Introduction
Oral cancer is the 6th most common malignancy
worldwide and oral squamous cell carcinoma (OSCC) represents 90% of
all cancer cases (1). The current
therapy is only effective for early stage OSCC, and the 5-year
overall survival rate was <50% in 2020 (2). Studies have indicated that tobacco
addiction, excessive alcohol consumption and human papillomavirus
(HPV) infection are the main risk factors for tumorigenesis and the
development of OSCC (3). Current
targeted therapies, such as targeting PD1/PDL1, are far from
satisfactory (4); therefore; new
predictive biomarkers and therapeutic targets are urgently
required.
Long non-coding (lnc)RNAs are 200 nucleotides in
length (5). With the application of
high-throughput sequencing, lncRNAs have been reported to be
associated with the progression of numerous types of cancer.
Rs6695584 single nucleotide polymorphism-induced lncRNA-SLCC1 drove
colorectal cancer by activating glycolysis signaling (6). lncRNA lnc030 maintained breast cancer
stemness by stabilizing SQLE mRNA and therefore promoted
cholesterol synthesis (7). lncRNA
CRNDE attenuated chemoresistance in gastric cancer by
SRSF6-regulated alternative splicing of PICALM (8). HOXB-AS3 was previously reported to
encode a micro-peptide and to suppress tumor progression in colon
cancer (9). However, biofunction
varies in different types of cancer (10,11). Its
potential function and underlying mechanism in OSCC are still
unknown.
In the present study, the expression level of
HOXB-AS3 in patients with OSCC was analyzed and its biological
functions and underlying mechanism in OSCC cell lines was also
investigated. The results demonstrated, to the best of knowledge,
for the first time that HOXB-AS3 and its encoded protein were
upregulated in OSCC tumors. Knocking down HOXB-AS3 repressed OSCC
cell proliferation and viability, and a Co-immunoprecipitation (IP)
assay indicated that the HOXB-AS3-encoded protein interacted with
IGF2BP2; therefore, stabilized c-Myc.
Materials and methods
Patient information and clinical
sample collection
Paired OSCC samples and adjacent normal tissues were
randomly collected from patients who underwent resection surgery at
the Affiliated Stomatological Hospital of Nanchang University
(Jiangxi, China) between January 2012 and December 2013. The
following inclusion criteria was used: Diagnosed with OSCC by
pathologists between January 2012 and December 2013 at the
Affiliated Stomatological Hospital of Nanchang University. Patients
with metastasis were excluded. A total of 20 cases (age range,
42–68 years; 15 male and 5 female) were divided into 4 groups,
according to their clinical features and the TNM stage system
(12). There were 5 patients with
stage I, 3 patients with stage II, 8 patients with stage III and 4
cases with stage 4. Adjacent normal tissues were collected >5 cm
from the tumor site and confirmed by a pathologist as normal
tissue. All the OSCC tissue samples were confirmed by pathology.
The present study was approved by the Ethics Committee of The
Affiliated Stomatological Hospital of Nanchang University (Jiangxi,
China). The clinical samples were collected from patients following
written informed consent.
Survival analysis
Using data from The Cancer Genome Atlas (TCGA)
database, the time of the end point of each patient was collected,
this was defined as the survival end point of each patient, all the
survival end points in of the whole cohort was defined as overall
survival. The overall survival analysis was thus automatically
determined using the Gene Expression Profiling Interactive Analysis
online tool by analyzing these data (gepia.cancer-pku.cn) by
inputing the gene name (HOXB-AS3) and cancer type (OSCC), 519
patients were enrolled and 44 normal tissues were collected. For
the patients recruited into the present study, the survival time
was collected between 2012 and 2018. From the patients recruited
into the present study, they were divided into 2 groups according
to the mean mRNA expression (3.56) and Kaplan-Meier survival
analysis was performed and the GraphPad software v.7.0 (GraphPad
Software, Inc.).
Reverse transcription-quantitative
analysis (RT-qPCR) analysis
Tissues and cells were harvested and lysed using
TRIzol® (cat. no. 93289; Sigma-Aldrich; Merck KGaA)
following the manufacturer's protocol. After purification and
reverse transcription (60°C for 5 min, 37°C for 15 min and 85°C for
5 sec) cDNA was collected. Specific primers were used for qPCR
using the SYBR-Green RT-PCR system with RT mix (cat. no. RR036B;
Takara Bio, Inc.) and SYBRGreen (cat. no. K0243; Themo Fisher
Scientific Inc.) with 2 step process. The following thermocycling
conditions were used: Initial denaturation at 95°C for 5 sec, then
40 cycles at 95°C for 5 sec, 95°C for 35 sec and 60°C for 30 sec,
then 60°C for 30 sec. GAPDH was used for normalization. The
following primers were used: HOXB-AS3 forward,
5′-CCCTCCAAGTCCAGTAAGAAGT-3′ and reverse
5′-AGATCCTAAGAGGTGCGAGTTTA-3′; and GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT−3′ and reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′ and the expression level was
normalized to the control (13).
Cell culture and transfection
The Cal-27 and UM2 cell lines were gifts from
Professor Wang from the Department of Oral and Maxillofacial
Surgery, Affiliated Hospital of Jiangxi University of Traditional
Chinese Medicine (Jiangxi, China). The cells were cultured with
complete DMEM (cat. no. D0697; Sigma-Aldrich; Merck KGaA)
containing 10% fetal bovine serum (cat. no. F8687; Invitrogen;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(cat. no. V900929; Sigma-Aldrich; Merck KGaA).
To establish a stable cell line, lentivirus was
obtained from GenScript, and the following short hairpin (sh)RNAs
were used in pCDH-CMV-MCS-EF1 + Puro (SBI pCD513B-1): Scrambled
negative control (NC) 5′-GTACCTACGTA-3′, shRNA1,
5′-CCATCCAAGTCTA-3′ and shRNA2, 5′-TAATGCAGCGTAAG-3′. ORF, the open
read frame of HOXB-AS3 encoding the protein but not the whole
lncRNA. Plasmids (4 µg) were co-transfected with packaging vectors
psPAX2 (Addgene, Inc.) (1 µg) and pMD2G (Addgene, Inc.) (3 µg) into
293T (cat. no. CRL-11268; ATCC) cells for lentivirus production
using Lipofectamine 3000® (Thermo Fisher Scientific
Inc.) in accordance to the manufacturer's instructions. After 72 h,
the suspension was collected. The lentivirus was stored at
1×108/ml. The cells were seeded into the 6-well plate
with 40% density and incubated overnight at 37°C, following which
10 µl (12 MOI) each lentivirus was added into the corresponding
well and incubated at 37°C for 3 days to successfully establish the
stable cell line. No selection method was used due to the high
transfection rate.
Western blot analysis
The cells were harvested and washed 3 times with ice
cold PBS and lysed with RIPA buffer (cat. no. 20-188;
Sigma-Aldrich; Merck KGaA). The protein was quantified using the
BCA kit (Beyotime Institute of Biotechnology), according to the
manufacturer's protocol. Equal amounts of protein (40 µg/lane) were
separated using 12% SDS-PAGE, then transferred onto a PVF membrane
and blocked with 5% skimmed milk for 1 h at room temperature. The
membranes were subsequently incubated with the primary antibodies
overnight at 4°C, washed with 0.1% TBS-Tween-20, then incubated
with the donkey anti mouse/rabbit secondary antibody (1:10,000;
cat. no. 5724; KPL, Inc.) for 1 h at room temperature. The target
proteins were detected using an ECL (EMD Millipore) kit. The
following primary antibodies were used: HOXB-AS3 (1:1,000; cat. no.
SBSN250; Genscript), β-actin (cat. no. 3700; 1:1,000; Cell
Signaling Technology, Inc.), IGF2BP2 (cat. no. ab109284; 1:1,000;
Abcam), and c-Myc (cat. no. ab32072; 1:1,000; Abcam).
Co-IP assay
The Cal-27 cell line was transfected with 2 µg Flag
tagged HOXB-AS3 (pCDH-CMV-MCS-EF1 + Puro (SBI pCD513B-1) (cat. no.
SBSN251; Genscript) using Lipofectamine 3000® (Thermo
Fisher Scientific Inc.) in accordance to the manufacturer's
instructions at 37°C for 1 day. Following incubation at 37°C for 3
days cells were harvested. After harvesting, cells were washed with
Phosphate-Buffered Saline (cat. no. C0221A; Beyotime Institute of
Biotechnology) the cells were lysed with 600 µl weak RIPA (cat. nο.
P0013D; Beyotime Institute of Biotechnology). After incubation at
4°C for 30 mins the suspension was collected. After centrifuging at
6×103 g for 10 mins at 4°C, the sediment was removed.
The 2 µg flag antibody (cat. no. 14793; Cell Signaling Technology,
Inc.) was added, and after incubation overnight at 4°C, the
suspension was collected, a 100 µl protein A + G beads (cat. no.
P2108; Beyotime Institute of Biotechnology) were added and after
rolling at room temperature for 30 mins, after centrifuging at
6×103 g for 10 mins at 4°C, the suspension was collected
and boiled at 100°C for 10 min and subjected to western blot
analysis as aforementioned. IGF2BP2 was detected.
Cell proliferation assay
A total of 500 cells per well were seeded into
96-well plates. After incubation at 37°C for 3 days, CCK-8 (cat.
no. ab228554; Abcam,) solution was added, after incubation at 37°C
for 4 h, the absorption at 450 nm was measured. The relative
absorption of sh-1,sh-2 and ORF group (experimental group) was
normalized to the control group(NC). The experiments were repeated
3 times and the data are presented as the mean ± SD.
Colony formation assay
To measure the long-term proliferation effects, 500
cells were seeded in a 6-well plate. After incubating for 14 days,
the colonies were fixed for 20 min with 4% paraformaldehyde and
stained with crystal violet (cat. no. C0775; Sigma-Aldrich; Merck
KGaA) at room temperature for 10 min, and the number of colonies
was counted under a light microscope (FV100; Olympus Corporation)
manually. The assay was performed 3 times and the data are
presented as the mean ± SD.
Statistical analysis
Data was shown in mean ± SD, all the experiments
were repeated at least 3 times. All data analysis was performed
with SPSS v20.0 (IBM, Corp.). The differences between 2 groups were
compared using a paired Student's t-test. For >2 groups, one-way
ANOVA was used, followed by Tukey's post hoc test. Kaplan-Meier
method were used to analyze overall survival (OS) time and the
curves were compared using the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
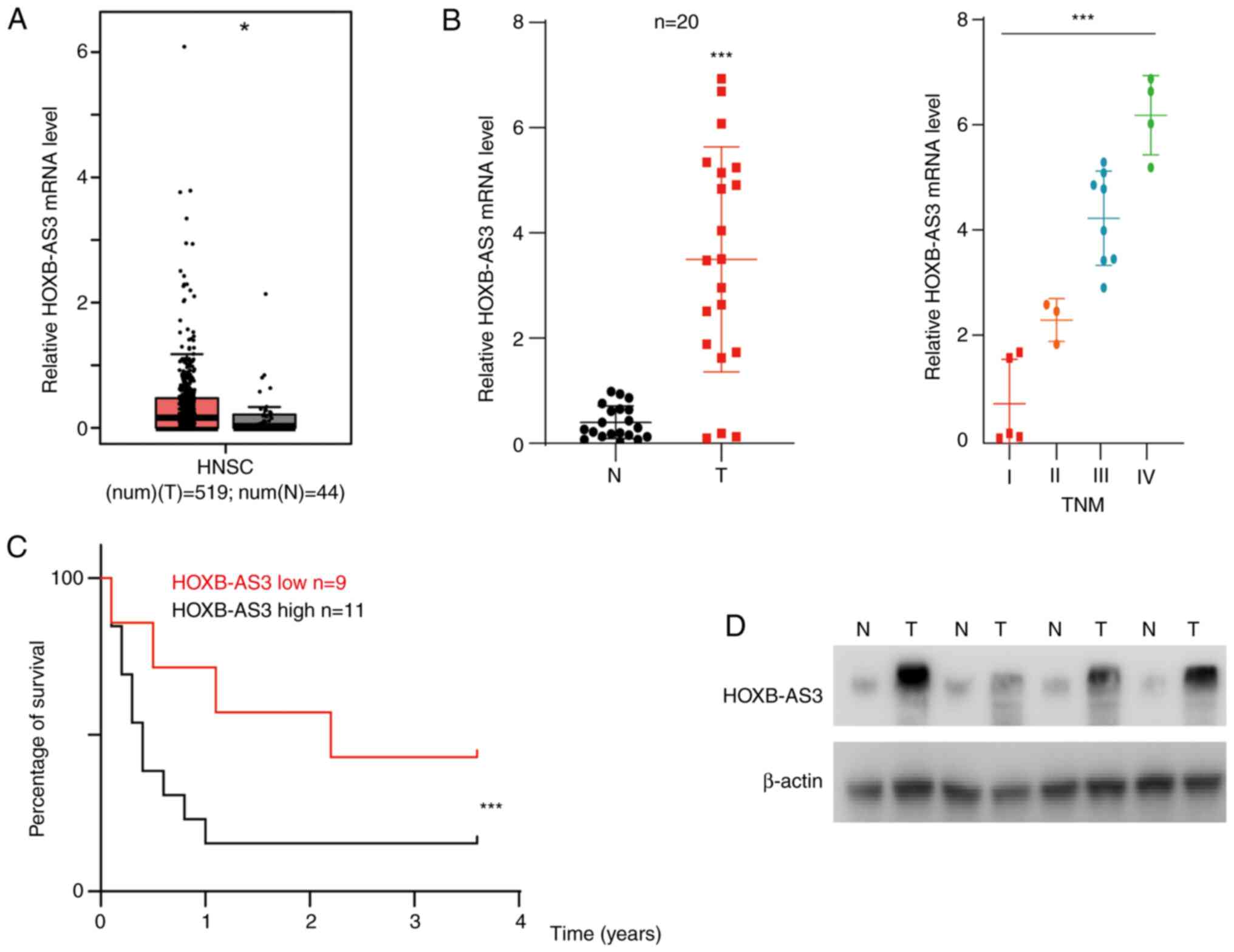
HOXB-AS3 is upregulated in human OSCC
tissues and is associated with poor prognosis
RNA sequencing datasets from TCGA were analyzed and
HOXB-AS3 was identified as one of the dysregulated genes in head
and neck cancer, which to the best of our knowledge has not been
investigated previously in head and neck cancer (P<0.05;
Fig. 1A). Next, the mRNA expression
level of HOXB-AS3 was analyzed in 20 randomly selected paired
adjacent normal and cancer tissues from patients recruited into the
present study. The results showed that HOXB-AS3 was upregulated in
cancer tissue compared with that in adjacent normal tissue
(P<0.001; Fig. 1B). The patients
were subsequently divided into 4 subtypes according to TNM stage.
The results indicated that patients with developed oral cancer had
higher mRNA expression levels of HOXB-AS3 (P<0.001; Fig. 1B). The Kaplan-Meier survival analysis
from TCGA database is shown in Fig.
S1, the overall survival showed no significance. From the
patients recruited into the present study, they were divided into 2
groups (high/low expression) according to the mean mRNA expression
level and the OS time was analyzed. The results showed that
HOXB-AS3 was associated with poor prognosis, with patients with low
expression levels having poor survival time (P<0.001; Fig. 1C). HOXB-AS3 was also reported to
encode a protein named HOXB-AS3 (8);
therefore, the protein expression level of HOXB-AS3 was analyzed in
4 randomly selected tumor and paired normal tissues. The results
showed that HOXB-AS3 protein expression level was increased in the
cancer tissue (Fig. 1D).
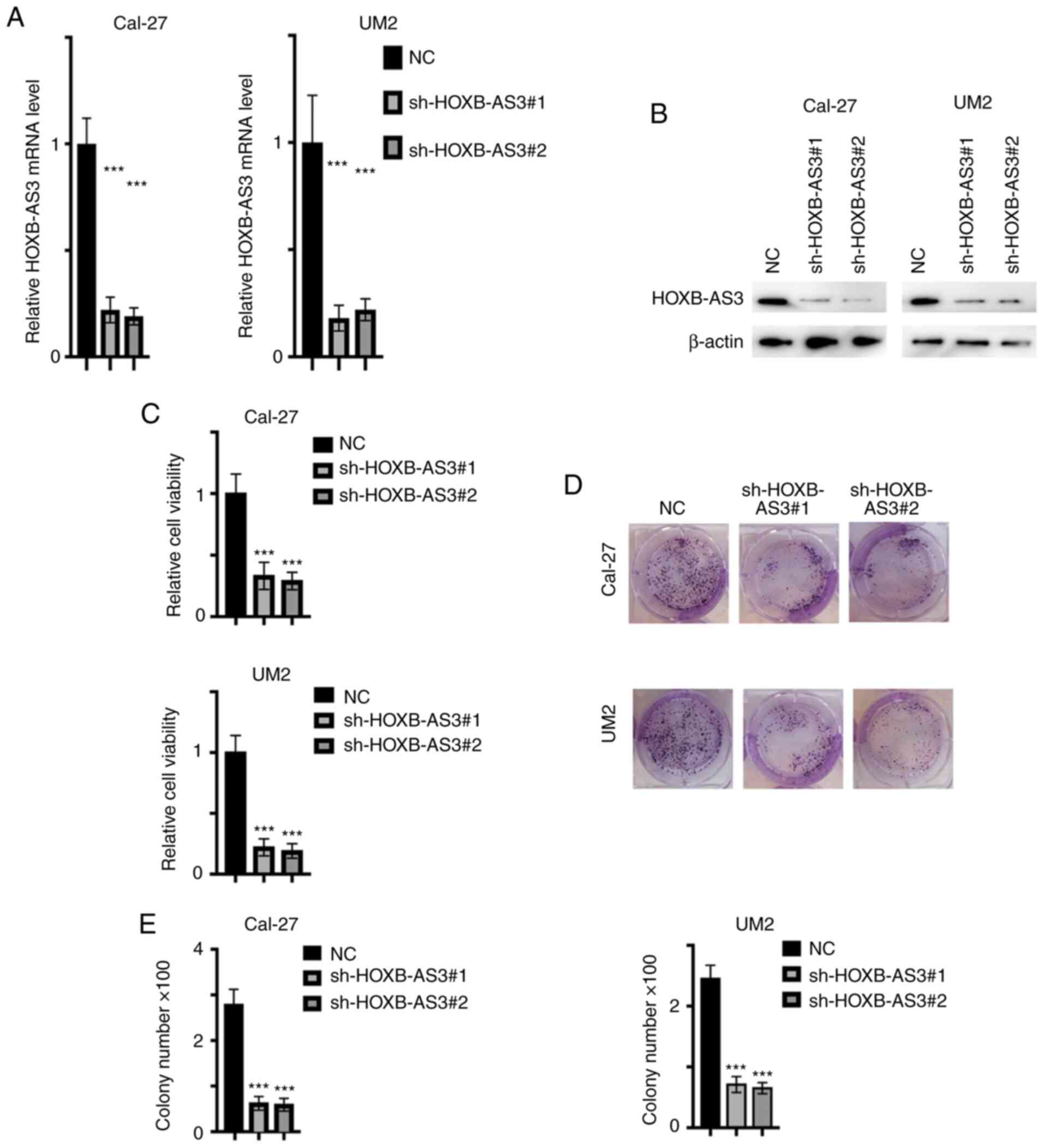
HOXB-AS3 promotes the proliferation of
OSCC
As aforementioned, HOXB-AS3 was found to be
upregulated in OSCC cancer tissue. To identify the biofunction of
HOXB-AS3 in the progression of the OSCC cell lines, HOXB-AS3 stable
knockdown cell lines were established using the Cal-27 and UM2 cell
lines. The relative mRNA (P<0.001; Fig. 2A) and protein (Fig. 2B) expression level of HOXB-AS3 was
decreased following transfection with sh-HOXB-AS3-1 and −2 in both
cell lines. Subsequently, the viability and proliferation ability
of the transfected cells was analyzed, and it was found that cells,
in which HOXB-AS3 mRNA expression was knocked down, had reduced
viability and proliferation ability from CCK-8 (P<0.001;
Fig. 2C) and colony formation assays
(P<0.001) (Fig. 2D and E),
respectively. This indicated that HOXB-AS3 promoted
proliferation.
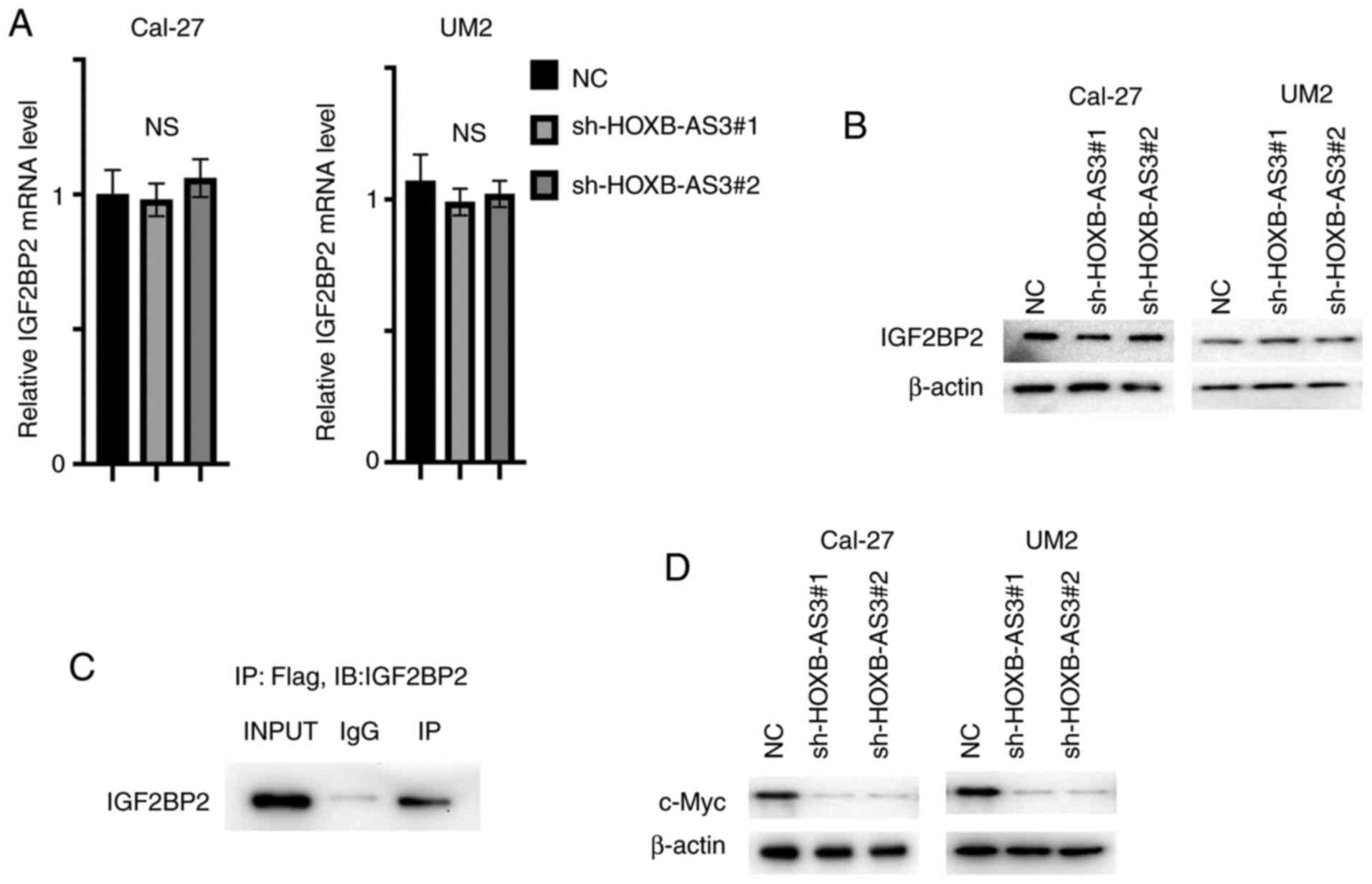
HOXB-AS3 directly binds with IGF2BP2
and thereby stabilizes c-Myc mRNA
HOXB-AS3 was reported to interact with hnRNPA1 and
affect the ratio of PKM1/PKM2 (8),
and IGF2BP2 was one of the potential interacting proteins (8). The mRNA and protein expression level of
IGF2BP2 was analyzed in the cell lines transfected with
sh-HOXB-AS3-1 and −2. The expression of IGF2BP2 was rarely changed,
the expression between groups showed no significance (Fig. 3A and B). Next Flag-tagged HOXB-AS3
was transfected into the 293T cell line and was then subjected to a
Co-IP assay. IGF2BP2 was detected and the results indicated that
HOXB-AS3 binds with IGF2BP2 as a whole complex (Fig. 3C). IGF2BP2 is a well-studied m6A
reader stabilizing c-Myc mRNA (14);
therefore, the protein expression level of c-Myc was analyzed in
cells transfected with sh-HOXB-AS3-1 and −2 and the results showed
that c-Myc protein expression level was decreased with the
knockdown of HOXB-AS3 in both cell lines (Fig. 3D).
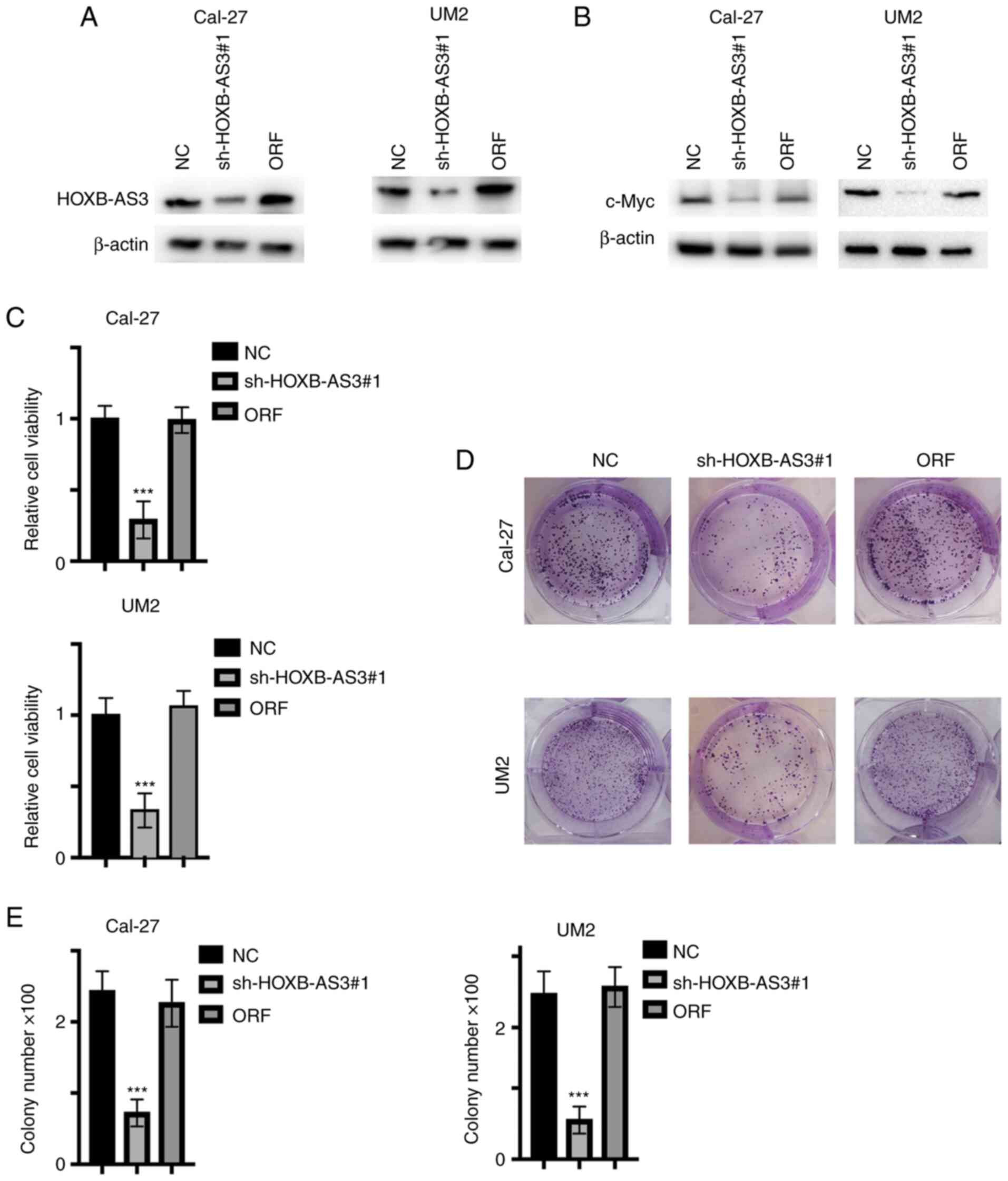
Micro-peptide HOXB-AS3, but not
HOXB-AS3 RNA promotes the proliferation of the OSCC cells
As aforementioned, it was found that HOXB-AS3
promoted the proliferation and viability of the OSCC cell lines by
stabilizing c-Myc mRNA. However, the mRNA and the protein were not
separated; therefore, to identify the potential function, the
HOXB-AS3 open reading frame was re-expressed in HOXB-AS3 stable
knockdown cell lines, the graphic illustration of ORF and HOXB-AS3
was shown in Fig. S2. Subsequently,
the protein expression levels of HOXB-AS3 and c-Myc were analyzed
(Fig. 4A and B, respectively). The
results showed that the protein expression level of c-Myc was
restored. In addition, cell viability and proliferation was
analyzed and the results also showed an increase in both (Fig. 4C-E), indicating that the HOXB-AS3
protein, but not HOXB-AS3 mRNA exerted its oncogenic function.
Discussion
OSCC is one of the most common malignancies
worldwide (1). Although many risk
factors, such as alcohol addiction and HPV infections, have been
reported to be involved in the tumourigenesis and progression of
OSCC (15), genomic mutations in
PTEN and AKT lead to the uncontrolled phosphorylation of AKT and
thus promote the proliferation of OSCC (16,17).
Oncogenic amplification, such as in the epidermal growth factor
receptor promotes the activation of downstream signaling and
eventually promotes OSCC survival (18). However, the exact mechanism of
tumourigenesis in OSCC remains unknown.
LncRNAs serve crucial roles in the tumourigenesis
and progression of OSCCs (19–24).
PDGF-BB regulates the transformation of fibroblasts into
cancer-associated fibroblasts via modulating the expression of
lncRNA LURAP1L-AS1 and this is mostly accomplished by affecting the
NF-κB signaling pathway (19).
lncRNA LINC01929 promotes the expression of Forkhead box protein C1
by sponging the microRNA(miR)-137-3p and serves oncogenic roles
(20). Tumor associated macrophages
secrete LncRNA LBX1-AS1 via the exosome, inhibiting OSCC
progression by targeting miR-182-5p/Forkhead box protein O3
(21). LncRNA PART1 directly binds
with the E2-ubiquitin ligating enzyme of EZH2 and promotes cell
proliferation (22). Taken together,
lncRNAs mostly exert their functions through sponging miRNA and RNA
binding protein partner (22).
However, lncRNAs are no longer absolutely non-coding. With the
application of high-throughput sequencing, an increasing number of
lncRNAs have been reported to be able to be translated into
proteins (23). HOXB-AS3 is one of
these translatable lncRNAs (9). The
biological function of HOXB-AS3 varies in colon cancer (9) and lung cancer (24). However, HOXB-AS3 is not only a coding
mRNA, but the lncRNA and its encoded protein exert their functions
separately (9,24). This may be the reason why conflicting
biological functions exist. In most studies, HOXB-AS3 exert its'
function as a ceRNA and the biological function was the overall
function of ceRNA and HOXB-AS3 protein (9,24).
However, in the study which first described HOXB-AS3 protein, it
was found that HOXB-AS3 protein exert its' tumor suppressing
function by switching the ratio of PKM1/Pyruvate Kinase M1/M2. In
the LC/MS mass spectrum analysis performed in the aforementioned
study, IGF2BP2 was one of the potential interacting protein
(9). IGF2BP2 is a novel m6A reader
and promotes the stability of mRNAs, such as Myc (9). The present study uncovered the role of
HOXB-AS3 in OSCC and to further uncover the mechanism, the lncRNA
and protein roles were separated. The present study reexpressed
HOXB-AS3 protein in stable knockdown cells and detected the
proliferation rate. Proliferation was completely restored with the
reexpression of HOXB-AS3 protein in the present study, indicating
that in OSCC, HOXB-AS3 promoted tumourigenesis and that this
oncogenic role is HOXB-AS3 protein dependent. The findings of the
present study indicated that HOXB-AS3 protein, but not the lncRNA
exert its' function in OSCC.
The present study analysed the expression pattern of
HOXB-AS3 in OSCC and its correlation with the prognosis of patients
in the TCGA database and our in-house database, however, the TCGA
database showed that the level of HOXB-AS3 didn't predict the
prognosis, it may because that the Head and Neck tumor in TCGA
database doesn't just contain data for OSCC. We found that HOXB-AS3
overexpression promoted the proliferation of OSCC by stabilizing
c-myc mRNA by directly binding to IGF2BP2. HOXB-AS3 binds with
IGF2BP2 and promotes its' function in stabilizing c-myc. To the
best of our knowledge, the present study is the first to have
demonstrated the potential biological function and underlying
mechanism of HOXB-AS3 in OSCC. A limitation of the present study
was the lack of in vivo experiments which should be
conducted by future studies to verify the findings of the present
study. Targeting HOXB-AS3 may be one of the potential therapy
strategies for patients with OSCC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Wang
(Department of Oral and Maxillofacial Surgery, Affiliated Hospital
of Jiangxi University of Traditional Chinese Medicine, Nanchang,
China) for the support with research design and providing
materials.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZGZ designed the article. FL, YYM, HL, XLL, HC, YZ
and ZGZ performed the experiments, data analysis and wrote the
manuscript. HC and ZGZ confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Studies were performed conforming to the Declaration
of Helsinki and approval was obtained from the Ethics Committee of
the Affiliated Stomatological Hospital of Nanchang University
(Jiangxi, China). Clinical samples were collected from patients
after written informed consent was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li P, Fang Q, Yang Y, Chen D, Du W, Liu F
and Luo R: Survival significance of number of positive lymph nodes
in oral squamous cell carcinoma stratified by p16. Front Oncol.
11:5454332021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Antonsson A, de Souza M, Wood ZC, Carroll
A, Van K, Paterson L, Pandeya N and Whiteman DC: Natural history of
oral HPV infection: Longitudinal analyses in prospective cohorts
from Australia. Int J Cancer. 148:1964–1972. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Panarese I, Aquino G, Ronchi A, Longo F,
Montella M, Cozzolino I, Roccuzzo G, Colella G, Caraglia M and
Franco R: Oral and Oropharyngeal squamous cell carcinoma:
Prognostic and predictive parameters in the etiopathogenetic route.
Expert Rev Anticancer Ther. 19:105–119. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang H, Zhang Z, Peng R, Zhang L, Liu H,
Wang X, Tian Y and Sun Y: RNA-Seq analysis reveals critical
transcriptome changes caused by sodium butyrate in DN mouse models.
Biosci Rep. 30:BSR202030052021. View Article : Google Scholar
|
|
6
|
Yan T, Shen C, Jiang P, Yu C, Guo F, Tian
X, Zhu X, Lu S, Han B, Zhong M, et al: Risk SNP-induced
lncRNA-SLCC1 drives colorectal cancer through activating glycolysis
signaling. Signal Transduct Target Ther. 6:702021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qin Y, Hou Y, Liu S, Zhu P, Wan X, Zhao M,
Peng M, Zeng H, Li Q, Jin T, et al: A novel long non-coding RNA
lnc030 maintains breast cancer stem cell stemness by stabilizing
SQLE mRNA and increasing cholesterol synthesis. Adv Sci (Weinh).
8:20022322020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang F, Wang H, Yu J, Yao X, Yang S, Li
W, Xu L and Zhao L: LncRNA CRNDE attenuates chemoresistance in
gastric cancer via SRSF6-regulated alternative splicing of PICALM.
Mol Cancer. 20:62021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang JZ, Chen M, Chen D, Gao XC, Zhu S,
Huang H, Hu M, Zhu H and Yan GR: A peptide encoded by a putative
lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell.
68:171–184, e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu S, Jia G, Zhang H, Wang L, Cong Y, Lv
M, Xu J, Ruan H, Jia X, Xu P and Wang Y: LncRNA HOXB-AS3 promotes
growth, invasion and migration of epithelial ovarian cancer by
altering glycolysis. Life Sci. 264:1186362021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Papaioannou D, Petri A, Dovey OM, Terreri
S, Wang E, Collins FA, Woodward LA, Walker AE, Nicolet D, Pepe F,
et al: The long non-coding RNA HOXB-AS3 regulates ribosomal RNA
transcription in NPM1-mutated acute myeloid leukemia. Nat Commun.
10:53512019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beltramini GA, Belloni LM, Fusco N,
Sacconi A, Muti P, Baj A, Bolzoni AR and Giannì AB: Comparing
prognostic utility between the 8th edition of TNM staging system
and the lymph node ratio for oral squamous cell carcinoma. Head
Neck. Jun 11–2021.(Epub ahead of Print). View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng J, Zhou X, Feng W, Jia M, Zhang X,
An T, Luan M, Pan Y, Zhang S, Zhou Z, et al: Risk stratification by
long non-coding RNAs profiling in COVID-19 patients. J Cell Mol
Med. 25:4753–4764. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Starzynska A, Adamska P, Sejda A,
Sakowicz-Burkiewicz M, Adamski ŁJ, Marvaso G, Wychowański P and
Jereczek-Fossa BA: Any role of PIK3CA and PTEN biomarkers in the
prognosis in oral squamous cell carcinoma? Life (Basel).
10:3252020.PubMed/NCBI
|
|
17
|
Manikandan M, Deva Magendhra Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: MicroRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saravani S, Parsamanesh N and
Miri-Moghaddam E: Role of EGFR gene polymorphisms in oral squamous
cell carcinoma patients of Southeast Iran: A case-control study.
Caspian J Intern Med. 11:391–397. 2020.PubMed/NCBI
|
|
19
|
Ren X, Li L, Wu J, Lin K, He Y and Bian L:
PDGF-BB regulates the transformation of fibroblasts into
cancer-associated fibroblasts via the lncRNA
LURAP1L-AS1/LURAP1L/IKK/IκB/NF-κB signaling pathway. Oncol Lett.
22:5372021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Che H, Che Y, Zhang Z and Lu Q: Long
non-coding RNA LINC01929 accelerates progression of oral squamous
cell carcinoma by targeting the miR-137-3p/FOXC1 axis. Front Oncol.
11:6578762021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ai Y, Wei H, Wu S, Tang Z, Li X and Zou C:
Exosomal lncRNA LBX1-AS1 derived from RBPJ
overexpressed-macrophages inhibits oral squamous cell carcinoma
progress via miR-182-5p/FOXO3. Front Oncol. 11:6058842021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu Q, Du Y, Wang S and Zheng X: LncRNA
PART1 promotes cell proliferation and inhibits apoptosis of oral
squamous cell carcinoma by blocking EZH2 degradation. J Biochem.
Mar 16–2021.(Epub ahead of Print). View Article : Google Scholar
|
|
23
|
Lu S, Zhang J, Lian X, Sun L, Meng K, Chen
Y, Sun Z, Yin X, Li Y, Zhao J, et al: A hidden human proteome
encoded by ‘non-coding’ genes. Nucleic Acids Res. 47:8111–8125.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang W, Kai J, Li D, Wei Z, Wang Y and
Wang W: lncRNA HOXB-AS3 exacerbates proliferation, migration, and
invasion of lung cancer via activating the PI3K-AKT pathway. J Cell
Physiol. 235:7194–7203. 2020. View Article : Google Scholar : PubMed/NCBI
|