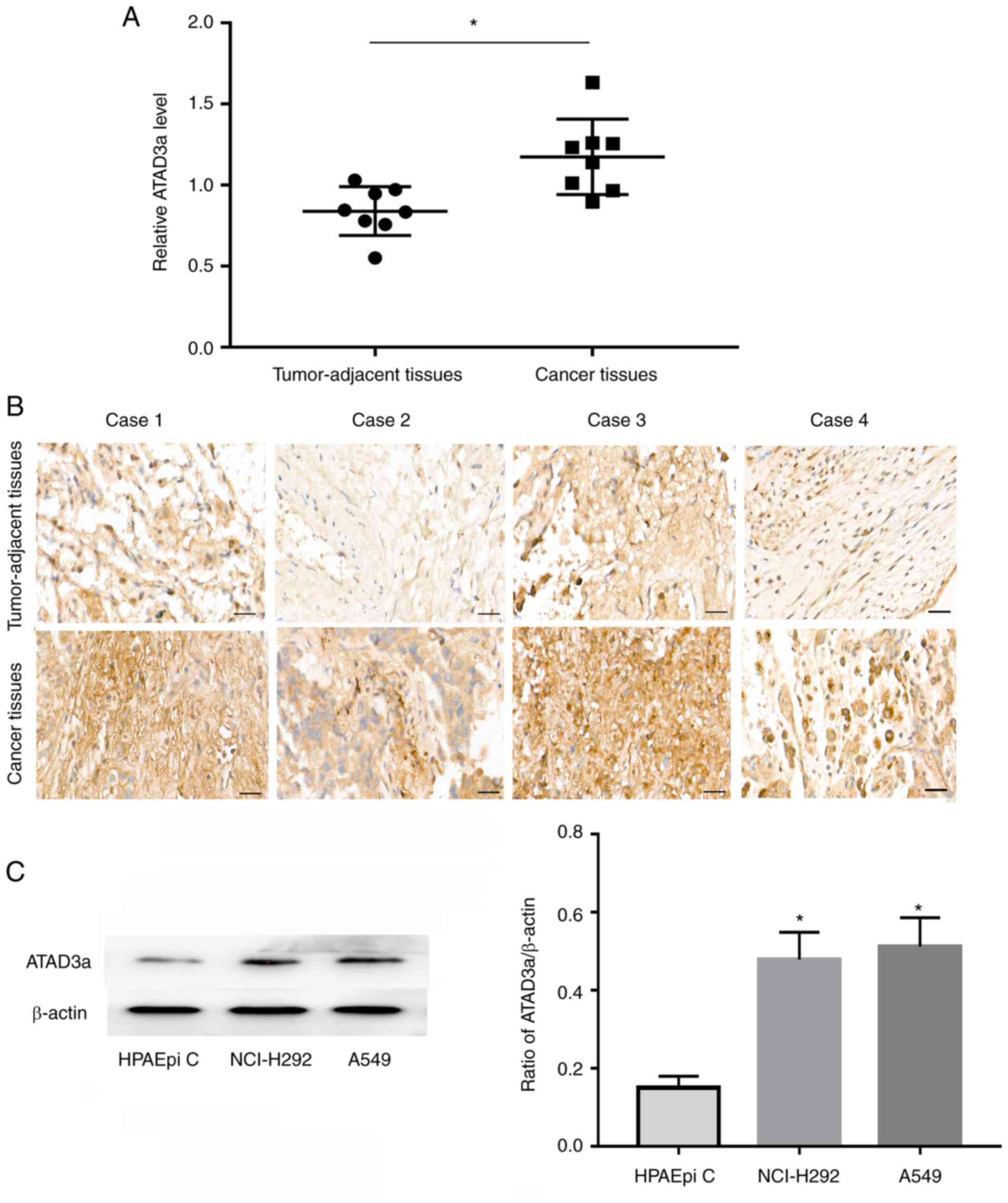
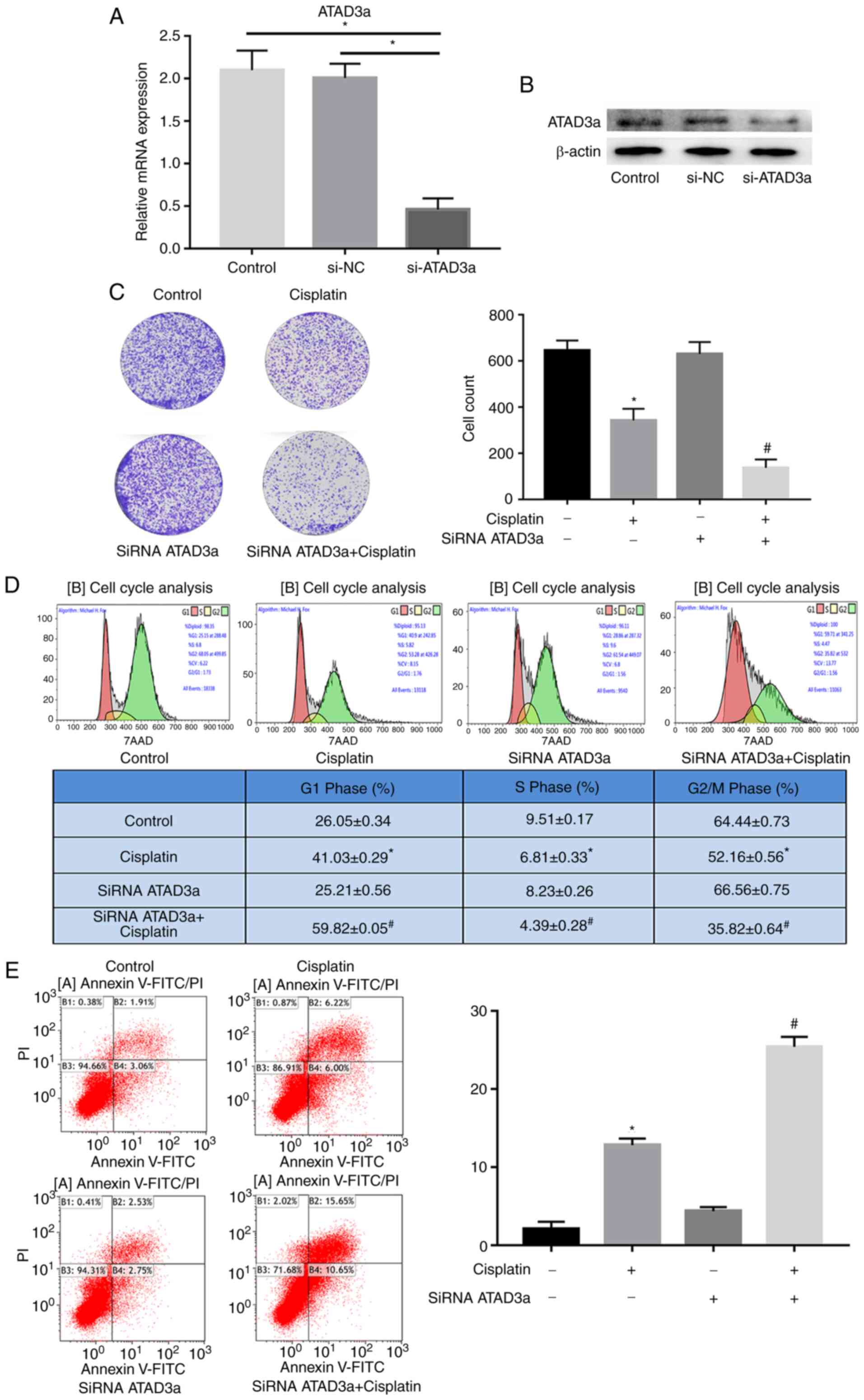
|
1
|
Bezzecchi E, Ronzio M, Semeghini V,
Andrioletti V, Mantovani R and Dolfini D: NF-YA overexpression in
lung cancer: LUAD. Genes (Basel). 11:1982020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu M, Wang D, Wang X and Zhang Y:
Correlation between mucin biology and tumor heterogeneity in lung
cancer. Semin Cell Dev Biol. 64:73–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haragan A, Field JK, Davies MPA, Escriu C,
Gruver A and Gosney JR: Heterogeneity of PD-L1 expression in
non-small cell lung cancer: Implications for specimen sampling in
predicting treatment response. Lung Cancer. 134:79–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Böttge F, Schaaij-Visser TB, de Reus I,
Piersma SR, Pham T, Nagel R, Brakenhoff RH, Thunnissen E, Smit EF
and Jimenez CR: Proteome analysis of non-small cell lung cancer
cell line secretomes and patient sputum reveals biofluid biomarker
candidates for cisplatin response prediction. J Proteomics.
196:106–119. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Tian M, Chang Y, Xue C and Li Z:
Investigation of structural proteins in sea cucumber
(Apostichopus japonicus) body wall. Sci Rep. 10:187442020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bradshaw RA, Hondermarck H and Rodriguez
H: Cancer proteomics and the elusive diagnostic biomarkers.
Proteomics. 19:e18004452019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang C, Yang S, Jin L, Dai G, Yao Q, Xiang
H, Zhang Y, Liu X and Xue B: Biological and clinical significance
of GATA3 detected from TCGA database and FFPE sample in bladder
cancer patients. Onco Targets Ther. 13:945–958. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong J, Ye Z, Clark CR, Lenz SW, Nguyen
JH, Yan H, Robertson KD, Farrugia G, Zhang Z, Ordog T and Lee JH:
Enhanced and controlled chromatin extraction from FFPE tissues and
the application to ChIP-seq. BMC Genomics. 20:2492019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greytak SR, Engel KB, Bass BP and Moore
HM: Accuracy of molecular data generated with FFPE biospecimens:
Lessons from the Literature. Cancer Res. 75:1541–157. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zakrzewski F, Gieldon L, Rump A, Seifert
M, Grützmann K, Krüger A, Loos S, Zeugner S, Hackmann K, Porrmann
J, et al: Targeted capture-based NGS is superior to multiplex
PCR-based NGS for hereditary BRCA1 and BRCA2 gene analysis in FFPE
tumor samples. BMC Cancer. 19:3962019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coscia F, Doll S, Bech JM, Schweize L,
Mund A, Lengyel E, Lindebjerg J, Madsen GI, Moreira JM and Mann M:
A streamlined mass spectrometry-based proteomics workflow for
large-scale FFPE tissue analysis. J Pathol. 251:100–112. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marchione DM, Ilieva I, Devins K, Sharpe
D, Pappin DJ, Garcia BA, Wilson JP and Wojcik JB: HYPERsol:
High-quality data from archival FFPE tissue for clinical
proteomics. J Proteome Res. 19:973–983. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lang L, Loveless R and Teng Y: Emerging
links between control of mitochondrial protein ATAD3A and cancer.
Int J Mol Sci. 21:79172020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Slizen MV and Galzitskaya OV: Comparative
analysis of proteomes of a number of nosocomial pathogens by KEGG
modules and KEGG pathways. Int J Mol Sci. 21:78392020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Camidge DR, Doebele RC and Kerr KM:
Comparing and contrasting predictive biomarkers for immunotherapy
and targeted therapy of NSCLC. Nat Rev Clin Oncol. 16:341–355.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han Z, Feng Y, Gu B, Li Y and Chen H: The
post-translational modification, SUMOylation, and cancer (Review).
Int J Oncol. 52:1081–1094. 2018.PubMed/NCBI
|
|
17
|
Lu Z and Hunter T: Metabolic kinases
moonlighting as protein kinases. Trends Biochem Sci. 43:301–310.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barton MK: Structured palliative care
program found to be helpful for caregivers of patients with lung
cancer. CA Cancer J Clin. 66:5–6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu J, Zhang C, Wang X, Zhai L, Ma Y, Mao
Y, Qian K, Sun C, Liu Z, Jiang S, et al: Integrative proteomic
characterization of human lung adenocarcinoma. Cell.
182:245–261.e17. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Keskin O, Tuncbag N and Gursoy A:
Predicting protein-protein interactions from the molecular to the
proteome level. Chem Rev. 116:4884–4909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Newton Y, Sedgewick AJ, Cisneros L,
Golovato J, Johnson M, Szeto CW, Rabizadeh S, Sanborn JZ, Benz SC
and Vaske C: Large scale, robust, and accurate whole transcriptome
profiling from clinical formalin-fixed paraffin-embedded samples.
Sci Rep. 10:175972020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu Y, Weiss T, Zhang Q, Sun R, Wang B, Yi
X, Wu Z, Gao H, Cai X, Ruan G, et al: High-throughput proteomic
analysis of FFPE tissue samples facilitates tumor stratification.
Mol Oncol. 13:2305–2328. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park C, Na KJ, Choi H, Ock CY, Ha S, Kim
M, Park S, Keam B, Kim TM, Paeng JC, et al: Tumor immune profiles
noninvasively estimated by FDG PET with deep learning correlate
with immunotherapy response in lung adenocarcinoma. Theranostics.
10:10838–10848. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu F, Zhan X, Zheng X, Xu H, Li Y, Huang
X, Lin L and Chen Y: A signature of immune-related gene pairs
predicts oncologic outcomes and response to immunotherapy in lung
adenocarcinoma. Genomics. 112:4675–4683. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Jiang W, Li T, Li M, Li X, Zhang Z,
Zhang S, Liu Y, Zhao W, Gu Y, et al: Identification of a small
mutation panel of coding sequences to predict the efficacy of
immunotherapy for lung adenocarcinoma. J Transl Med. 18:252020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marinelli D, Mazzotta M, Scalera S,
Terrenato I, Sperati F, D'Ambrosio L, Pallocca M, Corleone G,
Krasniqi E, Pizzuti L, et al: KEAP1-driven co-mutations in lung
adenocarcinoma unresponsive to immunotherapy despite high tumor
mutational burden. Ann Oncol. 12:1746–1754. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu C and Hwang MJ: Risk stratification for
lung adenocarcinoma on EGFR and TP53 mutation status, chemotherapy,
and PD-L1 immunotherapy. Cancer Med. 8:5850–5861. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du P and Xu C: Predicting multisite
protein subcellular locations: Progress and challenges. Expert Rev
Proteomics. 10:227–237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Laursen L, Karlsson E, Gianni S and Jemth
P: Functional interplay between protein domains in a supramodular
structure involving the postsynaptic density protein PSD-95. J Biol
Chem. 295:1992–2000. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dawson N, Sillitoe I, Marsden RL and
Orengo CA: The classification of protein domains. Methods Mol Biol.
1525:137–164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng W, Zhou X, Wu Q, Pearce R, Li Y and
Zhang Y: FUpred: Detecting protein domains through
deep-learning-based contact map prediction. Bioinformatics.
12:3749–3757. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Libiad M, Motl N, Akey DL, Sakamoto N,
Fearon ER, Smith JL and Banerjee R: Thiosulfate
sulfurtransferase-like domain-containing 1 protein interacts with
thioredoxin. J Biol Chem. 293:2675–2686. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu Y, Mehta-D'souza P, Biswas I,
Villoutreix BO, Wang X, Ding Q and Rezaie AR: Ile73Asn mutation in
protein C introduces a new N-linked glycosylation site on the first
EGF-domain of protein C and causes thrombosis. Haematologica.
105:1712–1722. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang F, Wang-Gou S, Cao H, Jiang N and Li
X: Proteomics identifies EGF-like domain multiple 7 as a potential
therapeutic target for epidermal growth factor receptor-positive
glioma. Cancer Commun (Lond). 40:518–530. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mizutani K, Guo X, Shioya A, Zhang J,
Zheng J, Kurose N, Ishibashi H, Motono N, Uramoto H and Yamada S:
The impact of PRDX4 and the EGFR mutation status on cellular
proliferation in lung adenocarcinoma. Int J Med Sci. 16:1199–1206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang S, Ko JC, Yang J and Shih JY:
Afatinib is effective in the treatment of lung adenocarcinoma with
uncommon EGFR p.L747P and p.L747S mutations. Lung Cancer.
133:103–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Neugut AI, MacLean SA, Dai W and Jacobson
JS: Physician characteristics and decisions regarding cancer
screening: A systematic review. Popul Health Manag. 22:48–62. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heldring N, Nyman U, Lönnerberg P,
Onnestam S, Herland A, Holmberg J and Hermanson O: NCoR controls
glioblastoma tumor cell characteristics. Neuro Oncol. 16:241–249.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang H, Gao J, Zhang R, Li M, Peng Z and
Wang H: Molecular and immune characteristics for lung
adenocarcinoma patients with CMTM6 overexpression. Int
Immunopharmacol. 83:1064782020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ni K, Wang D, Xu H, Mei F, Wu C, Liu Z and
Zhou B: miR-21 promotes non-small cell lung cancer cells growth by
regulating fatty acid metabolism. Cancer Cell Int. 19:2192019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Barton MK: Use of posttreatment imaging
and biomarker testing for survivors of breast cancer. CA Cancer J
Clin. 66:175–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
VanderLaan PA, Rangachari D, Majid A,
Parikh MS, Gangadharan SP, Kent MS, McDonald DC, Huberman MS,
Kobayashi SS and Costa DB: Tumor biomarker testing in
non-small-cell lung cancer: A decade of change. Lung Cancer.
116:90–95. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu Y, Jamal M, Xie T, Sun J, Song T, Yin
Q, Li J, Pan S, Zeng X, Xie S and Zhang Q: Uridine-cytidine kinase
2 (UCK2): A potential diagnostic and prognostic biomarker for lung
cancer. Cancer Sci. 110:2734–2747. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gu B and Zhu WG: Surf the
post-translational modification network of p53 regulation. Int J
Biol Sci. 8:672–684. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Teng Y, Lang L and Shay C: ATAD3A on the
path to cancer. Adv Exp Med Biol. 1134:259–269. 2019. View Article : Google Scholar : PubMed/NCBI
|