Introduction
According to the International Agency for Research
on Cancer, lung cancer was considered as the most common type of
cancer and the leading cause of cancer-associated mortality in
2018, accounting for 18.4% of cancer-related deaths worldwide
(1). Based on the histological
characteristics of tumor cells, lung cancer is mainly divided into
two categories, small cell lung cancer (SCLC) and non-small cell
lung cancer (NSCLC), and >80% of lung cancers belong to the
NSCLC category (2). Most cancer
patients are diagnosed with advanced cancer at the time of medical
treatment, and the five-year survival rate is <18% (3). Studying the pathogenesis of NSCLC and
exploring novel therapeutic targets that could be effective for
NSCLC treatment is therefore crucial.
Long non-coding RNAs (lncRNAs) are defined as
transcripts of >200 nucleotides in length (4). LncRNAs have attracted increasing
attention in the recent years due to their important roles in the
pathological mechanisms of malignant tumors (5). These functional lncRNAs have clinical
significance and biological functions in various types of cancer,
such as the role of MVIH in pancreatic ductal adenocarcinomas
(6), PCTST in pancreatic cancer
(7) and HOTAIR in gastric cancer
(8). The expression of LINC01272 has
been found to be abnormal in numerous diseases, such as gastric
cancer (9), inflammatory bowel
diseases (10) and unstable
atherosclerotic plaque (11). In
addition, LINC01272 was reported to have an important role in
promoting gastric cancer progression (9). However, the role of LINC01272 remains
unclear in NSCLC. The results from our preliminary bioinformatics
analysis using data from The Cancer Genome Atlas (TCGA) database
demonstrated that LINC01272 expression is significantly
downregulated in NSCLC tumor, and that patients with lung cancer
and with low LINC01272 expression have a significantly worse
overall survival than patients with high LINC01272 expression.
Exploring the clinical significance and function of LINC01272
expression in patients with NSCLC is therefore of great importance
to further understand the role of LINC01272 and improve the
treatment of NSCLC.
MicroRNAs (miRNAs) are a class of non-coding RNA
molecules of ~22 nucleotide in length that promote the degradation
of target mRNAs or inhibit mRNA translation by recognizing and
binding to the 3′-untranslated regions of target mRNAs (12). Previous studies have demonstrated
that microRNAs (miRNAs) play important roles in cancer progression
by promoting or inhibiting tumorigenesis (13,14). It
has been reported that miR-1303 plays a crucial role in various
types of cancer (15,16). For example, it was shown that
increased miR-1303 expression in prostate cancer tissues and cell
lines could promote the proliferation, migration and invasion of
prostate cancer cells (15). Zhang
et al (16) reported that
miR-1303 expression is decreased in gastric cancer tissues and cell
lines, promoting gastric cancer cell proliferation, migration and
invasion. Furthermore, a previous study demonstrated that increased
expression of miR-1303 in NSCLC tissues and cells could promote
NSCLC cell proliferation, migration and invasion and might
therefore serve as a potential prognostic biomarker for NSCLC
(17). In addition, our preliminary
bioinformatics analysis predicted the binding site of LINC01272 to
miR-1303. We therefore hypothesized that LINC01272 may have a role
in NSCLC by targeting miR-1303.
The present study aimed to analyze the expression
level of LINC01272 in patients with NSCLC, evaluate the prognostic
value of LINC01272 and analyze the role of LINC01272/miR-1303 axis
in NSCLC.
Materials and methods
Patients and sample collection
The experimental procedures were approved by the
Ethics Committee of Weifang People's Hospital (approval no. 011827)
and patients provided signed informed consents prior to sampling.
Tumor tissue samples and adjacent non-tumor tissue samples (>5
cm from the tumor margin) were collected from 108 patients with
NSCLC who underwent curative resection between July 2012 and June
2015 at Weifang People's Hospital. All tissue samples were
histopathologically verified and stored in liquid nitrogen.
Patients who had received any anti-tumor treatment prior to the
surgery were excluded from the study. All patients received a
5-year follow-up with monthly telephone or face-to-face appointment
for patient survival. The clinicopathological characteristics of
patients are presented in Table
I.
 | Table I.Association between LINC01272
expression and the clinicopathological characteristics of patients
with non-small cell lung cancer. |
Table I.
Association between LINC01272
expression and the clinicopathological characteristics of patients
with non-small cell lung cancer.
|
|
| LINC01272
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | Number, n=108 | Low (n=58) | High (n=50) | P-values |
|---|
| Age, years |
|
|
| 0.704 |
|
≤60 | 39 | 20 | 19 |
|
|
>60 | 69 | 38 | 31 |
|
| Sex |
|
|
| 0.971 |
|
Female | 43 | 23 | 20 |
|
|
Male | 65 | 35 | 30 |
|
| Smoking |
|
|
| 0.982 |
| No | 39 | 21 | 18 |
|
|
Yes | 69 | 37 | 32 |
|
| Tumor size, cm |
|
|
| 0.021 |
| ≤3 | 54 | 23 | 31 |
|
|
>3 | 54 | 35 | 19 |
|
| Lymph node
metastasis |
|
|
| 0.037 |
| No | 51 | 22 | 29 |
|
|
Yes | 57 | 36 | 21 |
|
|
Differentiation |
|
|
| 0.101 |
| Well
and moderate | 60 | 28 | 32 |
|
|
Poor | 48 | 30 | 18 |
|
| TNM stage |
|
|
| 0.009 |
|
I–II | 46 | 18 | 28 |
|
|
III | 62 | 40 | 22 |
|
Cell culture
The four NSCLC cell lines SK-MES-1 (cat. no.
TCHu110), A549 (cat. no. TCHu150), NCI-H460 (cat. no. TCHu205) and
NCI-H522 (cat. no. MZ-1314) and the normal lung cell line NHBE-T
(cat. no. MZ-2677) were purchased from The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences. All cells
were cultured in Dulbecco's modified Eagle medium (DMEM;
Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and placed at 37°C
in a humidified incubator containing 5% CO2.
Cell transfection
The pcDNA3.1, pcDNA3.1-LINC01272 expression vector,
miR-1303 mimic and mimic negative control (mimic NC) were
synthesized from Shanghai GenePharma Co., Ltd.. Briefly, for the
synthesis of pcDNA3.1-LINC01272, the PCR products of LINC01272 were
firstly cloned into pUM-T vector and transformed into Escherichia
coli DH5α to obtain LINC01272 clones. The recombinant expression
vector pcDNA3.1-LINC01272 was generated by subcloning the obtained
LINC01272 clones directly into pcDNA3.1 plasmid through digestion
and ligation. Then, 30 nM pcDNA3.1 and 30 nM pcDNA3.1-LINC01272
were transfected into A549 and H460 cells, and 50 nM miR-1303 mimic
(5′-UUUAGAGACGGGGUCUUGCUCU-3′) and 50 nM mimic NC
(5′-UUCUCCGAACGUGUCACGU-3′) were transfected into A549 cells using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturers' protocol. Cells were used for
subsequent experiments following 48 h transfection at 37°C.
Bioinformatics analysis
GEPIA 2.0 (http://gepia2.cancer-pku.cn/#index) (18) was used to analyze data from TCGA
database (https://cancergenome.nih.gov/) and compare the
expression levels of LINC01272 in lung adenocarcinoma (LUAD) and
lung squamous cell carcinoma (LUSC) with that of healthy controls.
Kaplan-Meier plotter (http://www.kmplot.com/analysis/) (19) was used to analyze the data of the
Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), the European
Genome-Phenome Archive (EGA; http://ega-archive.org/datasets) and TCGA database and
evaluate the association between LINC01272 expression and the
survival prognosis in patients with lung cancer. The lncBase v.2 of
DIANA (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index)
was used to predict the binding sites of LINC01272 to potential
target miRNAs.
RNA extraction and reverse
transcription quantitative (RT-q)PCR
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from tissue samples
and NSCLC cells. The purity and concentration of the extracted RNA
were evaluated by NanoDrop 2000 (Thermo Fisher Scientific, Inc.).
Then, cDNA was synthesized using the Revert Aid First Strand cDNA
Synthesis Kit (Thermo Fisher Scientific) according to the
manufacturers' instructions.
The levels of LINC01272 and miR-1303 were measured
using RT-qPCR, which was carried out using a SYBR Green PCR kit
(Bio-Rad Laboratories, Inc.) and the Applied Biosystems 7900
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). LINC01272 and miR-1303 expression were normalized to GAPDH
and U6, respectively. The PCR reaction conditions were as follows:
95°C for 10 min, followed by 39 cycles at 95°C for 10 sec and 60°C
for 30 sec. The sequences of the primers were as follows:
LINC01272, forward 5′-TGTTCACTGCTGTACACCCA-3′, reverse
5′-TGTGGAGAGGGGATTTCTGG-3′; GAPDH, forward
5′-TGTTCGTCATGGGTGTGAAC-3′, reverse 5′-ATGGCATGGACTGTGGTCAT-3′;
miR-1303, forward 5′-GCCGAGTTTAGAGACGGGGT-3′, reverse
5′-CTCAACTGGTGTCGTGGA-3′; and U6, forward 5′-CTCGCTTCGGCAGCACA-3′
and reverse 5′-AACGCTTCACGAATTTGCGT-3′. The relative expression
levels were calculated using the 2−ΔΔCq method (20).
Cell proliferation assay
The proliferation of A549 and H460 cells was
evaluated using a Cell Counting Kit-8 (CCK-8) assay (Beyotime
Institute of Biotechnology). NSCLC cells were seeded into 96 well
plates at the density of 3×103 cells/well and cultured
in a humidified incubator at 37°C. At 0, 24, 48 and 72 h, CCK-8 (10
µl) reagent was added to each well and cells were further incubated
for 2 h at 37°C. The optical density was evaluated at 450 nm using
a microplate analyzer (Bio-Rad Laboratories, Inc.).
Cell migration and invasion
assays
The migratory and invasive abilities of A549 and
H460 cells were determined using Transwell chambers (Corning,
Inc.). Chambers precoated with Matrigel (Corning, Inc.) were used
for invasion assay, whereas chambers not precoated with Matrigel
were used for migration assay. A549 and H460 cells
(3×104 cells/well) were seeded into the upper chamber
with serum-free DMEM medium, and the lower chamber was filled with
DMEM medium supplemented with 10% FBS. After incubation at 37°C for
48 h, cells in the lower chambers were stained with 0.1% crystal
violet for 20 min at room temperature. The number of migrated or
invaded cells in five randomly selected fields was counted under an
inverted light microscope (Olympus Corporation; magnification,
×200).
Dual-luciferase reporter assay
The binding sequence of LINC01272 to miR-1303 was
obtained using DIANA (http://starbase.sysu.edu.cn/) platform. The wild type
(WT) LINC01272 sequence containing the miR-1303-binding sequence
and the mutant (MUT) LINC01272 sequence were cloned into the
pmirGLO dual luciferase miRNA target expression vector (Promega
Corporation). Then, LINC01272 (WT) and LINC01272 (MUT) were
co-transfected with miR-1303 mimic or mimic NC, respectively, into
A549 cells using Lipofectamine 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). After 48 h transfection, the relative
luciferase activity was measured using the dual-luciferase reporter
assay system (Promega Corporation). Firefly luciferase activity was
normalized to Renilla luciferase activity. All procedures
followed the manufacturers' instructions.
Statistical analysis
Data were presented as the means ± standard
deviation. Differences between two groups were analyzed using
paired Student's t-test or χ2 test, and comparisons
between multiple groups were assessed using one-way ANOVA followed
by Tukey's post hoc test. Kaplan-Meier curves and log-rank test
were used to analyze the relationship between LINC01272 expression
and the overall survival of patients with NSCLC. The prognostic
value of LINC01272 in patients with NSCLC was confirmed by
univariate and multivariate Cox regression analysis. Each
experiment was repeated at least three times. SPSS 22.0 software
(IBM Corp.) and GraphPad Prism 7.0 software (GraphPad Software,
Inc.) were used to perform analyses. P<0.05 was considered to
indicate a statistically significant difference.
Results
LINC01272 expression and its
relationship with patient overall survival based on bioinformatics
analysis
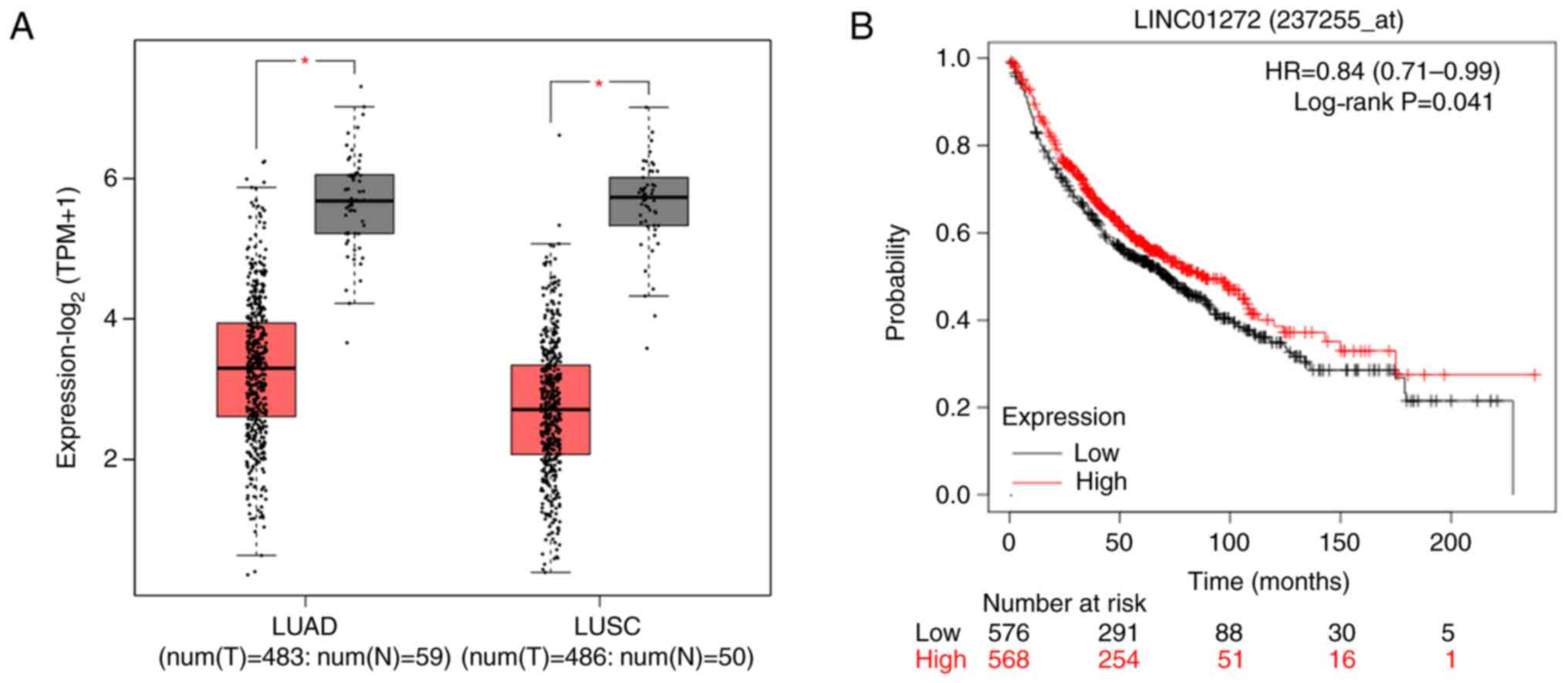
According to bioinformatics analysis, the results
from TCGA database analysis revealed that the expression of
LINC01272 was downregulated in NSCLC tumors compared with normal
tissues (Fig. 1A), and the results
from GEO, EGA and TCGA database analyses showed that low LINC01272
expression was associated with worse overall survival in patients
with lung cancer (Fig. 1B).
LINC01272 is downregulated in NSCLC
tissues and cell lines
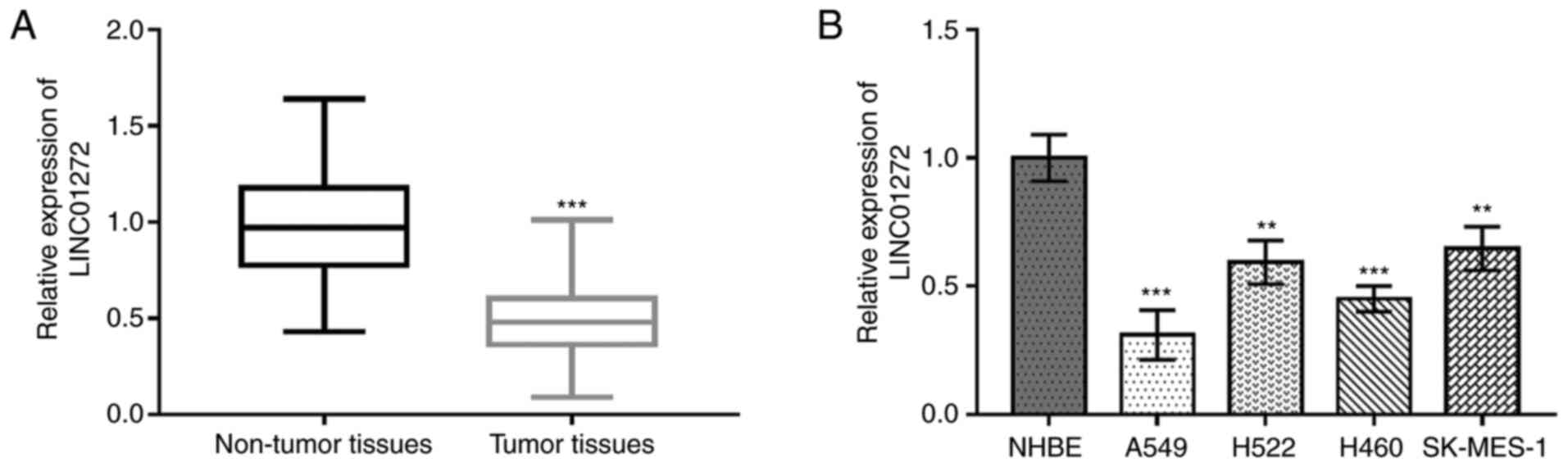
The expression level of LINC01272 in NSCLC tissues
and cell lines was evaluated using RT-qPCR. As presented in
Fig. 2A, LINC01272 expression was
significantly decreased in NSCLC tissues compared with adjacent
non-tumor tissues (P<0.001). In addition, LINC01272 expression
was significantly decreased in NSCLC cells compared with NHBE cells
(all P<0.01; Fig. 2B).
Furthermore, because A549 and H460 cells expressed the lowest
levels of LINC01272, these cells lines were used for subsequent
experiments.
Relationship between LINC01272
expression and the clinicopathological characteristics of patients
with NSCLC
Analysis of the relationship between LINC01272
expression and the clinicopathological characteristics of patients
with NSCLC suggested that LINC01272 might be involved in the
development of NSCLC. The median expression value of miR-1303
(0.50) was used as the cutoff value to divide the patients into low
and high miR-1303 expression groups. As presented in Table I, LINC01272 expression was associated
with tumor size (P=0.021), lymph node metastasis (P=0.037) and
Tumor-Node-Metastasis (TNM) stage (P=0.009). However, there were no
association between LINC01272 expression and the other variables,
including age, sex, smoking and differentiation (all
P>0.05).
Association between LINC01272
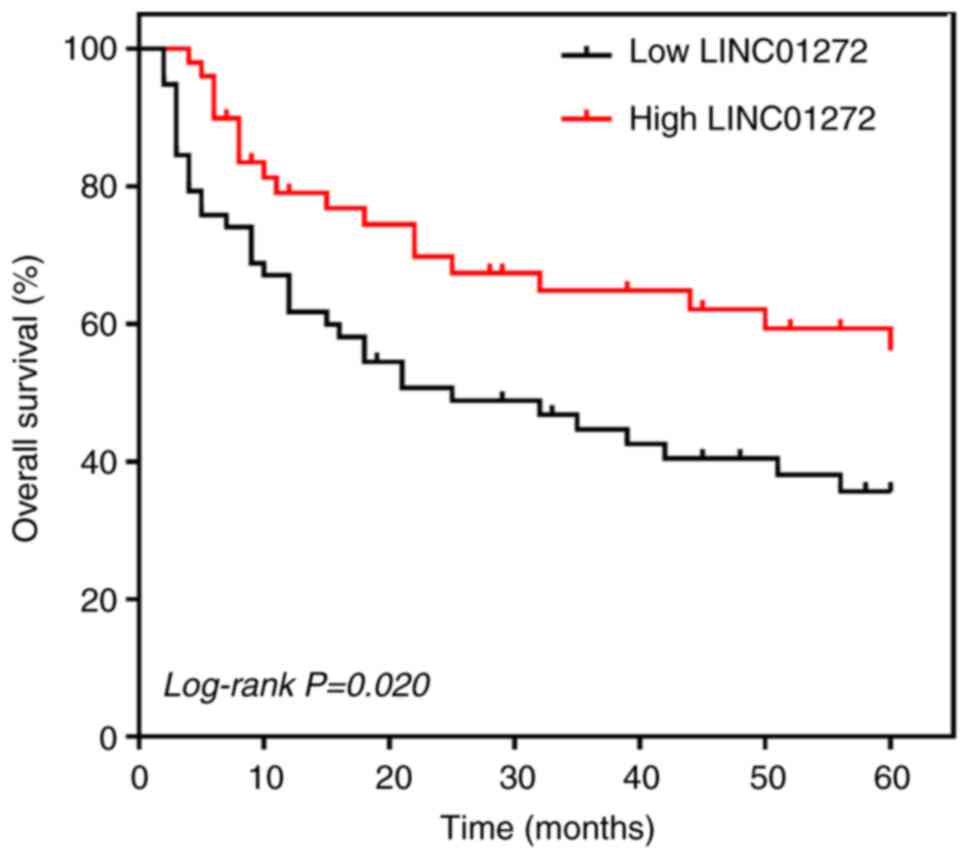
expression and the overall survival of patients with NSCLC
The prognostic value of LINC01272 expression in
patients with NSCLC was evaluated by Kaplan-Meier survival curves
and Cox regression analyses. Patients with NSCLC with high
LINC01272 expression had higher overall survival than those with
low LINC01272 expression (log-rank P=0.020; Fig. 3). In addition, the results from Cox
regression analysis (Table II)
indicated that LINC01272 expression, lymph node metastasis,
differentiation and TNM stage were associated with the overall
survival of patients with NSCLC, and that LINC01272 expression and
TNM stage could be used as two independent prognostic factors
[LINC01272, hazard ratio (HR)=1.988, 95% confidence interval
(CI)=1.290–3.078; P=0.009; TNM stage: HR=1.596, 95% CI=1.107–2.506;
P=0.027].
 | Table II.Cox regression analysis results for
patients with non-small cell lung cancer. |
Table II.
Cox regression analysis results for
patients with non-small cell lung cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age | 1.365 | 0.815–2.241 | 0.267 | – | – | – |
| Sex | 1.226 | 0.761–1.960 | 0.403 | – | – | – |
| Smoking | 1.872 | 0.758–2.955 | 0.181 | – | – | – |
| Tumor size | 1.376 | 0.862–2.103 | 0.159 | – | – | – |
| Lymph node
metastasis | 1.599 | 1.022–2.389 | 0.038a | 1.423 | 0.927–2.138 | 0.091 |
|
Differentiation | 1.285 | 1.018–2.654 | 0.042a | 1.253 | 0.906–1.966 | 0.128 |
| TNM stage | 1.941 | 1.385–3.159 | 0.012a | 1.596 | 1.107–2.506 | 0.027a |
| LINC01272 | 2.085 | 1.523–3.503 | 0.005b | 1.988 | 1.290–3.078 | 0.009b |
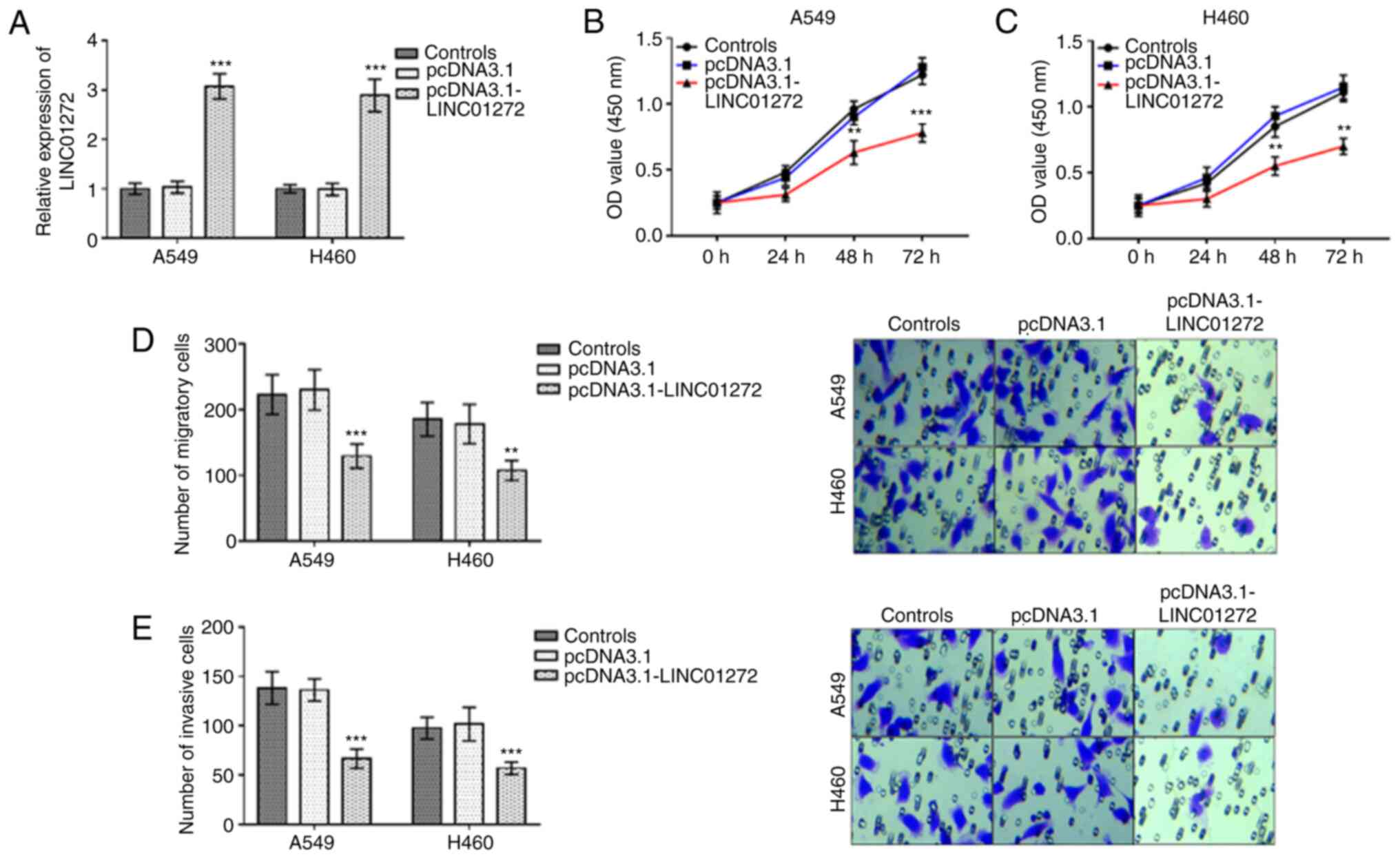
Inhibitory effects of LINC01272
overexpression on NSCLC cell proliferation, migration and
invasion
Following transfection with pcDNA3.1 or
pcDNA3.1-LINC01272, the expression level of LINC01272 was
significantly increased by pcDNA3.1-LINC01272 in A549 cells and
H460 cells (Fig. 4A; P<0.001).
Furthermore, the proliferation of A549 cells (Fig. 4B; P<0.01) and H460 cells (Fig. 4C; P<0.01) was significantly
inhibited following LINC01272 overexpression. In A549 cells and
H460 cells, the migratory (Fig. 4D;
P<0.01) and invasive (Fig. 4E;
P<0.001) abilities were significantly inhibited by LINC01272
overexpression.
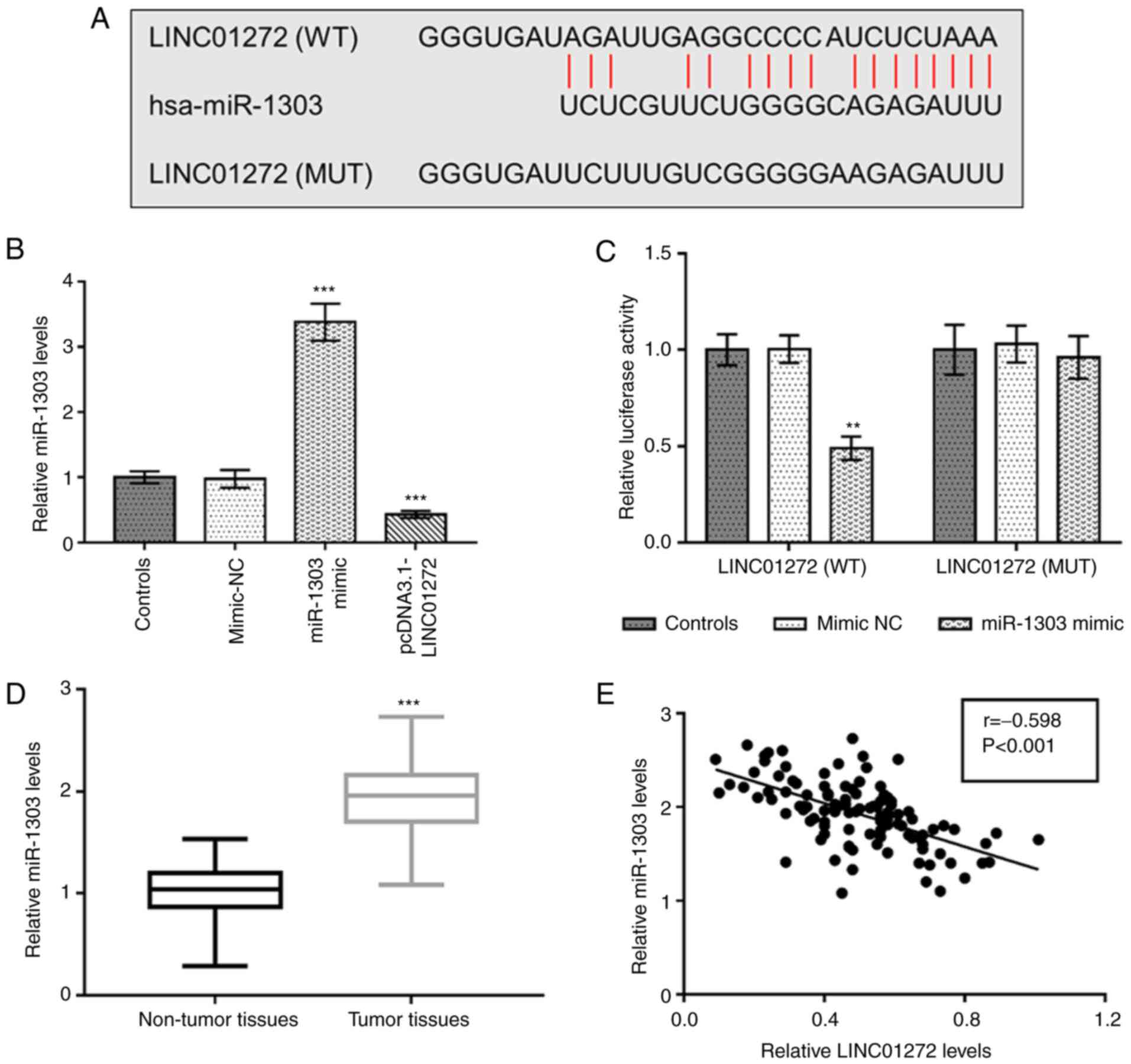
Negative relationship between
LINC01272 and miR-1303 in NSCLC
The complementary binding sequences between
LINC01272 and miR-1303 were seen in Fig.
5A. As presented in Fig. 5B,
miR-1303 was significantly upregulated following transfection with
miR-1303 mimic but downregulated after LINC01272 overexpression in
A549 cells (P<0.001). Furthermore, a dual-luciferase reporter
assay was conducted to confirm the interaction between LINC01272
and miR-1303 in NSCLC cells. The results demonstrated that
overexpression of miR-1303 inhibited the relative luciferase
activity of LINC01272 (WT) group (P<0.01), whereas no changes
were observed in the luciferase activity of LINC01272 (MUT) group
(P>0.05; Fig. 5C). As presented
in Fig. 5D, miR-1303 expression was
significantly increased in tumor tissues compared with adjacent
non-tumor tissues (P<0.001). In addition, a negative correlation
was observed between miR-1303 expression and LINC01272 expression
in NSCLC tissues (Fig. 5E; r=−0.598,
P<0.001).
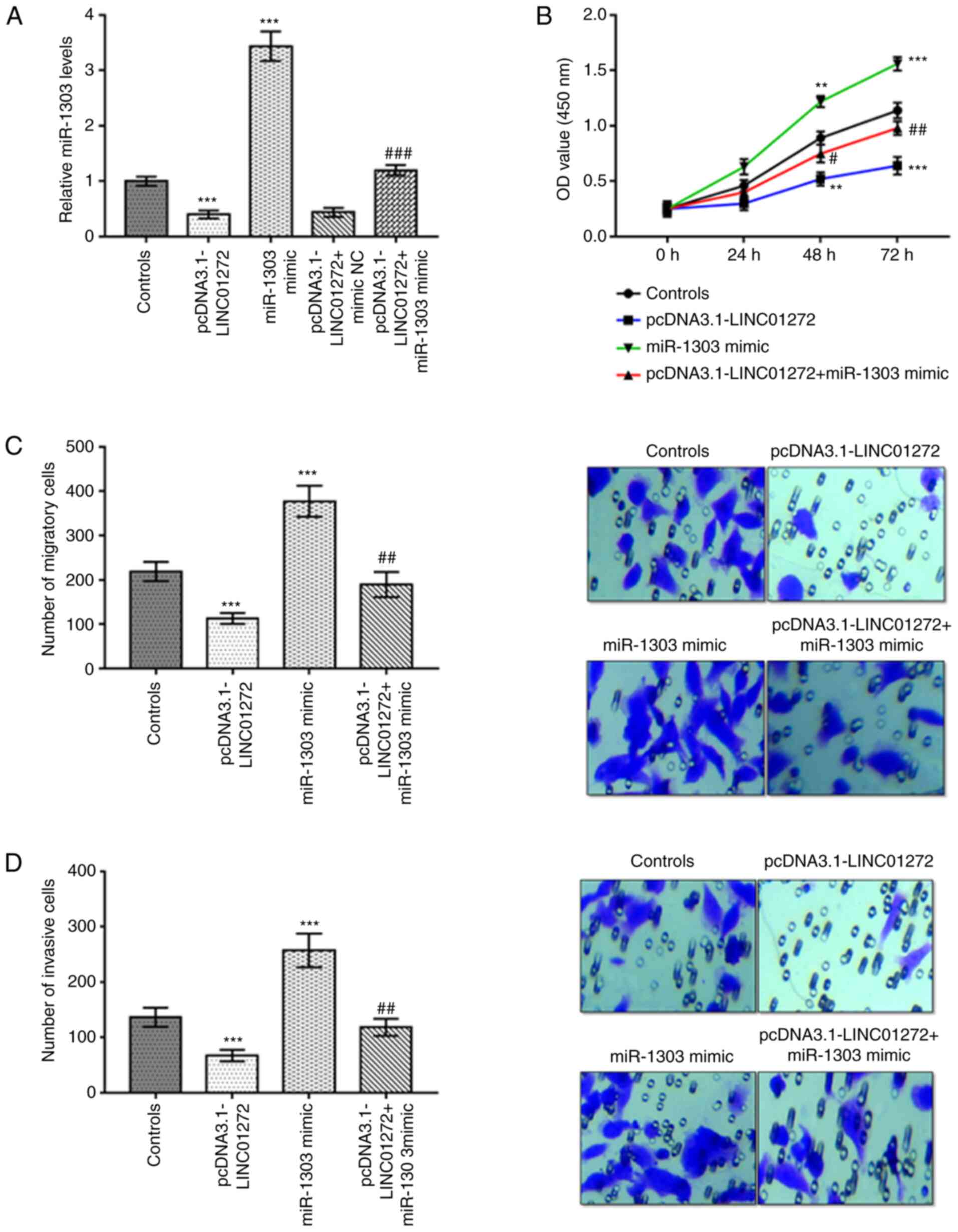
miR-1303 overexpression reverses the
function of LINC01272 in NSCLC cells
As presented in Fig.
6A, miR-1303 expression was significantly decreased after
transfection with pcDNA3.1-LINC01272, whereas it was significantly
increased by miR-1303 mimic in A549 cells (all P<0.001). In
addition, miR-1303 mimic reversed the suppressive effect of
pcDNA3.1-LINC01272 on the expression of miR-1303 in A549 cells
(P<0.001). Overexpression of LINC01272 inhibited, while miR-1303
overexpression promoted the proliferation, migration and invasion
of A549 cells (Fig. 6B-D;
P<0.01). Furthermore, miR-1303 overexpression reversed the
inhibitory effects of LINC01272 overexpression on the function of
A549 cells (Fig. 6B-D;
P<0.05).
Discussion
It has been demonstrated that lncRNAs aberrant
expression is closely related to the progression of numerous types
of cancer (6,7,21),
suggesting that lncRNAs might be involved in the progression of
cancer. In addition, numerous lncRNAs, including PTAR (22), NBR2 (23) and DLEU2 (24), have been reported to be abnormally
expressed in NSCLC. According to bioinformatics analysis of data
from TCGA database, LINC01272 expression level was found to be
significantly downregulated in NSCLC. The findings from the present
study confirmed the downregulation of LINC01272 in NSCLC tumor
tissues and cells. Furthermore, LINC01272 expression was
significantly correlated with tumor size, lymph node metastasis and
TNM stage in patients with NSCLC. In addition, LINC01272 has been
found to serve as an important regulator in other diseases. For
example, LINC01272 is upregulated in gastric cancer and its
expression is associated with tumor stage and lymph node metastasis
(9). Wang et al (10) reported an increased expression of
LINC01272 in tissues and plasma samples of patients with
inflammatory bowel disease compared with healthy volunteers. The
present study hypothesized therefore that LINC01272 may be involved
in the progression of NSCLC.
Increasing evidence has demonstrated that certain
lncRNAs can function as potential biomarkers for NSCLC prognosis
(25), such as SLC16A1-AS1 (26), XIST (27) and LINC-PINT (28). Our preliminary bioinformatics
analysis using GEO, EGA and TCGA database data reported that
patients with lung cancer and with low LINC01272 expression had a
significantly worse overall survival than patients with high
LINC01272 expression. Furthermore, in the present study, decreased
expression of LINC01272 was found in NSCLC tissues. The present
study analyzed the prognostic significance of LINC01272 in NSCLC.
It has been reported that SLC16A1-AS1 (HR=3.351, 95%
CI=2.027–5.541, P<0.001) (26),
XIST (HR=2.645, 95% CI=1.672–7.393, P=0.029) (27) and LINC-PINT (HR=2.628, 95%
CI=1.589–4.348, P<0.001) (28)
can be used as prognostic biomarkers in patients with NSCL. The
prognostic value of LINC01272 was evaluated according to the 5-year
survival information of patients with NSCLC in the present study.
The overall survival of patients with low LINC01272 expression was
worse compared with that of patients with high LINC01272
expression. In addition, LINC01272 was independently related to
overall survival, suggesting that LINC01272 may be considered as a
potential prognostic biomarker for NSCLC. Since the HR of LINC01272
is lower than the HRs of SLC16A1-AS1, XIST and LINC-PINT, the
overall mortality risk of patients with NSCLC and with low
LINC01272 expression was relatively low.
Increasing evidence indicates that lncRNAs can
regulate NSCLC tumor biological functions (29–31). For
example, LINC00673 can regulate the proliferation, migration and
invasion of NSCLC cells by sponging miR-150-5p (29). Furthermore, PCAT7 can promote the
proliferation, migration and invasion, and inhibit the apoptosis of
NSCLC cells by inhibiting miR-134-5p (30). A study by An et al (31) reported that LINC00668 downregulation
inhibits NSCLC cell proliferation, migration and invasion, and
stimulates NSCLC cell apoptosis. The results from the present study
demonstrated that LINC01272 overexpression could inhibit NSCLC cell
proliferation, migration and invasion. In addition, LINC01272
knockdown has been found to inhibit the biological function of
gastric cancer cells (9). LINC01272
may therefore serve an anticancer role in NSCLC.
Previous studies have demonstrated that miRNA-lncRNA
interactions play important roles in the occurrence and development
of cancer (32–34). According to bioinformatics
prediction, miR-1303 was identified as a target gene of LINC01272
in the present study, and this interaction was confirmed through a
luciferase reporter assay. miR-1303 is a key molecule involved in
various cancers, such as prostate cancer (15) and gastric cancer (16). The present study reported a higher
miR-1303 expression in tumor tissues compared with non-tumor
tissues, which was in accordance with a study from Chen et
al (35). Furthermore, miR-1303
expression was negatively correlated with LINC01272 expression in
tumor tissues of patients with NSCLC. In addition, overexpression
of LINC01272 inhibited miR-1303 expression in NSCLC cells, and
miR-1303 overexpression reversed the effects of LINC01272 on NSCLC
cell proliferation, migration and invasion, suggesting that
LINC01272 may inhibit NSCLC cell function by targeting miR-1303.
Certain lncRNAs have been found to exert their biological functions
by regulating miR-1303 in several diseases. For example, BCRT1 was
reported to contribute to breast cancer cell proliferation,
migration and invasion by targeting miR-1303/PTBP3 axis (36). In addition, LINC01433 upregulation
can promote cell proliferation and migration by sponging miR-1301
in esophageal squamous cell carcinoma (37).
A study by Liu et al (15) reported that miR-1303 can enhance the
cell proliferation, migration and invasion via targeting DKK3 in
prostate cancer. Cheng et al (38) demonstrated that miR-1303-p can
regulate the cell proliferation and apoptosis of clear cell renal
cell carcinoma by targeting STARD9. Therefore, we speculated that
miR-1303 may regulate NSCLC cell function by targeting DKK3 or
STARD. However, whether DKK3 and STARD9 are targets of miR-1303 in
NSCLC remains unclear. Thus, the lack of miR-1303 target is a major
limitation of the present study, and further investigation is
therefore required to validate the results. In addition, the sample
size was small, which is also a limitation of this study, and
future study including a large research cohort is needed.
In conclusion, the present study demonstrated that
LINC01272 expression was decreased in NSCLC tumor tissues and NSCLC
cells compared with normal tissues and cells, suggesting that
LINC01272 may serve as an independent prognostic biomarker for
patients with NSCLC. In addition, LINC01272 overexpression could
inhibit NSCLC cell proliferation, migration and invasion by
inhibiting miR-1303 in NSCLC. The novel LINC01272/miR-1303 axis may
therefore be considered as a new biomarker and therapeutic target
for NSCLC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SZ and JZ designed the study, performed clinical
studies and analyzed data. JZ performed the cell experiments. SZ
and JZ wrote and revised the manuscript. SZ and JZ confirmed the
authenticity of all raw data. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
A signed written informed consent was obtained from
each patient and the experimental procedures were all in accordance
with the guideline of the Ethics Committee of Weifang People's
Hospital (approval no. 011827).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goebel C, Louden CL, McKenna R Jr, Onugha
O, Wachtel A and Long T: Diagnosis of non-small cell lung cancer
for early stage asymptomatic patients. Cancer Genomics Proteomics.
16:229–244. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang Q, Tang Y, Tang C, Cong H, Wang X,
Shen X and Ju S: Diminished LINC00173 expression induced miR-182-5p
accumulation promotes cell proliferation, migration and apoptosis
inhibition via AGER/NF-κB pathway in non-small-cell lung cancer. Am
J Transl Res. 11:4248–4262. 2019.PubMed/NCBI
|
|
4
|
Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X
and Tai S: LncRNA HOXA-AS2 and its molecular mechanisms in human
cancer. Clin Chim Acta. 485:229–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu S, Zheng Q, Xiong J, Wu H, Wang W and
Zhou W: Long non-coding RNA MVIH promotes cell proliferation,
migration, invasion through regulating multiple cancer-related
pathways, and correlates with worse prognosis in pancreatic ductal
adenocarcinomas. Am J Transl Res. 12:2118–2135. 2020.PubMed/NCBI
|
|
7
|
Wang Y, Ding X, Hu H, He Y, Lu Z, Wu P,
Tian L, Xia T, Yin J, Yuan H, et al: Long non-coding RNA lnc-PCTST
predicts prognosis through inhibiting progression of pancreatic
cancer by downregulation of TACC-3. Int J Cancer. 143:3143–3154.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leng X, Liu G, Wang S, Song J, Zhang W,
Zhang X, Rong L, Ma Y and Song F: LINC01272 promotes migration and
invasion of gastric cancer cells via EMT. Onco Targets Ther.
13:3401–3410. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang S, Hou Y, Chen W, Wang J, Xie W,
Zhang X and Zeng L: KIF9-AS1, LINC01272 and DIO3OS lncRNAs as novel
biomarkers for inflammatory bowel disease. Mol Med Rep.
17:2195–2202. 2018.PubMed/NCBI
|
|
11
|
Hung J, Scanlon JP, Mahmoud AD, Rodor J,
Ballantyne M, Fontaine MAC, Temmerman L, Kaczynski J, Connor KL,
Bhushan R, et al: Novel plaque enriched long noncoding RNA in
atherosclerotic macrophage regulation (PELATON). Arterioscler
Thromb Vasc Biol. 40:697–713. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Z, Sha HH and Li HJ: Functions and
mechanisms of miR-186 in human cancer. Biomed Pharmacother.
119:1094282019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iacona JR and Lutz CS: miR-146a-5p:
Expression, regulation, and functions in cancer. Wiley Interdiscip
Rev RNA. 10:e15332019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mamoori A, Gopalan V and Lam AK: Role of
miR-193a in cancer: Complexity and factors control the pattern of
its expression. Curr Cancer Drug Targets. 18:618–628. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu B, Zhou W, Jiang H, Xiang Z and Wang
L: miR-1303 promotes the proliferation, migration and invasion of
prostate cancer cells through regulating the Wnt/β-catenin pathway
by targeting DKK3. Exp Ther Med. 18:4747–4757. 2019.PubMed/NCBI
|
|
16
|
Zhang SJ, Feng JF, Wang L, Guo W, Du YW,
Ming L and Zhao GQ: miR-1303 targets claudin-18 gene to modulate
proliferation and invasion of gastric cancer cells. Dig Dis Sci.
59:1754–1763. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv QL, Wang LC, Li DC, Lin QX, Shen XL,
Liu HY, Li M, Ji YL, Qin CZ and Chen SH: Knockdown lncRNA DLEU1
inhibits gliomas progression and promotes temozolomide
chemosensitivity by regulating autophagy. Front Pharmacol.
11:5605432020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar
|
|
20
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feng J, Li J, Qie P, Li Z, Xu Y and Tian
Z: Long non-coding RNA (lncRNA) PGM5P4-AS1 inhibits lung cancer
progression by up-regulating leucine zipper tumor suppressor
(LZTS3) through sponging microRNA miR-1275. Bioengineered.
12:196–207. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu W, Sun Z, Yang L, Han Y, Yue L, Deng L
and Yao R: lncRNA PTAR promotes NSCLC cell proliferation, migration
and invasion by sponging microRNA-101. Mol Med Rep. 20:4168–4174.
2019.PubMed/NCBI
|
|
23
|
Gao YP, Li Y, Li HJ and Zhao B: LncRNA
NBR2 inhibits EMT progression by regulating Notch1 pathway in
NSCLC. Eur Rev Med Pharmacol Sci. 23:7950–7958. 2019.PubMed/NCBI
|
|
24
|
Zhou Y, Shi H, Du Y, Zhao G, Wang X, Li Q,
Liu J, Ye L, Shen Z, Guo Y and Huang Y: lncRNA DLEU2 modulates cell
proliferation and invasion of non-small cell lung cancer by
regulating miR-30c-5p/SOX9 axis. Aging (Albany NY). 11:7386–7401.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song JY, Li XP, Qin XJ, Zhang JD, Zhao JY
and Wang R: A fourteen-lncRNA risk score system for prognostic
prediction of patients with non-small cell lung cancer. Cancer
Biomark. 29:493–508. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu HY, Lu SR, Guo ZH, Zhang ZS, Ye X, Du
Q, Li H, Wu Q, Yu B, Zhai Q and Liu JL: lncRNA SLC16A1-AS1 as a
novel prognostic biomarker in non-small cell lung cancer. J
Investig Med. 68:52–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fang H, Yang L, Fan Y, Mo C, Luo L, Liang
D and Jiang Y: Upregulation of tissue long noncoding RNA X inactive
specific transcript predicts poor postoperative survival in
patients with non-small cell lung cancer. Medicine (Baltimore).
99:e217892020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang C, Gong C, Li J and Tang J:
Downregulation of long non-coding RNA LINC-PINT serves as a
diagnostic and prognostic biomarker in patients with non-small cell
lung cancer. Oncol Lett. 21:2102021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H,
Kong J, Ding K, Shen HM, Wu H, et al: Long non-coding RNA linc00673
regulated non-small cell lung cancer proliferation, migration,
invasion and epithelial mesenchymal transition by sponging
miR-150-5p. Mol Cancer. 16:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Q, Wu Y, Xiao J and Zou J: Long
non-coding RNA prostate cancer-associated transcript 7 (PCAT7)
induces poor prognosis and promotes tumorigenesis by inhibiting
mir-134-5p in non-small-cell lung (NSCLC). Med Sci Monit.
23:6089–6098. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
An YX, Shang YJ, Xu ZW, Zhang QC, Wang Z,
Xuan WX and Zhang XJ: STAT3-induced long noncoding RNA LINC00668
promotes migration and invasion of non-small cell lung cancer via
the miR-193a/KLF7 axis. Biomed Pharmacother. 116:1090232019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luo H, Xu C, Le W, Ge B and Wang T: lncRNA
CASC11 promotes cancer cell proliferation in bladder cancer through
miRNA-150. J Cell Biochem. 120:13487–13493. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luan X and Wang Y: LncRNA XLOC_006390
facilitates cervical cancer tumorigenesis and metastasis as a ceRNA
against miR-331-3p and miR-338-3p. J Gynecol Oncol. 29:e952018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao W, Geng D, Li S, Chen Z and Sun M:
LncRNA HOTAIR influences cell growth, migration, invasion, and
apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer
Med. 7:842–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen J, Jiang T, Yu B, Li T, Zhao P, Yuan
L and Qi J: Upregulation of microRNA-1303 is a potential prognostic
marker of non-small cell lung cancer. Cancer Biomark. 28:439–446.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang Y, Song X, Li Y, Chen B, Zhao W,
Wang L, Zhang H, Liu Y, Han D, Zhang N, et al: LncRNA BCRT1
promotes breast cancer progression by targeting miR-1303/PTBP3
axis. Mol Cancer. 19:852020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng L, Liu YT, Wu CP, Jiang JT, Zhang L,
Wang ZL and Wang QY: Long non-coding RNA linc01433 promotes
tumorigenesis and progression in esophageal squamous cell carcinoma
by sponging miR-1301. Eur Rev Med Pharmacol Sci. 24:4785–4792.
2020.PubMed/NCBI
|
|
38
|
Cheng T, Shuang W, Ye D, Zhang W, Yang Z,
Fang W, Xu H, Gu M, Xu W and Guan C: SNHG16 promotes cell
proliferation and inhibits cell apoptosis via regulation of the
miR-1303-p/STARD9 axis in clear cell renal cell carcinoma. Cell
Signal. 84:1100132021. View Article : Google Scholar : PubMed/NCBI
|