Introduction
Esophageal cancer (EC), accompanied by high
morbidity and mortality, has become one of the most common
malignancies in the world (1).
Esophageal squamous cell carcinoma (ESCC) accounts for ~80% of EC
cases (1,2). ESCC is highly aggressive and is
characterized by a poor prognosis (1,2). There
is a high incidence of EC in China due to numerous factors,
including inappropriate diets (low fruit and vegetable intake and
low fruit intake), drinking alcohol and tobacco smoking (3). The 5-year survival rate of patients
with EC is 15–25% (4), and patients
with advanced EC have a poor prognosis (4). Therefore, it is essential to identify
new biomarkers for early diagnosis and to explore the possible
molecular mechanisms for target-specific drug development.
Long non-coding RNAs (lncRNAs) are a group of RNA
molecules >200 nucleotides in length (5). Dysregulation of lncRNAs is often
observed in several types of cancer, such as EC (6). For example, lncRNA PVT1 promotes the
proliferation, invasion, colony formation and tumor sphere
formation of EC cells by regulating Yes-associated protein 1
signaling (7). lncRNA EIF3J-AS1
increases the invasion of EC cells by mediating AKT1 expression
through interaction with microRNA (miRNA/miR)-373-3p (8). lncRNA opa-interacting protein 5
antisense transcript 1 (OIP5-AS1) is derived from the anti-sense of
the OIP5 gene, and it promotes epithelial-mesenchymal transition
(EMT), migration and invasion in cisplatin-resistant oral squamous
cell carcinoma cells (9). OIP5-AS1
may effectively facilitate the progression of ovarian cancer by
mediating CCNG1 expression through sponging of miR-128-3p (10). Recently, OIP5-AS1 has been reported
to promote the development of EC (11). However, the underlying mechanism
remains unclear.
The expression levels of vesicular overexpressed in
cancer prosurvival protein 1 (VOPP1) are often increased in cancer
(12). VOPP1 enhances cell
proliferation, migration and invasion in lung adenocarcinoma
(13). Additionally, it has become a
putative oncogene in gastric cancer (14). VOPP1 also interacts with cytoplasmic
NF-κB protein (15). Recently, VOPP1
has been identified as one of the key factors associated with EC
development (1). However, the
pathological roles of VOPP1 in EC remain unknown. The present study
aimed to investigate the biological activities of OIP5-AS1 and
VOPP1 in the progression of EC.
Materials and methods
Tissue collection
A total of 32 pairs of human EC tissue samples and
corresponding adjacent non-tumor tissues (5 cm away from the tumor
tissue) were collected from newly diagnosed patients (mean age,
61.6±4.8 years; range, 54 to 75 years) at The First Affiliated
Jiujiang Hospital of Nanchang University (Jiujiang, China) between
March 2016 and July 2019. The patients had not received any
chemotherapy. The clinicopathological data of patients with EC is
shown in Table SI. The EC tissue
samples were histologically confirmed as ESCC by two pathologists.
The tissue samples were immediately frozen in liquid nitrogen and
stored at −80°C until further use. All patients signed the consent
forms prior to the use of their tissues in the present study. The
experimental protocols have been approved by the Ethics Committee
of The First Affiliated Jiujiang Hospital of Nanchang University
according to the Declaration of Helsinki Principles.
Cell culture
Human esophageal carcinoma cells (EC109, EC9706 and
TE-1) and human immortalized normal esophageal epithelial cells
(Het-1A) were obtained from the American Type Culture Collection.
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin and streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) was employed for cell culture. All cells were cultured in a
humidified incubator containing 5% CO2 at 37°C. For
ensuring the phenotype of Het-1A, the first or second generation
was used for investigation.
Cell transfection
Three short hairpin RNAs (shRNAs; sh-OIP5-AS1-#1,
sh-OIP5-AS1-#2 and sh-OIP5-AS1-#3) and a scrambled shRNA negative
control (sh-NC) were obtained from Shanghai GeneChem Co., Ltd. The
shRNAs (100 nM) were inserted into pGPH1/Neo (40 nM; Shanghai
GenePharma Co., Ltd.). Then, 75 pmol of constructed pGPH1/Neo were
transfected into EC cells using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. Briefly, the plasmid pGPH1/Neo/shRNA
and Lipofectamine® 3000 were diluted with Opti-MEM
(serum-reduced medium) and incubated for 5 min at room temperature.
The two transfection mixtures were mixed and incubated at room
temperature for 20 min. The transfection mixture was then gently
added to each well, and the cells were incubated at 37°C (5%
CO2). The cells were transfected with sh-OIP5-AS1 or
sh-NC for 48 h prior to further experiments. Neomycin (cat. no.
1405-10-3; Sigma-Aldrich; Merck KGaA) at 400 µg/µl was used to
select the stable transfected cells for 4 weeks.
miR-30a mimics (sense, 5′-UGUAAACAUCCUCGACUGCAAG-3′
and anti-sense, 5′-CUUCCAGUCGAGGAUCUUUACA-3′) and miR-NC (sense,
5′-UCACAACCUCCUAGAUAGAGUAGA-3′ and anti-sense,
5′-UACUCUUUCUAGGAGGUAGUGAUU-3′) were purchased from Guangzhou
RiboBio Co., Ltd. EC cells at 60% confluence (1×105
cells/well) were used for transfection using
Lipofectamine® 3000 according to the manufacturer's
instructions. The concentrations of miR-30a mimics and miR-NC in
the final transfection system were 50 nM. The transfected cells
were incubated at 37°C (5% CO2), and collected 48 h
later for the following experiments.
Furthermore, 2000 ng pcDNA3.1-VOPP1 vector
(Guangzhou RiboBio Co., Ltd.) and the empty vector pcDNA3.1
(negative control) were prepared and then transfected into EC
cells, respectively, using Lipofectamine 3000 according to the
manufacturer's instructions. The transfected cells were incubated
with 5% CO2 at 37°C, and 48 h after transfection for the
further experimentation.
MTT assays
Transfected EC cells (5×103/well) in
96-well plates were cultured at 37°C for 48 h. The MTT assay was
conducted according to the instructions of the MTT Cell
Proliferation and Cytotoxicity Assay kit (cat. no. C0009S; Beyotime
Institute of Biotechnology). Specifically, MTT (0.5 mg/m) was added
to each well and incubated at 37°C for 4 h. Then, the formazan
crystals were dissolved in 150 µl DMSO in the dark. The wavelength
of 490 nm was used for measurement using a microplate reader
(Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from EC tissues and cultured cells was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. Specifically, 2 µg RNA was reverse transcribed using
random primers and M-MLV Reverse Transcriptase RNase H Minus (both
Promega Corporation), according to the manufacturer's protocols.
qPCR assays were conducted on Power SYBRs Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) to detect the
mRNA expression levels of OIP5-AS1 and VOPP1. miR-30a expression
was detected using the Taqman MicroRNA Reverse Transcription kit
and Taqman Universal Master Mix II kit (both Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. The procedure was carried out as follows: 95°C for 6
min, followed by 40 cycles at 95°C for 40 sec and 65°C for 30 sec,
and finally 75°C for 8 min. Electrophoresis in 1.5% (wt/v) agarose
gel was used to identify and confirm the PCR results. GAPDH and U6
were used as the endogenous reference genes for mRNA and miRNA,
respectively. All the primers for the sense and anti-sense chains
were obtained from Biomics. The primer sequences were as follows:
OIP5-AS1 forward, 5′-TGCGAAGATGGCGCAGTAAG-3′ and reverse,
5′-TAGTTCCTCTCGTCTGGCCG-3′; VOPP1 forward,
5′-GATGAACCCTGTCGGGAAT-3′ and reverse, 5′-GGCCTTCACTACCTGTTCGTA-3′;
GAPDH forward, 5′-AGGTGAAGGTCGGAGTCAACG-3′ and reverse:
5′-AGGGGTCATTGATGGCAACA-3′; miR-30a forward,
5′-ACACTCCAGCTGGGTGTAAACATCCTCGAC-3′ and reverse,
5′-CAGTGCGTGTCGTGGAGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The gene expression of miRNA
and mRNA was indicated as fold-changes using the 2−∆∆Cq
method (16).
Western blotting
The total proteins were extracted from tissues and
cultured cells in ice-old RIPA lysis buffer (Beyotime Institute of
Biotechnology), and the protein concentrations were determined
using a BCA protein assay kit (Beyotime Institute of
Biotechnology). Total proteins (30 µg/lane) of each experimental
group were subjected to 10% SDS-PAGE and then transferred onto
polyvinylidene fluoride membranes (EMD Millipore). After blocking
in TBS containing 5% skimmed milk for 1 h at room temperature, the
membranes were incubated at 4°C overnight with primary antibodies
against VOPP1 (1:1,000; Sigma-Aldrich; Merck KGaA; cat. no.
HPA038371) and GAPDH (1:1,000; Sigma-Aldrich; Merck KGaA; cat. no.
SAB1410512). Subsequently, the membranes were incubated with the
secondary antibody conjugated with peroxidase (1:2,000;
Sigma-Aldrich; Merck KGaA; cat. no. AP510) for 1 h at 37°C. Protein
bands were detected using the enhanced chemiluminescence detection
system (Bio-Rad Laboratories, Inc.) and Quantity One software
v4.6.2 (Bio-Rad Laboratories, Inc.).
Transwell migration and invasion
assays
EC cells were prepared for cell suspensions using
serum-free medium to a density of 3×104 cells/ml. The
cell suspensions were added into the upper chambers of Transwell
plates at room temperature, while RPMI-1640 medium supplemented
with 20% FBS was added to the lower chambers. For the invasion
assay, Transwell membranes were pre-coated with Matrigel (EMD
Millipore) overnight at room temperature. Following incubation for
24 h at 37°C, the migratory and invasive cells were collected,
washed and stained with 0.5% crystal violet (Sigma-Aldrich; Merck
KGaA) for 15 min at room temperature. Finally, the cells were
counted under an inverted light microscope (Olympus CK-40; Olympus
Corporation) at a magnification of ×100.
Dual-luciferase reporter assays
The online predicted system StarBase v2.0
(http://starbase.sysu.edu.cn) and
TargetScan7.2 (http://www.targetscan.org) were employed to seek the
targets of OIP5-AS1 and miR-30a, respectively. The recombinant
luciferase plasmids were constructed by cloning the sequences of
wild-type (WT) OIP5-AS1 and 3′-UTR of VOPP1, respectively, into the
pGL-3 luciferase basic vector (Promega Corporation). In addition,
their mutant-types (MUT) were also constructed as MUT-OIP5-AS1 and
MUT-VOPP1, respectively. Each constructed plasmid was transfected
into EC cells with miR-30a mimics or miR-NC using Lipofectamine
3000, as aforementioned. Following incubation for 48 h at 37°C,
firefly and Renilla luciferase activities were detected
using the Glomax 96 luminometer (Promega Corporation) according to
the manufacturer's instructions. Firefly luciferase activity was
normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP)
assays
RIP assays were conducted to further investigate the
direct interaction between OIP5-AS1 and miR-30a using the Magna RNA
immunoprecipitation kit (EMD Millipore), according to the
manufacturer's instructions. Specifically, EC cells
(2×107 cells) were harvested using trypsin and lysed in
RIP cell lysis buffer (MilliporeSigma). The obtained supernatant
was used for the RIP assays using the EZ-Magna RIPTM RNA-Binding
Protein Immunoprecipitation Kit (MilliporeSigma); 10% cell extract
was used as the input, and 100 µl cell extract was incubated at 4°C
with 50 µl protein A/G-conjugated magnetic beads with antibodies
against Argonaute2 (Ago2; Sigma-Aldrich; Merck KGaA; cat. no.
MABE56) and anti-IgG (Sigma-Aldrich; Merck KGaA; cat. no. I5131)
(the negative control) overnight. The magnetic beads were harvested
by centrifugation at 1,000 × g at room temperature for 2 min, and
then rinsed with 1 ml RIP buffer. After detachment with proteinase
K at 55°C for 30 min, the immunoprecipitated RNA was extracted and
analyzed by RT-qPCR, as aforementioned. Finally, the expression
levels of OIP5-AS1 and miR-30a in anti-IgG and anti-Ago2 groups
were compared.
Statistical analysis
All experiments were performed in triplicate and
data are presented as the mean ± standard deviation. SPSS 20.0
software (IBM Corp.) was used for statistical analysis. Pearson's
χ2 test or Fisher's exact test were used to analyze the
clinicopathological data of patients. Differences between EC and
normal tissues were analyzed by paired Student's t-test. One-way
ANOVA and Tukey's post-hoc test were used to compare differences
among multiple groups. An unpaired Student's t-test was used to
make statistical comparisons between two groups. Pearson's
correlation coefficient was used to calculate the correlation.
P<0.05 was considered to indicate a statistically significant
difference.
Results
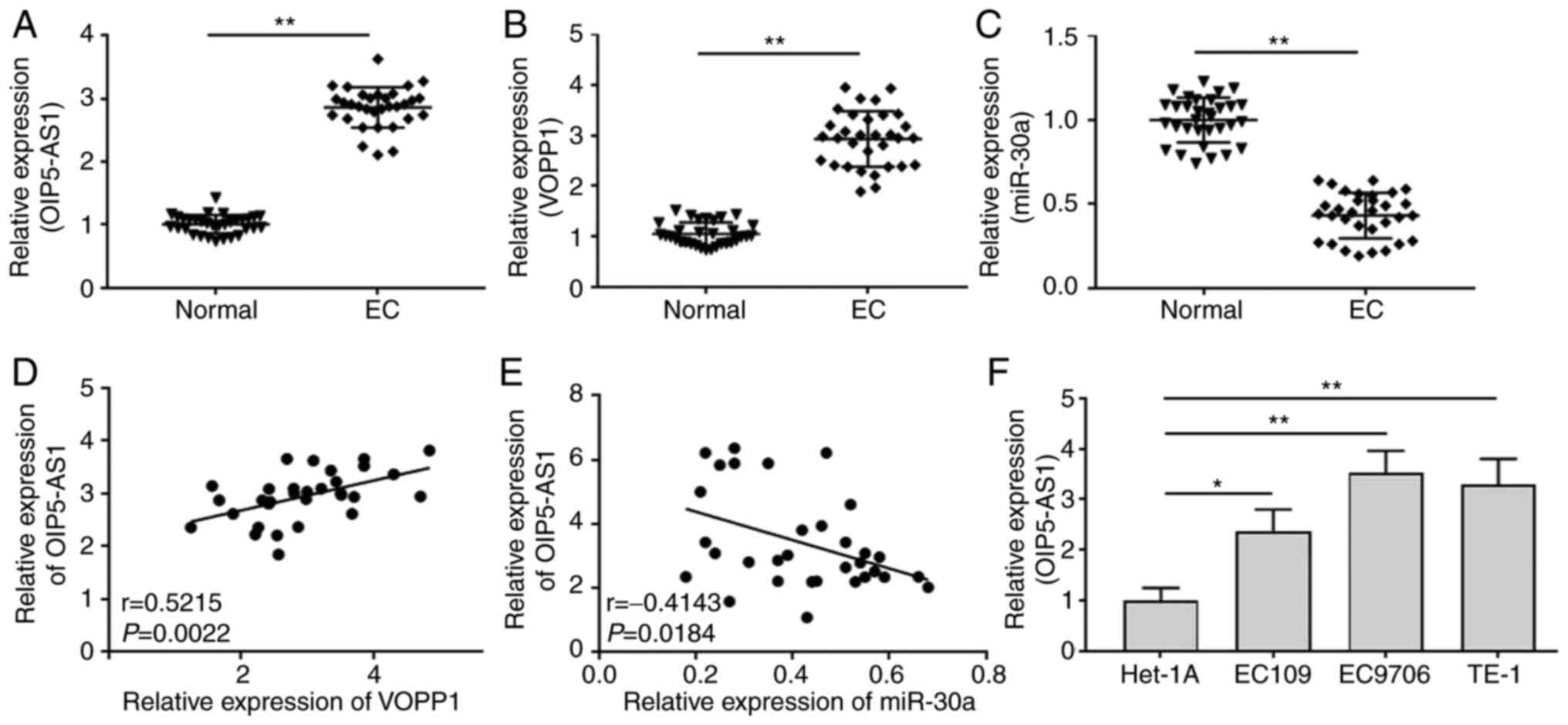
OIP5-AS1 expression is increased in EC
tissues and cultured cells
To investigate the roles of OIP5-AS1 in EC, the
expression levels of OIP5-AS1, VOPP1 and miR-30a in EC tissues and
cultured cells were detected by RT-qPCR. The clinicopathological
data of patients with EC showed that OIP5-AS1 expression was
significantly associated with differentiation, tumor invasion depth
and histological grade (Table SI).
In addition, OIP5-AS1 (Fig. 1A) and
VOPP1 (Fig. 1B) expression was
significantly increased, while miR-30a (Fig. 1C) expression was significantly
decreased in EC tissues compared with in adjacent normal tissues.
Pearson's correlation analysis indicated that there was a positive
correlation between OIP5-AS1 and VOPP1 expression (Fig. 1D) and a negative correlation between
OIP5-AS1 and miR-30a expression (Fig.
1E) in EC tissues (n=32). In the three cultured EC cells
(EC109, EC9706 and TE-1) and normal Het-1A cell, OIP5-AS1
expression was also significantly upregulated in EC cultured cells
than that in Het-1A cells (Fig.
1F).
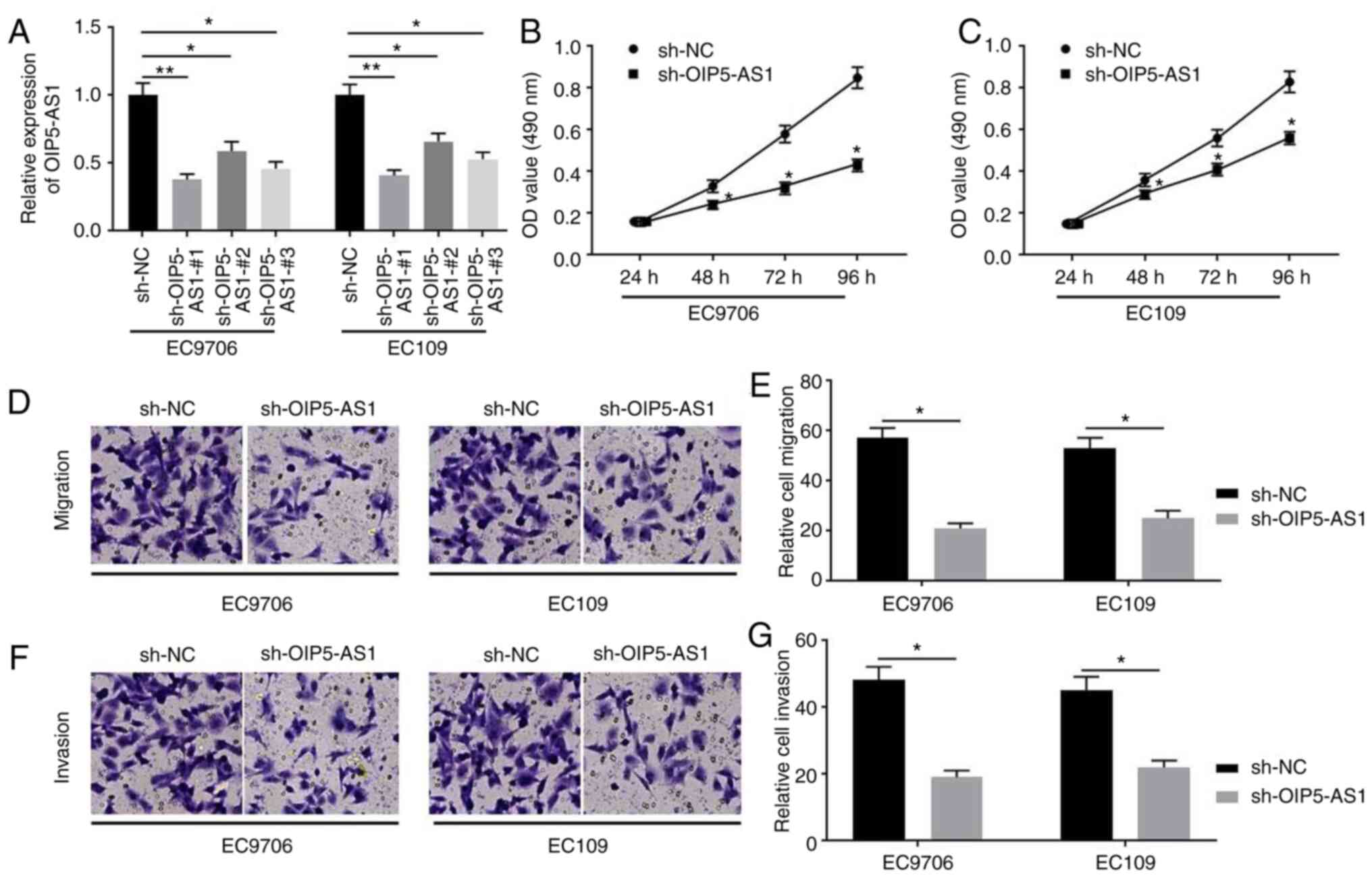
Knockdown of OIP5-AS1 suppresses cell
proliferation, migration and invasion in EC cells
To investigate the biological functions of OIP5-AS1
in EC cells, three shRNAs (sh-OIP5-AS1-#1, sh-OIP5-AS1-#2 and
sh-OIP5-AS1-#3) against OIP5-AS1 and sh-NC were constructed and
transfected into EC cells. RT-qPCR analysis was performed to
confirm the transfection efficiencies (Fig. 2A). All shRNAs exhibited inhibitory
effects on OIP5-AS1 expression in EC9706 and EC109 cells, and since
sh-OIP5-AS1-#1 exhibited the most effective activity, it was
selected for subsequent experiments. MTT assays indicated that
OIP5-AS1-knockdown by sh-OIP5-AS1 transfection resulted in
suppression of EC9706 (Fig. 2B) and
EC109 (Fig. 2C) cell proliferation
at 48–96 h. The effects of OIP5-AS1 on the migration and invasion
of EC9706 and EC109 cells were investigated. The results indicated
that knockdown of OIP5-AS1 expression significantly decreased the
capacities of migration (Fig. 2D and
E) and invasion (Fig. 2F and G)
in EC9706 and EC109 cells.
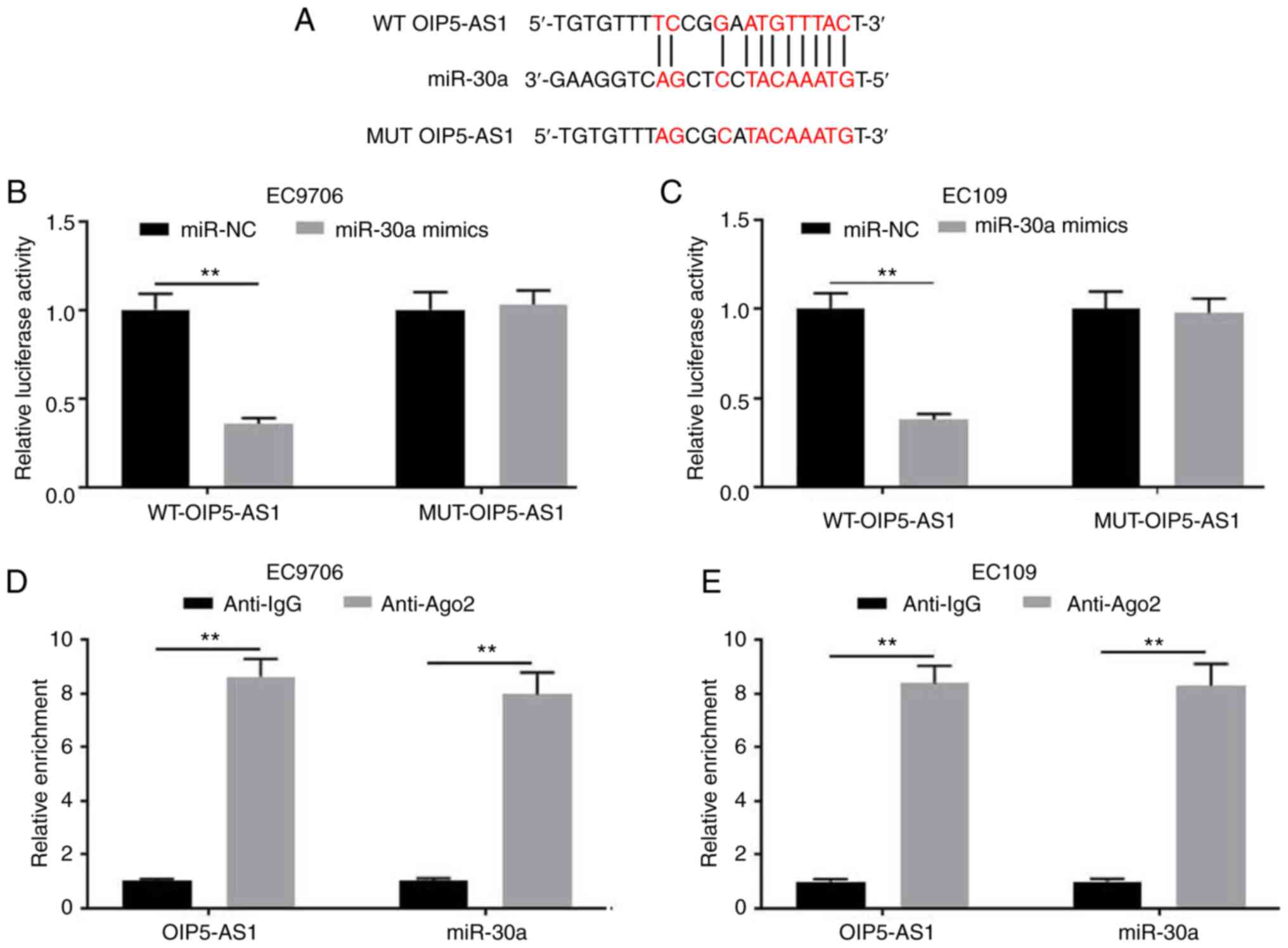
OIP5-AS1 interacts with miR-30a
To further investigate the possible underlying
mechanism of OIP5-AS1 in EC cells, the potential miRNAs that bind
to OIP5-AS1 were investigated, since lncRNAs exhibit regulatory
activity by sponging miRNAs (17).
Starbase2.0 software was employed to predict the target miRNAs that
interacted with OIP5-AS1. As a result, miR-30a was identified as a
possible target of OIP5-AS1 (Fig.
3A). The dual-luciferase reporter assays revealed that the
luciferase activity in the reporter containing the WT-OIP5-AS1 was
significantly using miR-30a mimics; by contrast, no significant
differences were observed in the relative luciferase activities
with the reporter containing the MUT-OIP5-AS1 (Fig. 3B and C). In addition, RIP assays
indicated that OIP5-AS1 could interact with miR-30a (Fig. 3D and E). Overall, miR-30a may be a
potential target of OIP5-AS1.
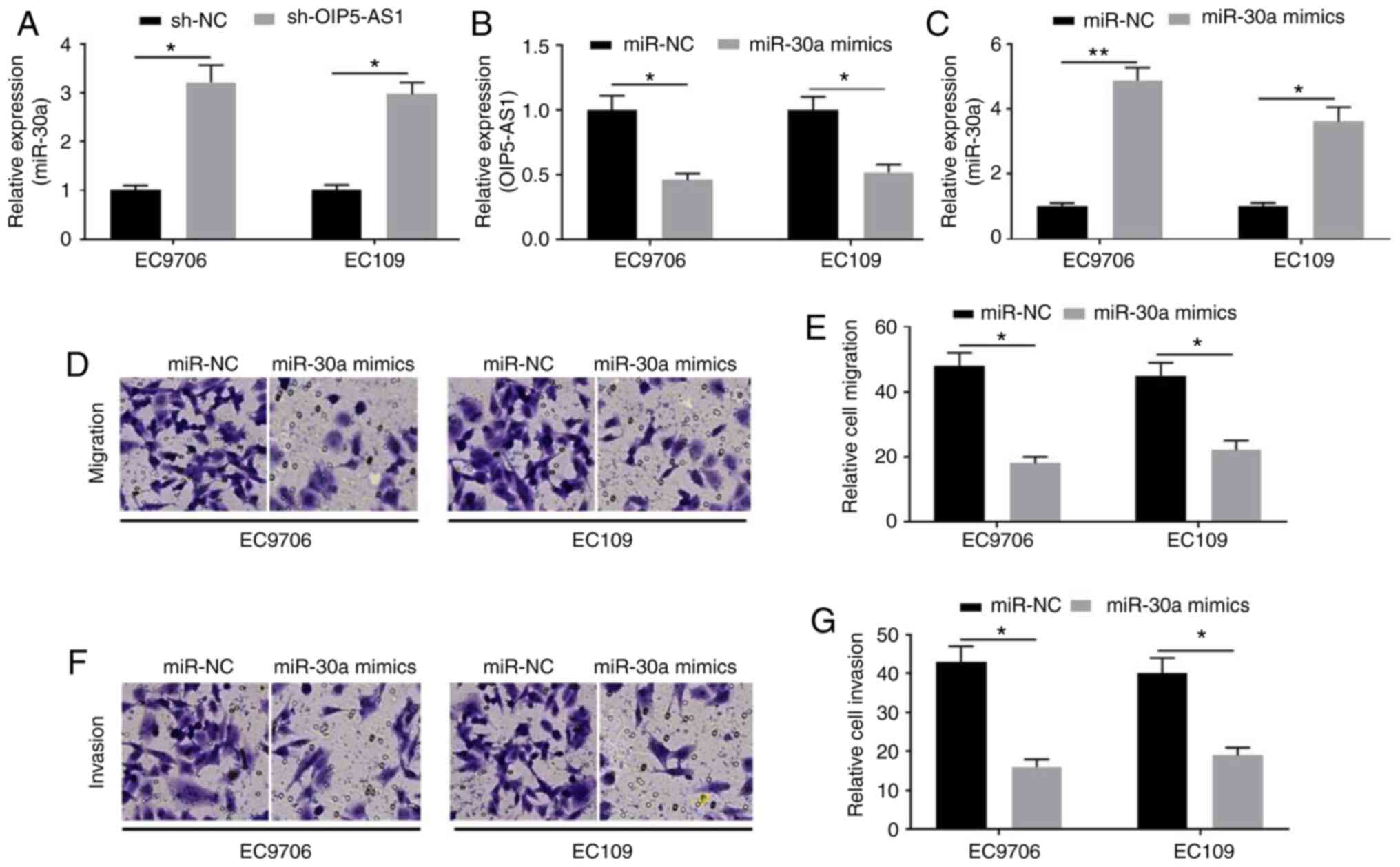
Overexpression of miR-30a ameliorated
the migration and invasion of EC cells
To investigate the roles of miR-30a in EC cells,
RT-qPCR assays were used to detect miR-30a expression. Suppression
of OIP5-AS1 significantly increased miR-30a expression (Fig. 4A). In turn, miR-30a mimics
significantly decreased OIP5-AS1 expression (Fig. 4B). miR-30a expression detected by
RT-qPCR suggested successful transfection (Fig. 4C). Furthermore, miR-30a
overexpression suppressed the migration (Fig. 4D and E) and invasion (Fig. 4F and G) of EC9706 and EC109
cells.
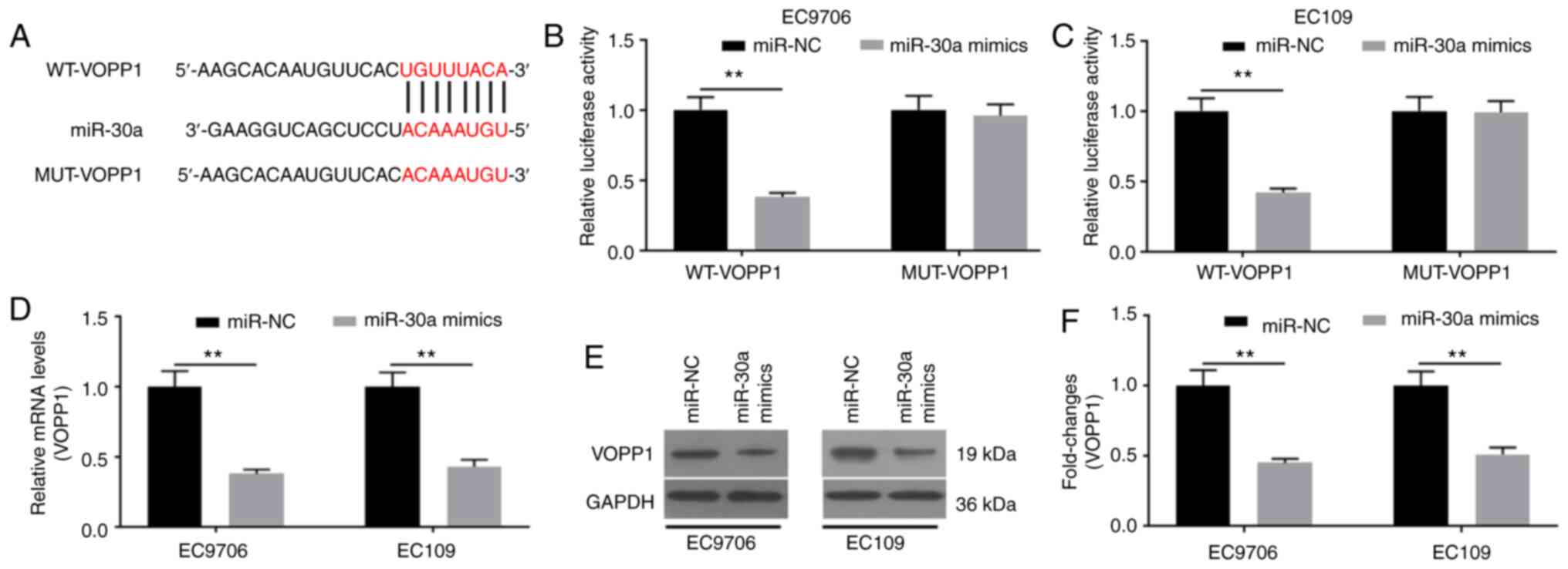
VOPP1 is a target of miR-30a
To further explore how miR-30a affected the
biological functions in EC cells, the target genes of miR-30a were
predicted by TargetScan7.2. As a result, VOPP1 was identified as a
potential target of miR-30a (Fig.
5A), which was verified using the dual-luciferase reporter
assay (Fig. 5B and C). The relative
luciferase activities did not show statistical difference with the
reporter containing the mutant site of VOPP1; by contrast, the
relative luciferase activities in the reporter containing the WT
binding site of VOPP1 were significantly decreased (Fig. 5B and C). The mRNA and protein
expression levels of VOPP1 were determined. It was revealed that
miR-30a mimics significantly downregulated VOPP1 mRNA (Fig. 5D) and protein (Fig. 5E and F) expression. Collectively,
miR-30a may specifically target VOPP1 to degrade it by binding to
its 3′-UTR.
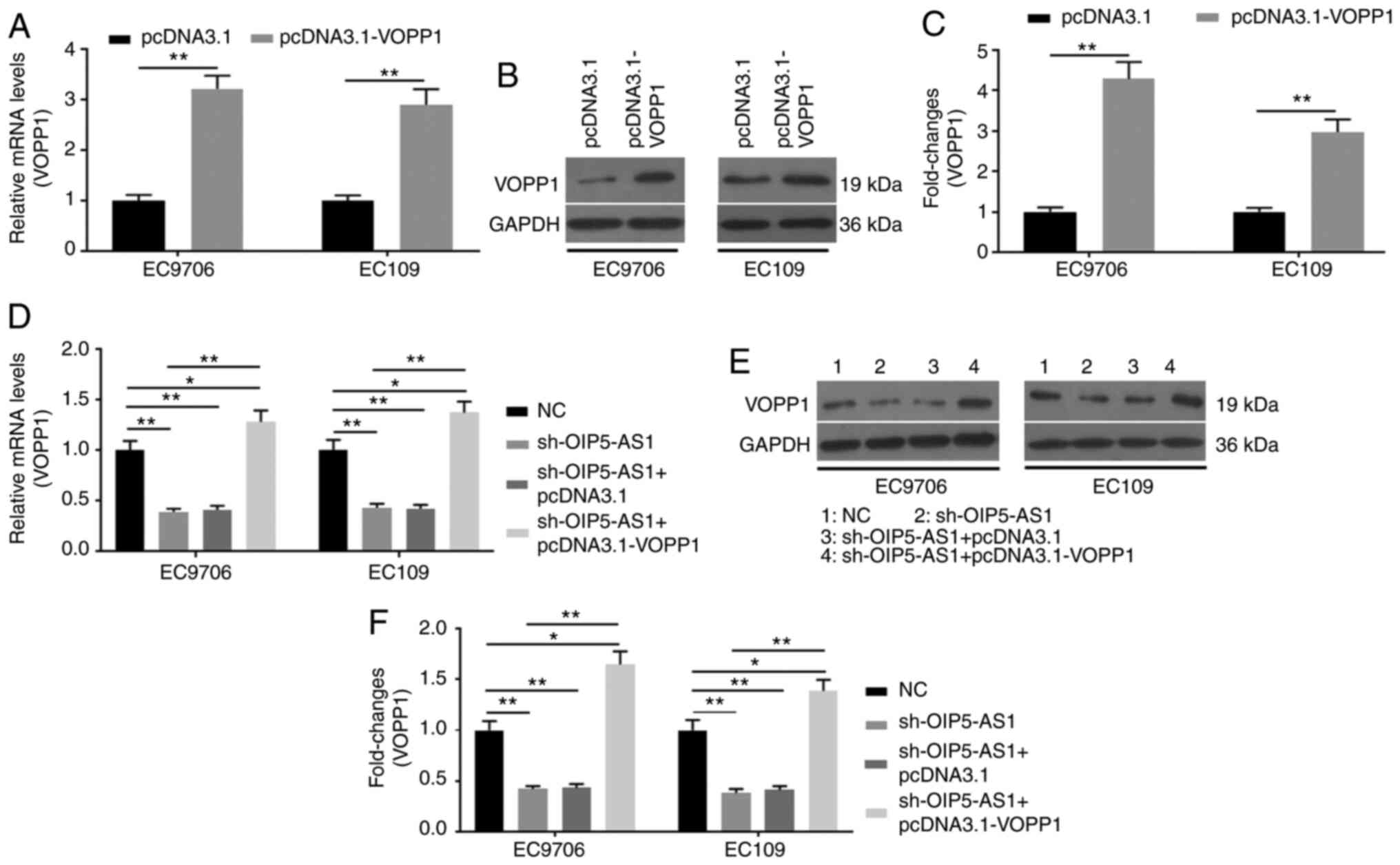
Overexpression of VOPP1 rescues the
biological functions induced by OIP5-AS1-knockdown in EC cells
Transfection of pcDNA3.1 and pcDNA3.1-VOPP1 into EC
cells was conducted. RT-qPCR (Fig.
6A) and western blot (Fig. 6B and
C) assays confirmed the successful transfection. To further
investigate the roles of the downstream factor VOPP1 in
sh-OIP5-AS1-transfected EC cells, pcDNA3.1-VOPP1 was prepared for
co-transfection. RT-qPCR (Fig. 6D)
and western blot (Fig. 6E and F)
assays were used to verify the successful co-transfection of
sh-OIP5-AS1 and pcDNA3.1-VOPP1, as showed by up regulation of VOPP1
expression at the mRNA and protein levels. VOPP1 overexpression
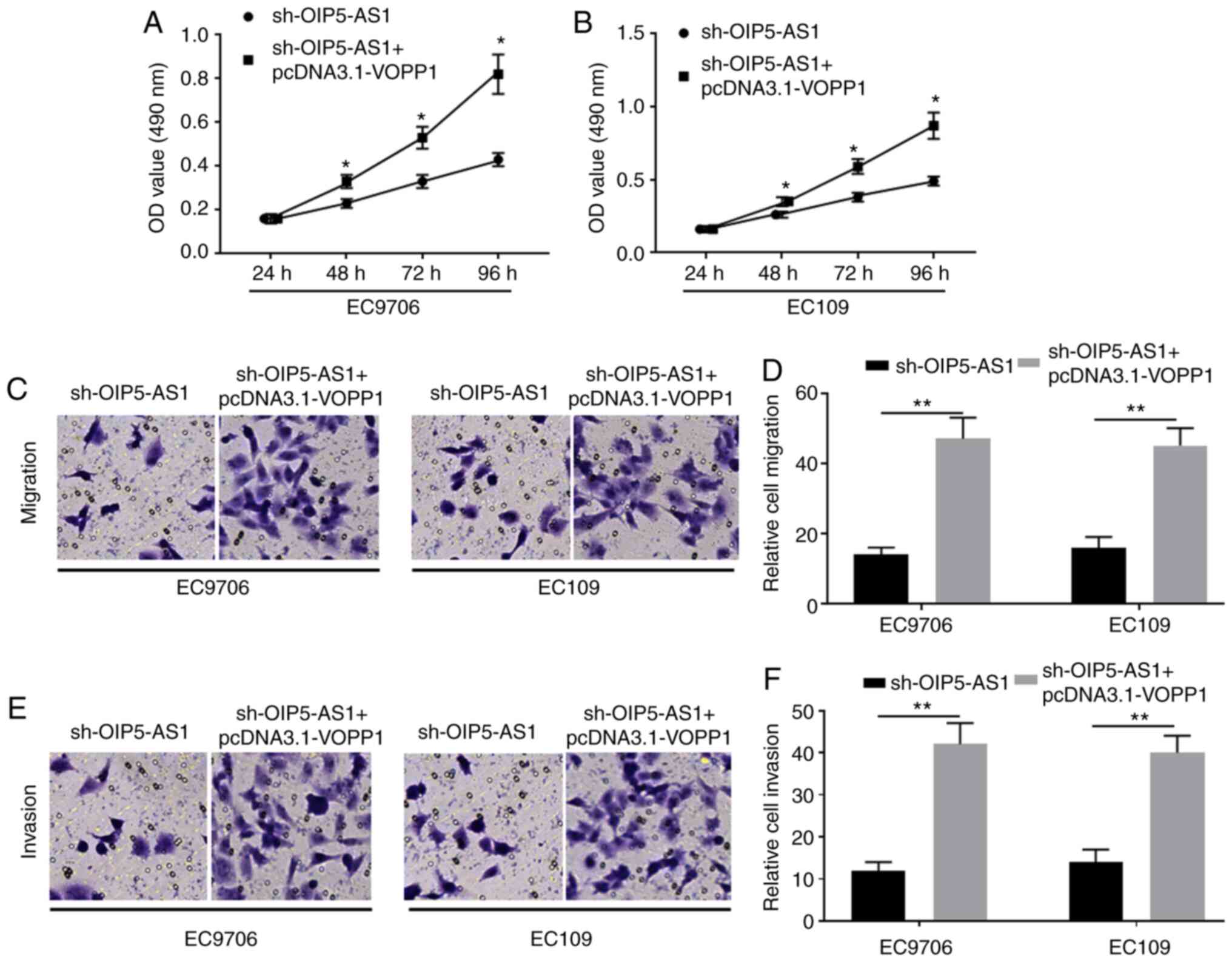
showed similar effects as OIP5-AS1 in EC9706 and EC109 cells.
Specifically, overexpression of VOPP1 ameliorated the decreased
proliferative activity of EC9706 (Fig.
7A) and EC109 (Fig. 7B) cells
induced by OIP5-AS1-knockdown. Similarly, VOPP1 overexpression also
improved cell migration (Fig. 7C and
D) and invasion (Fig. 7E and F),
which were attenuated by sh-OIP5-AS1 transfection in EC9706 and
EC109 cells. Collectively, overexpression of VOPP1 improved the
negative effects of OIP5-AS1-knockdown in EC cells.
Discussion
EC is the 8th most common type of cancer (18) and has complex pathological
mechanisms, which remain incompletely understood. The potential
post-transcriptional interactions between lncRNAs and mRNAs
generate an integrated lncRNA-mRNA network, orchestrating the
oncogenic/oncosuppressive development. Numerous advanced
technologies, such high-throughput sequencing and proteomics, have
been employed to explore the critical factors associated with the
pathological development of EC (19). In the present study, it was revealed
that OIP5-AS1 expression was upregulated in EC tissues and cultured
EC cells. Knockdown of OIP5-AS1 attenuated the proliferation,
migration and invasion of EC cells. miR-30a was identified to be a
target of OIP5-AS1, and miR-30a-mimics transfection produced
similar effects as those of OIP5-AS1-knockdown. VOPP1 was a direct
target of miR-30a, and overexpression of VOPP1 rescued the negative
effects of OIP5-AS1-knockdown in EC cells. Collectively, OIP5-AS1
may promote EC progression via mediating the miR-30a/VOPP1
axis.
Transcriptome sequencing has identified >58,000
lncRNAs in human cells (20). An
integrative bioinformatics method has been used to identify the key
functional lncRNAs involved in EC development (21). lncRNA625 has been found to promote
cell proliferation, migration and invasion in EC cells, and it
exhibits a specific prognostic value for clinical diagnosis
(21). lncRNA CCAT1 expression has
been reported to be upregulated in ESCC tissues, to promote cell
proliferation and migration by mediating the SPRY4/HOXB13 axis via
sponging miR-7, and to be associated with a poor survival in
patients with ESCC (22). OIP5-AS1
has been reported to regulate carcinogenesis in various types of
cancer (23). In cervical cancer,
silencing of OIP5-AS1 markedly decreases cell proliferation, colony
formation and invasion by increasing the expression of integrin
subunit a6 through sponging miR-143-3 (24). Additionally, OIP5-AS1 expression is
upregulated in trastuzumab-resistant breast cancer cells by
mediating the miR381-3p/HMGB3 axis, and knockdown of OIP5-AS1 may
rescue the sensitivity of cells to trastuzumab (25). The present study revealed that
OIP5-AS1 expression was upregulated in EC tissues and cultured
cells, and knockdown of OIP-AS1 expression decreased the
proliferation, migration and invasion of EC9706 and EC109
cells.
Generally, lncRNAs show biological functions by
sponging miRNAs, which normally degrade their target genes by
binding to their 3′-UTR (26).
miR-30a-5p expression is downregulated in renal cancer, and its
overexpression ameliorates the malignant phenotypes of renal
carcinoma cells by degrading SOX4 (27). miR-30a has been considered as a
potential prognostic biomarker for clear cell renal cell carcinoma,
and it inhibits cell migration and invasion of HCT116 cells
(28). In the present study, it was
revealed that miR-30a was downregulated in EC tissues and cultured
cells, and miR-30a-mimics transfection significantly ameliorated
cell migration and invasion. For further investigation of OIP5-AS1
biological functions in EC, the prediction by StarBase2.0 software
was conducted, revealing that OIP5-AS1 interacted with miR-30a and
inhibited its activity. Thus, it was suggested that OIP5-AS1
promoted the progression of EC by sponging miR-30a in EC9706 and
EC109 cells.
To further investigate the molecular mechanism of
OIP5-AS1/miR-30a in mediating EC progression, the target of miR-30a
was predicted by TargetScan7.2. VOPP1 was verified as the direct
target of miR-30a. VOPP1 expression has been reported to be
upregulated in various types of cancer, including colorectal
cancer, squamous cell carcinoma, gastric cancer and glioblastoma
(29). VOPP1 has been demonstrated
to promote cell proliferation and migration, and inhibit apoptosis
in SMMC-7721 and BEL-7404 cells (30) and human squamous cell carcinoma cell
lines (31), and suppression of
VOPP1 expression attenuates EMT and the subsequent intrusion and
metastasis of human lung adenocarcinoma cells (13). VOPP1 acts as an essential gene for
RPB3 for hepatocellular carcinoma proliferation (30). However, in breast cancer cells, VOPP1
may promote cellular transformation and enhance the growth of
transplanted tumors by inhibiting the antitumor effect of WW
domain-containing oxidoreductase (32). In the present study, VOPP1 expression
was upregulated in EC9706 and EC109 cells. VOPP1 overexpression
effectively rescued the effects of sh-OIP5-AS1 on EC cells. Thus,
VOPP1 acted as the downstream factor and the effector of OIP5-AS1.
However, there are limitations in the present study that should be
addressed in future studies. To further investigate the roles of
OIP5-AS1/miR-30a/VOPP1 in EC cells, OIP5-AS1- and/or VOPP1-knockout
animals or miR-30a transgenic animals should be used.
In conclusion, OIP5-AS1 promoted the proliferation,
migration and invasion of EC9706 and EC109 cells by enhancing VOPP1
expression through sponging miR-30a.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
National Science Foundation of China (grant no. 81960883) and the
Project of Jiangxi Provincial Department of Health (grant no.
20197133).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SJC designed the study and wrote the manuscript. JX,
ZC, ZF, SXC, YG, XL and KC performed the experiments, analyzed the
data, revised and finalized the manuscript. SJC and JX confirmed
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All patients signed the consent forms prior to the
use of their tissues in the present study. The experimental
protocols have been approved by the Ethics Committee of The First
Affiliated Jiujiang Hospital of Nanchang University (Jiujiang,
China) according to the Declaration of Helsinki Principles.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no completing
interests.
References
|
1
|
Chen N, Zhang G, Fu J and Wu Q:
Identification of key modules and hub genes involved in esophageal
squamous cell carcinoma tumorigenesis using WCGNA. Cancer Control.
27:10732748209788172020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang H, Si J, Yue J and Ma S: The
mechanisms and reversal strategies of tumor radioresistance in
esophageal squamous cell carcinoma. J Cancer Res Clin Oncol.
147:1275–1286. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ye H, Mulmi Shrestha S, Zhu J, Ding Y and
Shi R: Long non-coding RNA LINC00491 promotes proliferation and
inhibits apoptosis in esophageal squamous cell carcinoma. Int J Mol
Med. 47:472021. View Article : Google Scholar
|
|
5
|
Andrey G and Duboule D: SnapShot: Hox gene
regulation. Cell. 156:856–856.e1. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xue W, Zheng Y, Shen Z, Li L, Fan Z, Wang
W, Zhu Z, Zhai Y, Zhao J and Kan Q: Involvement of long non-coding
RNAs in the progression of esophageal cancer. Cancer Commun (Lond).
41:371–388. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu Y, Li Y, Jin J, Han G, Sun C, Pizzi MP,
Huo L, Scott A, Wang Y, Ma L, et al: LncRNA PVT1 up-regulation is a
poor prognosticator and serves as a therapeutic target in
esophageal adenocarcinoma. Mol Cancer. 18:1412019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wei WT, Wang L, Liang JX, Wang JF, Li Q
and Zeng J: LncRNA EIF3J-AS1 enhanced esophageal cancer invasion
via regulating AKT1 expression through sponging miR-373-3p. Sci
Rep. 10:139692020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao Z, Li J, Jin Q and Liu D: Long
non-coding RNA OIP5-AS1 contributes to cisplatin resistance of oral
squamous cell carcinoma through the miR-27b-3p/TRIM14 axis. Exp
Ther Med. 21:4082021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Fu X, Wang X, Liu Y and Song X:
Long non-coding RNA OIP5-AS1 facilitates the progression of ovarian
cancer via the miR-128-3p/CCNG1 axis. Mol Med Rep. 23:3882021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang L, Ren X, Ma X, Yin L, Niu X and Xing
S: LncRNA OIP5-AS1 promotes the development of esophageal squamous
cell carcinoma by binding to miR-1297. Panminerva Med. Jan
24–2020.(Epub ahead of print).
|
|
12
|
Liu F and Wen C: LINC01410 knockdown
suppresses cervical cancer growth and invasion via targeting
miR-2467-3p/VOPP1 Axis. Cancer Manag Res. 12:855–861. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li YJ, Zhang W, Xia H, Zhang BS, Chen P,
Zhao YL and Li J: miR-218 suppresses epithelial-to-mesenchymal
transition by targeting Robo1 and Ecop in lung adenocarcinoma
cells. Future Oncol. 13:2571–2582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao C, Pang M, Zhou Z, Long S, Dong D,
Yang J, Cao M, Zhang C, Han S and Li L: Epidermal growth factor
receptor-coamplified and overexpressed protein (VOPP1) is a
putative oncogene in gastric cancer. Clin Exp Med. 15:469–475.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baras A and Moskaluk CA: Intracellular
localization of GASP/ECOP/VOPP1. J Mol Histol. 41:153–164. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang
X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW and Liu XS:
Integrative analyses reveal a long noncoding RNA-mediated sponge
regulatory network in prostate cancer. Nat Commun. 7:109822016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng BZ, Liu TD, Chen G, Zhang JX and
Kang X: The effect of curcumin on cell adhesion of human esophageal
cancer cell. Eur Rev Med Pharmacol Sci. 22:551–560. 2018.PubMed/NCBI
|
|
19
|
Weng L, Shen S, Wu S, Yin X, Liu B, Shang
M, Zou X and Mao A: Identification of critical genes and proteins
for stent restenosis induced by esophageal benign hyperplasia in
esophageal cancer. Front Genet. 11:5639542020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li CQ, Huang GW, Wu ZY, Xu YJ, Li XC, Xue
YJ, Zhu Y, Zhao JM, Li M, Zhang J, et al: Integrative analyses of
transcriptome sequencing identify novel functional lncRNAs in
esophageal squamous cell carcinoma. Oncogenesis. 6:e2972017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang E, Han L, Yin D, He X, Hong L, Si X,
Qiu M, Xu T, De W, Xu L, et al: H3K27 acetylation activated-long
non-coding RNA CCAT1 affects cell proliferation and migration by
regulating SPRY4 and HOXB13 expression in esophageal squamous cell
carcinoma. Nucleic Acids Res. 45:3086–3101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ghafouri-Fard S, Dashti S, Farsi M, Hussen
BM and Taheri M: A review on the role of oncogenic lncRNA OIP5-AS1
in human malignancies. Biomed Pharmacother. 137:1113662021.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang J, Jiang B, Hai J, Duan S, Dong X and
Chen C: Long noncoding RNA opa-interacting protein 5 antisense
transcript 1 promotes proliferation and invasion through elevating
integrin α6 expression by sponging miR-143-3p in cervical cancer. J
Cell Biochem. 120:907–916. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu Q, Li Y, Peng S, Li J and Qin X:
Exosomal-mediated transfer of OIP5-AS1 enhanced cell
chemoresistance to trastuzumab in breast cancer via up-regulating
HMGB3 by sponging miR-381-3p. Open Med (Wars). 16:512–525. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eulalio A, Huntzinger E and Izaurralde E:
Getting to the root of miRNA-mediated gene silencing. Cell.
132:9–14. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen M, Wei X, Shi X, Lu L, Zhang G, Huang
Y and Hou J: LncRNA HIF1A-AS2 accelerates malignant phenotypes of
renal carcinoma by modulating miR-30a-5p/SOX4 axis as a ceRNA.
Cancer Biol Med. Mar 12–2021.(Epub ahead of print).
|
|
28
|
Yu D, Liu H, Qin J, Huangfu M, Guan X, Li
X, Zhou L, Dou T, Liu Y, Wang L, et al: Curcumol inhibits the
viability and invasion of colorectal cancer cells via miR-30a-5p
and Hippo signaling pathway. Oncol Lett. 21:2992021. View Article : Google Scholar
|
|
29
|
Hu P, Wang B, Chen T, Xu Y, Zheng G, Zhu Y
and Du X: RNA polymerase II subunit 3 regulates vesicular,
overexpressed in cancer, prosurvival protein 1 expression to
promote hepatocellular carcinoma. J Int Med Res.
49:3000605219905122021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fang Z, Wu L, Dai H, Hu P, Wang B, Han Q,
Xu Y, Lv S, Zhu Y, Gan M, et al: The role of vesicular
overexpressed in cancer pro-survival protein 1 in hepatocellular
carcinoma proliferation. Cancer Biomark. 28:9–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baras AS, Solomon A, Davidson R and
Moskaluk CA: Loss of VOPP1 overexpression in squamous carcinoma
cells induces apoptosis through oxidative cellular injury. Lab
Invest. 91:1170–1180. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bonin F, Taouis K, Azorin P, Petitalot A,
Tariq Z, Nola S, Bouteille N, Tury S, Vacher S, Bièche I, et al:
VOPP1 promotes breast tumorigenesis by interacting with the tumor
suppressor WWOX. BMC Biol. 16:1092018. View Article : Google Scholar : PubMed/NCBI
|