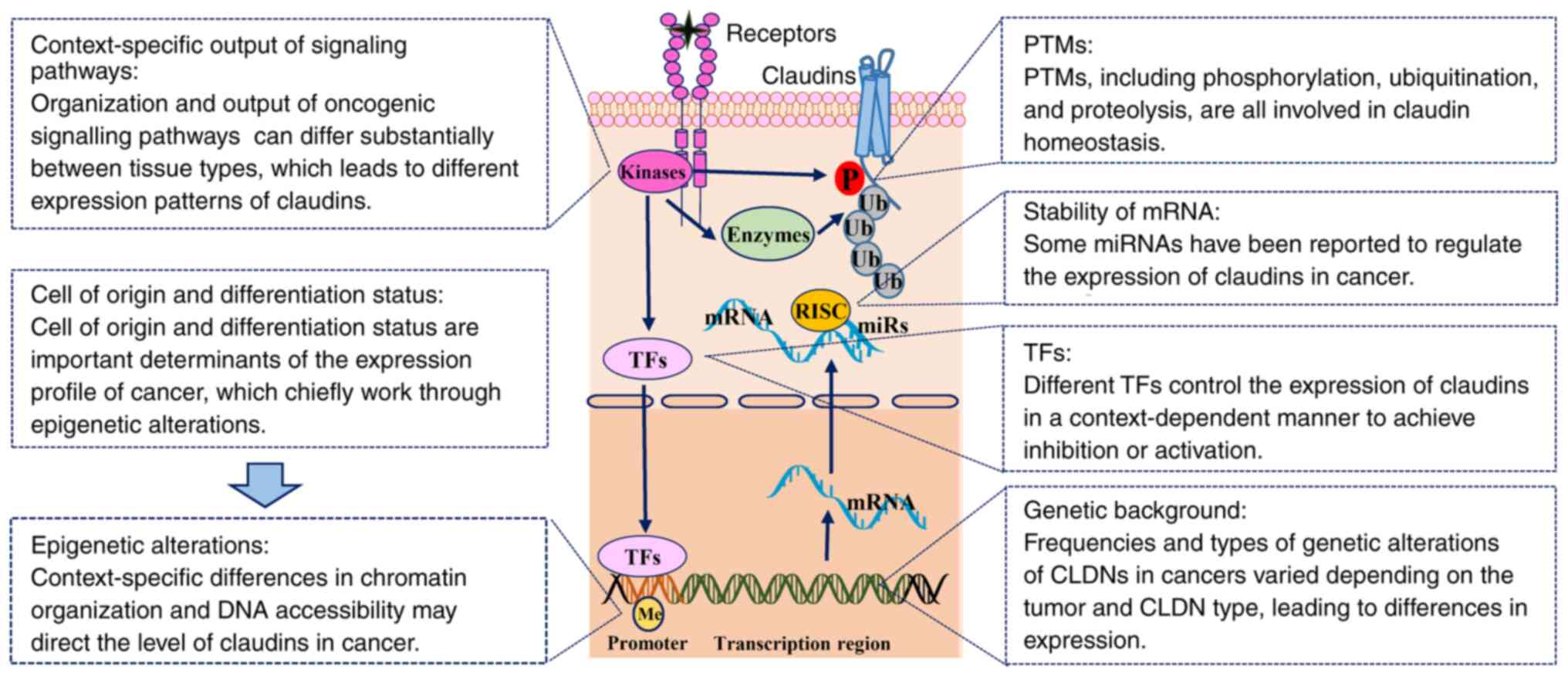 | Figure 4.Regulation of claudin expression in
cancer. The expression of claudins is tightly controlled via a wide
range of regulatory mechanisms, including genetic background,
epigenetic alterations, transcriptional changes and PTMs, which are
commonly modulated by oncogenic signaling pathways. Extrinsic
factors include the cancer cell of origin, differentiation status,
pre-existing or acquired tumor microenvironment and environmental
factors, which also determine the expression profile of the
claudins. CLDN, claudin; Me, methylation; miRNA, microRNA; P,
phosphorylation; PTM, post-translational modification; RISC,
RNA-induced silencing complex; TF, transcription factor; Ub,
ubiquitination. |
 | Table I.Regulation of claudin expression in
cancer. a |
Table I.
Regulation of claudin expression in
cancer. a
| Type of claudin
regulation | Regulator | Cancer type | Mechanism | Function | (Refs.) |
|---|
| Genetic | Amplification | SCC |
Amplification→claudin-1 increased | – |
|
|
|
| Ovarian cancer |
Amplification→claudin-6 increased | – |
|
| Epigenetic | DNA
methylation | Breast cancer |
DOCK1→RRP1B→DNMT→claudin-1 decreased | Viability and
motility increased | (52) |
|
| Histone
modifications | HCC | EZH2→H3K27ME3
increased→ claudin-14 decreased→Wnt/β-catenin signaling activity
increased | Motility, invasive
and EMT increased | (53) |
| Signaling pathways
and | ERK, Akt
and/or | Breast cancer |
ADAM15→PI3K/Akt/mTOR increased→claudin-1
increased | Cell clustering
increased | (88) |
| TFs | β-catenin | Lung cancer |
PI3K/Akt/NF-κB→claudin-1 increased | Anticancer drugs
penetration decreased | (82) |
|
|
| Cervical
cancer | Estrogen→GPR30→ERK
and/or Akt increased→claudin-1 increased | Survival,
proliferation, migration and invasion increased | (85) |
|
|
| Pancreatic
cancer | PKCα→Snail and
MAPK/ERK increased→claudin-1 decreased | EMT increased | (83) |
|
|
| CRC | β-catenin→claudin-7
decreased | EMT increased | (89) |
|
| TGF-β/S MAD | Lung cancer |
TGF-β→Smad→claudin-4 increased | Motility and
tumorigenicity increased | (90) |
|
| JAK/STAT | HCC | hGH→STAT3→claudin-1
decreased | Invasive and
CSC-like properties increased | (91) |
|
| EMT-TFs | SCC | Snail→claudin-11
increased | Collective cell
migration and invasion increased | (61) |
|
|
| Pancreatic
cancer | ZIP4→ZEB1→claudin-1
decreased→ FAK and Paxill increased | Migration and
invasion increased | (62) |
|
| Others | Gastric cancer | RUNX3→claudin-1
increased | Tumorigenicity
decreased | (92) |
|
|
| Prostate
cancer | AR→claudin-8
increased | Proliferation and
migration increased | (93) |
|
|
| Gastric cancer |
NSAIDs→intracellular Ca2+
increased→claudin-4 increased |
Anchorage-independent growth and cell
migration decreased | (94) |
|
|
| Lung cancer |
TNF-α→PKCδ/iPLA2/PGE2/PPARγ
increased→claudin-1 increased | Morphology changes
and migration increased | (95) |
| mRNA stability | miRs | CRC | miR-488→claudin-2
decreased→ MAPK decreased | Cell proliferation,
EMT, and lymph node metastasis decreased | (64) |
|
|
| HCC | miR-486→claudin-10
decreased | Growth, colony
formation and migration decreased | (65) |
|
|
| Lung cancer |
miR-146-5p→claudin-12
decreased→Wnt/β-catenin and PI3K/AKT/MAPK increased | Cell viability,
migration and invasion increased; apoptosis decreased | (63) |
|
|
| Gastric cancer | miR-1303→claudin-18
decreased | Proliferation,
migration and invasion increased | (66) |
|
| LncRNA | Colon cancer |
LINC00662→miR-340-5p
decreased→claudin-8/IL22/ERK increased | Proliferation,
invasion and migration increased; apoptosis decreased | (67) |
|
|
| Gastric cancer | LncRNA
PCAT18→miR-135b decreased→claudin-11 increased | Proliferation,
migration and invasion decreased | (68) |
|
| Others | Colon cancer |
HDACs→3′-UTR→claudin-1 increased | dedifferentiation
and invasion increased | (69) |
| PTMs |
Phospharylation | SCC | Claudin-11 tyrosine
phosphorylation→ Src increased→p190RhoGAP increased→RhoA activity
decreased→stable cell-cell contacts increased | Collective cell
migration and invasion increased | (61) |
|
|
| RCC | EphA2/Ephrin
A1→claudin-4 tyrosine phosphorylation increased→ cytoplasmic
translocation increased; PKC-ε→claudin-4 serine phosphorylation
increased +YAP→ nuclear translocation increased | EMT increased | (72) |
|
| Palmitoylation | RCC | Palmitoylated
claudin-7→GEM→integrin and EpCAM recruitment increased→cytoskeletal
linker proteins and MMP14, CD147 and TACE association
increased | Motility, matrix
degradation and EpCAM cleavage increased | (73) |
|
|
| Ovarian cancer | ZDHHC12→claudin-3
palmitoylation increased→accurate plasma membrane localization and
protein stability increased | Tumorigenic
promotion effect increased | (74) |
Genetic background
Currently, cancer is regarded as a disease
characterized by uncontrolled cellular growth caused primarily by
genetic alterations, which mainly occur in a set of cancer driver
genes, conferring transformed cells with certain selective
advantages over neighboring cells (45). Although no claudin gene mutations
were identified as drivers in any tumor type in an analysis using
the IntOGen platform, genetic alteration patterns can be informed
from sequencing data from a number of large tumor sequencing
initiatives, such as TCGA (46,47). In
the present review, cBioPortal was searched using TCGA PanCancer
Atlas data (https://www.cbioportal.org/) and the frequencies and
types of genetic alterations of CLDNs in cancer were found
to vary depending on the tumor and CLDN type, and an
important degree of variability across cancer subtypes
(histological and molecular) was also observed. Some genetic
alterations occur across several cancer types, while others tend to
be more specific. Therefore, the differences in the expression of
claudins in cancer may result from this genetic heterogeneity. Two
examples are the expression patterns of claudin-1 and −6, which are
always overexpressed in squamous cancer types and ovarian cancer,
respectively, consistent with the high amplification rates of the
two genes in these cancer types (31,48).
However, in a number of cases, the impact of amplification on the
expression of an individual gene is relatively subtle, and the
common deep deletion of CLDN20-25 has not been validated at
the protein level. Therefore, except for genetic alterations, the
expression of claudins in cancer is regulated by other factors.
Epigenetic alterations
At the epigenetic level, at least some members of
the claudin family are subjected to extensive epigenetic
regulation. The context-specific differences in chromatin
organization and DNA accessibility may direct the level of claudins
in cancer (49). For example,
promoter hypomethylation and histone H3 hyperacetylation of
CLDN3 were shown to increase claudin-3 expression in ovarian
cancer cells, while DNA hypermethylation of the promoter of
CLDN1 was an important factor for the low expression of
claudin-1 in ER-positive breast cancer (50,51). In
breast cancer cells with low claudin expression, dedicator of
cytokinesis 1 mediates the downregulation of claudin-1 expression
through promoter activity suppression by DNA methyltransferase,
leading to cancer progression and metastasis (52). For some claudins, expression changes
with changes in methylation. For example, claudin-18.2 expression
decreases as the methylation of CpGs in promoter regions increases
in gastric cancer, but increases as methylation decreases in
pancreatic cancer (27). Histone
modifications such as H3K27ME3, catalyzed by enhancer of zeste
homolog 2, were also demonstrated to decrease the expression of
claudin-14, providing an explanation for the aggressive nature of
hepatocellular carcinoma (HCC) with downregulation of claudin-14
(53). Epigenetic alterations have a
greater impact on cancer cell phenotypes than genomic alterations,
and the epigenetic status can be highly plastic, suggesting that
epigenetic regulation have a dominant impact on claudin expression,
consistent with the diverse and dynamic expression patterns of
claudins.
Transcription factors
Expression regulation of claudins is also tightly
controlled by different transcription factors (TFs). Different TFs
control the expression of claudins in a context-dependent manner to
achieve inhibition or activation, which reflects the diverse
expression patterns of the claudins. Acting at the onset and in the
progression of cancer, neoplastic conversion profoundly alters the
claudin repertoire in cancer cells to achieve neoplastic
transformation. TFs involved in epithelial-mesenchymal transition
(EMT) are major drivers of these alterations (54–56).
Studies have shown that claudin-1 expression is inhibited by Slug
and Snail in breast cancer, but activated by caudal homeobox
proteins (Cdx1 and Cdx2), GATA binding protein 4 and β-catenin in
colon cancer via interactions with different elements in the
promoter (54,55). Claudin-7 expression is negatively
regulated by Snail by the direct binding of Snail to the E-box of
promoter regions, but is positively regulated by embryonic liver
fodrin 3 and hepatocyte nuclear factor 4, whose binding sites are
all also located in the promoter region (56–58).
Some claudins also exhibit high pairwise sequence homology, so
coordinated gene expression is possible, and the same TF can
regulate the expression of different claudins simultaneously; for
example, Sp1 is known to regulate claudin-3 and −4 promoter
activity in ovarian cancer (50,59,60). The
effects of TFs on claudin expression are context-dependent; for
example, the EMT-TF Snail induces claudin-11 expression and prompts
collective migration for tumor progression in squamous cell
carcinoma, whereas another EMT-TF, zinc finger E-box binding
homeobox 1 (ZEB1), promotes pancreatic cancer progression by
repressing claudin-1 expression, reflecting the complexity of the
roles of the claudins in cancer development (61,62).
Stability of mRNA
At the mRNA level, alternative or mis-splicing can
change the expression levels of claudins. Several mis-splicing
variants resulting in a premature stop codon and thus likely
leading to nonsense-mediated decay have been found for CLDN1
and CLDN18 (27,29). The stability of mRNA is one of the
most important determinants of protein expression. MicroRNAs
(miRs/miRNAs) are short non-coding RNAs (ncRNAs) that inhibit the
expression of target genes by directly binding to their target
mRNAs, and some miRNAs have been reported to regulate the
expression of claudins in cancer. For example, miR-146-5p promotes
cell viability, migration and invasion, inhibits apoptosis and
activates oncogenic pathways by decreasing claudin-2 expression in
lung cancer cells, while miR-488 could suppress the cell
proliferation, EMT and lymph node metastasis of CRC cells via
inhibition of claudin-2 expression (63,64).
miR-486 functions as a tumor suppressor in HCC by targeting
claudin-10, which decreases cancer cell proliferation, colony
formation and migration, while miR-1303 functions as an oncogenic
miRNA in gastric cancer. Downregulation of miR-1303 can inhibit
proliferation, migration and invasion of cancer cells by targeting
claudin-18 (65,66). Another type of ncRNA, long ncRNA, can
also affect the expression of claudins through effects on miRNAs,
with protumor or antitumor roles in a context-dependent manner
(67,68). Furthermore, one study revealed that
histone deacetylases (HDACs) modulate claudin-1 mRNA stability by
interacting with its 3′ untranslated region, indicating the
application of HDAC inhibitors in such settings with anticancer
promise (69).
PTMs
Apart from these transcriptional regulatory
mechanisms, claudin levels and functions are also known to be
regulated by PTMs that affect their protein interactions,
trafficking, subcellular localization, oligomer assembly and
overall turnover (25).
Dysregulation of the transportation of claudins, such as the
cytoplasmic accumulation of claudin-1 induced by inhibition of
endosomal sorting complexes required for transport, causes TJs to
disassemble and lose cell polarity (70). PTMs, including phosphorylation,
palmitoylation, ubiquitination, glycosylation and proteolysis, are
all involved in claudin homeostasis (25). Almost all signaling pathways involved
in cancer contain kinases; therefore, it is not surprising that the
phosphorylation of claudins is an important PTM that affects their
localization and function. In addition, some data have demonstrated
that claudin delocalization contributes to cancer hallmarks. For
instance, activated ephrin receptor A2 phosphorylates the tyrosine
residue of claudin-4, decreasing its association with ZO-1 and
leading to claudin-4 accumulation in the cytoplasm, which decreases
the integration of claudin-4 into TJs. Further phosphorylation of
the serine residue by protein kinase C (PKC) results in nuclear
import of claudin-4 (71,72). By contrast, tyrosine phosphorylation
of claudin-11 activates Src to maintain stable cell-cell contacts
by suppressing RhoA activity, which prompts the formation of
circulating tumor cell clusters (61). Palmitoylation of claudin-7 affects
the relative amounts of claudin-7 in the basolateral membrane by
leading to the incorporation of claudin-7 into glycolipid-enriched
membrane domains and inhibiting its integration into TJs, while
palmitoylation of claudin-3 by zinc finger DHHC-type
palmitoyltransferase 12 contributes to plasma membrane
localization, protein stability and ovarian cancer cell
tumorigenesis (73,74). Although evidence from humans is
scarce, ubiquitination and protease cleavage of claudins have been
observed in human cells under normal physiological or inflammatory
conditions, but their roles in cancer need further research
(75–77).
Cell of origin and differentiation
status
The cell of origin and differentiation status are
important determinants of the expression profile of cancer, which
is shaped during the development of the cancer, although
alterations always occur to some extent after malignant
transformation. For example, claudin-1 is especially crucial for
squamous epithelium such as the skin, whose germline mutations
mainly cause neonatal ichthyosis. Therefore, the expression of
claudin-1 is high in squamous cancer types, whereas cancer types
with adenocarcinomatous components are likely to lose claudin-1
expression (48). Another example is
claudin-6, which is an oncofetal cell surface antigen that is
completely silenced in normal human tissues, but reactivated in
germline cancers such as testicular, ovarian and uterine cancer
(31). Another mechanism
contributing to the different expression profiles of claudins in
cancer is the differentiation status of cancer cells. Key studies
have shown that some cancer types display a hierarchical nature
founded by a cancer stem cell or a malignant cell that has gained
stem cell properties, leading to reversal of the differentiation
signals put in place during development and acquisition of
characteristics of the undifferentiated state (78). In this regard, this phenomenon may
explain the dysregulated expression of claudins in some cancer
types. For example, claudin-2 expression is restricted to the
stem/progenitor cell compartment in the normal intestinal
epithelium; however, claudin-2 is widely expressed in human CRC
tissues (79,80). By contrast, the expression of
claudin-18.2 is absent from the stem cell zone of gastric glands,
but increased in differentiated gastric cells. When gastric cancer
arises from these stem cells or the cells transition into an
undifferentiated state, the expression of claudin-18.2
significantly decreases (81).
Context-specific output of signaling
pathways
In the majority of instances, oncogene activation
lies upstream of most protein expression dysregulation in cancer
cells. However, different cell and tissue types show profound
differences in their response to oncogenic driver mutations, and
the organization and output of oncogenic signaling pathways, such
as MAPK, PI3K/Akt, WNT/β-catenin and NF-κB signalling, can differ
substantially between tissue types, which leads to different
expression patterns of claudins (82–95). For
example, in lung cancer cells, claudin-1 is upregulated through
activation of the PI3K/Akt/NF-κB pathway, while in pancreatic
cancer cells, claudin-1 is downregulated by PKCα via Snail- and
MAPK/ERK-dependent pathways (82,83).
Moreover, the membrane zinc importer Zrt-/Irt-like protein 4 (ZIP4)
was found to be overexpressed in human pancreatic, and it was
demonstrated that ZIP4 represses claudin-1 through a ZEB1-dependent
transcriptional mechanism (84). The
contradictory roles of claudin-1 in different cancer types reflect
the fact that different oncogenic pathways can hijack different
roles of claudins to contribute to cancer progression. In addition,
signaling pathways also encode, process and integrate external and
internal signals, providing a specific and appropriate response to
external stimuli. Therefore, the expression of claudins is further
complicated by the output of these signals integrated from external
stimuli, such as spatial and temporal variability in nutrients,
oxygenation, growth factors and hormones. For example,
estrogen/GPR30 signaling induces claudin-1 expression in
ER-negative cervical adenocarcinoma (85). Glucose was found to regulate the
expression of claudin-2 in endometrial cancer (86). Additionally, some vitamins can
regulate the expression of claudins. For example, vitamin D can
regulate claudin-2 and −4 protein levels (87).
Future perspectives and conclusions
Consistent with the diversity of claudin expression
in cancer, numerous studies on the roles of claudins in
carcinogenesis have reported contradictory results. However, the
data suggest that even if it does not function as a cancer driver,
claudin dysregulation assists in the initiation and progression of
cancer and is involved in nearly all aspects of tumor biology and
all steps of tumor development. In addition to their various roles
in cancer, from a translational perspective, as classic cell
surface molecules, claudins may be ideal targets for therapy. As
aforementioned, claudin expression has tissue specificity and is
dysregulated in cancer. Furthermore, it has been observed that most
claudins are buried in the TJ complex in normal tissues, while
higher accessibility is caused by extrajunctional mislocalization
in malignant tissues (96). Owing to
this specific expression profile and difference between normal and
tumor cells, claudin-targeting strategies seem to hold substantial
promise, and this idea has been validated by clinical applications
using specific claudin-targeting therapies (targeting claudin-6 and
−18.2) (97). For the majority of
claudins, with the exception of claudin-6 and −18.2, although the
antitumor effect has been verified in proof-of-concept experiments,
they remain at the laboratory stage, and their translation into
clinical practice is eagerly awaited.
Overall, the dysregulated expression of claudins is
a common phenotype associated with a number of different cancer
types. The tissue-, cancer- and stage-specific expression patterns
of claudins are the result of the regulation of various factors at
different levels, from genetic background to tumor microenvironment
alterations. Along with the understanding of the dysregulation of
claudin expression in cancer, the exploration of their role in
tumorigenesis and thus in cancer treatment and prevention will be
advanced.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed during the
current study are available in TCGA repository and cBioPortal.
Authors' contributions
JL conceived the paper, wrote the initial draft,
generated the figures, edited and revised the original manuscript,
and read and approved the final manuscript. Data authentication is
not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The author declares that they have no competing
interests.
References
|
1
|
Furuse M, Sasaki H, Fujimoto K and Tsukita
S: A single gene product, claudin-1 or −2, reconstitutes tight
junction strands and recruits occludin in fibroblasts. J Cell Biol.
143:391–401. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singh AB, Uppada SB and Dhawan P: Claudin
proteins, outside-in signaling, and carcinogenesis. Pflugers Arch.
469:69–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hashimoto Y, Tachibana K, Krug SM,
Kunisawa J, Fromm M and Kondoh M: Potential for tight junction
protein-Directed drug development using claudin binders and
angubindin-1. Int J Mol Sci. 20:40162019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schneeberger EE and Lynch RD: The tight
junction: A multifunctional complex. Am J Physiol Cell Physiol.
286:C1213–C1228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Niessen CM: Tight junctions/adherens
junctions: Basic structure and function. J Invest Dermatol.
127:2525–2532. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meng W and Takeichi M: Adherens junction:
Molecular architecture and regulation. Cold Spring Harb Perspect
Biol. 1:a0028992009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kottke MD, Delva E and Kowalczyk AP: The
desmosome: Cell science lessons from human diseases. J Cell Sci.
119((Pt 5)): 797–806. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farquhar MG and Palade GE: Junctional
complexes in various epithelia. J Cell Biol. 17:375–412. 1963.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zihni C, Mills C, Matter K and Balda MS:
Tight junctions: From simple barriers to multifunctional molecular
gates. Nat Rev Mol Cell Biol. 17:564–580. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lal-Nag M and Morin PJ: The claudins.
Genome Biol. 10:2352009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsukita S, Tanaka H and Tamura A: The
Claudins: From tight junctions to biological systems. Trends
Biochem Sci. 44:141–152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suzuki H, Nishizawa T, Tani K, Yamazaki Y,
Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O and Fujiyoshi
Y: Crystal structure of a claudin provides insight into the
architecture of tight junctions. Science. 344:304–307. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Suzuki H, Tani K, Tamura A, Tsukita S and
Fujiyoshi Y: Model for the architecture of claudin-based
paracellular ion channels through tight junctions. J Mol Biol.
427:291–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saitoh Y, Suzuki H, Tani K, Nishikawa K,
Irie K, Ogura Y, Tamura A, Tsukita S and Fujiyoshi Y: Tight
junctions. Structural insight into tight junction disassembly by
Clostridium perfringens enterotoxin. Science. 347:775–778.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shinoda T, Shinya N, Ito K, Ohsawa N,
Terada T, Hirata K, Kawano Y, Yamamoto M, Kimura-Someya T, Yokoyama
S and Shirouzu M: Structural basis for disruption of claudin
assembly in tight junctions by an enterotoxin. Sci Rep.
6:336322016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakamura S, Irie K, Tanaka H, Nishikawa K,
Suzuki H, Saitoh Y, Tamura A, Tsukita S and Fujiyoshi Y:
Morphologic determinant of tight junctions revealed by claudin-3
structures. Nat Commun. 10:8162019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen L, Weber CR, Raleigh DR, Yu D and
Turner JR: Tight junction pore and leak pathways: A dynamic duo.
Annu Rev Physiol. 73:283–309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Otani T and Furuse M: Tight junction
structure and function revisited. Trends Cell Biol. 30:805–817.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ahmad R, Kumar B, Chen Z, Chen X, Müller
D, Lele SM, Washington MK, Batra SK, Dhawan P and Singh AB: Loss of
claudin-3 expression induces IL6/gp130/Stat3 signaling to promote
colon cancer malignancy by hyperactivating Wnt/β-catenin signaling.
Oncogene. 36:6592–6604. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou B, Flodby P, Luo J, Castillo DR, Liu
Y, Yu FX, McConnell A, Varghese B, Li G, Chimge NO, et al:
Claudin-18-mediated YAP activity regulates lung stem and progenitor
cell homeostasis and tumorigenesis. J Clin Invest. 128:970–984.
2018. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hagen SJ: Non-canonical functions of
claudin proteins: Beyond the regulation of cell-cell adhesions.
Tissue Barriers. 5:e13278392017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu Z, Kim DH, Fan J, Lu Q, Verbanac K,
Ding L, Renegar R and Chen YH: A non-tight junction function of
claudin-7-Interaction with integrin signaling in suppressing lung
cancer cell proliferation and detachment. Mol Cancer. 14:1202015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nübel T, Preobraschenski J, Tuncay H,
Weiss T, Kuhn S, Ladwein M, Langbein L and Zöller M: Claudin-7
regulates EpCAM-mediated functions in tumor progression. Mol Cancer
Res. 7:285–299. 2009. View Article : Google Scholar
|
|
24
|
Pope JL, Bhat AA, Sharma A, Ahmad R,
Krishnan M, Washington MK, Beauchamp RD, Singh AB and Dhawan P:
Claudin-1 regulates intestinal epithelial homeostasis through the
modulation of Notch-signalling. Gut. 63:622–634. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu F, Koval M, Ranganathan S, Fanayan S,
Hancock WS, Lundberg EK, Beavis RC, Lane L, Duek P, McQuade L, et
al: Systems proteomics view of the endogenous human claudin protein
family. J Proteome Res. 15:339–359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Milatz S and Breiderhoff T: One gene, two
paracellular ion channels-claudin-10 in the kidney. Pflugers Arch.
469:115–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Zhang Y, Hu D, Gong T, Xu R and Gao
J: Analysis of the expression and genetic alteration of CLDN18 in
gastric cancer. Aging (Albany NY). 12:14271–14284. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Milatz S: A novel claudinopathy based on
claudin-10 mutations. Int J Mol Sci. 20:53962019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Blanchard AA, Zelinski T, Xie J, Cooper S,
Penner C, Leygue E and Myal Y: Identification of claudin 1
transcript variants in human invasive breast cancer. PLoS One.
11:e01633872016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ben-David U, Nudel N and Benvenisty N:
Immunologic and chemical targeting of the tight-junction protein
Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat
Commun. 4:19922013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reinhard K, Rengstl B, Oehm P, Michel K,
Billmeier A, Hayduk N, Klein O, Kuna K, Ouchan Y, Wöll S, et al: An
RNA vaccine drives expansion and efficacy of claudin-CAR-T cells
against solid tumors. Science. 367:446–453. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Erdélyi-Belle B, Török G, Apáti Á, Sarkadi
B, Schaff Z, Kiss A and Homolya L: Expression of tight junction
components in hepatocyte-like cells differentiated from human
embryonic stem cells. Pathol Oncol Res. 21:1059–1070. 2015.
View Article : Google Scholar
|
|
33
|
Fujita H, Chiba H, Yokozaki H, Sakai N,
Sugimoto K, Wada T, Kojima T, Yamashita T and Sawada N:
Differential expression and subcellular localization of claudin-7,
−8, −12, −13, and −15 along the mouse intestine. J Histochem
Cytochem. 54:933–944. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
D'Souza T, Sherman-Baust CA, Poosala S,
Mullin JM and Morin PJ: Age-related changes of claudin expression
in mouse liver, kidney, and pancreas. J Gerontol A Biol Sci Med
Sci. 64:1146–1153. 2009. View Article : Google Scholar
|
|
35
|
Perdomo-Ramirez A, Aguirre M, Davitaia T,
Ariceta G, Ramos-Trujillo E; RenalTube Group, ; Claverie-Martin F:
Characterization of two novel mutations in the claudin-16 and
claudin-19 genes that cause familial hypomagnesemia with
hypercalciuria and nephrocalcinosis. Gene. 689:227–234. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ouban A: Claudin-1 role in colon cancer:
An update and a review. Histol Histopathol. 33:1013–1019.
2018.PubMed/NCBI
|
|
37
|
Wang K, Xu C, Li W and Ding L: Emerging
clinical significance of claudin-7 in colorectal cancer: A review.
Cancer Manag Res. 10:3741–3752. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dhawan P, Singh AB, Deane NG, No Y, Shiou
SR, Schmidt C, Neff J, Washington MK and Beauchamp RD: Claudin-1
regulates cellular transformation and metastatic behavior in colon
cancer. J Clin Invest. 115:1765–1776. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Blanchard AA, Skliris GP, Watson PH,
Murphy LC, Penner C, Tomes L, Young TL, Leygue E and Myal Y:
Claudins 1, 3, and 4 protein expression in ER negative breast
cancer correlates with markers of the basal phenotype. Virchows
Arch. 454:647–656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cherradi S, Ayrolles-Torro A, Vezzo-Vié N,
Gueguinou N, Denis V, Combes E, Boissière F, Busson M,
Canterel-Thouennon L, Mollevi C, et al: Antibody targeting of
claudin-1 as a potential colorectal cancer therapy. J Exp Clin
Cancer Res. 36:892017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matsuoka T, Mitomi H, Fukui N, Kanazawa H,
Saito T, Hayashi T and Yao T: Cluster analysis of claudin-1 and −4,
E-cadherin, and β-catenin expression in colorectal cancers. J Surg
Oncol. 103:674–686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bhat AA, Ahmad R, Uppada SB, Singh AB and
Dhawan P: Claudin-1 promotes TNF-α-induced epithelial-mesenchymal
transition and migration in colorectal adenocarcinoma cells. Exp
Cell Res. 349:119–127. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bornholdt J, Friis S, Godiksen S, Poulsen
SS, Santoni-Rugiu E, Bisgaard HC, Lothe IM, Ikdahl T, Tveit KM,
Johnson E, et al: The level of claudin-7 is reduced as an early
event in colorectal carcinogenesis. BMC Cancer. 11:652011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tanaka M, Shibahara J, Fukushima N,
Shinozaki A, Umeda M, Ishikawa S, Kokudo N and Fukayama M:
Claudin-18 is an early-stage marker of pancreatic carcinogenesis. J
Histochem Cytochem. 59:942–952. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stratton MR: Exploring the genomes of
cancer cells: Progress and promise. Science. 331:1553–1558. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gonzalez-Perez A, Perez-Llamas C, Deu-Pons
J, Tamborero D, Schroeder MP, Jene-Sanz A, Santos A and Lopez-Bigas
N: IntOGen-mutations identifies cancer drivers across tumor types.
Nat Methods. 10:1081–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Martínez-Jiménez F, Muiños F, Sentís I,
Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O,
Bonet J, Kranas H, et al: A compendium of mutational cancer driver
genes. Nat Rev Cancer. 20:555–572. 2020. View Article : Google Scholar
|
|
48
|
Paschoud S, Bongiovanni M, Pache JC and
Citi S: Claudin-1 and claudin-5 expression patterns differentiate
lung squamous cell carcinomas from adenocarcinomas. Mod Pathol.
20:947–954. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schaefer MH and Serrano L: Cell
type-specific properties and environment shape tissue specificity
of cancer genes. Sci Rep. 6:207072016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Honda H, Pazin MJ, D'Souza T, Ji H and
Morin PJ: Regulation of the CLDN3 gene in ovarian cancer cells.
Cancer Biol Ther. 6:1733–1742. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Di Cello F, Cope L, Li H, Jeschke J, Wang
W, Baylin SB and Zahnow CA: Methylation of the claudin 1 promoter
is associated with loss of expression in estrogen receptor positive
breast cancer. PLoS One. 8:e686302013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chiang SK, Chang WC, Chen SE and Chang LC:
DOCK1 regulates growth and motility through the RRP1B-claudin-1
pathway in claudin-low breast cancer cells. Cancers (Basel).
11:17622019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li CP, Cai MY, Jiang LJ, Mai SJ, Chen JW,
Wang FW, Liao YJ, Chen WH, Jin XH, Pei XQ, et al: CLDN14 is
epigenetically silenced by EZH2-mediated H3K27ME3 and is a novel
prognostic biomarker in hepatocellular carcinoma. Carcinogenesis.
37:557–566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Martínez-Estrada OM, Cullerés A, Soriano
FX, Peinado H, Bolós V, Martínez FO, Reina M, Cano A, Fabre M and
Vilaró S: The transcription factors Slug and Snail act as
repressors of Claudin-1 expression in epithelial cells. Biochem J.
(394(Pt 2)): 449–457. 2006. View Article : Google Scholar
|
|
55
|
Bhat AA, Sharma A, Pope J, Krishnan M,
Washington MK, Singh AB and Dhawan P: Caudal homeobox protein Cdx-2
cooperates with Wnt pathway to regulate claudin-1 expression in
colon cancer cells. PLoS One. 7:e371742012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Feng J, Cen J, Li J, Zhao R, Zhu C, Wang
Z, Xie J and Tang W: Histone deacetylase inhibitor valproic acid
(VPA) promotes the epithelial mesenchymal transition of colorectal
cancer cells via up regulation of Snail. Cell Adh Migr. 9:495–501.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kohno Y, Okamoto T, Ishibe T, Nagayama S,
Shima Y, Nishijo K, Shibata KR, Fukiage K, Otsuka S, Uejima D, et
al: Expression of claudin7 is tightly associated with epithelial
structures in synovial sarcomas and regulated by an Ets family
transcription factor, ELF3. J Biol Chem. 281:38941–38950. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Farkas AE, Hilgarth RS, Capaldo CT,
Gerner-Smidt C, Powell DR, Vertino PM, Koval M, Parkos CA and
Nusrat A: HNF4alpha regulates claudin-7 protein expression during
intestinal epithelial differentiation. Am J Pathol. 185:2206–2218.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Baltzegar DA, Reading BJ, Brune ES and
Borski RJ: Phylogenetic revision of the claudin gene family. Mar
Genomics. 11:17–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Honda H, Pazin MJ, Ji H, Wernyj RP and
Morin PJ: Crucial roles of Sp1 and epigenetic modifications in the
regulation of the CLDN4 promoter in ovarian cancer cells. J Biol
Chem. 281:21433–21444. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li CF, Chen JY, Ho YH, Hsu WH, Wu LC, Lan
HY, Hsu DS, Tai SK, Chang YC and Yang MH: Snail-induced claudin-11
prompts collective migration for tumour progression. Nat Cell Biol.
21:251–262. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu M, Yang J, Zhang Y, Zhou Z, Cui X,
Zhang L, Fung KM, Zheng W, Allard FD, Yee EU, et al: ZIP4 promotes
pancreatic cancer progression by repressing ZO-1 and claudin-1
through a ZEB1-dependent transcriptional mechanism. Clin Cancer
Res. 24:3186–3196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sun X, Cui S, Fu X, Liu C, Wang Z and Liu
Y: MicroRNA-146-5p promotes proliferation, migration and invasion
in lung cancer cells by targeting claudin-12. Cancer Biomark.
25:89–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang YB, Shi Q, Li G, Zheng JH, Lin J and
Qiu W: MicroRNA-488 inhibits progression of colorectal cancer via
inhibition of the mitogen-activated protein kinase pathway by
targeting claudin-2. Am J Physiol Cell Physiol. 316:C33–C47. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sun H, Cui C, Xiao F, Wang H, Xu J, Shi X,
Yang Y, Zhang Q, Zheng X, Yang X, et al: MiR-486 regulates
metastasis and chemosensitivity in hepatocellular carcinoma by
targeting CLDN10 and CITRON. Hepatol Res. 45:1312–1322. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang SJ, Feng JF, Wang L, Guo W, Du YW,
Ming L and Zhao GQ: MiR-1303 targets claudin-18 gene to modulate
proliferation and invasion of gastric cancer cells. Dig Dis Sci.
59:1754–1763. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cheng B, Rong A, Zhou Q and Li W: LncRNA
LINC00662 promotes colon cancer tumor growth and metastasis by
competitively binding with miR-340-5p to regulate CLDN8/IL22
co-expression and activating ERK signaling pathway. J Exp Clin
Cancer Res. 39:52020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang XZ, Mao HL, Zhang SJ, Sun L, Zhang
WJ, Chen QZ, Wang L and Liu HC: lncRNA PCAT18 inhibits
proliferation, migration and invasion of gastric cancer cells
through miR-135b suppression to promote CLDN11 expression. Life
Sci. 249:1174782020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Krishnan M, Singh AB, Smith JJ, Sharma A,
Chen X, Eschrich S, Yeatman TJ, Beauchamp RD and Dhawan P: HDAC
inhibitors regulate claudin-1 expression in colon cancer cells
through modulation of mRNA stability. Oncogene. 29:305–312. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhou B, Moodie A, Blanchard AA, Leygue E
and Myal Y: Claudin 1 in breast cancer: New insights. J Clin Med.
4:1960–1976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tanaka M, Kamata R and Sakai R: EphA2
phosphorylates the cytoplasmic tail of Claudin-4 and mediates
paracellular permeability. J Biol Chem. 280:42375–42382. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Owari T, Sasaki T, Fujii K, Fujiwara-Tani
R, Kishi S, Mori S, Mori T, Goto K, Kawahara I, Nakai Y, et al:
Role of nuclear claudin-4 in renal cell carcinoma. Int J Mol Sci.
21:83402020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Heiler S, Mu W, Zöller M and Thuma F: The
importance of claudin-7 palmitoylation on membrane subdomain
localization and metastasis-promoting activities. Cell Commun
Signal. 13:292015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yuan M, Chen X, Sun Y, Jiang L, Xia Z, Ye
K, Jiang H, Yang B, Ying M, Cao J and He Q: ZDHHC12-mediated
claudin-3 S-palmitoylation determines ovarian cancer progression.
Acta Pharm Sin B. 10:1426–1439. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Mandel I, Paperna T, Volkowich A, Merhav
M, Glass-Marmor L and Miller A: The ubiquitin-proteasome pathway
regulates claudin 5 degradation. J Cell Biochem. 113:2415–2423.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gong Y, Wang J, Yang J, Gonzales E, Perez
R and Hou J: KLHL3 regulates paracellular chloride transport in the
kidney by ubiquitination of claudin-8. Proc Natl Acad Sci USA.
112:4340–4345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Willemsen LE, Hoetjes JP, van Deventer SJ
and van Tol EA: Abrogation of IFN-gamma mediated epithelial barrier
disruption by serine protease inhibition. Clin Exp Immunol.
142:275–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lytle NK, Barber AG and Reya T: Stem cell
fate in cancer growth, progression and therapy resistance. Nat Rev
Cancer. 18:669–680. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Rahner C, Mitic LL and Anderson JM:
Heterogeneity in expression and subcellular localization of
claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut.
Gastroenterology. 120:411–422. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Paquet-Fifield S, Koh SL, Cheng L, Beyit
LM, Shembrey C, Mølck C, Behrenbruch C, Papin M, Gironella M,
Guelfi S, et al: Tight junction protein claudin-2 promotes
self-renewal of human colorectal cancer stem-like cells. Cancer
Res. 78:2925–2938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sahin U, Koslowski M, Dhaene K, Usener D,
Brandenburg G, Seitz G, Huber C and Türeci O: Claudin-18 splice
variant 2 is a pan-cancer target suitable for therapeutic antibody
development. Clin Cancer Res. 14:7624–7634. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Akizuki R, Maruhashi R, Eguchi H,
Kitabatake K, Tsukimoto M, Furuta T, Matsunaga T, Endo S and Ikari
A: Decrease in paracellular permeability and chemosensitivity to
doxorubicin by claudin-1 in spheroid culture models of human lung
adenocarcinoma A549 cells. Biochim Biophys Acta Mol Cell Res.
1865:769–780. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kyuno D, Kojima T, Yamaguchi H, Ito T,
Kimura Y, Imamura M, Takasawa A, Murata M, Tanaka S, Hirata K and
Sawada N: Protein kinase Cα inhibitor protects against
downregulation of claudin-1 during epithelial-mesenchymal
transition of pancreatic cancer. Carcinogenesis. 34:1232–1243.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H,
Wang X, Zhang S, Liuzzi JP, Chang SM, Cousins RJ, et al: Aberrant
expression of zinc transporter ZIP4 (SLC39A4) significantly
contributes to human pancreatic cancer pathogenesis and
progression. Proc Natl Acad Sci USA. 104:18636–18641. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Akimoto T, Takasawa A, Takasawa K, Aoyama
T, Murata M, Osanai M, Saito T and Sawada N: Estrogen/GPR30
signaling contributes to the malignant potentials of ER-negative
cervical adenocarcinoma via regulation of claudin-1 expression.
Neoplasia. 20:1083–1093. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Okada T, Konno T, Kohno T, Shimada H,
Saito K, Satohisa S, Saito T and Kojima T: Possibility of targeting
claudin-2 in therapy for human endometrioid endometrial carcinoma.
Reprod Sci. 27:2092–2103. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Domazetovic V, Iantomasi T, Bonanomi AG
and Stio M: Vitamin D regulates claudin-2 and claudin-4 expression
in active ulcerative colitis by p-Stat-6 and Smad-7 signaling. Int
J Colorectal Dis. 35:1231–1242. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Mattern J, Roghi CS, Hurtz M, Knäuper V,
Edwards DR and Poghosyan Z: ADAM15 mediates upregulation of
claudin-1 expression in breast cancer cells. Sci Rep. 9:125402019.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kim WK, Kwon Y, Jang M, Park M, Kim J, Cho
S, Jang DG, Lee WB, Jung SH, Choi HJ, et al: β-catenin activation
down-regulates cell-cell junction-related genes and induces
epithelial-to-mesenchymal transition in colorectal cancers. Sci
Rep. 9:184402019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Rachakonda G, Vu T, Jin L, Samanta D and
Datta PK: Role of TGF-β-induced Claudin-4 expression through c-Jun
signaling in non-small cell lung cancer. Cell Signal. 28:1537–1544.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen YJ, You ML, Chong QY, Pandey V,
Zhuang QS, Liu DX, Ma L, Zhu T and Lobie PE: Autocrine human growth
hormone promotes invasive and cancer stem cell-like behavior of
hepatocellular carcinoma cells by STAT3 dependent inhibition of
CLAUDIN-1 expression. Int J Mol Sci. 18:12742017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chang TL, Ito K, Ko TK, Liu Q,
Salto-Tellez M, Yeoh KG, Fukamachi H and Ito Y: Claudin-1 has tumor
suppressive activity and is a direct target of RUNX3 in gastric
epithelial cells. Gastroenterology. 138:255–265.e1-3. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ashikari D, Takayama KI, Obinata D,
Takahashi S and Inoue S: CLDN8, an androgen-regulated gene,
promotes prostate cancer cell proliferation and migration. Cancer
Sci. 108:1386–1393. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Mima S, Tsutsumi S, Ushijima H, Takeda M,
Fukuda I, Yokomizo K, Suzuki K, Sano K, Nakanishi T, Tomisato W, et
al: Induction of claudin-4 by nonsteroidal anti-inflammatory drugs
and its contribution to their chemopreventive effect. Cancer Res.
65:1868–1876. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Iitaka D, Moodley S, Shimizu H, Bai XH and
Liu M: PKCδ-iPLA2-PGE2-PPARγ signaling cascade mediates TNF-α
induced Claudin 1 expression in human lung carcinoma cells. Cell
Signal. 27:568–577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Morin PJ: Claudin proteins in human
cancer: Promising new targets for diagnosis and therapy. Cancer
Res. 65:9603–9606. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Singh P, Toom S and Huang Y: Anti-claudin
18.2 antibody as new targeted therapy for advanced gastric cancer.
J Hematol Oncol. 10:1052017. View Article : Google Scholar : PubMed/NCBI
|