Introduction
Osteosarcoma (OS), originating from osteoblasts, is
the most common primary malignant bone tumor, arising frequently in
the metaphysis of long bones (1).
Recently, the focus of research on OS has been on biological
macromolecules and genes, including activated oncogenes (2), tumor suppressor gene inactivation
(3), the role of tumor
microangiogenesis in promoting tumor development (4), imbalance of cell proliferation and
apoptosis (5), abnormal expression
of biological macromolecules (6,7) and
other factors (8) that jointly
promote the occurrence and development of OS. Findings on the
biological functions of OS may eventually be applied to the
clinic.
Long non-coding RNAs (lncRNAs), which are widely
distributed in the human genome to modulate gene expression, have a
variety of biological functions, such as signaling (9), acting as molecular decoys (10) and guiding (11). The molecular decoy function of lncRNA
is also called the ‘sponge’ function, which can sequester small
non-coding RNAs [microRNAs (miRNAs/miRs)] away from their targets
and suppress their function (10).
For example, lncRNA myocardin-induced smooth muscle lncRNA, inducer
of differentiation is significantly upregulated in gastric cancer,
and regulates the proliferation and apoptosis of gastric cancer by
regulating the miR-29c-3p/MCL1 apoptosis regulatory, BCL2 family
member axis (12). Furthermore, in
OS, the Alu-mediated p21 transcriptional regulator, as an oncogenic
lncRNA, can inhibit miR-132-3p expression and promote OS cell
proliferation and migration (13).
lncRNA XLOC_005950 is located on chromosome 6q27 (14), and to the best of our knowledge,
there are currently no studies regarding its function in OS. Thus,
the role of lncRNA XLOC_005950 in regulating cell metabolism in OS
needs to be clarified.
miRNAs in eukaryotic cells, which are 20–24
nucleotides in length, are important and abundant components of
gene regulatory networks, which contain lncRNA-regulated miRNAs
(15), miRNAs that reverse-regulate
lncRNA (16) and lncRNAs that
regulate mRNA expression by targeting miRNAs (17). miR-542-3p expression is downregulated
in colorectal cancer, non-small cell lung cancer and hepatocellular
carcinoma (18–20). In OS, Luo et al (21) reported that lncRNA ADP dependent
glucokinase-antisense RNA 1 regulates OS cell proliferation and
apoptosis by modulating miR-542-3p expression. However, the
regulation of glucose metabolism by miR-542-3p in OS still requires
further investigation.
Aberrant metabolism is a hallmark of human cancer
(22). Malignant tumor cells use
glucose via glycolysis even when oxygen is sufficient (23), which is called aerobic glycolysis or
more commonly, the Warburg effect (24). The Warburg effect is controlled by
glycolytic enzymes, including glucose transporter, hexokinase (HK),
phosphofructokinase (PFK) and pyruvate kinase (PK) (25). PFK is a key rate-limiting enzyme in
glycolysis (26). It has been
reported that PFK activity significantly increases in certain tumor
cells and tissues (27–29). PFK, muscle (PFKM) is a known isoform
of PFK in humans (26). A previous
study demonstrated that reduced glycolysis induced by miR-21 is
mediated through targeting of the PFKM isoform at the committed
step of glycolysis (30). Thus,
altering the metabolism by interfering with the aerobic glycolytic
pathway of tumor cells is important for studying the biological
behavior of tumors. Previous studies have reported that lncRNAs
regulate glycolysis by sponging miRNAs (31,32).
The present study aimed to investigate the effect of
the regulation of the lncRNA-miRNA-mRNA network on the energy
metabolism pathway of aerobic glycolysis in OS cells, and
investigate the effects of this regulatory axis on cell
proliferation and apoptosis.
Materials and methods
Patients and tissue samples
Tumor tissues and corresponding adjacent normal
tissues (3 cm away from the tumor tissues) were collected from 25
patients (age range, 7–65 years; mean age, 24.76 years; 14 men and
11 women) with OS at the Department of Orthopedic Surgery of The
First Affiliated Hospital of Zhengzhou University (Zhengzhou,
China) between September 2015 and March 2017. All tissue samples
collected during tumor resection were immediately frozen in liquid
nitrogen and stored at −80°C until subsequent experimentation. The
present study was approved by the Medical Ethics Committee of
Zhengzhou University (Zhengzhou, China; approval no. 2015-03) and
patients provided written informed consent prior to the study
start. Patient characteristics are listed in Table I. The mean lncRNA XLOC_005950
expression in 25 OS tissues was regarded as the cut-off point.
 | Table I.Association between lncRNA
XLOC_005950 expression and the clinicopathological characteristics
of patients with osteosarcoma (n=25). |
Table I.
Association between lncRNA
XLOC_005950 expression and the clinicopathological characteristics
of patients with osteosarcoma (n=25).
|
|
| lncRNA XLOC_005950
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Number of
cases | Low (n=15) | High (n=10) | P-value |
|---|
| Sex |
|
|
| 0.7422 |
|
Male | 14 | 8 | 6 |
|
|
Female | 11 | 7 | 4 |
|
| Age, years |
|
|
| 0.6098 |
|
≤15 | 16 | 9 | 7 |
|
|
>15 | 9 | 6 | 3 |
|
| Primary site |
|
|
| 0.8702 |
|
Femur | 13 | 8 | 5 |
|
|
Tibia/Humerus | 12 | 7 | 5 |
|
| Tumor size, cm |
|
|
| 0.2936 |
| ≤4 | 8 | 6 | 2 |
|
|
>4 | 17 | 9 | 8 |
|
| Clinical stage |
|
|
| 0.0270a |
|
I+II | 9 | 8 | 1 |
|
|
III | 16 | 7 | 9 |
|
| Metastasis |
|
|
| 0.0124a |
|
Negative | 10 | 9 | 1 |
|
|
Positive | 15 | 6 | 9 |
|
Cell culture and transfection
The human osteoblastic cell line, hFOB1.19, and
three human OS cell lines, MG63, U2OS and Saos-2, were purchased
from The Cell Bank of Type Culture Collection of The Chinese
Academy of Sciences. MG63, U2OS and Saos-2 cells were maintained in
DMEM (Biological Industries) supplemented with 10% fetal bovine
serum (FBS, Biological Industries) and 1% penicillin/streptomycin
(Beijing Solarbio Science & Technology Co., Ltd.), at 37°C with
5% CO2. Human hFOB 1.19 cells were maintained in
DMEM/F12 medium (Biological Industries) supplemented with 15% FBS
(Gibco; Thermo Fisher Scientific, Inc.), at 33.5°C with 5%
CO2. Cells were harvested for subsequent experimentation
after 48 h.
hsa-miR-542-3p mimics negative control (NC),
hsa-miR-542-3p mimics, hsa-miR-542-3p inhibitor NC and
hsa-miR-542-3p inhibitor were purchased from Shanghai GenePharma
Co., Ltd. The sequences were as follows: hsa-miR-542-3p inhibitor,
5′-UUCAGUUAUCAAUCUGUCACA-3′; hsa-miR-542-3p inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′; hsa-miR-542-3p mimics,
5′-UGUGACAGAUUGAUAACUGAAA-3′; hsa-miR-542-3p mimics NC,
5′-UUCUCCGAACGUGUCACGUTT-3′. MG63 cells were transfected with 50 nM
miR-542-3p mimics or miR-542-3p NC using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C with 5%
CO2. Following incubation for 6 h at 37°C, the medium
was replaced with fresh complete DMEM and cultured for 36–48 h.
Similarly, MG63 cells were transfected with 50 nM miR-542-3p
inhibitor using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Transfected cells were immediately used for subsequent
experiments.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated from cells (hFOB1.19, MG63,
U2OS and Saos-2 cells) and tissues using TransZol reagent (TransGen
Biotech, Co., Ltd.). The RNA concentration was measured using a
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.).
lncRNA and mRNA were reverse transcribed into cDNA using the
PrimeScript™ RT Reagent kit with gDNA Eraser (Perfect Real Time)
(Takara Biotechnology Co., Ltd.), according to the manufacturer's
instructions. miRNA RT was performed with a stem-loop primer at
42°C for 60 min and 70°C for 5 min, using the Revert Aid First
Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). qPCR
was subsequently performed on an ABI 7500 Fast system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR Premix
Ex Taq II kit (Takara Biotechnology Co., Ltd.). The following
thermocycling conditions were used for qPCR: Initial denaturation
at 95°C for 30 sec; followed by 40 cycles of 95°C for 5 sec and
60°C for 34 sec. The primer sequences used for qPCR were designed
by Primer Premier 6.0 and synthesized by Sangon Biotech Co., Ltd.
(Table II). Relative expression
levels were calculated using the 2−ΔΔCq method (33). β-actin and U6 were used as endogenous
controls for mRNA and miRNA expression levels, respectively. All
experiments were performed in triplicate.
 | Table II.Primer sequences used for
quantitative PCR. |
Table II.
Primer sequences used for
quantitative PCR.
| Gene | Primer sequence
(5′-3′) |
|---|
| hsa-miR-542-3p |
TCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTTCAG |
| U6 |
AACGCTTCACGAATTTGCGT |
| lncRNA
XLOC_005950 | F:
GTTCAGGACATAGGTGGATT |
|
| R:
CGACACTATCAGAGGCATT |
| PFKM | F:
ACCCGTGGTTCTCGTCTC |
|
| R:
AAAGGCTGATGGCGTCCC |
| β-actin | F:
CTCCATCCTGGCCTCGCTGT |
|
| R:
GCTGTCACCTTCACCGTTC |
Plasmid construction
sgRNA was designed (http://crispr.mit.edu), synthesized (Sangon Biotech
Co., Ltd.) and inserted into a PX459 vector (Addgene, Inc.)
digested by BbsI. The sgRNA sequences directed against lncRNA
XLOC_005950 were annealed and cloned into the PX459 vector. A total
of two pairs of independent sgRNA sequences targeting lncRNA
XLOC_005950 were designed: Forward oligo1,
5′-CACCGGGAGTGCGTTCGGTGTGCCG-3′ and reverse oligo1,
5′-AAACCGGCACACCGAACGCACTCCC-3′; and forward oligo2,
5′-CACCGCAGGTGCATGCCAATATACC-3′ and reverse oligo2,
5′-AAACGGTATATTGGCATGCACCTGC-3′. Successful insertion of each sgRNA
was verified by sequencing.
CRISPR/Cas9 gene editing
A total of 4×105 MG63 cells were seeded
into 6-well plates and cultured for 24 h. After reaching 70–80%
confluency, cells were transfected with PX459 plasmid vectors
carrying sgRNAs using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 6 h post-incubation, the medium was replaced
with fresh complete DMEM. After 48 h, puromycin (0.5 µg/µl) was
added to the medium, and the transfected cells were subjected to
puromycin selection. After 2 weeks of transfection, the surviving
cells were expanded in the presence of 5 µg/ml puromycin. The
expression of lncRNA XLOC_005950 in the cells was detected by
genomic PCR analysis using the following primer sequences: Forward,
5′-CCTGCCAGTGTCTCCGCCGGTT-3′ and reverse,
5′-GCCTGACCAACATGGTGAAGC-3′. The following thermocycling conditions
were used for the PCR: Initial denaturation at 94°C for 5 min;
followed by 35 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C
for 30 sec; and a final step at 72°C for 10 min. The corresponding
PCR-amplified fragments were ligated into pGEM-T vectors (pGEM-T
Easy; Promega Corporation). The genome deletions were verified by
sequencing prior to subsequent experimentation. CRISPR/Cas9 gene
editing was used to knock out the genomic DNA fragments of lncRNA
XLOC_005950 containing the binding sites of miR-542-3p in MG63
cells. The knockout MG63 cell line was labeled as
MG63−/− in the present study.
Glucose uptake and lactate production
assay
Glucose, lactate and PFK assay kits were used to
determine glucose content, lactic acid content and activities of
PFK, respectively, following the manufacturer's instructions (cat.
nos. BC2500; BC2230; BC0530; Beijing Solarbio Science &
Technology Co., Ltd.). Briefly, the cultured cells (hFOB1.19, MG63
and MG63−/− cells) were digested with 0.25% trypsin and
centrifuged at 960 × g for 5 min at room temperature to obtain the
supernatant. Subsequently, 5×106 cells were lysed in 1
ml assay buffer for each kit, and the cells were sonicated by
ultrasound and centrifuged to obtain the supernatant for detection.
The detection reagents of the three kits were added to the
supernatant sample of each respective group following the
corresponding instructions. Following incubation at 37°C (for 15
min for glucose detection, 20 min for lactate detection and 10 min
for PFK activity detection), the absorbance of the samples was
measured using a spectrophotometer: The glucose content was
measured at a wavelength of 505 nm; the lactic acid content at a
wavelength of 570 nm and the PFK activity at a wavelength of 340
nm. The standard curve of lactic acid content detection was
determined according to the standard product. The results in each
group were normalized to the cell number, and independent
experiments were performed in triplicate.
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8, Dojindo Molecular
Technologies, Inc.) assay was performed to assess cell
proliferation. Briefly, hFOB1.19, MG63 and knockout cells were
seeded into 96-well plates (five replicate wells/group), with
2×103 cells/well. MG63 cells were transfected with 40 nM
miR-542-3p mimics (or miR-542-3p NC) and liposomes, and the medium
was changed after 5 h. Subsequently, at 0, 24, 48 and 72 h, CCK-8
solution (10 µl/well) was added to each well. Following incubation
for 1 h at 37°C with 5% CO2, the absorbance of each well
was measured at a wavelength of 450 nm, using a microplate reader
(Bio-Rad Laboratories, Inc.).
Cell apoptosis assay
Annexin V-FITC/propidium iodide (PI) apoptosis assay
reagent (cat. no. C1062L; Beyotime Institute of Biotechnology) was
used to determine the apoptotic rate. Following transfection for 48
h, MG63 cells were transfected with 50 nM miR-542-3p mimics or
miR-542-3p NC and washed twice with PBS at room temperature for 30
sec. The cells (1×105 cells/ml) were centrifuged at
1,000 × g for 5 min at room temperature to discard the supernatant
and resuspended in 195 µl binding buffer (cat. no. C1062L-2;
Beyotime Institute of Biotechnology). Subsequently, 5 µl Annexin
V-FITC and 10 µl PI were added for double staining for 15 min at
room temperature in the dark. Following incubation, centrifuge
tubes containing the stained cells were placed in an ice water
bath, avoiding light. The rate of apoptosis was analyzed using a
FACSCalibur flow cytometer (BD Biosciences). On the flow cytometry
scattergrams, cells in the upper-left quadrant labeled as Annexin
V-FITC−/PI+ were designated as mechanically
damaged cells and necrotic cells; cells in the upper-right quadrant
tagged as Annexin V-FITC+/PI+ were designated
as late-apoptotic cells; cells in the lower-right quadrant labeled
as Annexin V-FITC+/PI− were designated as
early apoptotic cells; and cells in the lower-left quadrant labeled
as Annexin V-FITC−/PI− were designated as
viable cells.
Western blotting
Total protein was extracted from hFOB1.19 and MG63
cells using RIPA lysis buffer (Beijing Solarbio Science &
Technology Co., Ltd.). Protein concentration was quantified using
the BCA protein assay kit (Beijing Solarbio Science &
Technology Co., Ltd.). The extracted proteins (50 µg/lane) were
separated via SDS-PAGE (8%) and transferred onto PVDF membranes
(EMD Millipore), using the wet transfer method. The membranes were
blocked with 5% non-fat milk for 1.5 h at room temperature and then
incubated with primary antibodies against PFKM, pyruvate kinase
M1/2 (PKM2), HK2 (all diluted 1:200; cat nos. sc-166722; sc-365684;
sc-374091, respectively) and β-actin (diluted 1:1,000; cat. no.
sc-8432) (all from Santa Cruz Biotechnology, Inc.), overnight at
4°C. After washing with TBST, membranes were incubated with
secondary antibodies (diluted 1:2,000; cat. no. sc-2031; Santa Cruz
Biotechnology, Inc.) for 2 h at room temperature. Protein bands
were detected and visualized using an enhanced chemiluminescence
reagent kit (Beyotime Institute of Biotechnology). β-actin was used
as the loading control for normalization.
Bioinformatics predictions and
luciferase reporter assay
Potential miRNAs interacting with lncRNA XLOC_005950
were predicted using DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index).
Putative binding sites of hsa-miR-542-3p and PFKM were predicted
using TargetScan (http://www.targetscan.org/vert_71), miRBase
(http://www.mirbase.org) and miRWalk (http://mirwalk.umm.uni-heidelberg.de).
Dual-luciferase reporter assay
Genomic DNA was used as the template, and PCR was
applied to obtain the PFKM 3′-untranslated region (UTR) and lncRNA
XLOC_005950 wild-type (wt) sequences containing hsa-miR-542-3p
binding sites to construct pGL3-promoter-PFKM-wt and
pGL3-promoter-Lnc-wt vectors. pGL3-Promoter vector and pRL-TK
vector were purchased from Promega Corporation. The primer
sequences were as follows: PFKM forward,
5′-GCTCTACCTAATAAGTCCACATCTTCTC-3′ and reverse,
5′-AAAGGCCGGCCCAGACAGCCAGCAAGTAG-3′; and lncRNA XLOC_005950
forward, 5′-GCTCTAGATGTCTCCGCCGGTTGAAA-3′ and reverse,
5′-AAAGGCCGGCCCTGACCAACATGGTGAAGC-3′.
In addition, mutant (mut) segments of lncRNA
XLOC_005950 and PFKM mRNA 3′-UTR were constructed by
overlap-extension PCR, generating pGL3-promoter-Lnc-mut and
pGL3-promoter-PFKM-mut vectors. MG63 cells were seeded into 24-well
plates and co-transfected with plasmids and hsa-miR-542-3p mimics
or NC for 36 h using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 36 h, firefly luciferase and
Renilla luciferase activities were measured on a microplate
luminometer (Centro XS LB960; Titertek-Berthold) using a Dual
Luciferase Reporter Assay kit (Promega Corporation). Firefly
luciferase activity was normalized to Renilla luciferase
activity.
Statistical analysis
Statistical analysis was performed using SPSS 19.0
software (IBM Corp.) and GraphPad Prism 5.0 software (GraphPad
Software, Inc.). All experiments were performed in triplicate and
data are presented as the mean ± standard deviation. χ2
and Fisher's exact tests were used to assess the association
between lncRNA XLOC_005950 expression and the clinicopathological
characteristics of patients with OS. Unpaired Student's t-test was
used to compare differences between two groups, while one-way ANOVA
followed by Tukey's post hoc test were used to compare differences
between multiple groups. Pearson's correlation analysis was
performed to determine the correlations between lncRNA XLOC_005950
and miR-542-3p, and miR-542-3p and PFKM. P<0.05 was considered
to indicate a statistically significant difference.
Results
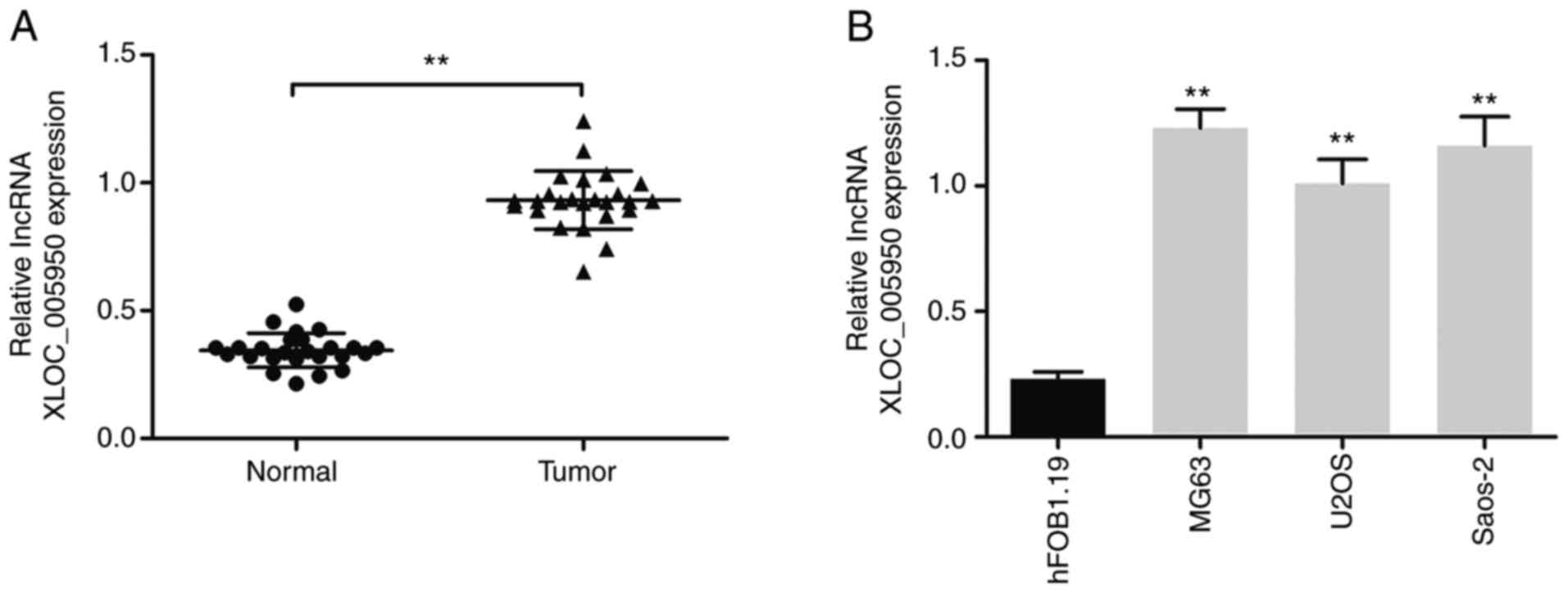
lncRNA XLOC_005950 expression is
upregulated in OS tissues and cell lines
lncRNA XLOC_005950 expression was detected in 25
paired OS tissues and matched adjacent normal tissues via RT-qPCR
analysis. The results demonstrated that lncRNA XLOC_005950
expression was upregulated in OS tissues and cell lines (MG63, U2OS
and Saos-2) compared with adjacent normal tissues and human normal
osteoblasts hFOB1.19, respectively (P<0.01; Fig. 1A and B). lncRNA XLOC_005950
expression was highest in MG63 cells compared with hFOB1.19 cells
(Fig. 1B). Thus, MG63 cells were
selected to construct lncRNA XLOC_005950 knockout cells.
Furthermore, the clinical data indicated that lncRNA XLOC_005950
expression was significantly associated with clinical stage and
metastasis (Table I). Taken
together, these results suggest that lncRNA XLOC_005950 expression
is upregulated in OS tissues and MG63 cells, which may be involved
in the progression of OS.
lncRNA XLOC_005950 knockdown triggers
changes in aerobic glycolysis and inhibits PFKM expression in MG63
cells
To determine the potential biological significance
of lncRNA XLOC_005950 in OS cells, a loss-of-function experiment
was performed in MG63 cells. CRISPR/Cas9 gene editing was used to
knock out lncRNA XLOC_005950 expression in MG63 cells. Sequencing
demonstrated that the genomic DNA fragments of lncRNA XLOC_005950
containing the binding sites of miR-542-3p was knocked out in MG63
cells (Fig. 2A). Given that the
experimental period required for using CRISPR technology to
construct knockout cell lines was relatively long in the present
study, only MG63 knockout cells were constructed to observe the
effect of lncRNA XLOC_005950 knockout on the cell phenotype.
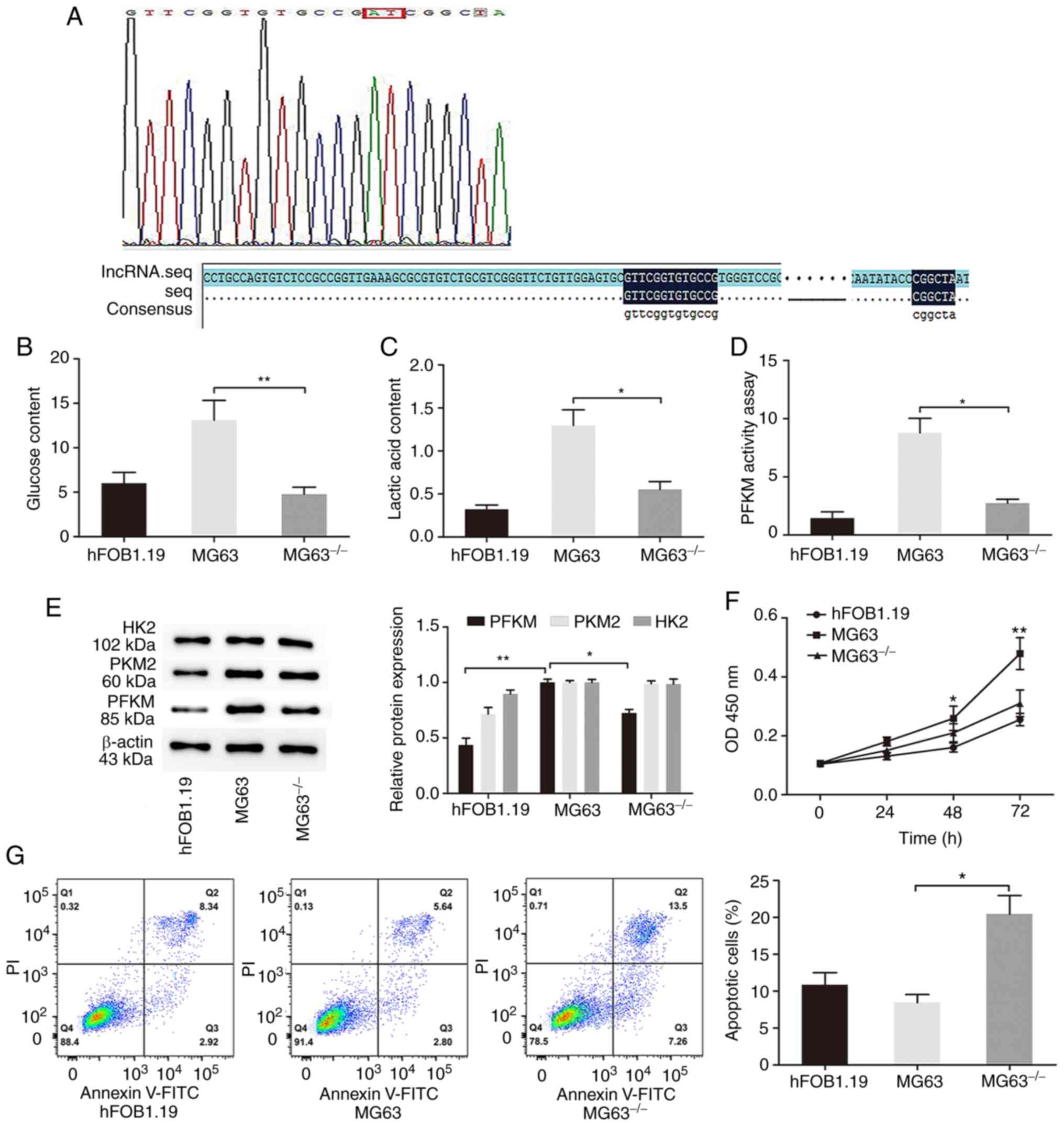 | Figure 2.lncRNA XLOC_005950 knockdown triggers
changes in osteosarcoma cells in terms of glycolysis and inhibits
proliferation while promoting cell apoptosis. (A) Following
CRISPR/Cas9 gene editing to knockout lncRNA XLOC_005950 expression
in MG63 cells, sequencing demonstrated that the specific fragments
of lncRNA XLOC_005950 had been knocked out. (B) Glucose
consumption, (C) lactate production and (D) PFKM activity were
detected in hFOB1.19, MG63 and knockout cells. (E) Western blot
analysis was performed to detect the protein expression levels of
hexokinase 2, PFKM and PKM2 in hFOB1.19, MG63 and knockout cells.
β-actin was used as the internal control for normalization. (F) The
Cell Counting Kit-8 assay was performed to assess cell
proliferation. (G) Flow cytometry was performed to detect the
apoptotic rates of cells. Data are presented as the mean ± standard
deviation. *P<0.05, **P<0.01. MG63−/−, MG63 cells
with lncRNAXLOC_005950 knocked out. lncRNA, long non-coding RNA;
PFKM, phosphofructokinase, muscle; PKM2, pyruvate kinase M1/2; OD,
optical density. |
Subsequently, the changes in aerobic glycolysis in
OS cells following lncRNA XLOC_005950 knockdown were investigated.
The results demonstrated that lncRNA XLOC_005950 knockdown via
transfection with PX459 with sgRNA in MG63 cells markedly decreased
glucose consumption and lactate production, and decreased PFKM
activity (P<0.05 and P<0.01; Fig.
2B-D). Inhibition of lncRNA XLOC_005950 in the
MG63−/− group decreased PFKM rather than HK2 and PKM2
expression (Fig. 2E). Collectively,
these results suggest that knockout of lncRNA XLOC_005950 can
decrease glucose and lactic content, as well as decrease PFKM
activity and protein expression, thus impairing aerobic
glycolysis.
lncRNA XLOC_005950 knockdown inhibits
MG63 cell proliferation and promotes cell apoptosis
Aerobic glycolysis, the main energy supply pattern
of OS cells (24), was partially
inhibited by knocking out lncRNA XLOC_005950 specific fragments.
The present study investigated whether it could affect the
proliferation and apoptotic abilities of OS cells. The results of
the CCK-8 assay demonstrated that the proliferation of the
MG63−/− group markedly decreased compared with that of
the MG63 group (P<0.01; Fig. 2F).
Furthermore, flow cytometry analysis was performed to detect cell
apoptosis. The results demonstrated that the apoptotic rate of the
MG63−/− group markedly increased compared with the MG63
group, and the difference was statistically significant (P<0.05;
Fig. 2G). Taken together, these
results suggest that lncRNA XLOC_005950 knockdown suppresses OS
cell proliferation and induces apoptosis in vitro.
lncRNA XLOC_005950 is a target of
hsa-miR-542-3p
To determine the potential molecular mechanism by
which lncRNA XLOC_005950 is involved in OS progression, the
correlations between lncRNA XLOC_005950 and miRNAs were
investigated. According to the results of DIANA tools, lncRNA
XLOC_005950 contains binding sites for hsa-miR-542-3p (Fig. 3A). RT-qPCR analysis demonstrated that
hsa-miR-542-3p expression was significantly downregulated in OS
tissues compared with adjacent normal tissues (P<0.05; Fig. 3B). Similarly, the data indicated that
hsa-miR-542-3p expression was downregulated in MG63 cells
(P<0.01; Fig. 3C). Notably,
lncRNA XLOC_005950 knockdown upregulated hsa-miR-542-3p expression
(P<0.05; Fig. 3C). In addition,
RT-qPCR analysis revealed a negative correlation between lncRNA
XLOC_005950 and hsa-miR-542-3p expression (r=−0.5318, P<0.01;
Fig. 3D). The dual-luciferase
reporter assay was performed to verify the direct interaction
between lncRNA XLOC_005950 and hsa-miR-542-3p. As presented in
Fig. 3E, overexpression of
hsa-miR-542-3p decreased the fluorescence intensity of the vector
carrying pGL3-promoter-Lnc-wt. Collectively, these results suggest
that recognition sites in hsa-miR-542-3p can directly bind to the
lncRNA XLOC_005950 sequence.
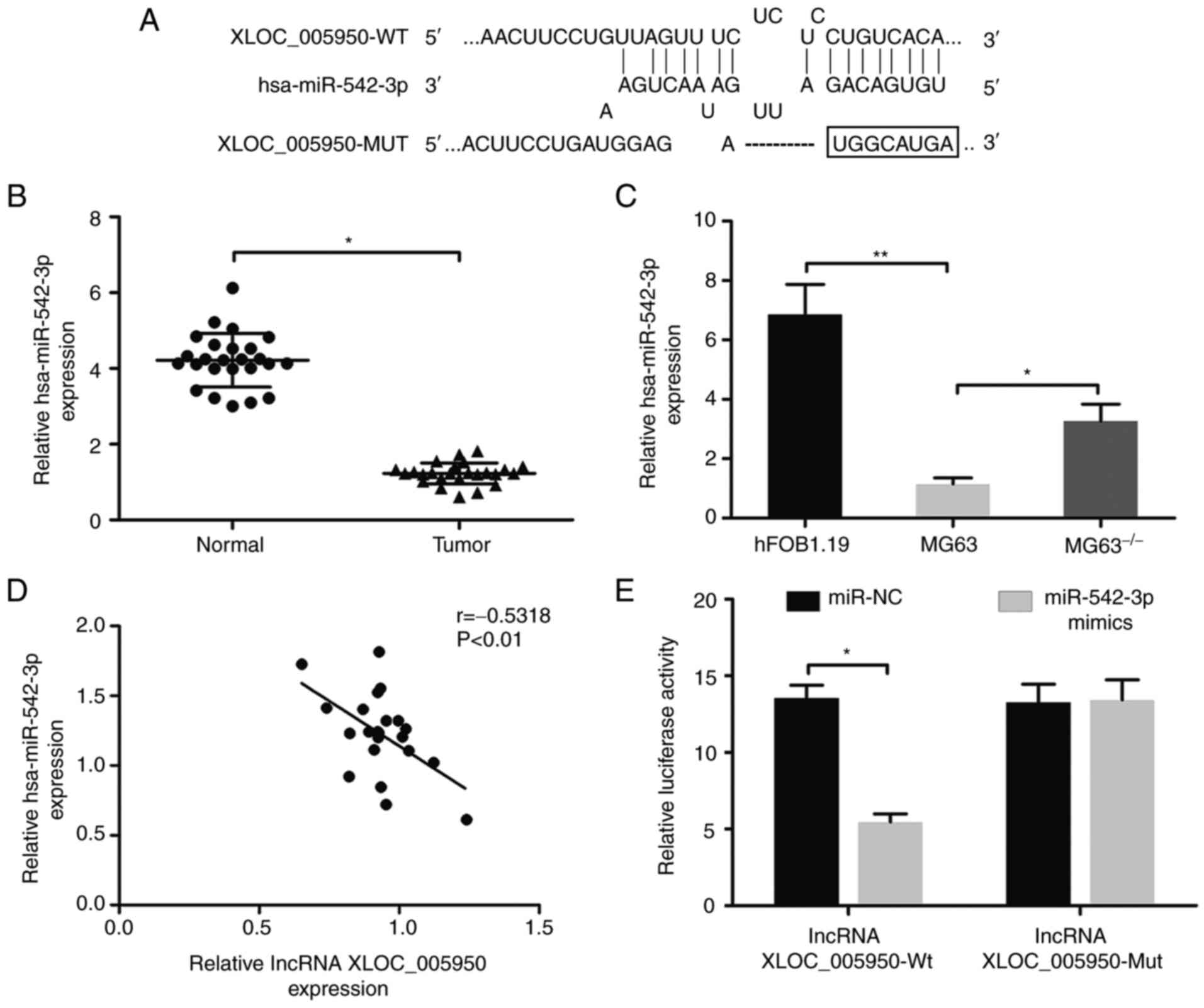 | Figure 3.lncRNA XLOC_005950 is a target of
hsa-miR-542-3p. (A) Binding sites of hsa-miR-542-3p and lncRNA
XLOC_005950. (B) RT-qPCR analysis was performed to detect
hsa-miR-542-3p expression in 25 paired OS tissues and adjacent
normal tissues. (C) RT-qPCR analysis was performed to detect
hsa-miR-542-3p expression in MG63, MG63−/− and human
osteoblast hFOB1.19 cells. (D) Correlation analysis in OS specimens
revealed a negative correlation between lncRNA XLOC_005950 and
hsa-miR-542-3p expression. (E) MG63 cells were co-transfected with
hsa-miR-542-3p mimics and pGL3-promoter-lnc-wt vector or
pGL3-promoter-lnc-mut vector. The relative luciferase activity was
detected via the dual-luciferase reporter assay. Data are presented
as the mean ± standard deviation (n=3). *P<0.05, **P<0.01.
MG63−/−, MG63 cells with lncRNA XLOC_005950 knocked out.
lncRNA, long non-coding RNA; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; OS, osteosarcoma; wt, wild-type;
mut, mutant; NC, negative control. |
hsa-miR-542-3p impairs glycolysis and
suppresses PFKM expression in MG63 cells
The present study investigated the role of
hsa-miR-542-3p in lncRNA XLOC_005950 knockout-induced glycolysis
inhibition of OS cells. The results demonstrated that
hsa-miR-542-3p expression was upregulated following transfection
with hsa-miR-542-3p mimics in MG63 cells (P<0.05; Fig. 4A). Overexpression of hsa-miR-542-3p
markedly decreased glucose and lactate content, as well as PFKM
activity in MG63 cells (P<0.05; Fig.
4D-F).
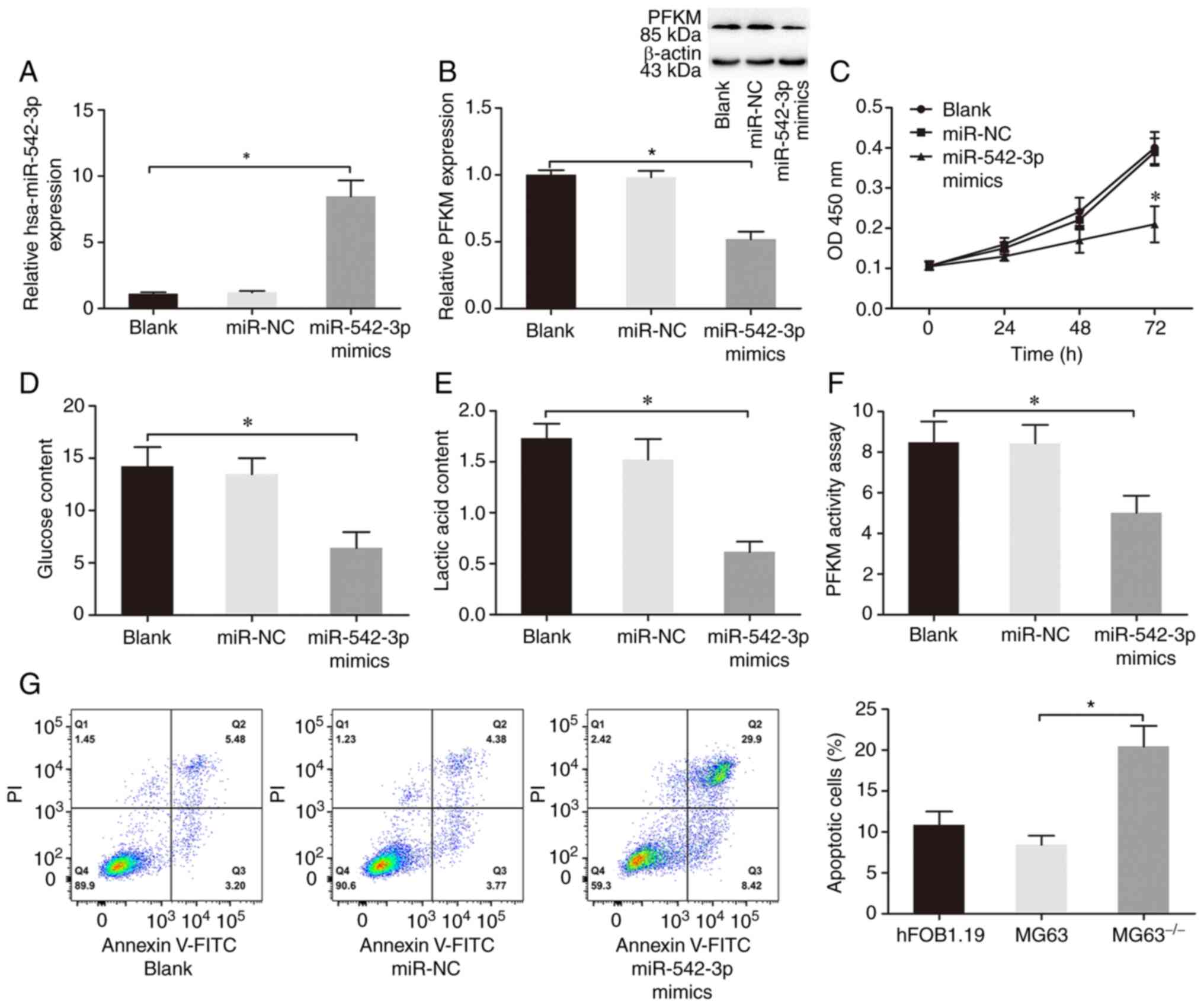 | Figure 4.hsa-miR-542-3p impairs glycolysis,
suppresses proliferation and induces apoptosis in OS cells. (A)
MG63 cells were transfected with miR-NC or hsa-miR-542-3p mimics,
and reverse transcription-quantitative PCR analysis was performed
to detect hsa-miR-542-3p expression. (B) Western blot analysis was
performed to detect PFKM protein expression in MG63 cells
transfected with miR-NC or hsa-miR-542-3p mimics. β-actin was used
as the internal control for normalization. (C) The Cell Counting
Kit-8 assay was performed to assess cell proliferation. (D) Glucose
and (E) lactic acid content, and (F) PFKM activity were determined
in the blank, miR-NC and miR-542-3p groups. (G) Flow cytometric
analysis was performed to detect the apoptotic rates of cells. Data
are presented as the mean ± standard deviation. *P<0.05. Blank,
MG63 cells transfected with Lipofectamine® 2000; miR-NC,
MG63 cells transfected with hsa-miR-542-3p NC and liposomes;
miR-542-3p mimics (hsa-miR-542-3p mimics), MG63 cells transfected
with hsa-miR-542-3p mimics and liposomes. miR, microRNA; OS,
osteosarcoma; NC, negative control; PFKM, phosphofructokinase,
muscle; OD, optical density. |
Western blot analysis was performed to determine the
regulatory mechanism of hsa-miR-542-3p on PFKM expression in OS
cells. The results demonstrated that overexpression of
hsa-miR-542-3p markedly decreased PFKM protein expression
(P<0.05; Fig. 4B). Taken
together, these results suggest that overexpression of
hsa-miR-542-3p impairs glycolysis and suppresses PFKM protein
expression in MG63 cells.
hsa-miR-542-3p suppresses
proliferation and induces apoptosis in MG63 cells
Given that hsa-miR-542-3p inhibited glycolysis in
MG63 cells, the roles of hsa-miR-542-3p in the proliferation and
apoptosis of OS cells were investigated. The results of the CCK-8
assay demonstrated that overexpression of hsa-miR-542-3p
significantly suppressed cell proliferation (P<0.05; Fig. 4C). In addition, overexpression of
hsa-miR-542-3p significantly induced MG63 cell apoptosis compared
with the control and blank groups (P<0.05; Fig. 4G). Collectively, these results
suggest that hsa-miR-542-3p, as a tumor suppressor in OS (18–20), can
arrest cell proliferation and promote apoptosis in MG63 cells.
hsa-miR-542-3p mediates the effect of
lncRNA XLOC_005950 on the metabolism and proliferation of MG63
cells
To determine whether hsa-miR-542-3p plays an
important role in the development of lncRNA XLOC_005950-mediated
OS, a hsa-miR-542-3p inhibitor was transfected into lncRNA
XLOC_005950-knockout MG63 cells. The upregulation of miR-542-3p
mediated by lncRNA XLOC_005950 knockout was reversed following
transfection with miR-542-3p inhibitor (P<0.01; Fig. 5A). In addition, rescue experiments
indicated that the inhibitory effect caused by lncRNA XLOC_005950
knockout on cell glucose and lactate content, as well as on PFKM
activity, were reversed following transfection with miR-542-3p
inhibitor (P<0.05 and P<0.01; Fig.
5B-D). Taken together, these results suggest that lncRNA
XLOC_005950 regulates MG63 cell metabolism and proliferation by
targeting hsa-miR-542-3p.
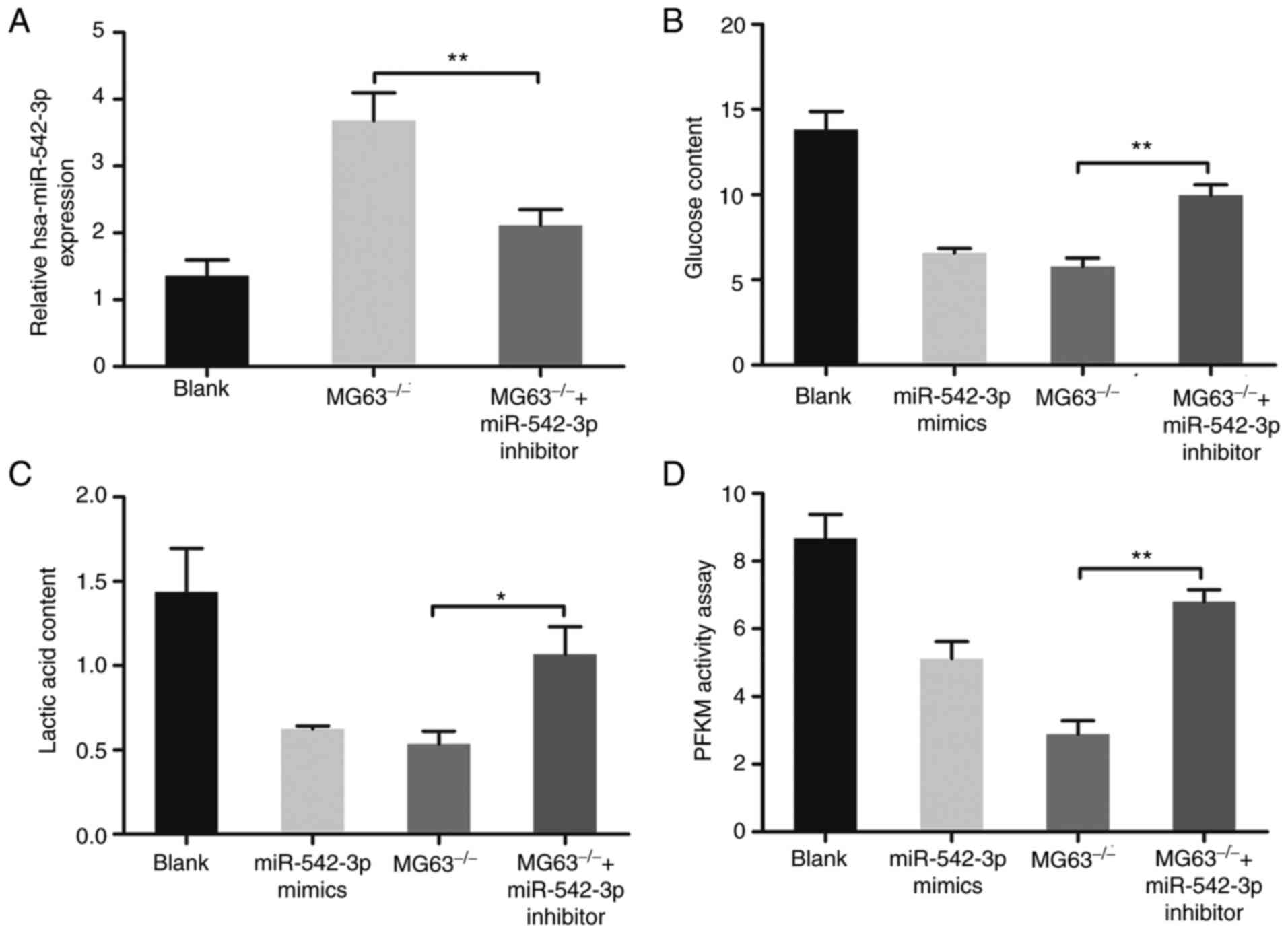 | Figure 5.hsa-miR-542-3p mediates the effect of
lncRNA XLOC_005950 on the metabolism and proliferation of MG63
cells. (A) Reverse transcription-quantitative PCR analysis was
performed to detect miR-542-3p expression in MG63 cells, knockout
MG63 cells and knockout MG63 cells transfected with miR-542-3p
inhibitor. (B) Glucose consumption, (C) lactate production and (D)
PFKM activity were detected in MG63 cells, knockout MG63 cells and
knockout cells transfected with miR-542-3p inhibitor. *P<0.05,
**P<0.01. Blank, MG63 cells transfected with
Lipofectamine® 2000; miR-542-3p mimics (hsa-miR-542-3p
mimics), MG63 cells transfected with hsa-miR-542-3p mimics and
liposomes; MG63−/−, MG63 cells with lncRNAXLOC_005950
knocked out; MG63−/− + miR-542-3p inhibitor, knocked out
MG63 cells transfected with miR-542-3p inhibitor and liposomes.
miR, microRNA; lncRNA, long non-coding RNA; PFKM,
phosphofructokinase, muscle. |
lncRNA XLOC_005950 controls the
hsa-miR-542-3p target, PFKM
To further determine whether hsa-miR-542-3p is
involved in lncRNA XLOC_005950-regulated PFKM expression, its
potential targets were predicted using the TargetScan, miRBase and
miRWalk databases. The results revealed that a hsa-miR-542-3p
binding site existed in the PFKM 3′-UTR (Fig. 6A). The luciferase activity of the
pGL3-promoter-PFKM-wt reporter plasmid significantly decreased in
the presence of hsa-miR-542-3p mimics (Fig. 6B). These results suggest that PFKM is
a downstream target of hsa-miR-542-3p in OS.
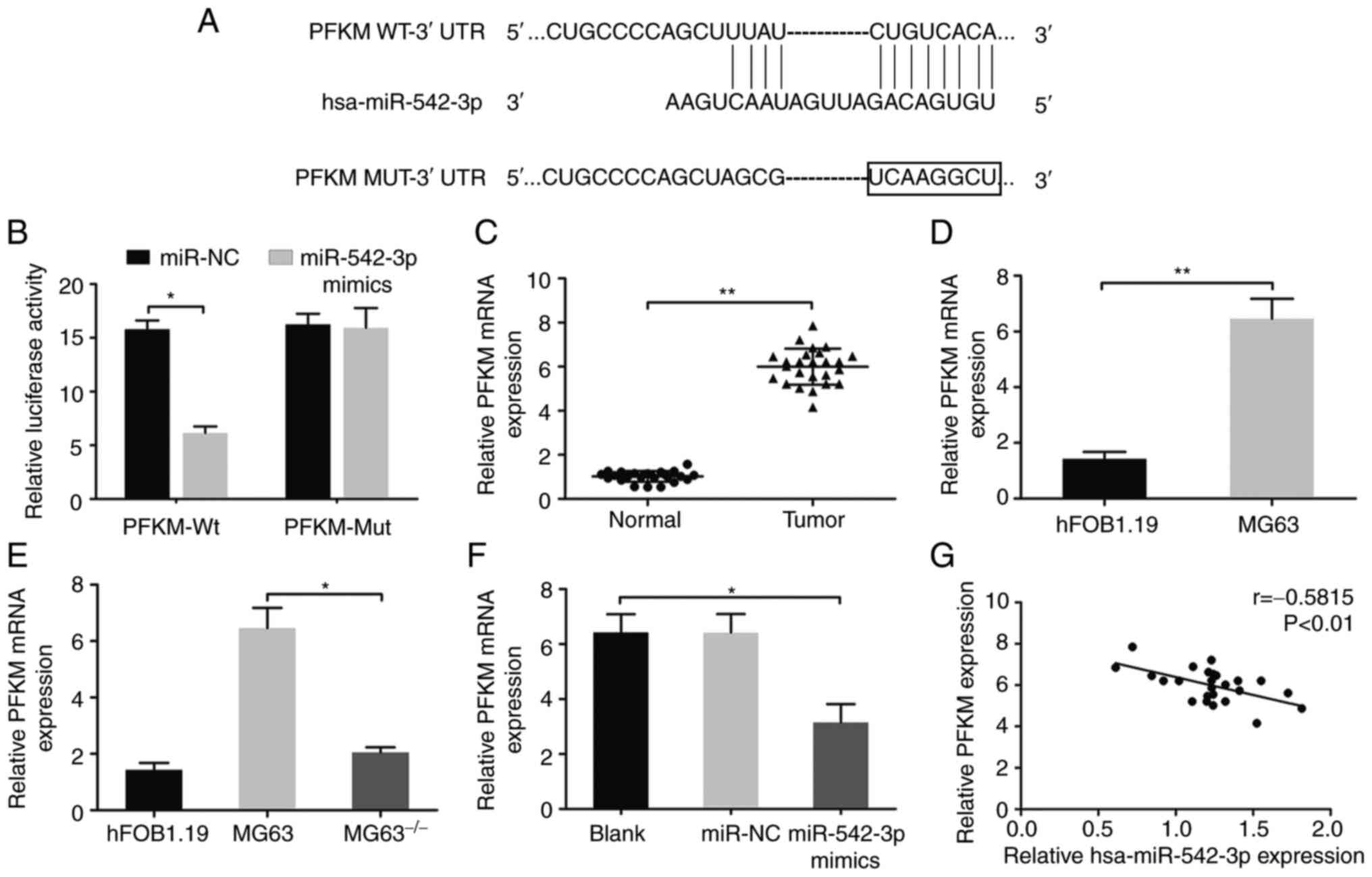 | Figure 6.lncRNA XLOC_005950 regulates the
expression of the hsa-miR-542-3p target, PFKM. (A) A predicted
hsa-miR-542-3p binding site was identified to exist in the PFKM
3′-UTR. (B) MG63 cells were co-transfected with hsa-miR-542-3p
mimics and PFKM 3′-UTR reporter construct. (C) Relative expression
of PFKM in 25 paired OS tissues and adjacent normal tissues. (D)
Relative expression of PFKM in MG63 cells and human osteoblast
hFOB1.19 cells. Reverse transcription-quantitative PCR analysis was
performed to detect PFKM mRNA expression in MG63 cells following
(E) lncRNA XLOC_005950 knockdown or (F) overexpression of
hsa-miR-542-3p. (G) A negative correlation was observed between
hsa-miR-542-3p and PFKM expression in OS specimens. Data are
presented as the mean ± standard deviation (n=3). *P<0.05,
**P<0.01. Blank, MG63 cells transfected with
Lipofectamine® 2000; miR-NC, MG63 cells transfected with
hsa-miR-542-3p NC and liposomes; miR-542-3p mimics (hsa-miR-542-3p
mimics), MG63 cells transfected with hsa-miR-542-3p mimics and
liposomes. MG63−/−, MG63 cells with lncRNA XLOC_005950
knocked out. lncRNA, long non-coding RNA; miR, microRNA; PFKM,
phosphofructokinase, muscle; UTR, untranslated region; OS,
osteosarcoma; NC, negative control; wt, wild-type; mut, mutant. |
RT-qPCR analysis demonstrated that PFKM expression
was upregulated in OS tissues and MG63 cells compared with adjacent
normal tissues and normal osteoblast hFOB1.19 cells, respectively
(P<0.01; Fig. 6C and D).
Furthermore, lncRNA XLOC_005950 knockdown inhibited PFKM protein
expression, and lncRNA XLOC_005950 bound to hsa-miR-542-3p to
inhibit hsa-miR-542-3p expression. To analyze the regulatory
pathway, RT-qPCR analysis was performed, and the results
demonstrated that lncRNA XLOC_005950 knockout or overexpression of
hsa-miR-542-3p significantly decreased PFKM expression in MG63
cells (P<0.05; Fig. 6E and F). In
addition, lncRNA XLOC_005950 and PFKM expression were highly
expressed in OS tissues and cells (Figs.
1A and B, 6C and D), while
hsa-miR-542-3p expression decreased (Fig. 3B and C). Correlation analysis
revealed a negative correlation between hsa-miR-542-3p and PFKM
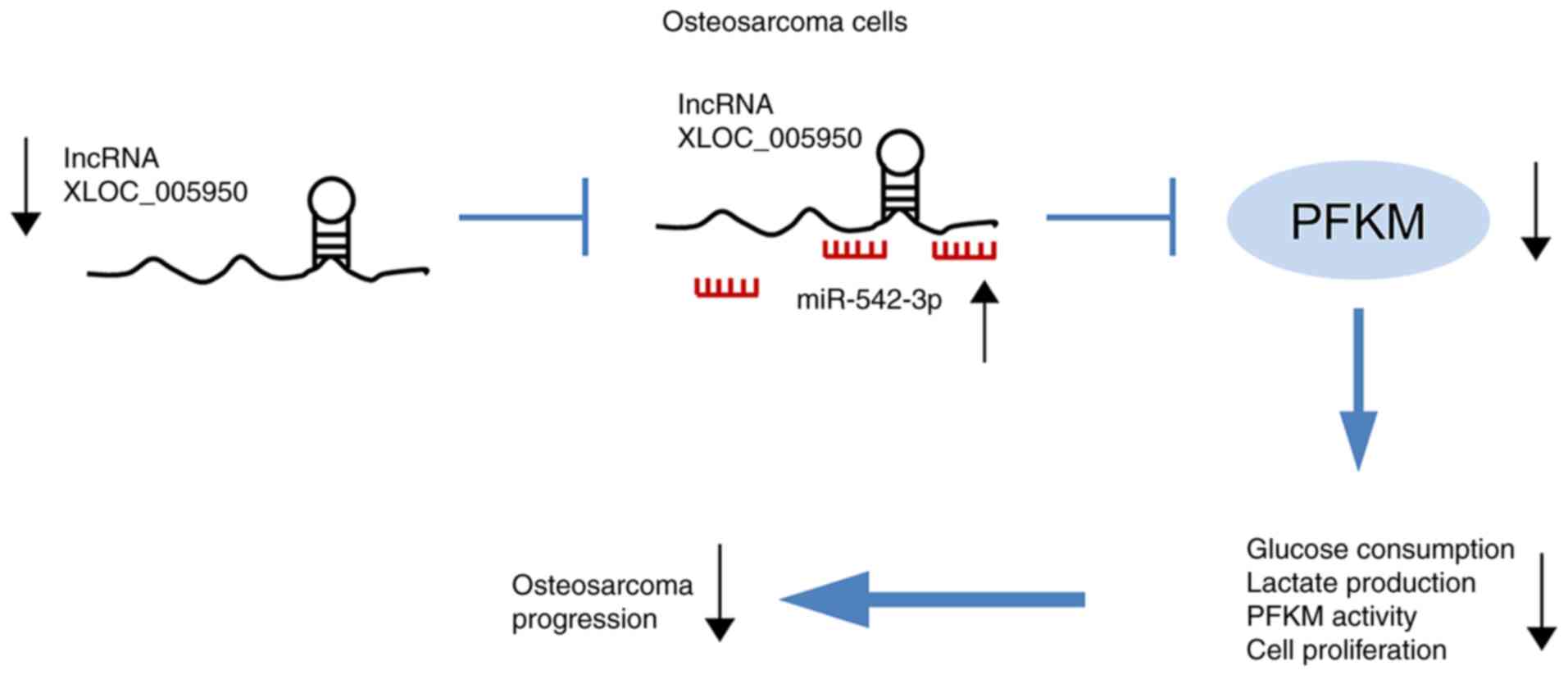
expression in OS tissues (r=−0.5815, P<0.01; Fig. 6G). Notably, lncRNA XLOC_005950
knockdown increased hsa-miR-542-3p expression and suppressed PFKM
expression, thus preventing glycolysis in OS cells, inhibiting the
main energy supply of tumor cells and inhibiting the proliferation
of OS cells (Fig. 7). Collectively,
these results suggest that lncRNA XLOC_005950 regulates PFKM
expression via hsa-miR-542-3p, affecting the glycolytic pathway of
MG63 cells, and regulating cell proliferation and apoptosis.
Discussion
Previous studies have reported a variety of complex
roles for lncRNAs in the regulation of gene expression, involving
several cellular processes (15,17,34).
Aberrant expression of lncRNA is associated with the occurrence and
development of several diseases, including cancer (35,36).
Zheng and Min (37) demonstrated
that HOX transcript antisense RNA (HOTAIR) is highly expressed in
OS cell lines. HOTAIR knockdown (using small interfering RNA)
decreased the proliferation rate of OS cells and promoted cell
apoptosis. In a previous study, lncRNA XLOC_005950 was reported in
gene chip analysis by Cabili et al (14). In the preliminary experiment of this
research, based on analysis results by Cabili et al, RT-qPCR
was performed to detect the expression of multiple lncRNAs in
hFOB1.19 and MG63 cells. lncRNA XLOC_005950 was the highest
expression in MG63 cells compared with normal osteoblast hFOB1.19,
and was selected as the research target. The present study used
CRISPR/Cas9-based genome editing system to analyze the effects of
lncRNA XLOC_005950 deletion in MG63 cells. The results demonstrated
that the efficiency of glycolysis metabolism of knockout cells
decreased, cell proliferation was inhibited and the apoptosis rate
increased. These results demonstrated that CRISPR/Cas9-mediated
knockout of lncRNA inhibited the metabolism and proliferation of
MG63 cells in vitro. Furthermore, increased lncRNA
XLOC_005950 expression promoted malignant proliferation of OS
cells, and clinicopathological analyses identified that patients
with OS with high lncRNA XLOC_005950 expression exhibited a higher
clinical stage compared with those with low expression. lncRNA
XLOC_005950 may serve as an oncogene in OS progression, which may
provide a new target for diagnosis and treatment of OS.
Several studies have suggested that miRNAs may exert
functions by targeting mRNAs, and negatively regulating gene
expression (38–40). miRNAs are abnormally expressed in OS.
For example, miR-381 expression (41) is upregulated in OS, while the
expression levels of miR-126 (42)
and miR-143 (43) are downregulated.
Zhong et al (44) reported
that lncRNA small nucleolar RNA host gene 8 promotes the
proliferation of OS cells by downregulating miR-542-3p expression.
Based on bioinformatics analysis and the dual-luciferase reporter
assay, hsa-miR-542-3p has a specific binding site for lncRNA
XLOC_005950, and PFKM is a target gene of hsa-miR-542-3p. The
present study demonstrated that hsa-miR-542-3p expression markedly
decreased in OS cells and tissues. Overexpression of hsa-miR-542-3p
inhibited the biological processes of MG63 cells, which was
consistent with the effect of knocking out lncRNA XLOC_005950 in
MG63 cells. Thus, lncRNA XLOC_005950 may act as a molecular sponge
to repress the function of hsa-miR-542-3p.
Glycolysis provides energy for the rapid
proliferation of tumor cells. It has been reported that metabolic
reprogramming promotes tumor cell survival and tumor development,
suggesting that tumor cells may depend on specific metabolic
pathways (45). Zancan et al
(46) reported that PFKM and PFKL
are highly expressed in breast cancer cells. Ahsan et al
(47) identified that PFKM gene
region 12q13.11 as a breast cancer susceptibility locus, is
associated with breast cancer risk. PFK activity is associated with
changes in intracellular metabolism (27,29). The
results of the present study demonstrated that PFKM was highly
expressed in OS cells and tissues. After gene editing aimed to
knockout lncRNA XLOC_005950 in MG63 cells, or after hsa-miR-542-3p
mimics transfection, the activity and expression of PFKM decreased,
and lactic acid and glucose content also decreased, which
subsequently decreased cell proliferation compared with that of
untreated MG63 cells. These results suggest that lncRNA XLOC_005950
may promote tumorigenesis and tumor development in OS, which may be
caused by enhanced expression of PFKM and promotion of the aerobic
glycolysis pathway in OS cells.
In conclusion, the results of the present study
demonstrated that the loss of lncRNA XLOC_005950 or overexpression
of hsa-miR-542-3p attenuated PFKM expression, PFKM activity,
glucose and lactic content and cell proliferation by modulating the
lncRNA XLOC_005950/hsa-miR-542-3p/PFKM axis, which potentially
reveals the underlying molecular mechanism of OS. However, the
present study is not without limitations. Prospective studies are
required to validate these results in more cell lines. In addition,
whether lncRNA XLOC_005950 can target other miRNAs, and whether
other factors are involved in the regulatory network to influence
OS progression require investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Henan Science
and Technology Research Project (grant no. 182102310340).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
YL, YDW and ZJ conceived and designed the present
study. YZ and YSW and YDW collected the samples. ZJ, YDW, XS, XZ
and SX performed the experiments. ZJ, YDW, XS, and YZ analyzed the
data. ZJ drafted the initial manuscript. YDW, ZJ and YL confirmed
the authenticity of all the raw data. All authors have read and
approved the final manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Zhengzhou University (Zhengzhou, China; approval no.
2015-03). Written informed consent was provided by all patients
prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Klein MJ and Siegal GP: Osteosarcoma:
Anatomic and histologic variants. Am J Clin Pathol. 125:555–581.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tian Z, Yang G, Jiang P, Zhang L, Wang J
and Sun S: Long non-coding RNA Sox4 promotes proliferation and
migration by activating Wnt/beta-catenin signaling pathway in
osteosarcoma. Pharmazie. 72:537–542. 2017.PubMed/NCBI
|
|
3
|
Min X, Heng H, Yu HL, Dan M, Jie C, Zeng
Y, Ning H, Liu ZG, Wang ZY and Lin W: Anticancer effects of
10-hydroxycamptothecin induce apoptosis of human osteosarcoma
through activating caspase-3, p53 and cytochrome c pathways. Oncol
Lett. 15:2459–2464. 2018.PubMed/NCBI
|
|
4
|
Lei Z, Duan H, Zhao T, Zhang Y, Li G, Meng
J, Zhang S and Yan W: PARK2 inhibits osteosarcoma cell growth
through the JAK2/STAT3/VEGF signaling pathway. Cell Death Dis.
9:3752018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miao J, Wang W, Wu S, Zang X, Li Y, Wang
J, Zhan R, Gao M, Hu M, Li J and Chen S: MiR-194 suppresses
proliferation and migration and promotes apoptosis of osteosarcoma
cells by targeting CDH2. Cell Physiol Biochem. 45:1966–1974. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu G, Du L, Guo Y, Xing B, Lu J and Wei Y:
Expression and role of microRNA-1271 in the pathogenesis of
osteosarcoma. Exp Ther Med. 15:1934–1940. 2018.PubMed/NCBI
|
|
7
|
Wang H, He H, Meng H, Cui Y and Wang W:
Effects of Grb2-associated binding protein 2-specific siRNA on the
migration and invasion of MG-63 osteosarcoma cells. Oncol Lett.
15:926–930. 2018.PubMed/NCBI
|
|
8
|
Liu DD and Kang Y: Ets2 anchors the
prometastatic function of mutant p53 in osteosarcoma. Genes Dev.
31:1823–1824. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Klattenhoff C, Bratu DP, McGinnis-Schultz
N, Koppetsch BS, Cook HA and Theurkauf WE: Drosophila
rasiRNA pathway mutations disrupt embryonic axis specification
through activation of an ATR/Chk2 DNA damage response. Dev Cell.
12:45–55. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun Y, Hu B, Wang Q, Ye M, Qiu Q, Zhou Y,
Zeng F, Zhang X, Guo Y and Guo L: Long non-coding RNA HOTTIP
promotes BCL-2 expression and induces chemoresistance in small cell
lung cancer by sponging miR-216a. Cell Death Dis. 9:852018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han Y, Wu N, Jiang M, Chu Y, Wang Z, Liu
H, Cao J, Liu H, Xu B and Xie X: Long non-coding RNA MYOSLID
functions as a competing endogenous RNA to regulate MCL-1
expression by sponging miR-29c-3p in gastric cancer. Cell Prolif.
52:e126782019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guan H, Shang G, Cui Y, Liu J, Sun X, Cao
W, Wang Y and Li Y: Long noncoding RNA APTR contributes to
osteosarcoma progression through repression of miR-132-3p and
upregulation of yes-associated protein 1. J Cell Physiol.
234:8998–9007. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lv L, Li T, Li X, Xu C, Liu Q, Jiang H, Li
Y, Liu Y, Yan H, Huang Q, et al: The lncRNA Plscr4 controls cardiac
hypertrophy by regulating miR-214. Mol Ther Nucleic Acids.
10:387–397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang BF, Cai W and Chen B: LncRNA SNHG12
regulated the proliferation of gastric carcinoma cell BGC-823 by
targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci.
22:1297–1306. 2018.PubMed/NCBI
|
|
18
|
Yuan L, Yuan P, Yuan H, Wang Z, Run Z,
Chen G, Zhao P and Xu B: MiR-542-3p inhibits colorectal cancer cell
proliferation, migration and invasion by targeting OTUB1. Am J
Cancer Res. 7:159–172. 2017.PubMed/NCBI
|
|
19
|
Liu B, Li J, Zheng M, Ge J, Li J and Yu P:
MiR-542-3p exerts tumor suppressive functions in non-small cell
lung cancer cells by upregulating FTSJ2. Life Sci. 188:87–95. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang T, Liu W, Meng W, Zhao H, Yang Q, Gu
SJ, Xiao CC, Jia CC and Fu BS: Downregulation of miR-542-3p
promotes cancer metastasis through activating TGF-β/Smad signaling
in hepatocellular carcinoma. Onco Targets Ther. 11:1929–1939. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo XF, Wu XJ, Wei X, Wang AG, Wang SH and
Wang JL: LncRNA ADPGK-AS1 regulated cell proliferation, invasion,
migration and apoptosis via targeting miR-542-3p in osteosarcoma.
Eur Rev Med Pharmacol Sci. 23:8751–8760. 2019.PubMed/NCBI
|
|
22
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, He ZC, Liu Q, Zhou K, Shi Y, Yao XH,
Zhang X, Kung HF, Ping YF and Bian XW: Large intergenic Non-coding
RNA-RoR inhibits aerobic glycolysis of glioblastoma cells via Akt
pathway. J Cancer. 9:880–889. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu L, Chen X, Sun X, Wang L and Chen S:
The glycolytic switch in tumors: How many players are involved? J
Cancer. 8:3430–3440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Houddane A, Bultot L, Novellasdemunt L,
Johanns M, Gueuning MA, Vertommen D, Coulie PG, Bartrons R, Hue L
and Rider MH: Role of Akt/PKB and PFKFB isoenzymes in the control
of glycolysis, cell proliferation and protein synthesis in
mitogen-stimulated thymocytes. Cell Signal. 34:23–37. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan S, Wei X, Xu S, Sun H, Wang W, Liu L,
Jiang X, Zhang Y and Che Y:
6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform 3
spatially mediates autophagy through the AMPK signaling pathway.
Oncotarget. 8:80909–80922. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu W, Ye L, Zhang J, Yu P, Wang H, Ye Z
and Tian J: PFK15, a small molecule inhibitor of PFKFB3, induces
cell cycle arrest, apoptosis and inhibits invasion in gastric
cancer. PLoS One. 11:e01637682016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi L, Pan H, Liu Z, Xie J and Han W:
Roles of PFKFB3 in cancer. Signal Transduct Target Ther.
2:170442017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hackett EE, Charles-Messance H, O'Leary
SM, Gleeson LE, Muñoz-Wolf N, Case S, Wedderburn A, Johnston DGW,
Williams MA, Smyth A, et al: Mycobacterium tuberculosis limits host
glycolysis and IL-1β by restriction of PFK-M via MicroRNA-21. Cell
Rep. 30:124–136.e4. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y,
Xue C, Ren F, Ren Z, Li J, et al: Long non-coding RNA PVT1 promotes
tumor progression by regulating the miR-143/HK2 axis in gallbladder
cancer. Mol Cancer. 18:332019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song J, Wu X, Liu F, Li M, Sun Y, Wang Y,
Wang C, Zhu K, Jia X, Wang B and Ma X: Long non-coding RNA PVT1
promotes glycolysis and tumor progression by regulating miR-497/HK2
axis in osteosarcoma. Biochem Biophys Res Commun. 490:217–224.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen F, Mo J and Zhang L: Long noncoding
RNA BCAR4 promotes osteosarcoma progression through activating
GLI2-dependent gene transcription. Tumour Biol. 37:13403–13412.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xia WK, Lin QF, Shen D, Liu ZL, Su J and
Mao WD: Clinical implication of long noncoding RNA 91H expression
profile in osteosarcoma patients. Onco Targets Ther. 9:4645–4652.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng H and Min J: Role of long noncoding
RNA HOTAIR in the growth and apoptosis of osteosarcoma cell MG-63.
Biomed Res Int. 2016:57576412016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pillai RS: MicroRNA function: Multiple
mechanisms for a tiny RNA? RNA. 11:1753–1761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang L, Liu ZM, Rao YW, Cui SQ, Wang H and
Jia XJ: Downregulation of microRNA-586 inhibits proliferation,
invasion and metastasis and promotes apoptosis in human
osteosarcoma U2-OS cell line. Cytogenet Genome Res. 146:268–278.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen L, Wang Q, Wang GD, Wang HS, Huang Y,
Liu XM and Cai XH: MiR-16 inhibits cell proliferation by targeting
IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS
Lett. 587:1366–1372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y, Zhao C, Yu Z, Chen J, She X, Li P,
Liu C, Zhang Y, Feng J, Fu H, et al: Low expression of miR-381 is a
favorite prognosis factor and enhances the chemosensitivity of
osteosarcoma. Oncotarget. 7:68585–68596. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiang R, Zhang C, Liu G, Gu R and Wu H:
MicroRNA-126 inhibits proliferation, migration, invasion, and EMT
in osteosarcoma by targeting ZEB1. J Cell Biochem. 118:3765–3774.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu H, Wang H, Liu H and Chen Y: Effect of
miR-143 on the apoptosis of osteosarcoma cells. Int J Clin Exp
Pathol. 8:14241–14246. 2015.PubMed/NCBI
|
|
44
|
Zhong GB, Jiang CQ, Yu XS, Liu ZD, Wang WL
and Xu RD: Long noncoding RNA SNHG8 promotes the proliferation of
osteosarcoma cells by downregulating miR-542-3p. J Biol Regul
Homeost Agents. 34:517–524. 2020.PubMed/NCBI
|
|
45
|
Clementino M, Shi X and Zhang Z: Oxidative
stress and metabolic reprogramming in Cr(VI) carcinogenesis. Curr
Opin Toxicol. 8:20–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zancan P, Sola-Penna M, Furtado CM and Da
Silva D: Differential expression of phosphofructokinase-1 isoforms
correlates with the glycolytic efficiency of breast cancer cells.
Mol Genet Metab. 100:372–378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ahsan H, Halpern J, Kibriya MG, Pierce BL,
Tong L, Gamazon E, McGuire V, Felberg A, Shi J, Jasmine F, et al: A
genome-wide association study of early-onset breast cancer
identifies PFKM as a novel breast cancer gene and supports a common
genetic spectrum for breast cancer at any age. Cancer Epidemiol
Biomarkers Prev. 23:658–669. 2014. View Article : Google Scholar : PubMed/NCBI
|