Introduction
Sulforaphane is an isothiocyanate compound derived
from cruciferous vegetables. Previous studies have shown that
sulforaphane exhibits inhibitory effects on the progression of
promyelocytic leukemia, skin, bladder, prostate, colon, pancreatic,
liver, lung, nasopharyngeal, ovarian, breast and cervical cancer
(1–12). Sulforaphane may inhibit the
proliferation and malignant transformation of cancer cells by
producing reactive oxygen species, inhibiting cytochrome P450 3A4
and phase-I metabolism enzymes, inhibiting G1 to S-phase
progression and G2/M phase arrest, as well as activating
the intrinsic and extrinsic apoptotic pathways (13–17).
Moreover, sulforaphane can achieve efficient glutathione depletion
to improve the accumulation of cisplatin in cancer cells (18).
Sulforaphene exhibits a similar structure to
sulforaphane with the exception of one carbon-carbon double bond.
Both molecules are isothiocyanates derived from cruciferous
vegetables (Table I). This compound
inhibits the activity of phase-I enzymes, such as cytochrome P450
enzymes. It also promotes the production of reactive oxygen
species, regulates the cell cycle and serves as a robust anticancer
component derived from various vegetables (15,19,20).
Sulforaphene exerts anticancer effects on lung, esophageal, colon,
gastric, liver, breast, cervical and thyroid cancer by inducing
apoptosis and blocking the cell cycle (19–26).
Moreover, sulforaphane and sulforaphene possess similar structures,
anticancer mechanisms and inhibitory effects (promoting cancer cell
apoptosis and inhibiting the cell cycle) on colon cancer
progression (15–17,20).
However, their underlying mechanisms of action are different. In
addition, the two compounds are effective on different types of
cancer and it is unknown whether they can complement each other or
act synergistically. Therefore, the aim of the present study was to
assess the effects of sulforaphane and sulforaphene on colon cancer
cells and examine on the differences in gene regulation mediated by
these two compounds.
 | Table I.Differences between sulforaphane and
sulforaphene. |
Table I.
Differences between sulforaphane and
sulforaphene.
| Property | Sulforaphane | Sulforaphene |
|---|
| Molecular
formula |
C6H11NOS2 |
C6H9NOS2 |
| Structural
formula |  |  |
| Chemical name |
1-isothiocyanato-4-[(S)-methylsulfinyl]
butane |
(E)-4-isothiocyanato-1-methylsulfinylbut-1-ene |
| Molecular mass,
g/mol | 177.3 | 175.264 |
| SMILES |
C[S@](=O)CCCCN=C=S |
CS(=O)C=CCCN=C=S |
| CAS Number | 155320-20-0 | 592-95-0 |
| Purity, % | >98 | >98 |
| Form/State | Solid | Solid |
| Solubility | Soluble in water,
in ethanol and in DMSO | Soluble in
water |
| XLogP3 | 1.4 | 1.5 |
| Number of hydrogen
bond donors | 0 | 0 |
| Number of hydrogen
bond acceptors | 4 | 4 |
| Number of rotatable
bonds | 5 | 4 |
| Source | Brassica
oleracea | Brassica
oleracea |
| Biological
characterization | Non-competitive
antagonist of the aryl hydrocarbon receptor, natural
isothiocyanate, antitumor agent, active in vivo and in
vitro (https://www.abcam.cn/s-sulforaphane-ah-receptor-antagonist-ab141970.html) | Carcinogenesis
inhibitor, anticancer, antidiabetic and antioxidant effects, active
in vivo and in vitro (https://www.abcam.cn/s-sulforaphene-carcinogenesis-inhibitor-ab141972.html) |
| Type of anti-cancer
effect | Promyelocytic
leukemia (1), skin cancer (2), bladder cancer (3), prostate cancer (4), colon cancer (5), pancreatic cancer (6), liver cancer (7), lung cancer (8), nasopharyngeal cancer (9), ovarian cancer (10), breast cancer (11), cervical cancer (12) | Lung cancer
(21), esophageal cancer (20), colon cancer (22), gastric cancer (23), liver cancer (24), breast cancer (19), cervical cancer (25), thyroid cancer (26) |
Materials and methods
Materials
The human colorectal adenocarcinoma cell line,
SW480, was purchased from the American Type Culture Collection
(CCL-228™) and cultured in DMEM/high glucose medium (HyClone;
Cytiva; cat. no. SH30022.01B) containing 10% fetal bovine serum
(HyClone; Cytiva; cat. no. SH30087.01) at 37°C in a 5%
CO2 incubator. Sulforaphane (Abcam; cat. no. ab141970)
and sulforaphene (Abcam; cat. no. ab141972) were purchased from
Abcam (purity >98%), solubilized in double-distilled water and
diluted to 25 µmol/l in culture medium. The RNA TRIzol®
extraction kit was purchased from Invitrogen, Thermo Fisher
Scientific, Inc.. The GN-genechip Clariom™ S Array next-generation
sequencing (NGS) chip was purchased from Affymetrix, Thermo Fisher
Scientific, Inc. (cat. no. 902927; human).
Comparison of physical and chemical
properties
Based on the reagents sulforaphane and sulforaphene
presented on the supplier's website (sulforaphane: https://www.abcam.cn/s-sulforaphane-ah-receptor-antagonist-ab141970.html;
sulforaphene: http://www.abcam.cn/s-sulforaphene-carcinogenesis-inhibitor-ab141972.html),
the physicochemical properties (e.g. molecular structure, chemical
name, molecular mass, form, purity and solubility) of the two
compounds were compared and compiled. Literature surrounding their
biological activity was reviewed in order to evaluate the different
effects of the two compounds on cancer, from a physicochemical
perspective.
Cell treatment
SW480 colon cancer cells were maintained in
DMEM/high glucose medium containing 10% fetal bovine serum, at 37°C
in a 5% CO2 incubator, and were cultured to >90%
confluence. A total of three groups were used, namely sulforaphane,
sulforaphene and blank control. The cells were treated at the
logarithmic growth phase, and each group was plated in
triplicate.
Measurement of cell viability
IC50 values were determined by assessing
cell viability at 48 h, using the MTT method (Fig. S1). Drug treatment was performed
using 25 µmol/l sulforaphane or sulforaphene. An equivalent volume
(10 µl in a 100 µl culture system) of double-distilled water was
added to the blank control group. Formazan was dissolved in
dimethyl sulfoxide, and the OD value was measured at 490 nm.
NGS
Following 48 h of incubation, total RNA was
extracted using TRIzol® reagent (Thermo Fisher
Scientific). The quality of the RNA was assessed using a Nanodrop™
2000 spectrophotometer (Thermo Fisher Scientific, Inc.) and an
Agilent 2100 Bioanalyzer (Agilent Technologies Inc.). The samples
met the following conditions prior to NGS: i) A260/A280 ratio,
1.7–2.2; ii) RNA integrity number ≥7.0; and iii) 28S/18S ratio
>0.7. Biotinylated cRNAs were prepared from 6 µg total RNA
according to the standard Affymetrix protocol (Expression Analysis
Technical Manual, 2001, Affymetrix) (27). Following fragmentation, 10 µg of cRNA
were hybridized for 16 h at 45°C on GN-GeneChip Clariom S Array,
human (Thermo Fisher Scientific; cat. no. 902926). GeneChips were
washed and stained in the GeneChip Fluidics Station 450 (Thermo
Fisher Scientific, Inc.) and scanned using the GeneChip Scanner
3000 (Thermo Fisher Scientific, Inc.). Paired-end sequencing was
performed, resulting in short sequence reads of ~150 bp.
Data analysis
The data were analyzed with Microarray Suite version
5.0 (MAS 5.0) using Affymetrix default analysis settings and global
scaling as the normalization method. The trimmed mean target
intensity of each array was arbitrarily set to 100. Pearson's
correlation and principal component analysis were performed on the
NGS data to determine the homogeneity of the three replicate
samples in the group. Data passed the quality control threshold if
the correlation coefficient was >0.8 with ‘cor()’ and
‘princomp()’. The limma package (version 3.40.2; http://www.bioconductor.org/packages/release/bioc/html/limma.html)
was used to analyze differential gene expression between the
groups, and differentially expressed genes were defined as
fold-change ≥2.0, false discovery rate <0.05, P<0.05 and
adjusted P<0.05. Based on these results, differential gene and
cluster analyses were performed and the data were evaluated by Gene
Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analysis using the DAVID 6.8 database
(https://david.ncifcrf.gov/). The
RIdeogram package was used to map the location of differentially
expressed genes across the chromosomes (https://cran.r-project.org/web/packages/RIdeogram/index.html).
All statistical analyses were carried out using R software version
3.5.1 (https://www.r-project.org/).
Results
Comparison of the differences in the
physicochemical properties of sulforaphane and sulforaphene
Sulforaphane and sulforaphene are both single-chain
compounds derived from cruciferous vegetables and share the same
chiral center, resulting in the formation of specific isomers
(15). Interestingly, sulforaphane
[Chemical Abstracts Service (CAS) no. 155320-20-0] and sulforaphene
(CAS no. 592-95-0) are distinguished by the presence of only one
carbon-carbon double bond. The physicochemical properties of the
two compounds with regard to their molecular structure, chemical
name, molecular mass, form, purity and solubility were evaluated in
order to identify the specific differences between them (Table I). Notably, sulforaphane and
sulforaphene underwent specific changes in soluble solvents,
resulting in a 2 g/mol difference in molecular mass. Based on the
XLogP3 values, sulforaphene exhibited a higher lipid-water
partition coefficient. Its solubility in organic solvents has not
been previously examined, as opposed to sulforaphane, which
exhibits a slightly lower XLogP3 value and is soluble in ethanol
and DMSO. In addition, the carbon-carbon double bonds resulted in a
reduced number of rotatable bonds in sulforaphene.
RNA and chip assay quality
control
The viability of the cells was measured after 48 h
of treatment with the two compounds, at a concentration of 25
µmol/l, and the results showed that both reached effective
inhibitory concentrations (Fig.
S1). The quality of the total RNA extracted using the
TRIzol® method was assessed in each sample (Table SI). The signal intensity of the
sequencing chip probes for each sample was tested and analyzed
using Pearson's correlation and principal component analyses. The
results indicated that the correlation coefficient for each sample
pair was >0.85 in three treatment groups (Fig. S2). In the principal component 1 and
principal component 2 dimensions, the aggregation and separation
trends of the samples were distinct in the three groups (Fig. S3).
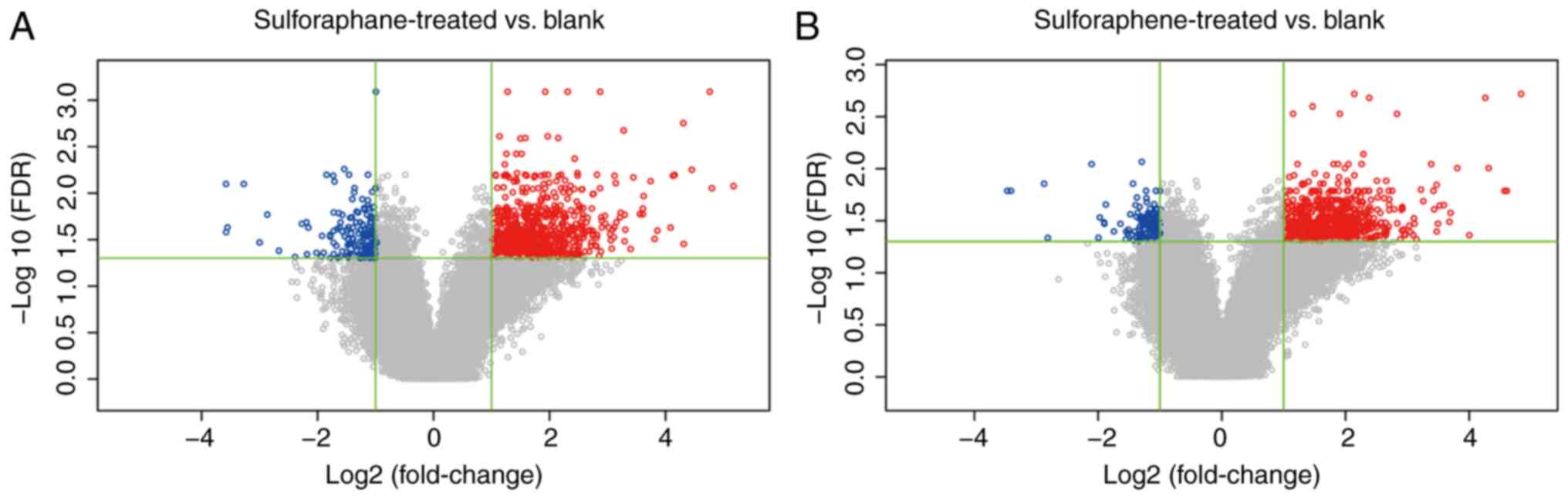
Differentially expressed gene analysis
of the sulforaphene and sulforaphane groups
A total of 873 probes were differentially expressed
in the sulforaphene group compared with the blank control group and
862 genes were obtained, of which 130 were downregulated and 732
were upregulated in the sulforaphene group. A total of 959 probes
in the sulforaphane group were differentially expressed compared
with the blank control group, and 949 genes were obtained. Among
these, 161 genes were downregulated and 788 genes were upregulated
in the sulforaphane group (Fig. 1).
In addition, differential gene expression analysis was performed,
and no genes were differentially expressed between the three
groups.
Gene position distribution and cluster
analysis of differential expression between the two groups
The genetic loci of the genes which were
differentially expressed in the sulforaphene group were identified.
The distribution position and number of differentially expressed
genes on the 22 pairs of autosomes and the X and Y sex chromosomes
did not markedly differ (Fig.
2).
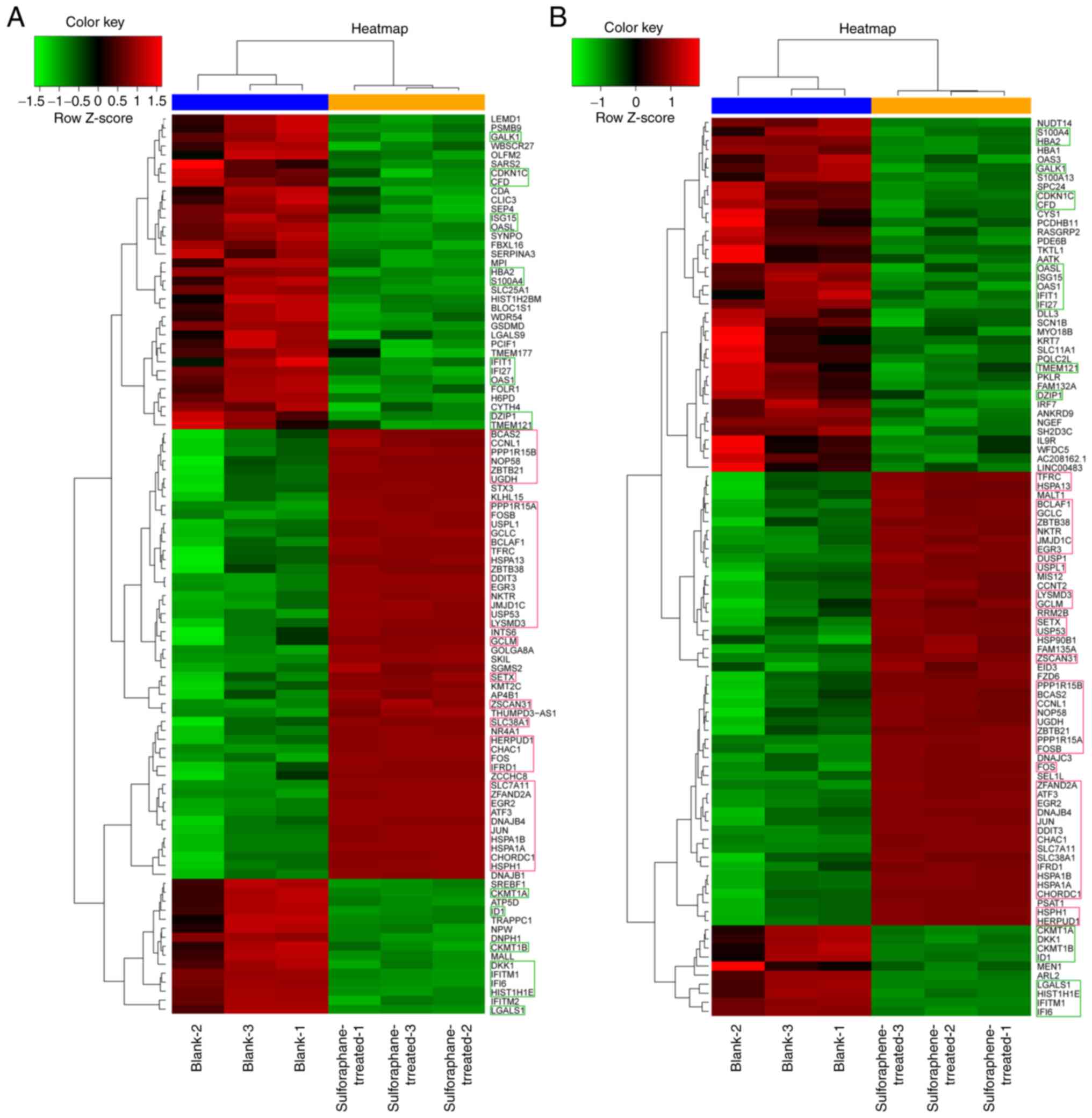
Hierarchical clustering analysis indicated that the
clustering of 58 genes (about 3% of the total differentially
expressed genes) was similar between the two groups. A total of 20
genes (GALK1, OAS1, CKMT1A, DKK1, CKMT1B, ID1, LGALS1, HIST1H1E,
IFITM1, IFI6, S100A4, HBA2, CDKN1C, CFD, OASL, ISG15, IFIT1, IFI27
TMEM121 and DZIP1) exhibited lower Z scores than the
blank control group. The Z scores of the remaining 38 genes
(BCAS2, CCNL1, PPP1R15B, NOP58, ZBTB21, UGDH, PPP1R15A, FOSB,
USPL1, GCLC, BCLAF1, TFRC, HSPA13, ZBTB38, DDIT3, EGR3, NKTR,
JMJD1C, USP53, LYSMD3, GCLM, SETX, ZSCAN31, SLC38AC, HERPUD1,
CHAC1, FOS, IFRD1, SLC7A11, ZFAND2A, EGR2, ATF3, DNAJB4, JUN,
HSPA1B, HSPA1A, CHORDC1 and HSPH1) were higher than
those noted in the blank control group (Fig. 3).
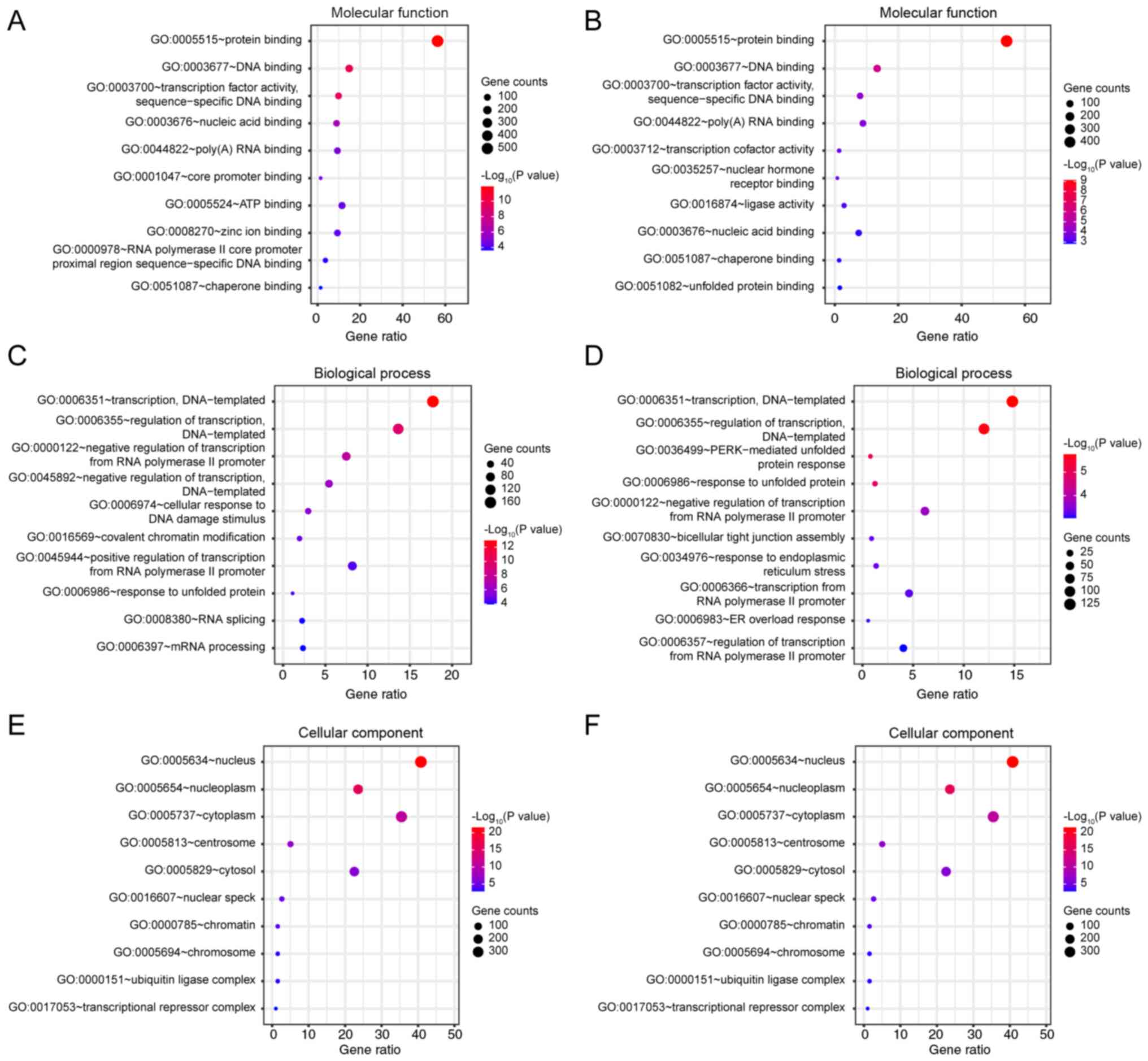
GO analysis
GO analysis of differentially expressed genes was
performed using the DAVID 6.8 database. Gene enrichments were
identified with regard to molecular function, biological processes
and cellular components (Fig. 4).
The majority of the genes in the sulforaphane and the
sulforaphene-treated groups were enriched in the same functions,
relative to the blank group, notably in ‘protein binding’
(GO:0005515, Fig. 4A and B),
‘transcription, DNA-templated’ (GO:0006351) and ‘regulation of
transcription, DNA-templated’ (GO:0006355, Fig. 4C and D), ‘nucleus’ (GO:0005634) and
‘nucleoplasm’ (GO:0005654, Fig. 4E and
F). However, subtle differences were noted in ‘ATP binding’
(GO:0005524, in sulforaphane group, Fig.
4A), ‘negative regulation of transcription, DNA-templated’
(GO:0045892, in sulforaphane group, Fig.
4A) and ‘positive regulation of transcription from RNA
polymerase II promoter’ (GO:0045944, in sulforaphane group,
Fig. 4A). Under ‘cellular
component’, the two groups of annotations to terms are the same,
while the unique sulforaphene group is ‘transcription cofactor
activity’ (GO:003712, Fig. 4B) and
‘transaction from RNA polymerase II promoter’ (GO:0006366, Fig. 4D).
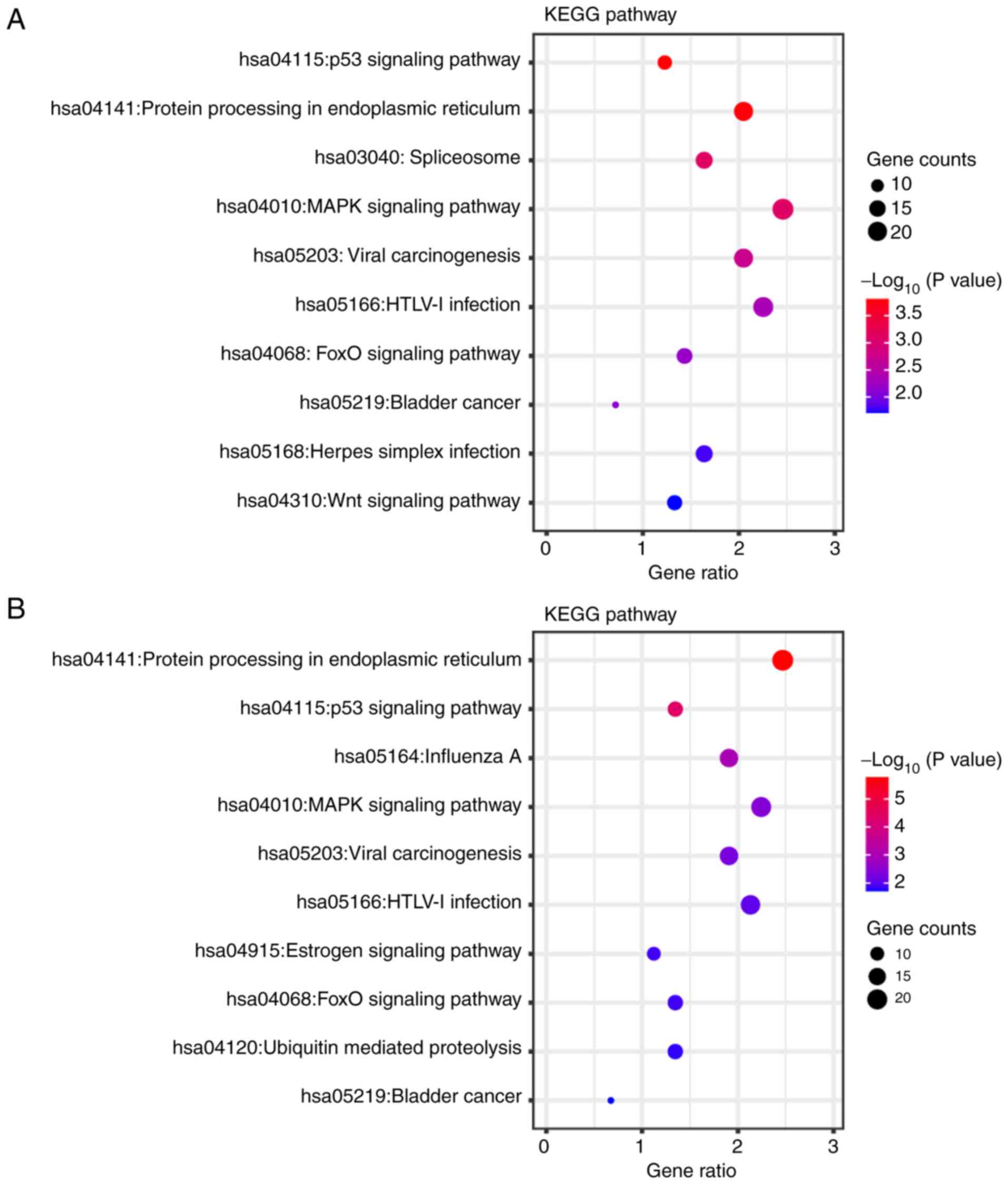
KEGG pathway annotations
The differentially expressed genes identified in the
sulforaphane and the sulforaphene-treated groups were annotated
using KEGG to assess the specific effects of these compounds on
SW480 cells (Fig. 5). Overlapping
pathways were identified, such as ‘p53 signaling pathway’
(hsa:04115), ‘protein processing in endoplasmic reticulum’
(hsa:04141), ‘spliceosome’ (hsa:03040), ‘MAPK signaling pathway’
(hsa:04010), ‘viral carcinogenesis’ (hsa:05203), ‘HTLV–I infection’
(hsa:05166), ‘FOXO signaling pathway’ (hsa:04068) and ‘bladder
cancer’ (hsa:05219). In the sulforaphane group, the differentially
expressed genes were uniquely found to be associated with the
‘spliceosome’ (hsa:03040), ‘herpes simplex infection’ (hsa:05168)
and ‘Wnt signaling pathway’ (hsa:04310), while in the sulforaphene
group the differentially expressed genes were associated with the
‘estrogen signaling pathway’ (hsa:04915) and ‘ubiquitin mediated
proteolysis’ (hsa:04120). What's more, a deeper visualization of
the pathways regulated by these two compounds are displayed
(Fig. S4). In the p53 signaling
pathway, both compounds upregulated MDM2 (Fig. S4A and B), while in the MAPK
signaling pathway both upregulated RafB (Fig. S4C and D). The FOXO pathway was
inhibited by sulforaphene, and sulforaphane directly upregulated
FOXO (Fig. S4E and F). The
WNT pathway was up-regulated by sulforaphane and core molecules
such as Wnt and APC (Fig.
S4G), while sulforaphene can upregulate some members of the
estrogen signaling pathway (Fig.
S4H).
Discussion
Colorectal cancer is one of the most common
gastrointestinal malignancies. The Global Cancer Observatory 2018
data suggested that the incidence of colon cancer ranked third
among men and second among women (28). In 2018, a total of 1,096 million new
cases and 551,000 deaths from colorectal cancer were reported
(28). The National Cancer Center of
China released the status of malignant tumors in 2016, which stated
that the number of new patients and deaths from colorectal cancer
ranked fourth among other cancer types, accounting for 376,000 and
191,000 new cases, respectively (29). Therefore, the identification of
practical and effective therapeutic drugs has become an urgent
medical requisite. Recent years have witnessed the development of
new technologies and treatment methods in the field of cancer
research. Moreover, the application of NGS has facilitated research
into the pathogenesis of colorectal cancer.
Cruciferous plants, such as cabbage, broccoli,
cauliflower, kale, lettuce and white radish contain >120 types
of glucosinolates that are similar in structure, consisting of one
β-D glucosinolate, one sulfonated oxime group and a side chain.
When glucosinolates are hydrolyzed by acid or specific enzymes
(such as myrosinase), they produce structural homologs and
isothiocyanates (14–16). Previous studies have shown that
isothiocyanates can improve the immunity of the body and enhance
antioxidant (in the liver), anti-inflammatory (in the prostate,
oral cavity and glial cells) and anticancer (in the skin, bladder,
colon, pancreas, liver, lung, nasopharynx, ovary, breast,
esophagus, stomach, cervix and thyroid) activities (2–11,19,20,23–26,30,31).
Sulforaphene is the isothiocyanate with the most potent anticancer
ingredients found in vegetables (15). Phase-I and -II enzymes are involved
in the metabolism of chemical carcinogens. Phase-I enzymes activate
chemical carcinogens to electrophiles, whereas phase-II enzymes
convert electrophiles to low-toxicity metabolites and readily
excreted substances (15).
Sulforaphene inhibits phase-I enzymes, such as cytochrome P450
enzymes and induces the expression levels of phase-II
detoxification enzymes, such as glutathione sulfur transferase,
epoxide hydrolase and uridine 5′-diphospho-
glucuronosyltransferase-glucuronyltransferase (15). Due to the lipophilic nature of
sulforaphene, the inhibition of the biosynthesis of phospholipids
and other lipid substances is a vital mechanism that can induce
growth arrest and apoptosis of cancer cells (32–34).
Previous studies have shown that sulforaphane and
sulforaphene have the same active center and similar chemical
structures (15–17). Despite their similarities, they have
been reported to be effective against distinct cancer types, as
aforementioned. This may be attributed to the limited number of
studies performed on these two compounds or could be due to their
limited efficacy against certain cancer types.
In the present study, the effects of sulforaphane
and sulforaphene were evaluated in colorectal cancer cells using
NGS in order to assess the differences between the anticancer
efficacy of the two compounds. Sulforaphene increased and decreased
the expression levels of 732 and 130 genes, respectively, while
sulforaphane increased and decreased the expression levels of 788
and 161 genes, respectively. Notably, none of these differentially
expressed genes were simultaneously detected in all three groups,
which indicated that sulforaphene and sulforaphane exhibited
distinct roles in colon cancer cells. However, these effects need
to be confirmed using multiple colon cancer cell lines. Although
the SW480 cell line is widely for the study of colon cancer, it
still does not fully represent the physiological characteristics of
colon cancer, and it is imperative to validate it using a wider
variety of colon cancer cell lines or by extracting primary cells
from tumor tissues of colon cancer patients in a sufficient sample
size. This is one of the limitations of the present study.
Hierarchical clustering analysis of differentially expressed genes
indicated that the Z score of 20 genes, such as GALK1, was
lower than that of the control group, whereas the cluster analysis
of the 38 genes, such as BCAS2, was higher than that of the
control group. This was observed in both sulforaphene and
sulforaphane groups. These 58 genes may be the common targets of
sulforaphane and sulforaphene. Sulforaphene and sulforaphane may
induce anticancer effects by affecting these 58 genes. GO and KEGG
analyses indicated that the functions and pathways enriched by the
differentially expressed genes produced by sulforaphene and
sulforaphane were similar. These pathways and functions included
‘protein binding’ (GO:0005515), ‘transcription, DNA-templated’
(GO:0006351), ‘regulation of transcription, DNA-templated’
(GO:0006355), ‘nucleus’ (GO:0005634), ‘nucleoplasm’ (GO:0005654),
‘p53 signaling pathway’ (hsa:04115), ‘MAPK signaling pathway’
(hsa:04010) and ‘FOXO signaling pathway’ (hsa:04068), which are
common cancer-associated functions or pathways. Sulforaphane may
also act on the Wnt pathway, suggesting a potential role in
epithelial-to-mesenchymal transition (9). Furthermore, sulforaphene is
specifically enriched in the estrogen signaling pathway, indicating
that it is associated with female endocrine cancers, such as those
of breast, cervical, endometrial and ovarian origin, which is
consistent with previous reports (19,25,35,36).
Based on the aforementioned results, it may be
hypothesized that sulforaphane and sulforaphene exert similar
anticancer effects. However, KEGG signaling pathway enrichment
suggested differences between the two molecules. For example, genes
with altered expression in both groups were enriched in the ‘p53
signaling pathway’, ‘MAPK signaling pathway’ and ‘FOXO signaling
pathway’ pathways. p53 is a key transcription factor required in
maintaining DNA integrity and is often referred to as the ‘guardian
of the genome’. Under steady-state conditions, the activity of p53
in colon cancer cells is inhibited by its binding to the negative
regulators mouse double minute (MDM) 2 and MDM4,
which induce p53 ubiquitination and protein degradation, while
maintaining the expression of tumor suppressors at low levels in
order to circumvent their tumor inhibitory function (37). The oncogenic activity of sulforaphane
and sulforaphene was presumably caused by the upregulation of MDM2
expression. The MAPK/PI3K signaling pathway is involved in cell
proliferation and survival. Changes in the expression levels of
proteins involved in this pathway cause an increase in the
proliferative potential of tumor cells (24,27,35,36).
KRAS, BRAF and PIK3CA mutations are common in
colorectal cancer (38). The
oncogenic properties of sulforaphane and sulforaphene may be due to
their ability to modulate RafB and MAPK kinase kinase 1 and
consequently affect the MAPK signaling pathway. FOXO family members
alter cell metabolism by inhibiting glycolysis and promoting
mitochondrial respiration (39,40),
which is crucial for cancer cells with a more active metabolism.
The results of KEGG pathway analysis demonstrated that sulforaphane
and sulforaphene affected the expression levels of AMPK, PI3K and
MDM2, which may in turn influence elements of the FOXO pathway. The
Wnt/β-catenin pathway regulates several cellular functions, such as
stem cell regeneration and organogenesis (41). The activation of this pathway occurs
at the bottom of intestinal crypts and is associated with the loss
of function of adenomatous polyposis coli (APC), a tumor
regulator (41). APC was one
of the genes significantly upregulated by sulforaphane treatment.
This may have been caused by changes in the expression of proteins
associated with specific pathways, such as WNT and MAPK, which may
also explain the differences in the anticancer efficacy of
sulforaphane and sulforaphene. Nevertheless, the changes caused by
sulforaphane on colon cancer crypts requires experimental
validation using immunohistochemical analysis and pathological
evaluation. The unique pathway for sulforaphene identified was the
‘ubiquitin-mediated proteolysis’ pathway, which may be associated
with ubiquitination and degradation of the p53 protein (37). In contrast to sulforaphane,
sulforaphene may regulate the ‘estrogen signaling pathway’ through
the regulation of genes, such as heparin-binding-EGF, which is
related to previous studies suggesting an association with breast
and cervical cancers (19,25). However, the effects of sulforaphene
on ovarian and endometrial cancer require further
investigation.
In the present study, only one colon cancer cell
line was used. The genes presented in this study were only obtained
using NGS and bioinformatics analysis and were not validated using
quantitative PCR at the RNA level or western blot analysis at the
protein level, which is the greatest limitation of the present
study. Due to modifications, such as RNA m6A
methylation, the sequencing results may differ from the actual RNA
and protein levels. Moreover, the analysis did not include
functional tests to assess the proliferative, invasive and
migratory activities of colon cancer cells treated with
sulforaphane and sulforaphene. The conclusions relating to the
anticancer effects of these two molecules are completely based on
previous reports; therefore, bias is likely. Several studies have
shown that different concentration levels of sulforaphane and
sulforaphene exert specific anticancer activities (15,19,23,25).
However, whether the concentration used in the present study was
optimal remains to be explored using a concentration gradient
test.
In conclusion, sulforaphane and sulforaphene have
similar biological activities and exert similar effects on colon
cancer cells. However, a slight difference was observed in the
functional annotation of the genes and in pathway enrichment
analysis. Both of these compounds may affect the p53, MAPK and FOXO
signaling pathways in colon cancer cells. Sulforaphane may be
associated with the Wnt pathway, while sulforaphene with the
estrogen pathway.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by The Research on
Evaluation and Control Technology of Characteristic Agricultural
Products Quality Safety and Nutritional Function (grant no.
2017JHZ010).
Availability of data and materials
The datasets generated and/or analysed during the
current study are available in the Gene Expression Omnibus under
accession no. GSE174444 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174444).
Authors' contributions
LG and JSW carried out the experiments. YHZ and JHL
analyzed the experimental results. FYD, DC and XZ reviewed the
results and graphed the experimental results. YTW and SQZ designed
and managed the experiments. YTW and SQZ confirm the authenticity
of all the raw data. FYD and JSW wrote the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Alhazmi N, Pai CP, Albaqami A, Wang H,
Zhao X, Chen M, Hu P, Guo S, Starost K, Hajihassani O, et al: The
promyelocytic leukemia protein isoform PML1 is an oncoprotein and a
direct target of the antioxidant sulforaphane (SFN). Biochim
Biophys Acta Mol Cell Res. 1867:1187072020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li S, Yang Y, Sargsyan D, Wu R, Yin R, Kuo
HD, Yang I, Wang L, Cheng D, Ramirez CN, et al: Epigenome,
transcriptome and protection by sulforaphane at different stages of
UVB-induced skin carcinogenesis. Cancer Prev Res (Phila).
13:551–562. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mastuo T, Miyata Y, Yuno T, Mukae Y,
Otsubo A, Mitsunari K, Ohba K and Sakai H: Molecular mechanisms of
the anti-cancer effects of isothiocyanates from cruciferous
vegetables in bladder cancer. Molecules. 25:5752020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Z, Garzotto M, Davis EW II, Mori M,
Stoller WA, Farris PE, Wong CP, Beaver LM, Thomas GV, Williams DE,
et al: Sulforaphane bioavailability and chemopreventive activity in
men presenting for biopsy of the prostate gland: A randomized
controlled trial. Nutr Cancer. 72:74–87. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gasparello J, Gambari L, Papi C, Rozzi A,
Manicardi A, Corradini R, Gambari R and Finotti A: High levels of
apoptosis are induced in the human colon cancer HT-29 cell line by
co-administration of sulforaphane and a peptide nucleic acid
targeting miR-15b-5p. Nucleic Acid Ther. 30:164–174. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Desai P, Wang KZ, Ann D, Wang J and Prabhu
S: Efficacy and pharmacokinetic considerations of loratadine
nanoformulations and its combinations for pancreatic cancer
chemoprevention. Pharm Res. 37:212020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dos Santos PWDS, Machado ART, De Grandis
RA, Ribeiro DL, Tuttis K, Morselli M, Aissa AF, Pellegrini M and
Antunes LMG: Transcriptome and DNA methylation changes modulated by
sulforaphane induce cell cycle arrest, apoptosis, DNA damage, and
suppression of proliferation in human liver cancer cells. Food Chem
Toxicol. 136:1110472020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Y, Chen JQ, Ge MM, Zhang Q, Wang XQ,
Zhu JY, Xie CF, Li XT, Zhong CY and Han HY: Sulforaphane inhibits
epithelial-mesenchymal transition by activating extracellular
signal-regulated kinase 5 in lung cancer cells. J Nutr Biochem.
72:1082192019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen L, Chan LS, Lung HL, Yip TTC, Ngan
RKC, Wong JWC, Lo KW, Ng WT, Lee AWM, Tsao GSW, et al: Crucifera
sulforaphane (SFN) inhibits the growth of nasopharyngeal carcinoma
through DNA methyltransferase 1 (DNMT1)/Wnt inhibitory factor 1
(WIF1) axis. Phytomedicine. 63:1530582019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tian M, Tian D, Qiao X, Li J and Zhang L:
Modulation of Myb-induced NF-kB-STAT3 signaling and resulting
cisplatin resistance in ovarian cancer by dietary factors. J Cell
Physiol. 234:21126–21134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li S, Chen M, Wu H, Li Y and Tollefsbol
TO: Maternal epigenetic regulation contributes to prevention of
estrogen receptor-negative mammary cancer with broccoli sprout
consumption. Cancer Prev Res (Phila). 13:449–462. 2020.PubMed/NCBI
|
|
12
|
Hussain A, Priyani A, Sadrieh L,
Brahmbhatt K, Ahmed M and Sharma C: Concurrent sulforaphane and
eugenol induces differential effects on human cervical cancer
cells. Integr Cancer Ther. 11:154–165. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li D, Shao R, Wang N, Zhou N, Du K, Shi J,
Wang Y, Zhao Z, Ye X, Zhang X and Xu H: Sulforaphane activates a
lysosome-dependent transcriptional program to mitigate oxidative
stress. Autophagy. 17:872–887. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Juge N, Mithen RF and Traka M: Molecular
basis for chemoprevention by sulforaphane: A comprehensive review.
Cell Mol Life Sci. 64:1105–1127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y and Talalay P: Mechanism of
differential potencies of isothiocyanates as inducers of
anticarcinogenic phase 2 enzymes. Cancer Res. 58:4632–4639.
1998.PubMed/NCBI
|
|
16
|
Kamal MM, Akter S, Lin CN and Nazzal S:
Sulforaphane as an anticancer molecule: Mechanisms of action,
synergistic effects, enhancement of drug safety, and delivery
systems. Arch Pharm Res. 43:371–384. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clarke JD, Dashwood RH and Ho E:
Multi-targeted prevention of cancer by sulforaphane. Cancer Lett.
269:291–304. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Y, Han X, Li Y, Min H, Zhao X, Zhang Y,
Qi Y, Shi J, Qi S, Bao Y and Nie G: Sulforaphane mediates
glutathione depletion via polymeric nanoparticles to restore
cisplatin chemosensitivity. ACS Nano. 13:13445–13455. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pocasap P, Weerapreeyakul N and Thumanu K:
Structures of isothiocyanates attributed to reactive oxygen species
generation and microtubule depolymerization in HepG2 cells. Biomed.
Pharmacother. 101:698–709. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang C, Zhang J, Wu Q, Xu B, Jin G, Qiao
Y, Zhao S, Yang Y, Shang J, Li X and Liu K: Sulforaphene induces
apoptosis and inhibits the invasion of esophageal cancer cells
through MSK2/CREB/Bcl-2 and cadherin pathway in vivo and in vitro.
Cancer Cell Int. 19:3422019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang M, Wang H, Zhou M, Liu W, Kuang P,
Liang H and Yuan Q: The natural compound sulforaphene, as a novel
anticancer reagent, targeting PI3K-AKT signaling pathway in lung
cancer. Oncotarget. 7:76656–76666. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Byun S, Shin SH, Park J, Lim S, Lee E, Lee
C, Sung D, Farrand L, Lee SR, Kim KH, et al: Sulforaphene
suppresses growth of colon cancer-derived tumors via induction of
glutathione depletion and microtubule depolymerization. Mol Nutr
Food Res. 60:1068–1078. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mondal A, Biswas R, Rhee YH, Kim J and Ahn
JC: Sulforaphene promotes Bax/Bcl2, MAPK-dependent human gastric
cancer AGS cells apoptosis and inhibits migration via EGFR,
p-ERK1/2 down-regulation. Gen Physiol Biophys. 35:25–34.
2016.PubMed/NCBI
|
|
24
|
Pawlik A, Wała M, Hać A, Felczykowska A
and Herman-Antosiewicz A: Sulforaphene, an isothiocyanate present
in radish plants, inhibits proliferation of human breast cancer
cells. Phytomedicine. 29:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Biswas R, Mondal A, Chatterjee S and Ahn
JC: Evaluation of synergistic effects of sulforaphene with
photodynamic therapy in human cervical cancer cell line. Lasers Med
Sci. 31:1675–1682. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chatterjee S, Rhee Y, Chung PS, Ge RF and
Ahn JC: Sulforaphene enhances the efficacy of photodynamic therapy
in anaplastic thyroid cancer through Ras/RAF/MEK/ERK pathway
suppression. J Photochem Photobiol B. 179:46–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baechler EC, Batliwalla FM, Karypis G,
Gaffney PM, Moser K, Ortmann WA, Espe KJ, Balasubramanian S, Hughes
KM, Chan JP, et al: Expression levels for many genes in human
peripheral blood cells are highly sensitive to ex vivo incubation.
Genes Immun. 5:347–353. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ko MO, Kim MB and Lim SB: Relationship
between chemical structure and antimicrobial activities of
isothiocyanates from cruciferous vegetables against oral pathogens.
J Microbiol Biotechnol. 26:2036–2042. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim KH, Moon E, Kim SY, Choi SU, Lee JH
and Lee KR: 4-Methylthio-butanyl derivatives from the seeds of
raphanus sativus and their biological evaluation on
anti-inflammatory and antitumor activities. J Ethnopharmacol.
151:503–508. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang H, Kang MJ, Hur G, Lee TK, Park IS,
Seo SG, Yu JG, Song YS, Park JHY and Lee KW: Sulforaphene
suppresses adipocyte differentiation via induction of
post-translational degradation of CCAAT/enhancer binding protein
beta (C/EBPβ). Nutrients. 12:7582020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Feng C, Tan X, Hagedoorn PL, Gu
C, Xu H and Zhou X: Effect of aliphatic diamine spacer length on
enzymatic performance of myrosinase immobilized on chitosan
microsphere and its application for sulforaphene production. J
Biotechnol. 299:79–85. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen J, Bao C, Kim JT, Cho JS, Qiu S and
Lee HJ: Sulforaphene inhibition of adipogenesis via Hedgehog
signaling in 3T3-L1 adipocytes. J Agric Food Chem. 66:11926–11934.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rai R, Gong Essel K, Mangiaracina Benbrook
D, Garland J, Daniel Zhao Y and Chandra V: Preclinical efficacy and
involvement of AKT, mTOR, and ERK kinases in the mechanism of
sulforaphane against endometrial cancer. Cancers (Basel).
12:12732020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Biswas R, Ahn JC and Kim JS: Sulforaphene
synergistically sensitizes cisplatin via enhanced mitochondrial
dysfunction and PI3K/PTEN modulation in ovarian cancer cells.
Anticancer Res. 35:3901–3908. 2015.PubMed/NCBI
|
|
37
|
Farooqi AA, de la Roche M, Djamgoz MBA and
Siddik ZH: Overview of the oncogenic signaling pathways in
colorectal cancer: Mechanistic insights. Semin Cancer Biol.
58:65–79. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mármol I, Sánchez-de-Diego C, Pradilla
Dieste A, Cerrada E and Rodriguez Yoldi MJ: Colorectal carcinoma: A
general overview and future perspectives in colorectal cancer. Int
J Mol Sci. 18:1972017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chiacchiera F, Matrone A, Ferrari E,
Ingravallo G, Lo Sasso G, Murzilli S, Petruzzelli M, Salvatore L,
Moschetta A and Simone C: p38alpha blockade inhibits colorectal
cancer growth in vivo by inducing a switch from HIF1alpha-to
FoxO-dependent transcription. Cell Death Differ. 16:1203–1214.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Q, Tang H, Hu F and Qin C: Silencing of
FOXO6 inhibits the proliferation, invasion, and glycolysis in
colorectal cancer cells. J Cell Biochem. 120:3853–3860. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Krishnamurthy N and Kurzrock R: Targeting
the Wnt/beta-catenin pathway in cancer: Update on effectors and
inhibitors. Cancer Treat Rev. 62:50–60. 2018. View Article : Google Scholar : PubMed/NCBI
|