Introduction
Lung cancer is one of the most frequently occurring
malignant tumors in women and men. Non-small cell lung cancer
(NSCLC) is a major type of lung cancer, accounting for four fifths
of lung cancer cases worldwide, and can be further divided into
lung adenocarcinoma (LUAD) and squamous cell carcinoma (1). LUAD is more likely to occur in women
and nonsmokers, and patients are more prone to metastasis. The
primary challenges in LUAD therapy are diagnosis at the
intermediate or advanced stages, as well as tumor heterogeneity,
since high genetic heterogeneity exists between different LUAD
cells. Therefore, increasing efforts have been made to identify
different ‘driver mutations,’ primarily comprising the epidermal
growth factor receptor (EGFR), anaplastic lymphoma kinase, ROS
proto-oncogene 1, receptor tyrosine kinase and KRAS (2,3). KRAS
mutations are associated with the expression of programmed cell
death 1 ligand 1, which suggests that it may predict
immunotherapeutic benefits. However, none of the patients in the
present study exhibited KRAS mutations.
Understanding cellular heterogeneity is important in
precision oncology. Proteomics research, the study of all the
proteins in a biological system, has provided a crucial
understanding of cellular heterogeneity, and major advances in
proteomics have been applied to clinical usage and personalized
therapy (4–6). The use of formalin-fixed,
paraffin-embedded (FFPE) samples represents the gold standard for
tissue preservation, and is the most common method for preserving
tissue morphology and the proteome of clinical specimens (7). In cancer research, FFPE samples have
attracted increasing attention as an alternative to fresh tissues
(8–10). Mass spectrometry (MS)-based
proteomics has become a formal research tool for the study of FFPE
human and animal tissue samples (11,12).
In the present study, ‘FFPE proteomics’ was adopted
for proteomics research on LUAD tissues and matched-adjacent
nontumor tissues. Tandem mass tags (TMTs), in combination with
high-resolution LC-MS/MS analysis, were used to identify >5,800
proteins. AAA domain containing 3A (ATAD3a), a member of the ATPase
family, plays a crucial role in metabolic activity and has been
shown to be a novel therapeutic target (13). However, the role and mechanism of
ATAD3a in LUAD remain unclear. The present study further confirmed
that ATAD3a downregulation enhanced the sensitivity of LUAD cells
to cisplatin. Collectively, the results suggested sensitive
diagnostic signatures and novel therapeutic targets for LUAD. From
the initial screening to treatment decisions through recurrence and
monitoring, proteomics plays an important role. The present study
aimed to identify similar markers for the presence or prognosis of
disease, or targeted proteins that can be found in LUAD through the
applied use of proteomics technologies.
Materials and methods
Cell culture
The LUAD cancer cell line (NCI-H292) was cultured in
RPMI-1640 supplemented with 10% FBS (both Gibco; Thermo Fisher
Scientific, Inc.) and a mixture of 1% penicillin (100
U/ml)/streptomycin (100 µg/ml) (Beijing Solarbio Science &
Technology Co., Ltd.). As an alternative to primary cultured lung
cells, immortalized normal alveolar epithelial cells (HPAEpi C), as
well as the LUAD cancer cell line A549, were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
and 1% penicillin/streptomycin. All cell lines were maintained at
37°C (5% CO2) in a humidified atmosphere.
Tissue samples
The present study was approved by the Research
Ethics Committee of Henan Provincial People's Hospital (HPPH), and
written informed consent was obtained from all patients. FFPE LUAD
specimens and adjacent nontumor lung tissues were acquired from
patients who underwent thoracic surgery at HPPH between May 2019
and February 2020; the basic patient information is presented in
Table I. The exclusion criteria were
as follows: i) Patients who received chemotherapy or radiotherapy;
and ii) patients who possessed other types of tumor. The inclusion
criteria were as follows: i) All clinicopathological diagnoses were
confirmed by two highly skilled pathologists; ii) none of the
patients received any treatments before surgery; iii) no history of
other synchronous malignancies; and iv) no death within the
perioperative period.
 | Table I.Basic information about the LUAD
patients. |
Table I.
Basic information about the LUAD
patients.
| Patient no. | Age (years) | Sex | TNM stage |
|---|
| 1 | 66 | F | II |
| 4 | 70 | M | III |
| 5 | 64 | F | IV |
| 6 | 59 | F | III |
| 7 | 69 | F | III |
| 9 | 46 | M | IV |
| 10 | 61 | F | III |
| 11 | 76 | M | II |
Sample preparation and TMT
labeling
The 5 µm-thick FFPE samples were dewaxed for 10 min
using xylene at room temperature, and then centrifuged for 10 min
at 12,000 × g at room temperature. The supernatant was discarded,
and the sediment was rehydrated with a descending ethanol series
(absolute, 85, 75% and ddH2O).
Samples were applied to 5 kD cutoff filters (Agilent
Technologies, Inc.) to perform buffer exchange. Then, 5X the sample
volume of 50 mm Hepes buffer (pH 7.6) was added to each sample. The
filters were subsequently centrifuged for 20 min at 2,000 × g at
room temperature and the flow through was discarded. This step was
repeated three times to ensure that a complete exchange was
achieved. Protein concentrations of the collected samples were
subsequently determined with a DC Protein assay (Bio-Rad
Laboratories, Inc.). The volume of each sample containing 30 µg
protein was adjusted to 120 µl with the addition of 50 mm HEPES (pH
7.6). The samples were subsequently denatured at 99°C for 5 min.
Reduction and alkylation were performed by adding 13 µl of 100 mm
dithiothreitol and 20 µl of 100 mm iodoacetamide to each sample.
Tryptic digestion was performed overnight at 37°C (trypsin: Sample
ratio, 1:60), followed by TMT labeling, according to the
manufacturer's instructions (Thermo Fisher Scientific, Inc.).
Following digestion, 5 µl of each sample was taken off and run on a
short gradient LC-MS/MS for quality control. The samples were
subsequently dissolved in 15 µl of mobile phase A (95% water, 0.1%
formic acid) and 1 µl and subjected to LC-MS/MS analysis using a
hybrid Q-Exactive mass spectrometer (Thermo Fisher Scientific,
Inc.) as described in 2.5.3. To create an internal standard to link
the four TMT sets, a pooled internal standard from TIFs was
prepared by taking 4 µg from each sample. TIF samples for in-depth
analysis were subjected to TMT labeling according to the
manufacturer's instructions (Thermo Fisher Scientific, Inc.). The
four TMT-labeled sets were desalted and cleaned up by applying them
to Strata SCX cartridges, according to the manufacturer's
instructions (Phenomenex, http://www.phenomenex.com.cn), followed by
lyophilization. The samples were stored at −20°C until subsequent
experimentation.
HPLC fractionation and LC-MS/MS
analysis
For each LC-MS analysis, the auto sampler (Ultimate
3000 RSLC System; Thermo Fisher Scientific, Inc.) dispensed 15 µl
of mobile phase A (95% water, 5% dimethyl sulfoxide, 0.1% formic
acid) into the corresponding well of a 96-well V-bottom polystyrene
microtiter plate (Corning, Inc.). When mixed together, the samples
was added to the plate by aspirating/dispensing a 10 µl volume 10
times. Subsequently, a 7-µl aliquot was injected into a C18 guard
desalting column (Acclaim Pepmap 100, 75 µm × 2 cm, NanoViper;
Thermo Fisher Scientific, Inc.). After 5 min with the loading pump
at a flow rate of 5 µl·min−1, the 10-port valve switched
to analysis mode with the NC pump providing a flow rate of 250
nl·min−1 through the guard column. The curved gradient
(curve 6 in chromeleon software, Thermo Fisher Scientific, Inc.)
was subsequently applied with 3% mobile phase B (95% acetonitrile,
5% water, 0.1% formic acid) increased to 45% mobile phase B over 50
min, followed by a wash with 99% mobile phase B and
re-equilibration. The total LC-MS run time was 74 min. A nano
EASY-Spray column (Pepmap RSLC, C18, 2 µm bead size, 100 Å, 75 µm
internal diameter, 50 cm length; Thermo Fisher Scientific, Inc.)
was used on the nano electrospray ionization (NSI) EASY-Spray
source (Thermo Fisher Scientific, Inc.) at 60°C. Online LC-MS was
performed using a hybrid Q-Exactive mass spectrometer (Thermo
Fisher Scientific, Inc.). FTMS master scans with 70,000 resolution
and a mass range of 300–1,700 m/z were followed by data-dependent
MS/MS at 35,000 resolution for the top five ions by using higher
energy collision dissociation (HCD) at 30% normalized collision
energy. Precursors were isolated with a 2 m/z window. Automatic
gain control targets were 1e6 for MS1 and 1e5 for MS2. Maximum
injection times were 100 ms for MS1 and 450 ms for MS2. The entire
duty cycle lasted 2.5 sec. Dynamic exclusion was used with 60 sec
duration. Precursors with an unassigned charge state or a charge
state of 1 were excluded. An underfill ratio of 1% was used. MS/MS
data were searched by using Sequest HT of the proteome discoverer
1.4 software platform (Thermo Fisher Scientific, Inc.) against the
UniProt protein sequence database (140407) with a 1% peptide false
discovery rate (FDR) cut-off. A precursor mass tolerance of 10
p.p.m. and product mass tolerances of 0.02 Da were used. Additional
settings were as follows: Trypsin with 1 missed cleavage; IAA on
cysteine, TMT on lysine, N-terminal as fixed modification,
oxidation of methionine, and phosphorylation of serine, threonine,
or tyrosine as variable modifications. Quantitation of TMT 10-plex
reporter ions was performed using Proteome Discoverer (Thermo
Fisher Scientific, Inc.) on HCD-FTMS tandem mass spectra by using
an integration window tolerance of 20 p.p.m. FDR rate was estimated
by using percolator (part of PD 1.4). The mass spectrometry
proteomics data obtained have been deposited into the
ProteomeXchange Consortium2 via the PRIDE partner repository with
the dataset identifier, PXD001686.
Protein annotation and functional
enrichment
Gene Ontology (GO), protein domain, Kyoto
Encyclopedia of Genes and Genomes (KEGG) and subcellular
localization annotation was performed using the Database for
Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov). GO annotation at the
proteomics level was carried out using the UniProt-GOA database.
InterProScan is based on protein sequence algorithms, and the
InterPro homologous domain database annotates the domains of
identified proteins. The KEGG Automatic Annotation Server
identifies and then annotates proteins, and matches them with
corresponding pathways via KEGG mapper. WoLF PSORT software was
used to determine the subcellular localization of proteins. For
functional enrichment, Fisher's exact test was used to identify
significantly differentially expressed proteins between LUAD and
adjacent nontumor lung tissues, and P<0.05 was considered to
indicate a statistically significant difference.
Western blot analysis
Cells were lysed using cell lysis buffer (Abcam).
The BCA method was used to examine the concentrations of different
protein samples. The protein samples (20 µg/lane) were separated
via 12% SDS-PAGE and transferred to PVDF membranes (Bio-Rad
Laboratories, Inc.). After incubating with 5% skimmed milk for 1 h
at room temperature, the membranes were incubated overnight at 4°C
with the following primary antibodies: β-actin (Abcam; cat. no.
ab8226; 1:1,000 dilution) and ATAD3a (Abcam; cat. no. ab112572;
1:1,000 dilution). The bound antibodies were visualized with a
horseradish peroxidase-conjugated secondary antibody (Abcam; cat.
no. ab6708; 1:1000 dilution, and Abcam; cat. no. ab7090; 1:1,000
dilution) using enhanced chemiluminescence (Clarity Western ECL;
Bio-Rad Laboratories, Inc.) and the Ez-Capture MG system (Atto
Corp.). Image Lab analysis software (v4.0; Bio-Rad Laboratories,
Inc.) was used to analyze the intensities of protein bands.
Reverse transcription-quantitative
(RT-q) PCR and transfection
RNA samples were isolated from tissues and cells
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. To obtain cDNA, RT
was conducted using the iScript kit (Bio Rad Laboratories, Inc.)
and the TaqMan RT kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturers' protocol. The
amplification of ATAD3 (forward primer, 5′-GCGAGCCACCGAGAAGATAAG-3′
and reverse primer, 5′-TGGACCATCTCATTGATGCGG-3′) and GAPDH (forward
primer, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse primer,
5′-GGCTGTTGTCATACTTCTCATGG-3′) was performed using iQSYBR Green
SuperMix (Bio Rad Laboratories, Inc.). The following thermocycling
conditions were used for qPCR: 95°C for 5 min, followed by 40
cycles at 95°C for 30 sec, 60°C for 40 sec and 72°C for 30 sec. The
quantification was performed using the 2−ΔΔCq method
(1,9).
Transfections were performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instruction. The
cancer cells were plated into a 6-well plate at a density of
4×105 cells/well, and were transfected with the negative
control (si-NC) or ATAD3a-targeting siRNA (si-ATAD3a). The siRNA
sequences are as follows: si-NC, AAGTCGGGTCAAGAGAAGC; and
si-ATAD3a, AGAUGGAGCUGAGGCAUAA. Plasmid (1 µg) was transfected into
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Following transfection for 7 h at 37°C,
the culture medium was replaced with fresh complete culture medium.
Subsequent experimentation were performed 24 h
post-transfection.
Colony formation assay
Cells were seeded into 6-well culture dishes and
incubated at 37°C in a humidified incubator for two weeks. Colonies
were fixed with 4% paraformaldehyde for 15 min and stained with
0.1% crystal violet for 15 min at room temperature in 20% ethanol
and counted. The colony forming cells were measured at a wavelength
of 590 nm, using a microplate reader (Molecular Devices, LLC), and
the total number was counted. The average and formation rates of
colony forming cells were measured.
Cell cycle and apoptosis assays
The Cell Cycle Analysis Kit (cat. no. FXP0311-100)
and Annexin V/PI apoptosis kit (cat. no. FXP018-100; both Beijing
4A Biotech Co., Ltd.) were used to assess cell cycle distribution
and apoptosis, respectively, per the manufacturer's instructions.
Cell apoptosis was measured via PI/Annexin V-APC staining and
analyzed via flow cytometry. Briefly, cells were harvested and
washed with PBS. After staining with PI and Annexin V, cell
apoptosis was detected on flow cytometry. Cell cycle was measured
via PI staining and analyzed via flow cytometry. Briefly, cells
were washed three times with iced PBS and fixed in 70% ethanol
overnight at 4°C. After staining with PI, cell cycle was detected
via flow cytometry. Flow cytometric data were analyzed using Kaluza
analysis software kaluza C Beckman analysis software (version 2.1,
Beckman Coulter, Inc.).
Immunohistochemistry
Tissue sections (3-µm-thick) were fixed with 10%
formaldehyde and embedded in paraffin, then they were heated at
60°C for 2 h, dewaxed for 20 min in xylene twice, and rehydrated in
gradient ethanol. Antigen repair was performed in citrate buffer
(pH 6.0, 10 mM) in the microwave (95°C for 15 min), and blocked
with 3% H2O2 for 15 min at 28°C. After
washing with PBS, the slices were incubated with 5% bovine serum
(Gene Tech) to block the nonspecific binding, and the anti-ATAD3a
antibody (Abcam; cat. no. ab112572; 1:200 dilution) was incubated
overnight at 4°C. After washing three times, the tissue slices were
incubated with horseradish peroxidase coupled double antibody
(dilution 1:300; Gene Tech; cat. no. 2017-416029) at room
temperature for 30 min, according to the manufacturer's
instructions. Tissue sections were immersed in
3,3-o-diamino-benzidine (Gene Tech) for 2 min, stained with
hematoxylin, dehydrated and observed under a light microscope
(Olympus BX51, ×400 magnification).
Statistical analysis
Data analysis was carried out using GraphPad Prism 5
software (GraphPad Software, Inc). The data were analyzed using the
Kruskal-Wallis test (non-parametric) followed by a suitable post
hoc multiple comparisons test (Tukey's or Bonferroni's) in the
event of a significant result. The data were expressed as the mean
± standard deviation. Pearson's correlation analysis was performed
to compare pairwise comparisons among all the samples. For
functional enrichment, Fisher's exact test was used to assess the
differences in expressed proteins between LUAD and adjacent
nontumor lung tissues. P<0.05 was considered to indicate a
statistically significant difference.
Results
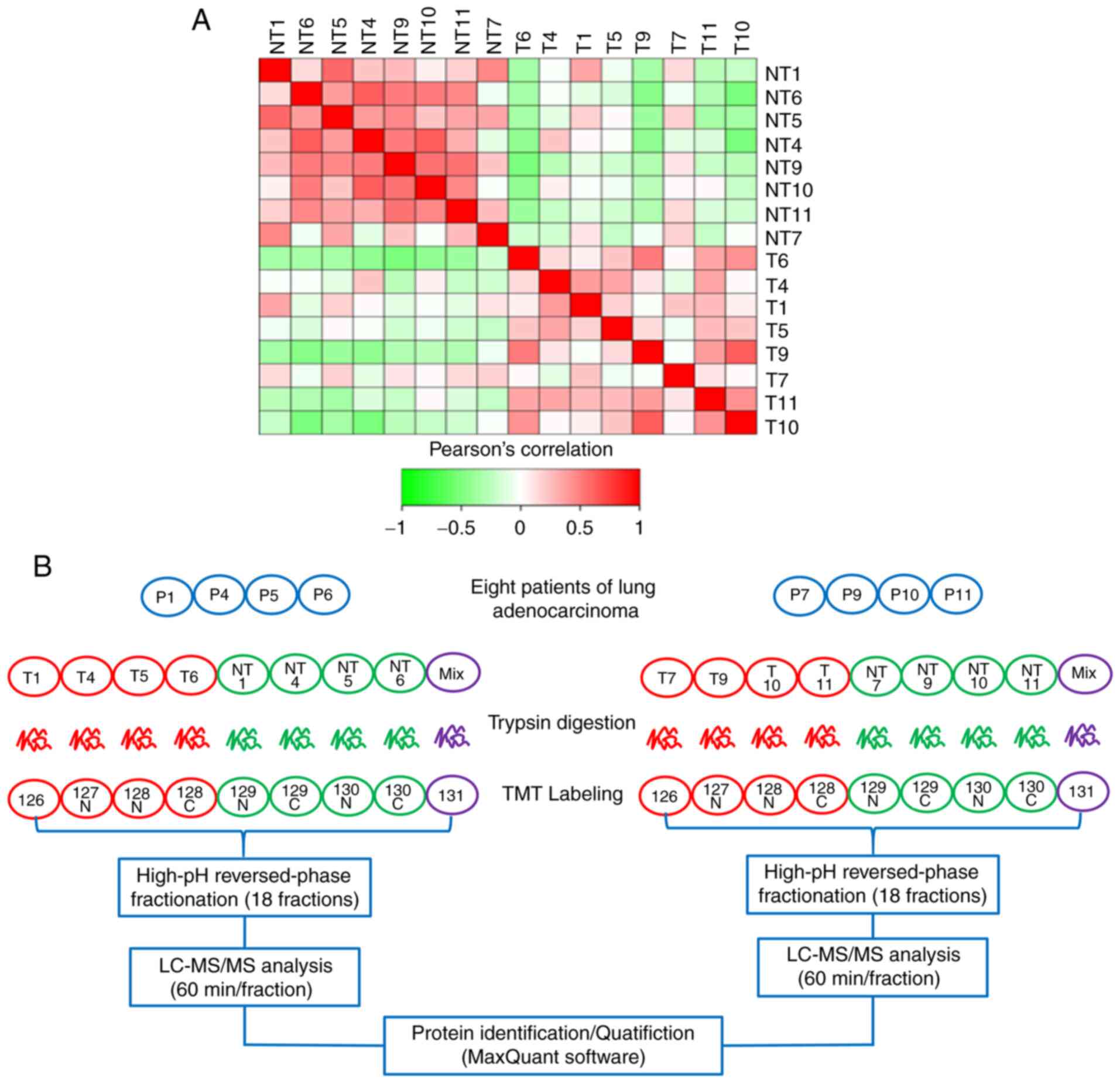
Experimental flow graph of
quantitative proteomics analysis
The quantitative results of biological repeat
samples were examined to determine whether they were statistically
consistent. Fig. 1A shows a heat map
of the Pearson's correlation coefficients of pairwise comparisons
among all the samples. The results showed a positive association
among all cancer samples and a negative association between
cancerous and normal samples, suggesting that the samples accorded
with statistical consistency. Next, to obtain the protein profile
of LUAD, cancer tissues and matched-adjacent nontumor tissues from
eight FFPE LUAD tissues were selected for proteomics analysis
according to strict standards. Tissue specimens from the eight
patients with LUAD were successfully analyzed by tandem mass
spectrometry combined with LC-MS/MS. Table II showed that mapping table of
sample name and labeling reagent. Two parallel TMT-10 PLEX
experiments were carried out; every four pairs of LUAD FFPE tissue
samples were mixed in each experiment (Fig. 1B).
 | Table II.Mapping table of sample name and
labeling reagent. |
Table II.
Mapping table of sample name and
labeling reagent.
| Sample name | Labeling
reagent | Sample name | Labeling
reagent |
|---|
| T1 | 126 | T7 | 126 |
| NT1 | 127N | NT7 | 127N |
| T4 | 128N | T9 | 128N |
| NT4 | 128C | NT9 | 128C |
| T5 | 129N | T10 | 129N |
| NT5 | 129C | NT10 | 129C |
| T6 | 130N | T11 | 130N |
| NT6 | 130C | NT11 | 130C |
| Mix | 131 | Mix | 131 |
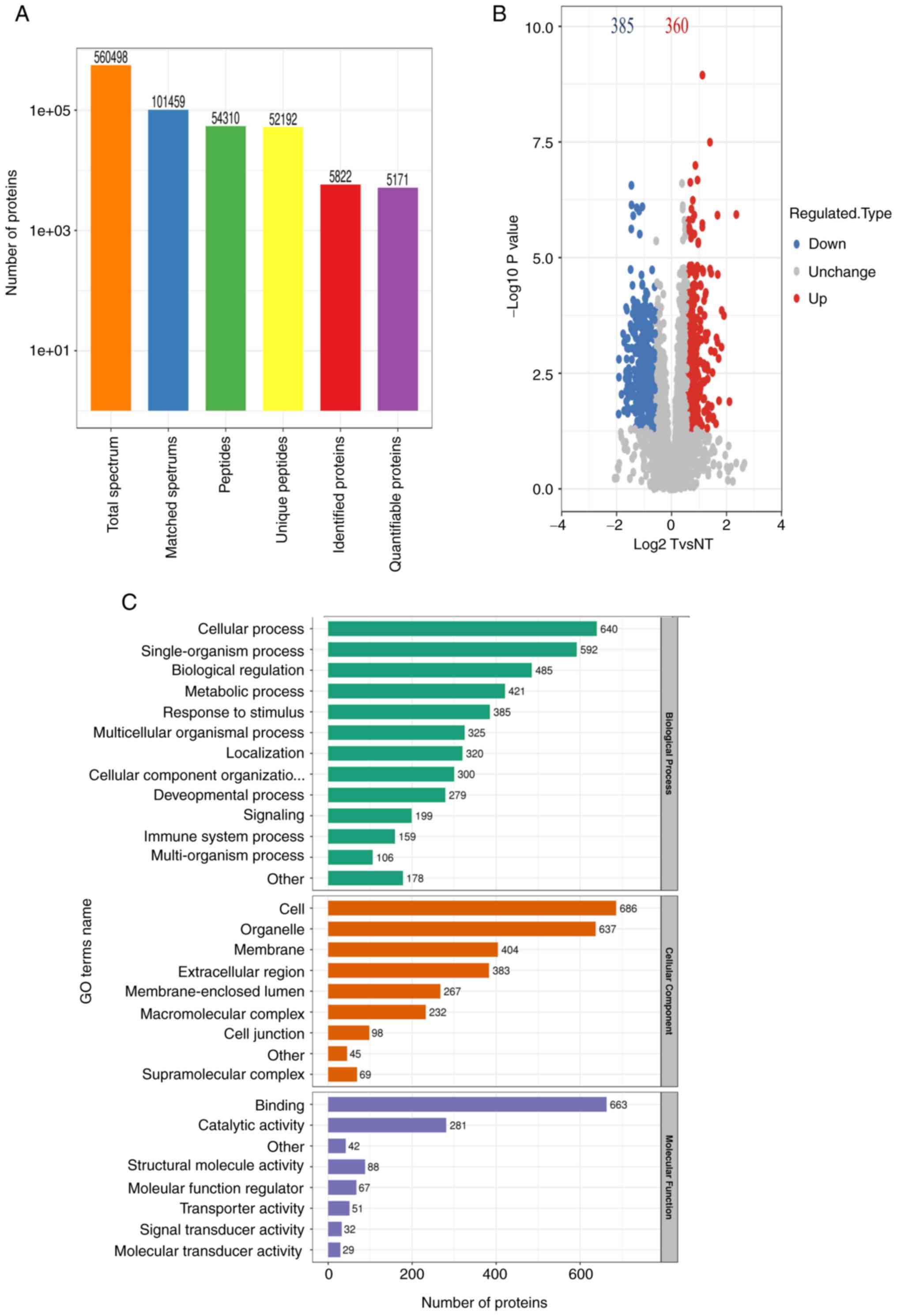
Identification and quantification of
proteins and GO functional annotation analysis of differentially
expressed proteins in LUAD
A total of 5,822 proteins were identified, and 5,171
proteins were quantified in eight pairs of LUAD FFPE tissue samples
(Fig. 2A). All proteins were
identified following false discovery rate correction. To identify
differentially expressed proteins, both a fold-change value and a
P-value were adopted for comparison, where P<0.05 and the
fold-change >1.5 or <0.67 were considered as significant.
Finally, 360 high-expression proteins and 385 low-expression
proteins were obtained in LUAD (Fig.
2B). Functional annotation of differentially expressed proteins
was performed by GO analysis. Identified proteins were assigned to
specific functions, including biological process (BP), cellular
component (CC) and molecular function (MF). BP annotation results
indicated that the differentially expressed proteins were primarily
involved in ‘cellular process’, ‘single-organism process’,
‘biological regulation’ and ‘metabolic process’, as well as
‘response to stimulus’. According to CC annotation, a large
majority of the differentially expressed proteins originated from
‘cell’, ‘organelle’, ‘membrane’ and ‘extracellular region’
(Fig. 2C). Using MF annotation, it
was found that differentially expressed proteins were primarily
associated with ‘binding’, ‘structural molecule activity’ and
‘catalytic activity’.
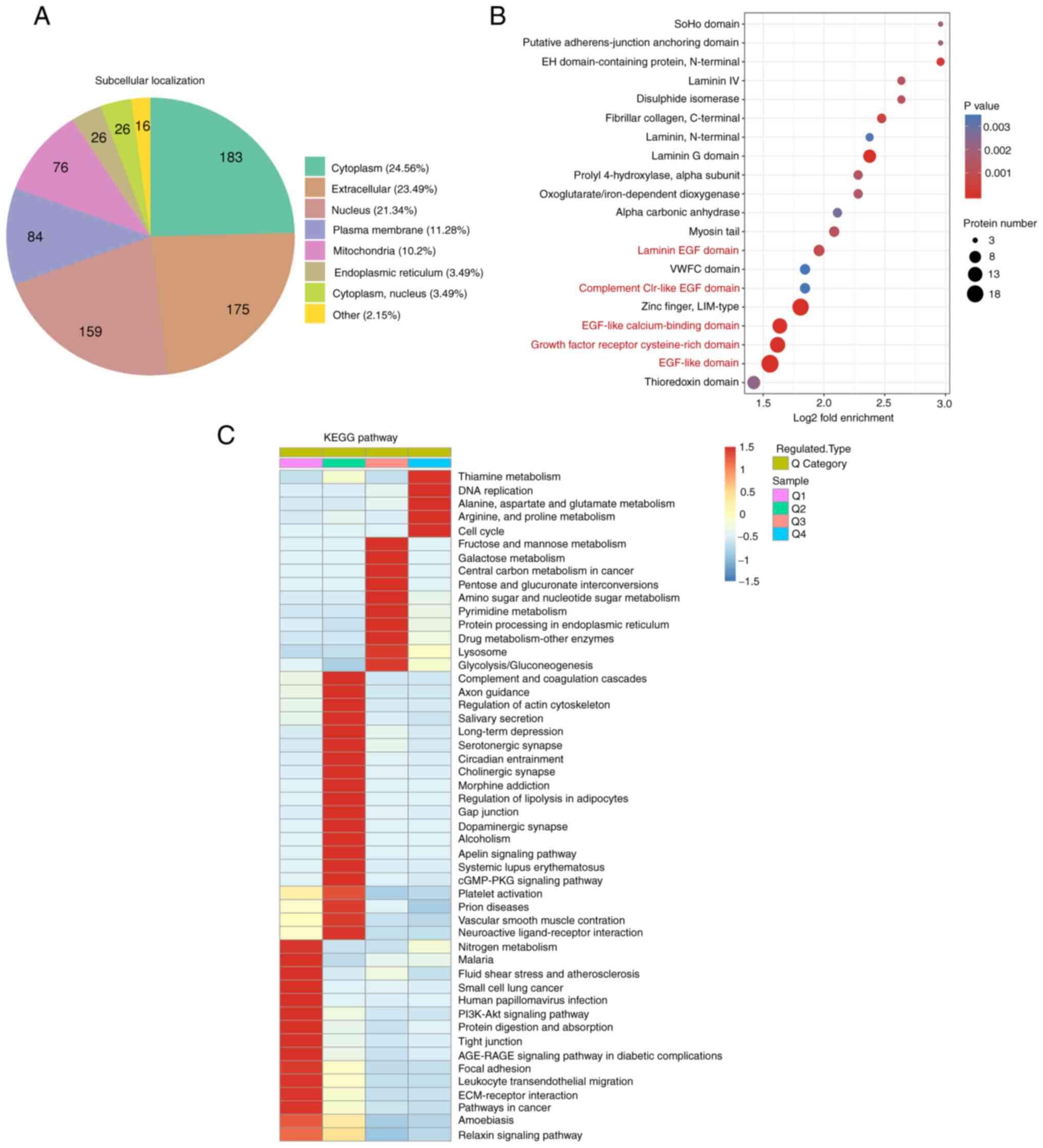
Subcellular location, protein domain
and KEGG enrichment analysis of differentially expressed proteins
in LUAD
Subcellular location analysis was conducted to
further investigate the differentially expressed proteins
associated with LUAD. In the present study, the most differentially
expressed proteins were localized to the cytoplasm, extracellular
matrix and nucleus (Fig. 3A).
Proteomics was also used to investigate the mechanisms and novel
targets of immunotherapy; 19 differentially expressed membrane
proteins were primarily involved in immune processes (Table III), indicating that these cell
membrane proteins may be potential immunotherapeutic targets.
 | Table III.Differentially expressed membrane
proteins in LUAD tissues and matched-adjacent nontumor tissues. |
Table III.
Differentially expressed membrane
proteins in LUAD tissues and matched-adjacent nontumor tissues.
| Protein
accession | Gene name | T/NT Ratio | Regulation
type | T/NT P-value |
|---|
| P53985 | SLC16A1 | 2.272 | Up | 0.021444 |
| P50281 | MMP14 | 1.944 | Up | 0.010844 |
| O15427 | SLC16A3 | 1.823 | Up | 0.010179 |
| P53634 | CTSC | 1.798 | Up | 0.009536 |
| Q07812 | BAX | 1.732 | Up | 0.000204 |
| O15260 | SURF4 | 1.619 | Up | 0.002158 |
| O96005 | CLPTM1 | 1.589 | Up | 0.006359 |
| Q03519 | TAP2 | 1.553 | Up | 0.018917 |
| Q8IWA5 | SLC44A2 | 0.568 | Down | 0.000458 |
| P08473 | MME | 0.553 | Down | 0.003921 |
| P56199 | ITGA1 | 0.541 | Down | 0.000276 |
| P04003 | C4BPA | 0.53 | Down | 0.003395 |
| P07204 | THBD | 0.517 | Down | 0.000996 |
| P98172 | EFNB1 | 0.486 | Down | 0.00156 |
| P08514 | ITGA2B | 0.48 | Down | 0.007982 |
| P56856 | CLDN18 | 0.463 | Down | 0.019284 |
| P04921 | GYPC | 0.367 | Down | 0.000904 |
| P16671 | CD36 | 0.338 | Down | 0.007999 |
Through fold enrichment, a number of differentially
expressed proteins were enriched in ‘EGF-like calcium-binding
domain’, ‘EGF-like domain’ and ‘laminin EGF domain’ (Fig. 3B and Table IV), suggesting that the EGF domain
and EGF-like domain play an important role in LUAD. KEGG is a
bioinformatics resource for identifying the functions and cells
types of organisms (14). KEGG
pathway analysis revealed that pathways involved in ‘thiamine
metabolism’ and ‘DNA replication’ were highly expressed in cancer
tissues (Fig. 3C). This was in
accordance with the strong proliferative capacity and high mutation
rates of cancer cells. The study also showed that ‘tight junction’,
‘PI3K-Akt signaling pathway’ and ‘nitrogen metabolism’ were
downregulated in cancer tissues (Fig.
3C). Furthermore, several metabolic pathways, including
‘glutamate metabolism’ and ‘fructose and galactose metabolism’,
were significantly enriched in cancer tissues (Fig. 3C). Furthermore, enrichment analysis
demonstrated that significantly upregulated proteins were
associated with the ‘endoplasmic reticulum lumen’, ‘protein
disulfide isomerase activity’, ‘vitamin binding’ and ‘cell cycle
G1/S phase transition’.
 | Table IV.Differentially expressed EGF
domain-related proteins in LUAD tissues and matched-adjacent
nontumor tissues. |
Table IV.
Differentially expressed EGF
domain-related proteins in LUAD tissues and matched-adjacent
nontumor tissues.
| Domain
description | Gene name | T/NT ratio | Regulation
type |
|---|
| Laminin EGF
domain | CRELD2 | 1.86 | Up |
| Laminin EGF
domain | LAMA3 | 0.4 | Down |
| Laminin EGF
domain | HSPG2 | 0.5 | Down |
| Laminin EGF
domain | LAMB2 | 0.39 | Down |
| Laminin EGF
domain | LAMC1 | 0.41 | Down |
| Laminin EGF
domain | LAMA5 | 0.41 | Down |
| Laminin EGF
domain | AGRN | 0.6 | Down |
| EGF-like domain,
extracellular | TNC | 1.94 | Up |
| EGF-like domain,
extracellular | TNXB | 0.54 | Down |
| EGF-like
domain | FBLN5 | 0.48 | Down |
| EGF-like
domain | LTBP4 | 0.54 | Down |
| EGF-like
domain | CRELD2 | 1.86 | Up |
| EGF-like
domain | LAMA3 | 0.4 | Down |
| EGF-like
domain | LTBP2 | 0.66 | Down |
| EGF-like
domain | NID2 | 0.58 | Down |
| EGF-like
domain | EFEMP1 | 0.55 | Down |
| EGF-like
domain | HSPG2 | 0.5 | Down |
| EGF-like
domain | FBLN2 | 0.65 | Down |
| EGF-like
domain | LAMB2 | 0.39 | Down |
| EGF-like
domain | THBS2 | 3.19 | Up |
| EGF-like
domain | TNC | 1.94 | Up |
| EGF-like
domain | TNXB | 0.54 | Down |
| EGF-like
domain | NID1 | 0.41 | Down |
| EGF-like
domain | LAMC1 | 0.41 | Down |
| EGF-like
domain | THBD | 0.52 | Down |
| EGF-like
domain | LDLR | 0.64 | Down |
| EGF-like
domain | F9 | 0.58 | Down |
| EGF-like
domain | LAMA5 | 0.41 | Down |
| EGF-like
domain | AGRN | 0.6 | Down |
| EGF-like
calcium-binding domain | FBLN5 | 0.48 | Down |
| EGF-like
calcium-binding domain | LTBP4 | 0.54 | Down |
| EGF-like
calcium-binding domain | CRELD2 | 1.86 | Up |
| EGF-like
calcium-binding domain | LTBP2 | 0.66 | Down |
| EGF-like
calcium-binding domain | NID2 | 0.58 | Down |
| EGF-like
calcium-binding domain | EFEMP1 | 0.55 | Down |
| EGF-like
calcium-binding domain | HSPG2 | 0.5 | Down |
| EGF-like
calcium-binding domain | FBLN2 | 0.65 | Down |
| EGF-like
calcium-binding domain | THBS2 | 3.19 | Up |
| EGF-like
calcium-binding domain | NID1 | 0.41 | Down |
| EGF-like
calcium-binding domain | THBD | 0.52 | Down |
| EGF-like
calcium-binding domain | F9 | 0.58 | Down |
| EGF-like
calcium-binding domain | AGRN | 0.6 | Down |
| EGF domain | NID2 | 0.58 | Down |
| EGF domain | THBS2 | 3.19 | Up |
| EGF domain | NID1 | 0.41 | Down |
Cluster analysis of GO enrichment of
differentially expressed proteins in LUAD
When further performing GO functional enrichment
analysis, it was found that 360 high-expression and 385
low-expression proteins in cancer tissues were considerably
differently expressed in CC, MF and BP. In CC, the upregulated
proteins in cancer tissues were mostly enriched in ‘endoplasmic
reticulum (ER) lumen’, while the downregulated proteins mostly
focused on ‘extracellular space’, ‘proteinaceous extracellular
matrix’ and ‘extracellular region’ (Fig.
4A). A large number of upregulated ER and Golgi proteins
indicated that protein synthesis and amino acid metabolism were
accelerated in patients with LUAD, and the large-scale
downregulation of extracellular matrix proteins may be associated
with the low production of extracellular mucus. In MF, the most
highly expressed proteins were associated with ‘isomerase activity’
and ‘procollagen-proline dioxygenase activity’, and most of the
low-expression proteins were associated with ‘structural molecule
activity’ and ‘actin binding’ (Fig.
4B). In BP, high-expression proteins were mostly associated
with metabolism, while low-expression proteins were mainly enriched
in multicellular processes and cell adhesion (Fig. 4C). The upregulation of metabolic
processes was consistent with most features of cancer.
Protein biomarker validation and
overview of kinases and post-translational modification enzymes in
LUAD
Cancer cells require profound remodeling of cellular
metabolism to meet the energy requirements for aerobic glycolysis
rather than mitochondrial oxidative phosphorylation (even under
aerobic conditions), a common phenomenon known as the Warburg
effect. In the present study, the expression of hexokinase 2, an
important rate-limiting enzyme in aerobic glycolysis, was
upregulated in cancer tissues (Fig.
5A). Moreover, the expression of some other important enzymes
in aerobic glycolysis was also upregulated in cancer tissues
(Fig. 5A).
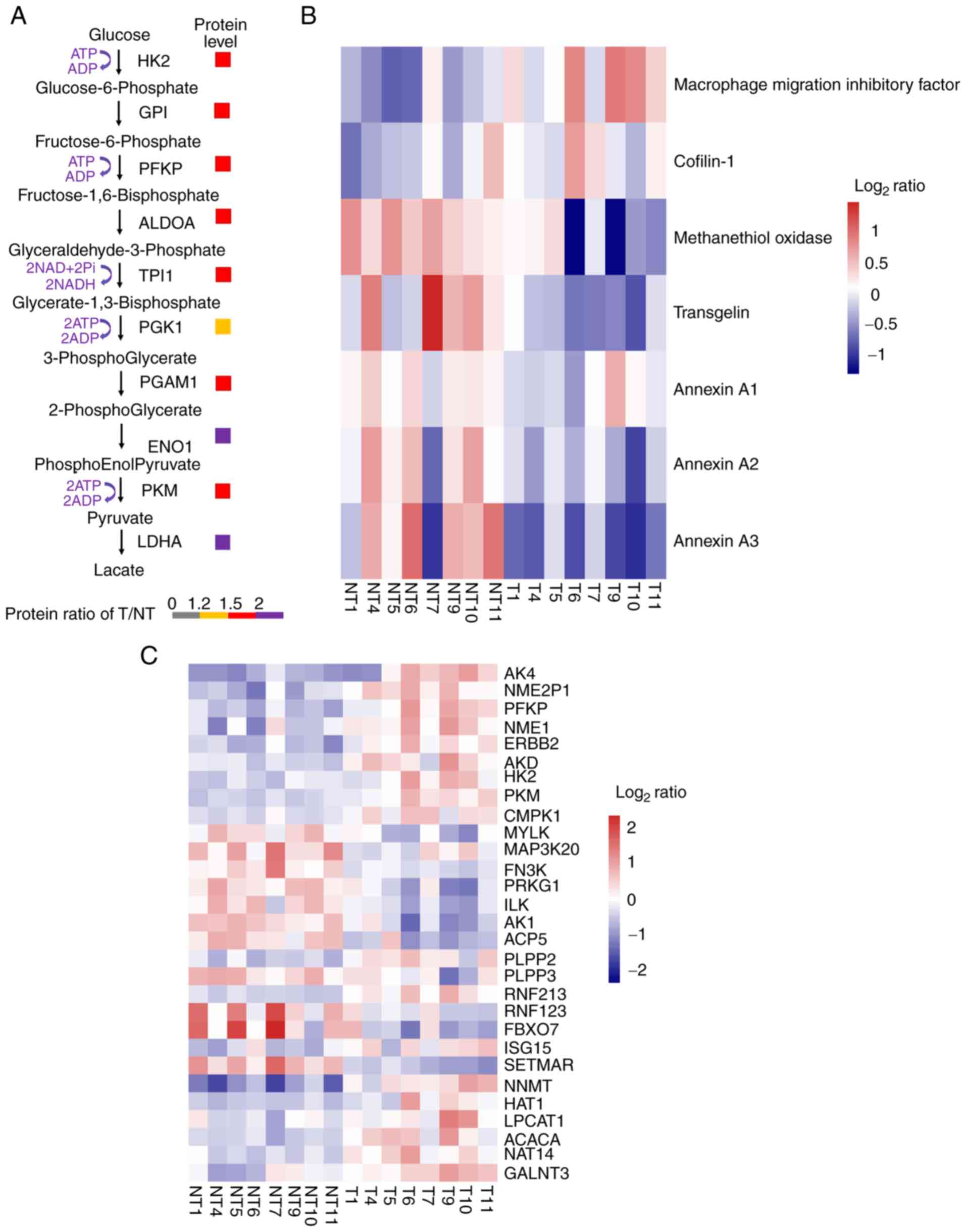 | Figure 5.Protein biomarker validation,
overview of kinases and post-translational modification enzymes in
lung adenocarcinoma. (A) Fold-change between tumor and
corresponding normal tissues. (B and C) Level of change of T/NT
(i.e., either increase or decrease) in protein expression was
determined as: i) Low (−1≤log2 ratio<0); ii) medium
(log2 ratio=0); or iii) high (0<log2
ratio≤1.0). (HK2, hexokinase 2; GPI, glucose 6 phosphate isomerase;
PFKP, phosphofructokinase; ALDOA, fructose-bisphosphate aldolase A;
TPI1, triosephosphate isomerase 1; PGK1, phosphoglycerate kinase 1;
PGAM1, phosphoglycerate mutase 1; ENO1, enolase 1α; PKM, pyruvate
kinase M; LDHA, lactate dehydrogenase A; AK4, adenylate kinase 4;
NME2P, non-metastatic2 pseudogene 1; PFKP, phosphofructokinase,
platelet; NME1, non-metastatic cells 1; ERBB2, V-erb-b2
erythroblastic leukemia viral oncogene homolog 2; ADK, adenylate
kinase; CMPK1, cytidine monophosphate kinase 1; MYLK, myosin light
chain kinase; MAP3K20, mitogen-activated protein kinase kinase
kinase 20; FN3K, fructosamine-3-kinase; PRKG1, protein kinase, cGMP
dependent type I; ILK, integrin-1inked kinase; AK1, adenylate
kinase 1; ACP5, acid phosphatase 5 tartrate resistant; PLPP2/3,
phospholipid phosphatase 2/3; RNF213/123, E3 ubiquitin-protein
ligase RNF213/123; FBXO7, F-box only protein 7; ISG15,
interferon-stimulated gene 15; SETMAR, SET domain and mariner
transposase fusion gene; NNMT, nicotinamide N-methyltransferase;
HAT1, histone acetyltransferase 1; LPCAT1,
lysophos-phatidylcholineacyltransferase 1; ACACA, acetylcoenzyme A
carboxylase α; NAT14, N-acetyltransferase 14; GALNT3, polypeptide
N-acetylgala-ctosaminyltransferase 3. |
Effective biomarkers are essential to improving
diagnosis and treatment in a number of human diseases, including
cancer, and some proteins were reported as biomarkers of LUAD in a
previous proteomics study (4,15), the
results of which are consistent with those of the present study. As
shown in Fig. 5B, the expression of
macrophage migration inhibitory factor and cofilin-1 was
upregulated, while the expression of methanethiol oxidase,
transgelin, annexin A1, A2 and A3 was downregulated in LUAD.
Post-translational modification (PTM) of proteins
plays a vital role in the signaling networks of cancer and the
regulation of cellular processes, including proliferation, cellular
localization of proteins, and protein complex formation (16). Protein kinases, the key regulators of
cellular function, not only regulate the process of
phosphorylation, but are themselves regulated by phosphorylation
and dephosphorylation processes (17). In the present study, enrichment
analysis results showed that the expression levels of some kinases
significantly changed across all eight samples. For example, the
expression of adenylate kinase 4 and phosphofructokinase platelet
were upregulated in cancer tissues (Fig.
5C). For ubiquitination regulatory proteins, the expression of
E3 ubiquitin-protein ligase ring finger protein 213 (RNF213) was
upregulated, while the expression of E3 ubiquitin-protein ligase
RNF123 was downregulated in LUAD. In addition, the expression of
F-box only protein 7 was increased in LUAD. For methylation, the
expression of recombinant myosin light chain kinase was
downregulated in LUAD, suggesting that kinases and PTM of proteins
plays a significant role in LUAD.
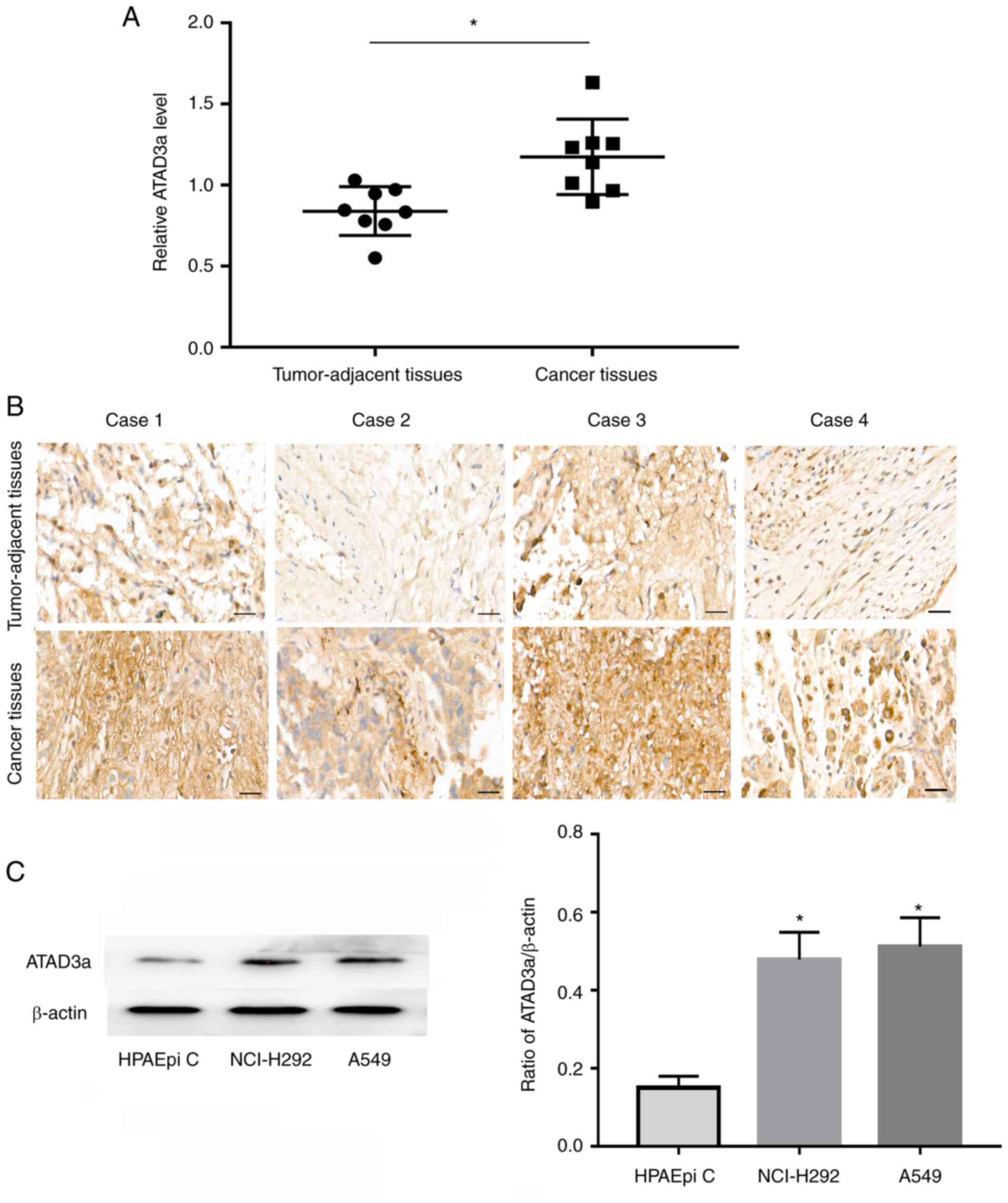
ATAD3a expression is upregulated in
LUAD
Proteomics analysis revealed that ATAD3a was more
highly expressed in LUAD than in adjacent nontumor tissues
(Fig. 6A). To verify the accuracy of
the proteomics analysis, immunohistochemical analysis of ATAD3a
expression was performed using FFPE tissue sections. The results
showed that ATAD3a expression was significantly higher in cancer
tissues than in adjacent nontumor lung tissues (Fig. 6B), which is consistent with the
results of the proteomics analysis. Western blot results also
showed that ATAD3a expression was significantly higher in LUAD
cancer cell lines (A549 and NCI-H292) than in immortalized normal
alveolar epithelial cells (HPAEpi C) (Fig. 6C and D).
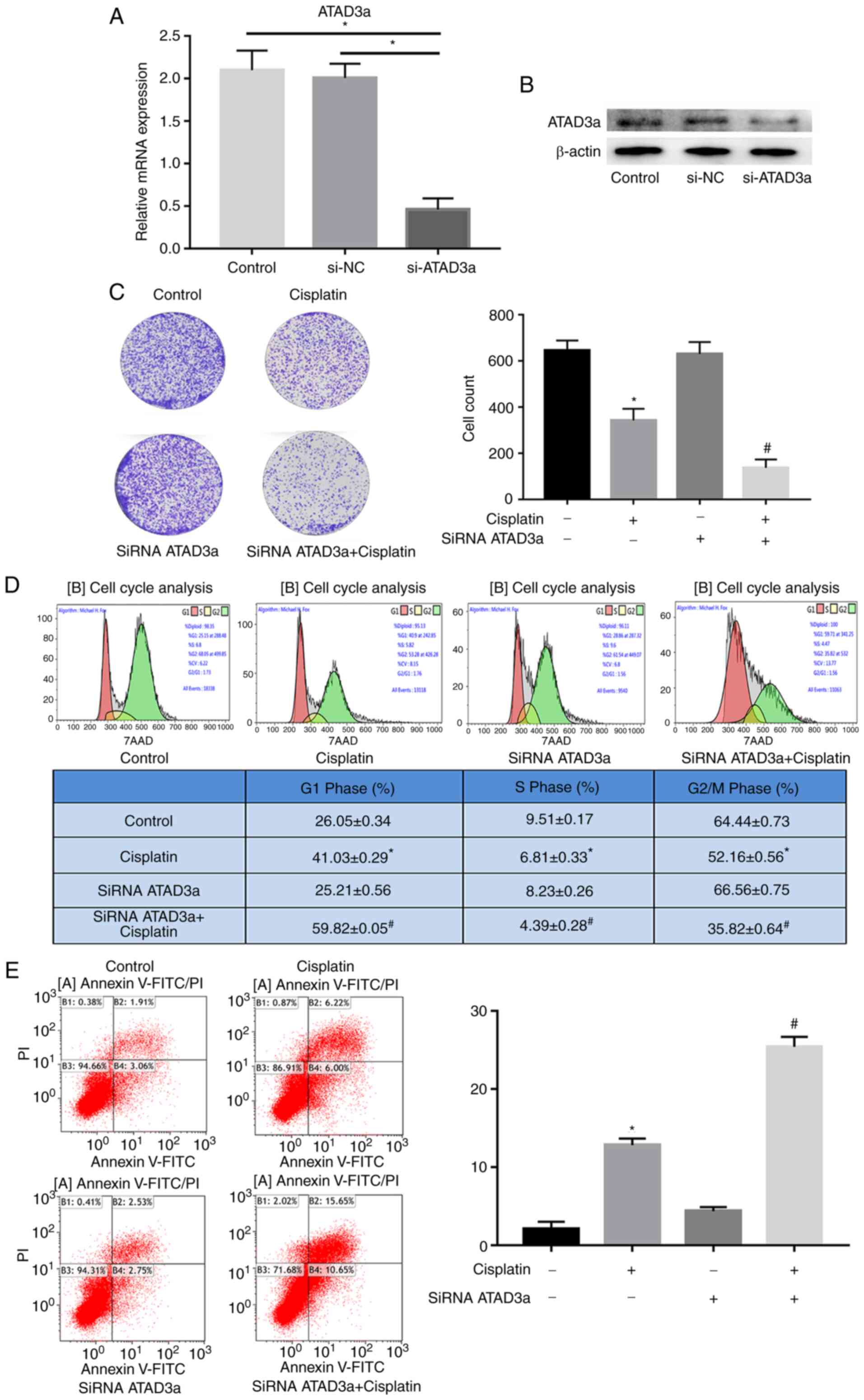
ATAD3a downregulation enhances the
sensitivity of LUAD cells to cisplatin by regulating cellular
proliferation, the cell cycle and apoptosis
To study the effects of ATAD3a expression on the
sensitivity of NCI-H292 cells to cisplatin, si-ATAD3a was used to
knockdown expression. RT-qPCR and western blot analyses showed that
ATAD3a mRNA and protein expression were significantly downregulated
in NCI-H292 cells transfected with si-ATAD3a, compared with the
control and si-NC groups (Fig. 7A and
B). As predicted, colony formation assays demonstrated that
si-ATAD3a increased cisplatin-induced growth inhibition in NCI-H292
cells (Fig. 7C). Flow cytometry
revealed that si-ATAD3a increased cisplatin-induced
G1-stage arrest and apoptosis in NCI-H292 cells
(Fig. 7D and E). These data
indicated that knocking down ATAD3a enhanced the sensitivity of
NCI-H292 cells to cisplatin by regulating proliferation, the cell
cycle and apoptosis.
Discussion
Lung cancer remains the leading cause of
cancer-related deaths among women and men worldwide. Moreover,
<15% of patients survive for 5 years following diagnosis at the
advanced stages of disease (1,18). Lung
cancer is categorized as NSCLC and SCLC based on histological
characteristics. NSCLC, which accounts for ~85% of all lung
cancers, can be further divided into two major histological
subtypes, LUAD and LUSC (19). LUAD
is found in 60% of this population. Despite efforts to improve
effective treatment, the prognosis of LUAD remains poor due to the
lack of specific biomarkers and personalized medical regimes
(3).
Proteomics not only gains insights into the overall
landscape of proteome-wide alterations of post-translational
modifications and signaling pathways, but also offers information
on protein-protein interactions and the associated underlying
mechanisms (20). FFPE tissue
samples offer comprehensive information for clinical and biomarker
research, and have been shown to be a rich resource for clinical
research and the most widely available tissue preservation method
(21,22). Using high-throughput proteomics
analysis, Zhu et al (22)
confirmed that FFPE tissue samples had great potential for the
identification of novel, promising protein biomarkers for prostate
carcinoma (22). In the present
study, FFPE LUAD and adjacent-nontumor lung tissues were selected
for proteomics analysis.
Immunotherapy can promote the anticancer properties
of the immune system, and hence plays an important role in LUAD
treatment (23–25). Compared with cytotoxic chemotherapy,
immunotherapy has a unique toxicity profile and the ability to
provide prolonged clinical benefit for patients (24). Marinelli et al (26) confirmed that the efficacy of
immunotherapy had notable interpatient heterogeneity in patients
with LUAD. In addition, immune checkpoint inhibitors were of
benefit to a great deal patients with LUAD, and a small mutation
panel of coding sequences could predict the long-term outcome of
immunotherapy (27). In the current
study, 19 differentially expressed membrane proteins were found to
be associated with immune processes, indicating that these cell
membrane proteins may be potential immunotherapeutic targets. The
subcellular location patterns of proteins provided crucial
information for understanding the underlying mechanism and
functions of specific proteins, which varied across different
pathological types and cellular conditions (28). In the present study, the most
differentially expressed proteins were localized to the cytoplasm,
followed by the extracellular matrix, nucleus and plasma
membrane.
A large number of recent studies have shown that
protein domains serve a vital role in tumorigenesis and progression
(29–32). Numerous proteins comprise more than
one domain, and each domain, a stable unit of the protein
structure, is responsible for a particular molecular function. In
the present study, ‘EGF-like calcium-binding domain’, ‘EGF-like
domain’ and ‘laminin EGF domain’ showed considerable differential
expression levels between LUAD and adjacent-nontumor lung tissues
(33). EGF is a growth factor that
stimulates cellular proliferation and differentiation (30). EGF binds to the extracellular domain
of EGFR and induces its dimerization, activating its intrinsic
kinase activity (34,35). EGF/EGFR signaling has been recognized
as an important molecular target in cancer therapy. The tyrosine
kinase inhibitors (TKIs) of EGFR are regarded as the first-line
standard option for patients with LUAD with EGFR mutations, and
EGFR mutation status determines the effectiveness of cancer
therapies. Liang et al (36)
confirmed that the EGFR mutation p.T790M was the most prominent
resistance mechanism to TKI therapy in LUAD. Moreover, they also
confirmed that after acquiring afatinib resistance, the p.T790M
mutation could not occur in patients with LUAD who also possessed
EGFR p.L747P or p.L747S mutations (36). In follow-up studies, further in
vivo and in vitro experiments are required to explore
the function or purpose of EGF-related domains and EGF-EGFR
signaling in LUAD.
The basic characteristics of cancer cells primarily
include strong proliferative capacity, high mutation rates and
changes in metabolic pathways. Cancer cells have longer telomerases
than normal cells, which are always in an active state, and allow
cancer cells to continuously divide (37–39).
Through KEGG analysis, pathways involved in cellular proliferation
and DNA replication were found to be highly expressed in LUAD
cancer tissues. Cancer is also a metabolic disease characterized by
metabolic reprogramming to support rapid cellular proliferation. Ni
et al (40) indicated that
LUAD cells favored the Warburg effect even under oxygen-rich
conditions, and that the metabolism of fatty acids and amino acids
was associated with the occurrence and progression of cancer
(40). In the present study, changes
in some of the most important metabolic pathways, such as thiamine,
alanine, aspartate and glutamate metabolism, occurred in patients
with LUAD.
Cluster analysis of GO functional enrichment
revealed that significantly upregulated proteins were primarily
associated with ‘ER lumen’, ‘protein disulfide isomerase activity’,
‘vitamin binding’ and ‘cell cycle G1/S phase
transition’. These results suggested that compared with
paracancerous tissues, the abnormal expression of multiple proteins
was related to ‘response’, ‘binding’ or ‘activity’ in LUAD. These
findings broaden the current understanding of LUAD. Tumor
biomarkers are defined as molecular or process-based changes that
reflect the status of an underlying malignancy. They can be divided
into three categories: i) Diagnostic biomarkers, (monitoring of
asymptomatic individuals and detection of early-stage cancer); ii)
prognostic biomarkers (predicting disease outcome and monitoring
disease recurrence); and iii) predictive biomarkers (monitoring
sensitivity to therapy and therapy response and aiding treatment
decisions) (41–43).
PTM enzymes perform >200 types of
post-translational modifications, including acetylation,
ubiquitination, phosphorylation, nitrosylation and methylation
(44), and affect nearly all aspects
of cancer biology and pathogenesis. A number of studies have
confirmed that PTMs may serve as biomarkers of cancer (44). The present study not only validated
seven LUAD-specific biomarkers by enrichment analysis, but also
found that the expression of numerous kinases and PTM enzymes was
significantly upregulated across all eight LUAD samples compared
with paracarcinoma tissues, suggesting that these proteins had the
potential to be novel biomarkers and may be beneficial to the
development of personalized treatments plans. Because the early
symptoms of LUAD are not apparent in the early stages of disease,
50–60% of LUAD is confirmed at the advanced stage. Cisplatin is the
most commonly used platinum-based drug, and plays an important role
in advanced LUAD. Patients are susceptible to cisplatin in early
treatment. However, resistance to LUAD has become a key obstacle in
the effective treatment of patients. ATAD3a, an integral
mitochondrial membrane protein, has been reported to be closely
associated with cancer cell metabolism, proliferation, apoptosis
and chemotherapy (45). Therefore,
the present study aimed to explore the role and mechanism of ATAD3a
in LUAD. ATAD3a expression was measured by immunohistochemical and
western blot analyses, and the results were in accordance with
those of the proteomics data analysis. The study also confirmed
that ATAD3a downregulation enhanced the sensitivity of LUAD cells
to cisplatin by regulating cellular proliferation, the cell cycle
and apoptosis. However, the specific molecular mechanisms
underlying ATAD3a upregulation in LUAD remain unknown. Hence,
further studies should be conducted to determine the potential
involvement of ATAD3a in LUAD. To conclude, the results of the
present study provided an improved understanding of potential
therapeutic targets for patients with LUAD.
Acknowledgements
The authors would like to thank Mr. Bin Wang (M.S.
Sales Representative at PTM Biolabs, Inc. Hangzhou, China) for TMT
labeling, HPLC fractionation and LC-MS/MS analysis.
Funding
The present study was supported by grants from the
Major Project of Science and Technology in Henan province (grant
no. 161100311400), the Medical Science and Technique Foundation of
Henan Provincial and ministry co-construction project (grant no.
SB201901084), and the Scientific Research Staring Foundation for
doctor of Henan Provincial People's Hospital (grant no.
201701).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QX performed most of the experiments and drafted the
initial manuscript. QX, ZL and LK were responsible for the study
conception and design. DW, XL and AH performed the experiments, and
analyzed and interpreted the data. HY, JT and PG performed
statistical and computational analyses. TS analyzed the data and
carried out the literature research. TS and LK supervised the
study, acquired funding, collected the tissues and edited the
manuscript. LK and TS confirmed the authenticity of the data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Henan Provincial People's Hospital (Zhengzhou,
China; approval no. 2019-050) and written informed consent was
provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bezzecchi E, Ronzio M, Semeghini V,
Andrioletti V, Mantovani R and Dolfini D: NF-YA overexpression in
lung cancer: LUAD. Genes (Basel). 11:1982020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu M, Wang D, Wang X and Zhang Y:
Correlation between mucin biology and tumor heterogeneity in lung
cancer. Semin Cell Dev Biol. 64:73–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haragan A, Field JK, Davies MPA, Escriu C,
Gruver A and Gosney JR: Heterogeneity of PD-L1 expression in
non-small cell lung cancer: Implications for specimen sampling in
predicting treatment response. Lung Cancer. 134:79–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Böttge F, Schaaij-Visser TB, de Reus I,
Piersma SR, Pham T, Nagel R, Brakenhoff RH, Thunnissen E, Smit EF
and Jimenez CR: Proteome analysis of non-small cell lung cancer
cell line secretomes and patient sputum reveals biofluid biomarker
candidates for cisplatin response prediction. J Proteomics.
196:106–119. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Tian M, Chang Y, Xue C and Li Z:
Investigation of structural proteins in sea cucumber
(Apostichopus japonicus) body wall. Sci Rep. 10:187442020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bradshaw RA, Hondermarck H and Rodriguez
H: Cancer proteomics and the elusive diagnostic biomarkers.
Proteomics. 19:e18004452019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang C, Yang S, Jin L, Dai G, Yao Q, Xiang
H, Zhang Y, Liu X and Xue B: Biological and clinical significance
of GATA3 detected from TCGA database and FFPE sample in bladder
cancer patients. Onco Targets Ther. 13:945–958. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong J, Ye Z, Clark CR, Lenz SW, Nguyen
JH, Yan H, Robertson KD, Farrugia G, Zhang Z, Ordog T and Lee JH:
Enhanced and controlled chromatin extraction from FFPE tissues and
the application to ChIP-seq. BMC Genomics. 20:2492019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greytak SR, Engel KB, Bass BP and Moore
HM: Accuracy of molecular data generated with FFPE biospecimens:
Lessons from the Literature. Cancer Res. 75:1541–157. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zakrzewski F, Gieldon L, Rump A, Seifert
M, Grützmann K, Krüger A, Loos S, Zeugner S, Hackmann K, Porrmann
J, et al: Targeted capture-based NGS is superior to multiplex
PCR-based NGS for hereditary BRCA1 and BRCA2 gene analysis in FFPE
tumor samples. BMC Cancer. 19:3962019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coscia F, Doll S, Bech JM, Schweize L,
Mund A, Lengyel E, Lindebjerg J, Madsen GI, Moreira JM and Mann M:
A streamlined mass spectrometry-based proteomics workflow for
large-scale FFPE tissue analysis. J Pathol. 251:100–112. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marchione DM, Ilieva I, Devins K, Sharpe
D, Pappin DJ, Garcia BA, Wilson JP and Wojcik JB: HYPERsol:
High-quality data from archival FFPE tissue for clinical
proteomics. J Proteome Res. 19:973–983. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lang L, Loveless R and Teng Y: Emerging
links between control of mitochondrial protein ATAD3A and cancer.
Int J Mol Sci. 21:79172020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Slizen MV and Galzitskaya OV: Comparative
analysis of proteomes of a number of nosocomial pathogens by KEGG
modules and KEGG pathways. Int J Mol Sci. 21:78392020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Camidge DR, Doebele RC and Kerr KM:
Comparing and contrasting predictive biomarkers for immunotherapy
and targeted therapy of NSCLC. Nat Rev Clin Oncol. 16:341–355.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han Z, Feng Y, Gu B, Li Y and Chen H: The
post-translational modification, SUMOylation, and cancer (Review).
Int J Oncol. 52:1081–1094. 2018.PubMed/NCBI
|
|
17
|
Lu Z and Hunter T: Metabolic kinases
moonlighting as protein kinases. Trends Biochem Sci. 43:301–310.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barton MK: Structured palliative care
program found to be helpful for caregivers of patients with lung
cancer. CA Cancer J Clin. 66:5–6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu J, Zhang C, Wang X, Zhai L, Ma Y, Mao
Y, Qian K, Sun C, Liu Z, Jiang S, et al: Integrative proteomic
characterization of human lung adenocarcinoma. Cell.
182:245–261.e17. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Keskin O, Tuncbag N and Gursoy A:
Predicting protein-protein interactions from the molecular to the
proteome level. Chem Rev. 116:4884–4909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Newton Y, Sedgewick AJ, Cisneros L,
Golovato J, Johnson M, Szeto CW, Rabizadeh S, Sanborn JZ, Benz SC
and Vaske C: Large scale, robust, and accurate whole transcriptome
profiling from clinical formalin-fixed paraffin-embedded samples.
Sci Rep. 10:175972020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu Y, Weiss T, Zhang Q, Sun R, Wang B, Yi
X, Wu Z, Gao H, Cai X, Ruan G, et al: High-throughput proteomic
analysis of FFPE tissue samples facilitates tumor stratification.
Mol Oncol. 13:2305–2328. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park C, Na KJ, Choi H, Ock CY, Ha S, Kim
M, Park S, Keam B, Kim TM, Paeng JC, et al: Tumor immune profiles
noninvasively estimated by FDG PET with deep learning correlate
with immunotherapy response in lung adenocarcinoma. Theranostics.
10:10838–10848. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu F, Zhan X, Zheng X, Xu H, Li Y, Huang
X, Lin L and Chen Y: A signature of immune-related gene pairs
predicts oncologic outcomes and response to immunotherapy in lung
adenocarcinoma. Genomics. 112:4675–4683. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Jiang W, Li T, Li M, Li X, Zhang Z,
Zhang S, Liu Y, Zhao W, Gu Y, et al: Identification of a small
mutation panel of coding sequences to predict the efficacy of
immunotherapy for lung adenocarcinoma. J Transl Med. 18:252020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marinelli D, Mazzotta M, Scalera S,
Terrenato I, Sperati F, D'Ambrosio L, Pallocca M, Corleone G,
Krasniqi E, Pizzuti L, et al: KEAP1-driven co-mutations in lung
adenocarcinoma unresponsive to immunotherapy despite high tumor
mutational burden. Ann Oncol. 12:1746–1754. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu C and Hwang MJ: Risk stratification for
lung adenocarcinoma on EGFR and TP53 mutation status, chemotherapy,
and PD-L1 immunotherapy. Cancer Med. 8:5850–5861. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du P and Xu C: Predicting multisite
protein subcellular locations: Progress and challenges. Expert Rev
Proteomics. 10:227–237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Laursen L, Karlsson E, Gianni S and Jemth
P: Functional interplay between protein domains in a supramodular
structure involving the postsynaptic density protein PSD-95. J Biol
Chem. 295:1992–2000. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dawson N, Sillitoe I, Marsden RL and
Orengo CA: The classification of protein domains. Methods Mol Biol.
1525:137–164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng W, Zhou X, Wu Q, Pearce R, Li Y and
Zhang Y: FUpred: Detecting protein domains through
deep-learning-based contact map prediction. Bioinformatics.
12:3749–3757. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Libiad M, Motl N, Akey DL, Sakamoto N,
Fearon ER, Smith JL and Banerjee R: Thiosulfate
sulfurtransferase-like domain-containing 1 protein interacts with
thioredoxin. J Biol Chem. 293:2675–2686. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu Y, Mehta-D'souza P, Biswas I,
Villoutreix BO, Wang X, Ding Q and Rezaie AR: Ile73Asn mutation in
protein C introduces a new N-linked glycosylation site on the first
EGF-domain of protein C and causes thrombosis. Haematologica.
105:1712–1722. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang F, Wang-Gou S, Cao H, Jiang N and Li
X: Proteomics identifies EGF-like domain multiple 7 as a potential
therapeutic target for epidermal growth factor receptor-positive
glioma. Cancer Commun (Lond). 40:518–530. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mizutani K, Guo X, Shioya A, Zhang J,
Zheng J, Kurose N, Ishibashi H, Motono N, Uramoto H and Yamada S:
The impact of PRDX4 and the EGFR mutation status on cellular
proliferation in lung adenocarcinoma. Int J Med Sci. 16:1199–1206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang S, Ko JC, Yang J and Shih JY:
Afatinib is effective in the treatment of lung adenocarcinoma with
uncommon EGFR p.L747P and p.L747S mutations. Lung Cancer.
133:103–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Neugut AI, MacLean SA, Dai W and Jacobson
JS: Physician characteristics and decisions regarding cancer
screening: A systematic review. Popul Health Manag. 22:48–62. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heldring N, Nyman U, Lönnerberg P,
Onnestam S, Herland A, Holmberg J and Hermanson O: NCoR controls
glioblastoma tumor cell characteristics. Neuro Oncol. 16:241–249.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang H, Gao J, Zhang R, Li M, Peng Z and
Wang H: Molecular and immune characteristics for lung
adenocarcinoma patients with CMTM6 overexpression. Int
Immunopharmacol. 83:1064782020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ni K, Wang D, Xu H, Mei F, Wu C, Liu Z and
Zhou B: miR-21 promotes non-small cell lung cancer cells growth by
regulating fatty acid metabolism. Cancer Cell Int. 19:2192019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Barton MK: Use of posttreatment imaging
and biomarker testing for survivors of breast cancer. CA Cancer J
Clin. 66:175–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
VanderLaan PA, Rangachari D, Majid A,
Parikh MS, Gangadharan SP, Kent MS, McDonald DC, Huberman MS,
Kobayashi SS and Costa DB: Tumor biomarker testing in
non-small-cell lung cancer: A decade of change. Lung Cancer.
116:90–95. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu Y, Jamal M, Xie T, Sun J, Song T, Yin
Q, Li J, Pan S, Zeng X, Xie S and Zhang Q: Uridine-cytidine kinase
2 (UCK2): A potential diagnostic and prognostic biomarker for lung
cancer. Cancer Sci. 110:2734–2747. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gu B and Zhu WG: Surf the
post-translational modification network of p53 regulation. Int J
Biol Sci. 8:672–684. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Teng Y, Lang L and Shay C: ATAD3A on the
path to cancer. Adv Exp Med Biol. 1134:259–269. 2019. View Article : Google Scholar : PubMed/NCBI
|