Introduction
Epithelial ovarian cancer (EOC) is the most fatal
gynecological cancer, with a survival rate of 46% at 5 years
post-diagnosis (1). The high
death-to-incidence rate is attributed to the advanced stage of the
disease at the time of diagnosis. Due to the asymptomatic nature of
EOC, ~75% of patients with ovarian cancer (OC) are diagnosed at an
advanced stage (2). In the late
stages of disease, there is a 5-year relative survival rate of 29%,
compared with 92% in the early stages of disease (3). It has also been revealed that 15% of
women diagnosed with EOC have a genomic predisposition to
developing the disease, including the presence of mutations in
causative breast cancer genes, BRCA1 and BRCA2, found in 65–75% of
hereditary EOC cases (2). Therefore,
the identification of further diagnostic and prognostic markers of
EOC is critical.
E74-like E26 transformation-specific (ETS)
transcription factor 3 (ELF3) is located on chromosome 1q32.1, and
encodes a protein 371 amino acids in length. ELF3 is an epithelial
cell-specific ETS transcription factor, and a member of the
epithelial cell-specific sub-group of the larger ETS transcription
factor family (4). The results of a
previous study demonstrated that inactivating and splice-site
mutations in ELF3 occur in ~6% of mucinous OCs (5). Yeung et al (6) suggested that ELF3 played a role in
tumor suppression, as loss of ELF3 mRNA and protein expression was
associated with a poor outcome in OC. On the other hand, ELF3 was
also found to promote the progression of colorectal cancer and
hepatocellular carcinoma (7,8).
In order to investigate its biological role in OC,
the aim of the present study was to analyze the expression of ELF3
in OC, and to determine the effects of ELF3 on OC cell
proliferation and cisplatin sensitivity. The mechanism by which
ELF3 promotes OC progression was also investigated.
Materials and methods
Bioinformatics analysis
The top 20 upregulated genes in OC, found using TCGA
and the Genotype-Tissue Expression Portal (GTEx), were analyzed for
their chromosomal distribution using the Gene Expression Profiling
Interactive Analysis (GEPIA) website (http://gepia.cancer-pku.cn/) (9), with a threshold of log2FoldChange>3;
q<0.01. The genetic alteration of ELF3, and the association
between ELF3 mRNA expression and genetic copy number alteration in
OC, was analyzed using cBioPortal (www.cbioportal.org) (10). GEPIA was also used to analyze the
expression of ELF3 in OC and non-cancerous control tissues, based
on TCGA and GTEx data. The protein expression of ELF3 in OC and
non-cancerous ovarian tissues was determined using The Human
Protein Atlas (https://www.proteinatlas.org/). The expression of ELF3
in OC, based on individual cancer stages and patient age, was
analyzed in UALCAN (http://ualcan.path.uab.edu/index.html) (11). Kaplan-Meier-plotter (http://kmplot.com/analysis/) (12) was used to analyze the overall and
progression-free survival rates of OC and ELF3 mRNA expression
based on the Kaplan-Meier-plotter cohort, and GEPIA was used to
perform survival analysis on TCGA cohort.
Cell lines and culture
A2780 and SKOV3 cell lines were purchased from the
American Type Culture Collection. A2780 cells were cultured in DMEM
and SKOV3 cells were cultured in McCoy's 5A medium, both
supplemented with 10% FBS (all Gibco; Thermo Fisher Scientific,
Inc.). A2780/DDP and SKOV3/DDP cell lines were established by
long-term exposure to cisplatin. Firstly, cells were treated with 5
µg/ml cisplatin for 1 h, and then cultured in medium without
cisplatin. After 3–5 days (at 70–80% confluency), the cells were
treated with 5 µg/ml cisplatin once more. When the cells were
resistant to the current cisplatin concentration, the concentration
was incrementally increased until reaching 200 µg/ml (5, 10, 20,
50, 100 and 200 µg/ml). The cells were than cultured in medium with
200 µg/ml cisplatin to retain resistance. All cells were maintained
at 37°C (5% CO2) in a humified incubator.
Plasmid construction and
transfection
To construct the overexpression vector, the open
reading frame sequence of ELF3 was amplified and cloned into the
pLenti-C-Myc-DDK-IRES-Puro (pCMV) vector (OriGene Technologies,
Inc.). 293T cells (ATCC) were transfected with the pCMV-ELF3 vector
(8 µg), along with pMD2.G (4 µg) and psPAX2 (8 µg) plasmids for
lentivirus production. Culture medium was collected 48 and 72 h
after transfection, and cell debris was removed by centrifugation
with 4,000 × g for 10 min at 4°C. The supernatants were clarified
with a 0.45-µm filter and stored at −80°C. A2780 cells were
infected with lentivirus for 48 h (multiplicity of infection, 10
for A2780 infection and 30 for SKOV3 infection) and subsequently
cultured for 2 weeks in DMEM medium (Gibco; Thermo Fisher
Scientific.) containing 2 µg/ml puromycin (Merck KGaA) to generate
stable expression cell lines. Stable cell line maintenance was
performed using 0.5 µg/ml puromycin. The negative control for
lentiviral transfection was the backbone vector without the ELF3
CDS.ELF3 small interfering (si)RNA was purchased from Shanghai
GenePharma Co., Ltd., with which SKOV3 cells were transfected using
Lipofectamine® 2000 (Invitrogen, Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
ELF3 siRNA sequence was 5′-GCCAUGAGGUACUACUACA-3′, and the si-NC
sequence was 5′-TTCTCCGAACGTGTCACGT-3′. For knockdown experiments,
cells were transfected with 50 nM siRNA at 37°C for 24 h. The time
interval between transfection and subsequent experimentation was 48
h.
Clonogenic assay
Cell suspensions were seeded into 6-well plates at a
density of 400 cells/well, and cultured at 37°C for 2 weeks. The
cell colonies were fixed with 100% methanol for 15 min, and stained
with 0.1% crystal violet for 30 min at room temperature. Colonies
of >50 cells were counted under a light microscope. The results
presented are the mean ± standard error and represent three
independent experiments.
Cellular proliferation assay
Cells were seeded into 96-well plates at a density
of 103 cells/well, and cultured at 37°C for 5–6 days.
Next, 10 µl Cell Counting Kit-8 (CCK-8) reagent was added to each
well at the same time every day, followed by incubation for 4 h at
37°C; the absorbance of each well was measured at 490 nm using a
microplate reader. The relative proliferation rate is presented as
the comparison of absorbance measured each day to the absorbance
measured on the first day of the experiment. All experiments were
repeated at least three times.
Xenograft experiments
The animal experiments were performed with the
approval of Shandong University Animal Care and Use committee
(approval no. KYLL-202011-130). A total of 10 female BALB/c-nude
mice (Charles River Laboratories, Inc.) were maintained in specific
pathogen-conditions (5 mice per cage) at 22°C (humidity, 60%;
ventilation rate, 15 times per hour) under a 12 h/12 h light/dark
cycle, with free access to food and water. Animal health was
monitored every 2 days. A2780 OC cells overexpressing ELF3 (and
controls cells overexpressing backbone vector) were subcutaneously
injected into the mice (age, 5 weeks; weight, 18–20 g; 5 mice per
group) at a density of 2×106 cells/mouse. The experiment
lasted for 24 days, and the mice were sacrificed by cervical
dislocation. The maximum tumor volume was 354.8 mm3, and
was calculated according to the following formula: Volume = 1/2 (a
× b2) (were a is the long axis of the tumor, and b is
the short axis of the tumor).
Western blotting
Cultured cells were harvested and lysed on ice for
30 min using RIPA buffer (Beyotime Institute of Biotechnology)
containing 1% PMSF and 1% sodium fluoride, with vortexing every 10
min. The protein concentration was determined using a BCA Assay kit
(Beyotime Institute of Biotechnology). A total of 30 µg
protein/lane was separated by SDS-PAGE (12%) and subsequently
transferred to a PVDF membrane. The membrane was blocked in 5%
skimmed milk solution at room temperature for 2 h, and then
incubated with primary antibodies at 4°C overnight. Following
washing with TBS-Tween-20 (0.1%), the membrane was incubated with
HRP-labeled secondary antibodies (1:10,000; cat. no. ab205718 and
ab6789; both Abcam), and the protein bands were visualized using an
ECL system (Cytiva). β-tubulin was used as the endogenous control.
ImageJ software (v6; National Institutes of Health) was used for
densitometric detection. The primary antibodies were as follows:
ELF3 (cat. no. A6371; ABclonal Biotech Co., Ltd.; 1:1,000), Bax
(cat. no. AF0120; Affinity Biosciences, Ltd.; 1:1,000), Bcl2 (cat.
no. 12789-1-AP; ProteinTech Group, Inc; 1:1,000), phosphorylated
(p-)mTOR (cat. no. 5536; CST Biological Reagents Co., Ltd.;
1:1,000), p-p70S6K (cat. no. 9234; CST Biological Reagents Co.,
Ltd.; 1:1,000), p-4EBP1 (cat. no. 9459; CST Biological Reagents
Co., Ltd.; 1:1,000), mTOR (cat. no. 2983; CST Biological Reagents
Co., Ltd.; 1:1,000), p70S6K (cat. no. 2708; CST Biological Reagents
Co., Ltd.; 1:1,000), 4EBP1 (cat. no. 19644; CST Biological Reagents
Co., Ltd.; 1:1,000) and β-tubulin (cat. no. ab6046; Abcam;
1:5,000)
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from cultured cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) per the manufacturer's protocol. Total RNA was reverse
transcribed into cDNA using the PrimeScript RT reagent kit (Takara
Bio, Inc.), and qPCR was subsequently performed using the SYBRGreen
qPCR master mix (Takara Bio, Inc.), according to the manufacturers'
instructions. The thermocycling conditions were as follows:
Denaturation at 95°C for 3 min, followed by 40 cycles at 95°C for
10 sec, 60°C for 30 sec and 72°C for 30 sec. The mRNA expression
levels of ELF3 were quantified using the 2−ΔΔCq method
(13). GAPDH was used as the
endogenous control. The primers used were as follows: ELF3 forward,
5′-AAGCGCCATTGACTTCTCAC-3′ and reverse, 5′-TCCAGCAGCTCAATGATCCA-3′;
and GAPDH forward, 5′-AGGTCGGAGTCAACGGATTT-3′ and reverse,
5′-TGACGGTGCCATGGAATTTG-3′.
Cisplatin sensitivity assay
Cells were seeded into 96-well plates at a density
of 1×103 cells/well and cultured at 37°C for 24 h. Then,
varying concentrations of cisplatin (0, 5,10, 20 and 40 µg/ml for
A2780 cells; and 0, 10,20,40 and 80 µg/ml for SKOV3 cells) were
added to the culture medium at the same time. After a further 24 h,
cell viability was assessed using CCK-8 reagent as
aforementioned.
Hematoxylin and eosin (H&E)
staining
Tumor xenografts tissues were fixed with formalin
(10%, at room temperature for 48 h), embedded in paraffin, and
subsequently cut into 4-µm sections. The tissue sections were
heated at 65°C for 30 min to melt the paraffin wax and then
immediately immersed in xylene. The sections were then rehydrated
through decreasing concentrations of ethanol, and washed in PBS.
The sections were stained with H&E (room temperature for 1
min), and then dehydrated using increasing concentrations of
ethanol and xylene. Finally, the sections were sealed with neutral
gum. Images were captured under a light microscope (magnification,
×400).
Statistical analysis
Statistical analysis was carried out using SPSS 25.0
(IBM). Unpaired Student's t-test and one-way ANOVA were used to
determine significant differences. Bonferroni's post hoc test was
used for pairwise comparisons following ANOVA. Survival curves were
analyzed using the log-rank test. Data are presented as the mean ±
standard deviation of three independent experiments, and P<0.05
was considered to indicate a statistically significant
difference.
Results
ELF3 is commonly upregulated in
OC
GEPIA was used to determine the distribution of
upregulated genes in OC, with a threshold of Log2FoldChange<3;
q<0.01. As demonstrated in Fig.
1A, a total of 374 genes were identified as upregulated in OC,
and were scattered on all chromosomes, excluding the Y chromosome.
The top 20 upregulated genes in OC are listed in Table I, which includes ELF3. Further
analysis of ELF3 mRNA expression using TCGA and GTEx revealed that
ELF3 was significantly upregulated in OC compared with
non-cancerous ovarian tissues (Fig.
1B). Genetic alterations of ELF3 were analyzed using
cBioPortal. As demonstrated in Fig.
1C, 8% of OC tissues displayed genetic amplification of ELF3,
though this was not associated with a significant increase in mRNA
expression (Fig. 1D), indicating a
more complex mechanism underlying ELF3 regulation in OC. Moreover,
ELF3 protein expression was analyzed using The Human Protein Atlas.
Immunohistochemical staining revealed an increase in ELF3 protein
expression in OC compared with non-cancerous ovarian tissue
(Fig. 1E). These results implicate
that ELF3 is commonly upregulated in OC.
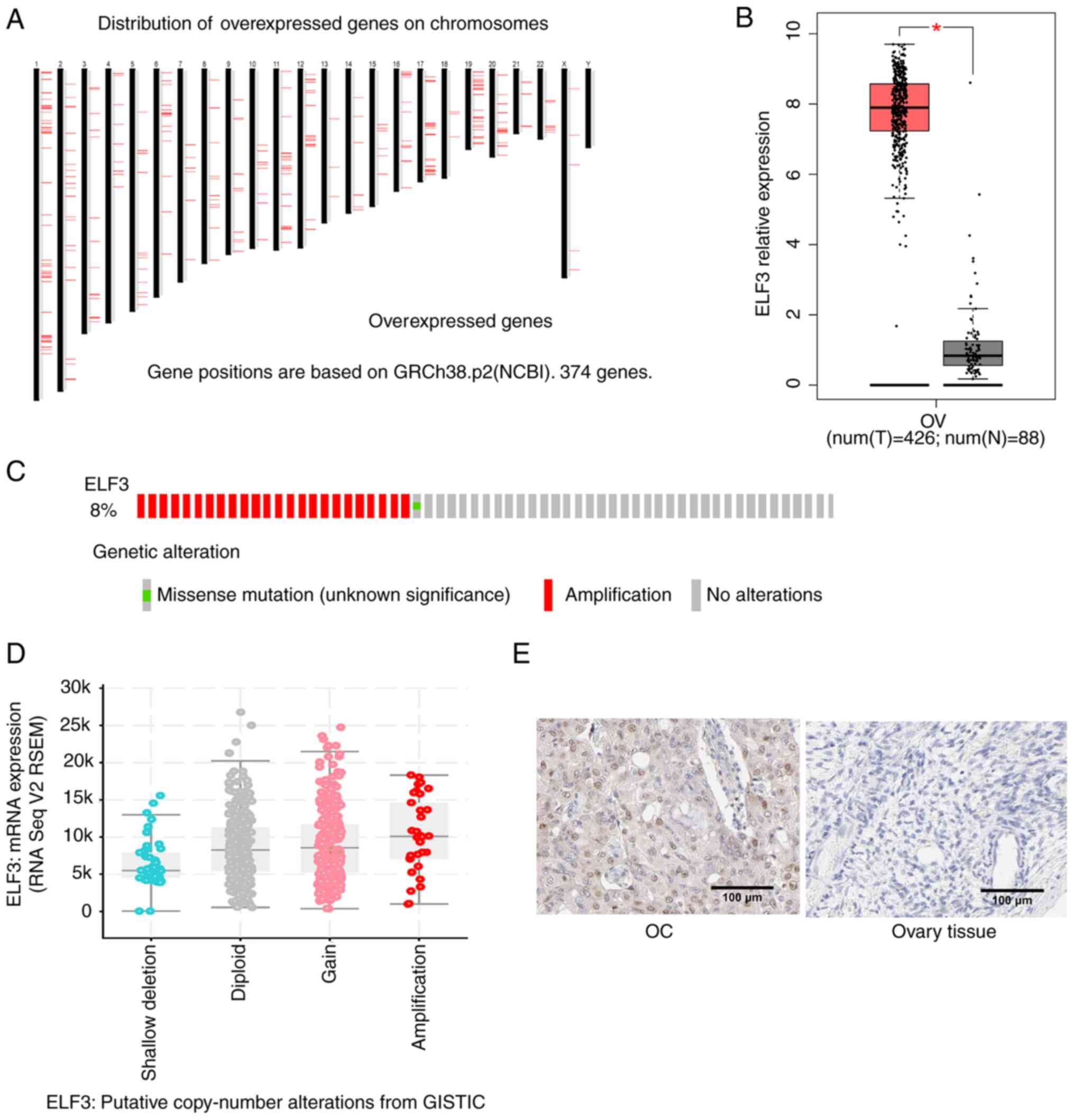 | Figure 1.ELF3 is overexpressed in OC. (A)
Chromosomal distribution of overexpressed genes based on TCGA OC
cohort using GEPIA; log2FoldChange>3, q<0.01 was considered
as the threshold. (B) Expression of ELF3 in OC tissue and normal
controls based on TCGA and GTEx data analyzed by GEPIA. (C) Genetic
alterations of ELF3 in OC based on the cBioPortal database; 8% of
patients with OC showed ELF3 genetic alternations, most of which
were amplifications. (D) Association between ELF3 mRNA expression
and genetic copy number alteration in OC, analyzed using
cBioPortal. (E) Representative images of ELF3 protein expression in
OC and normal ovary tissues, obtained from The Human Protein Atlas.
P-value was obtained by one-way ANOVA. *P<0.05. ELF3, E74-like
E26 transformation-specific transcription factor 3; OC, ovarian
cancer; TCGA, The Cancer Genome Atlas; GEPIA, Gene Expression
Profiling Interactive Analysis; OV, ovarian cancer (TCGA); T,
tumor; N, normal. |
 | Table I.Top 20 Upregulated genes in OC based
on TCGA cohort. |
Table I.
Top 20 Upregulated genes in OC based
on TCGA cohort.
| Gene | Gene ID | Median expression
(tumor) | Median expression
(normal) | Log2 (fold
change) | Adjusted
P-value |
|---|
| CLDN3 |
ENSG00000165215.6 | 532.873 | 0.100 | 8.923 | 8.49e-227 |
| WFDC2 |
ENSG00000101443.17 | 2341.633 | 4.850 | 8.645 | 9.12e-162 |
| SLPI |
ENSG00000124107.5 | 1338.266 | 2.718 | 8.493 | 1.01e-125 |
| FOLR1 |
ENSG00000110195.11 | 375.273 | 0.095 | 8.425 | 6.55e-143 |
| MSLN |
ENSG00000102854.14 | 428.721 | 0.425 | 8.236 | 4.64e-138 |
| CD24 |
ENSG00000272398.5 | 498.966 | 0.910 | 8.032 | 2.42e-158 |
| S100A1 |
ENSG00000160678.11 | 505.318 | 1.180 | 7.860 | 4.29e-105 |
| CLDN4 |
ENSG00000189143.9 | 265.56 | 0.460 | 7.512 | 6.91e-204 |
| LCN2 |
ENSG00000148346.11 | 255.202 | 0.470 | 7.445 | 3.75e-96 |
| KRT7 |
ENSG00000135480.14 | 364.872 | 1.130 | 7.425 | 3.60e-159 |
| SLC34A2 |
ENSG00000157765.11 | 176.227 | 0.070 | 7.372 | 9.83e-121 |
| CDKN2A |
ENSG00000147889.16 | 258.057 | 0.635 | 7.038 | 1.42e-83 |
| EPCAM |
ENSG00000119888.1 | 246.893 | 0.755 | 7.142 | 9.65e-199 |
| KLK7 |
ENSG00000169035.11 | 151.843 | 0.100 | 7.118 | 1.02e-112 |
| SMIM22 |
ENSG00000267795.5 | 165.581 | 0.210 | 7.105 | 1.12e-139 |
| ELF3 |
ENSG00000163435.15 | 237.617 | 0.785 | 7.063 | 3.96e-162 |
| KLK8 |
ENSG00000129455.15 | 154.326 | 0.165 | 7.059 | 3.74e-152 |
| RNVU1-7 |
ENSG00000206585.1 | 131.62 | 0.000 | 7.051 | 7.12e-27 |
| MAL2 |
ENSG00000147676.13 | 153.974 | 0.210 | 7.001 | 3.04e-182 |
| KLK6 |
ENSG00000167755.13 | 128.742 | 0.155 | 6.812 | 3.55e-94 |
ELF3 expression is associated with
cancer stage, patient age and prognosis of OC
To investigate the association between ELF3 mRNA
expression and the clinical characteristics of OC, UALCAN was used
to analyze OC data obtained from TCGA. As demonstrated in Fig. 2A and B, an increase in ELF3 mRNA
expression was associated with a higher OC cancer stage and lower
patient age, suggesting that upregulation of ELF3 is associated
with an increase in the malignant behavior of OC. Consistent with
these findings, Kaplan-Meier survival analysis indicated that high
expression of ELF3 is associated with poor prognosis in patients
with OC, based on Kaplan-Meier Plotter (Fig. 2C and D) and TCGA (Fig. S1B) cohort analysis. These results
suggested that high expression of ELF3 was associated with worse
clinicopathological features and poorer prognosis of patients with
OC.
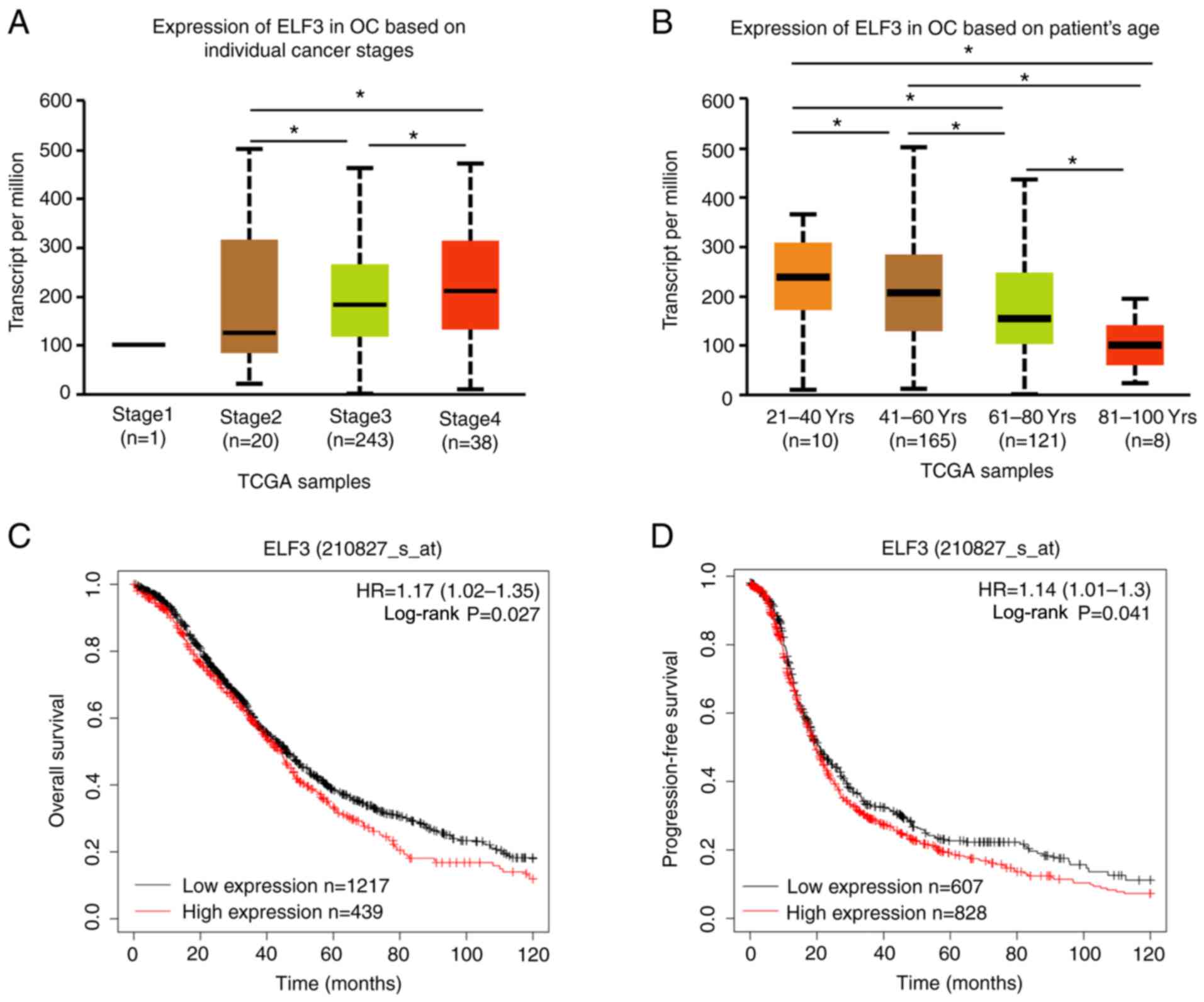 | Figure 2.Upregulation of ELF3 is associated
with higher individual cancer grade, lower patient age and poorer
prognosis. ELF3 expression in OC at (A) different individual cancer
stages and (B) different patient ages, based on TCGA cohort in
UALCAN. (C) Kaplan-Meier analysis of the effect of ELF3 mRNA
expression on the overall survival of patients with OC, based on
the Kaplan-Meier Plotter cohort. High and low expression groups
were separated based on the best cutoff; low ELF3 expression group,
n=1,217; and high ELF3 expression group, n=439. (D) Kaplan-Meier
analysis of the effect of ELF3 mRNA expression on the
progression-free survival of patients with OC, based on the
Kaplan-Meier Plotter online cohort. High and low expression groups
were separated based on the best cutoff; low ELF3 expression group,
n=607; and high ELF3 expression group, n=828. P-values were
obtained by one-way ANOVA or the log-rank test. *P<0.05.
E74-like E26 transformation-specific transcription factor 3; OC,
ovarian cancer; TCGA, The Cancer Genome Atlas; HR, hazard
ratio. |
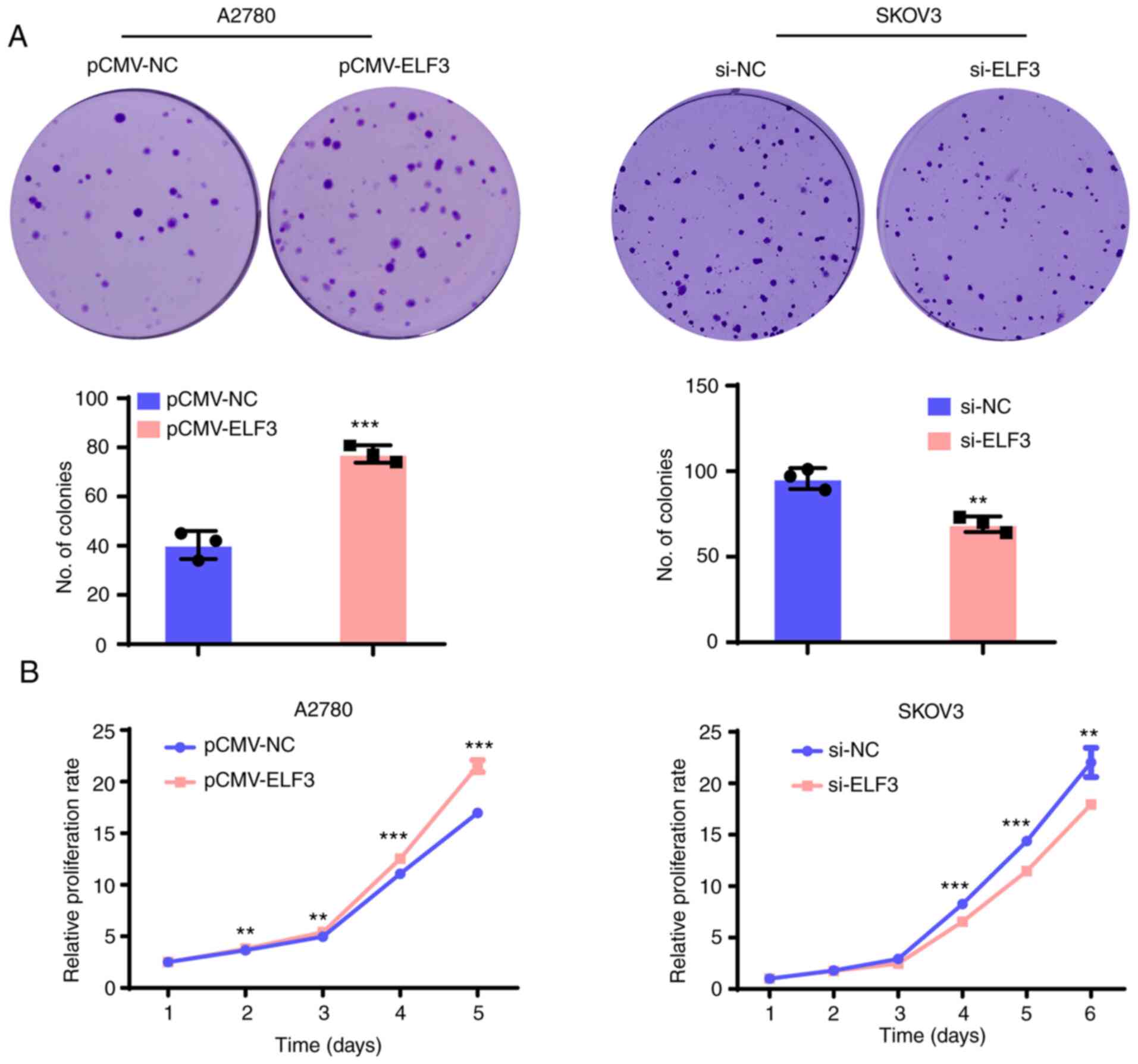
ELF3 promotes the proliferation of OC
cells and the growth of xenograft tumors in mice
To determine whether ELF3 was associated with the
proliferation of OC cells, ELF3 protein expression was evaluated in
six ovarian cancer cell lines by western blot analysis (Fig. S1A). A2780 cells (which express
endogenous ELF3 at low levels) were selected for ELF3
overexpression analysis (lentivirus infection), and SKOV3 cells
(which exhibit high levels of endogenous ELF3 expression) for ELF3
siRNA-knockdown. Clonogenic and proliferation assays were then
performed to evaluate the effects of ELF3 on OC cell proliferation.
As demonstrated in Fig. 3A,
overexpression of ELF3 increased the clonogenic ability of A2780
cells, while ELF3-knockdown reduced the clonogenic ability of SKOV3
cells. Growth curve assays demonstrated that ELF3 overexpression
increased the proliferation rate of A2780 cells, while knockdown
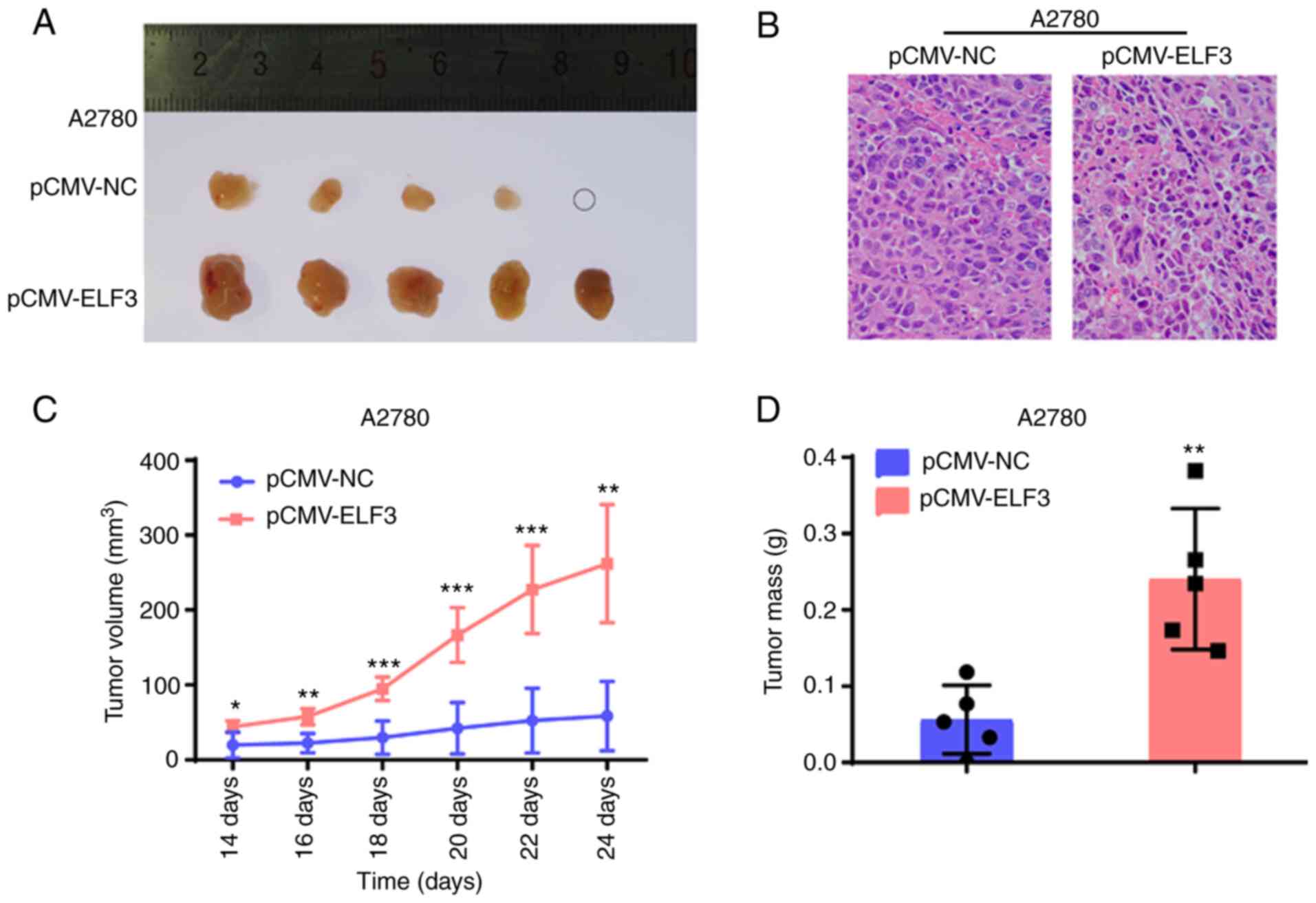
suppressed the proliferation rate of SKOV3 cells (Fig. 3B). Furthermore, a cell-derived
xenograft model was established to investigate the effect of ELF3
on tumor growth. A2780 cells with ELF3 overexpression (and the
corresponding control) were subcutaneously injected into nude mice
(n=5 per group), and overexpression significantly increased the
growth rate of the xenografted tumors (Fig. 4A); H&E staining confirmed OC cell
tumorigenesis (Fig. 4B).
Furthermore, overexpression of ELF3 in A2780 cells resulted in
increased tumor growth (Fig. 4C) and
tumor mass (Fig. 4D). Collectively,
these results revealed that ELF3 promoted OC cell proliferation and
tumor growth in vitro and in vivo.
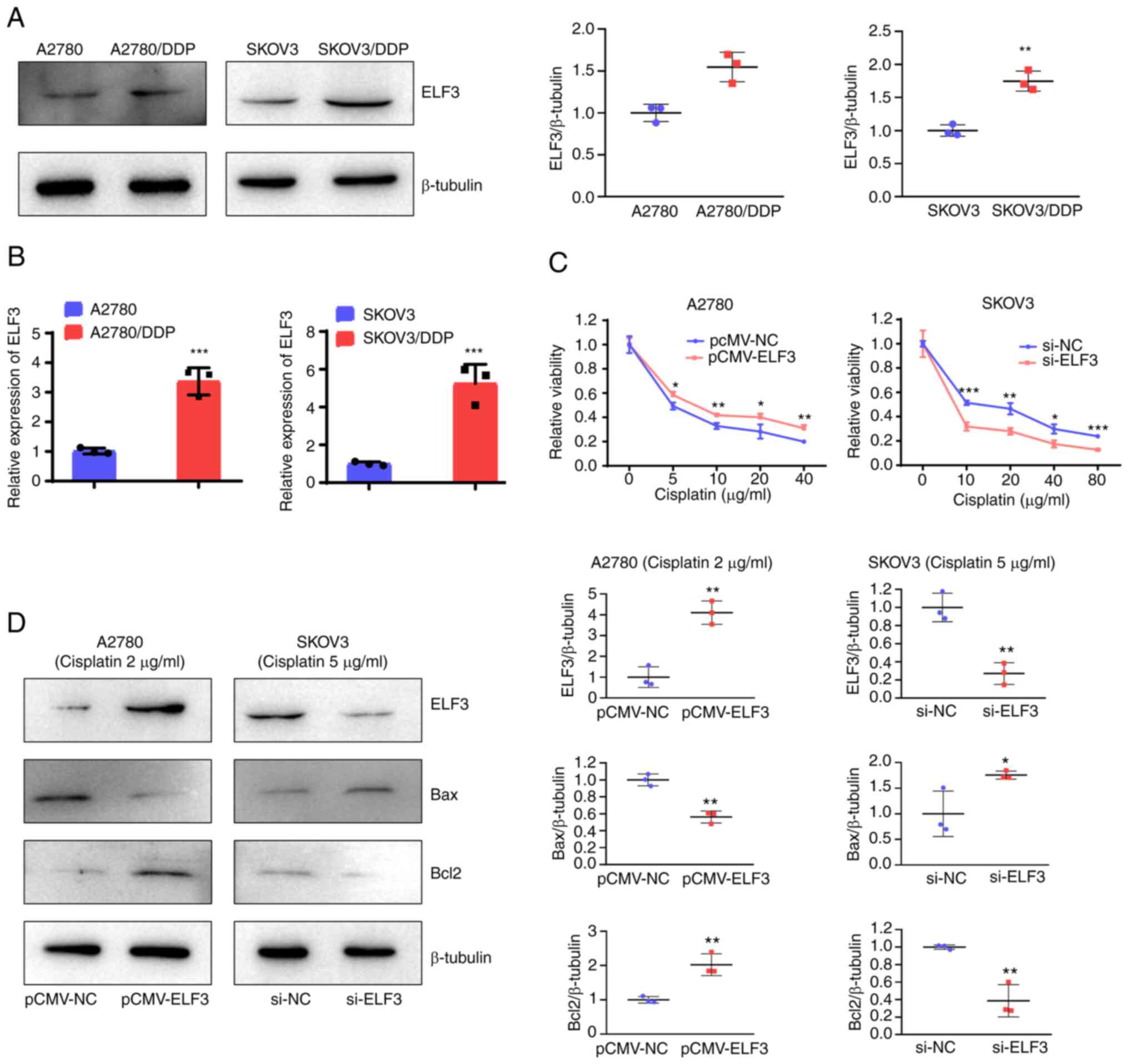
ELF3 overexpression reduces cisplatin
sensitivity of OC cells
To investigate the association between ELF3 and OC
cell sensitivity to cisplatin, the expression of ELF3 in
cisplatin-resistant A2780 (A2780/DDP) and SKOV3 (SKOV3/DDP) cells
was detected using western blotting and RT-qPCR. As demonstrated in
Fig. 5A and B, ELF3 expression was
upregulated in A2780/DDP and SKOV3/DDP cells compared with
cisplatin-sensitive A2780 and SKOV3 cells. A2780 cells with ELF3
overexpression and SKOV3 cells with ELF3-knockdown were used to
evaluate the effect of ELF3 on OC cell sensitivity to cisplatin. A
viability assay demonstrated that ELF3 overexpression reduced the
cisplatin sensitivity of A2780 cells, and ELF3-knockdown increased
the cisplatin sensitivity of SKOV3 cells (Fig. 5C). Western blotting results showed
that expression of Bax was decreased, and that of Bcl2 was
increased in ELF3-overexpressing A2780 cells compared with the
control cells. While the expression of Bax were increased and Bcl2
expression wA decreased in ELF3-knockdown SKOV3 cells, compared
with the control cells. These results indicated that ELF3 reduced
cisplatin-induced apoptosis (Fig.
5D). Collectively, ehe results suggest that ELF3 reduces
cisplatin sensitivity of OC cells.
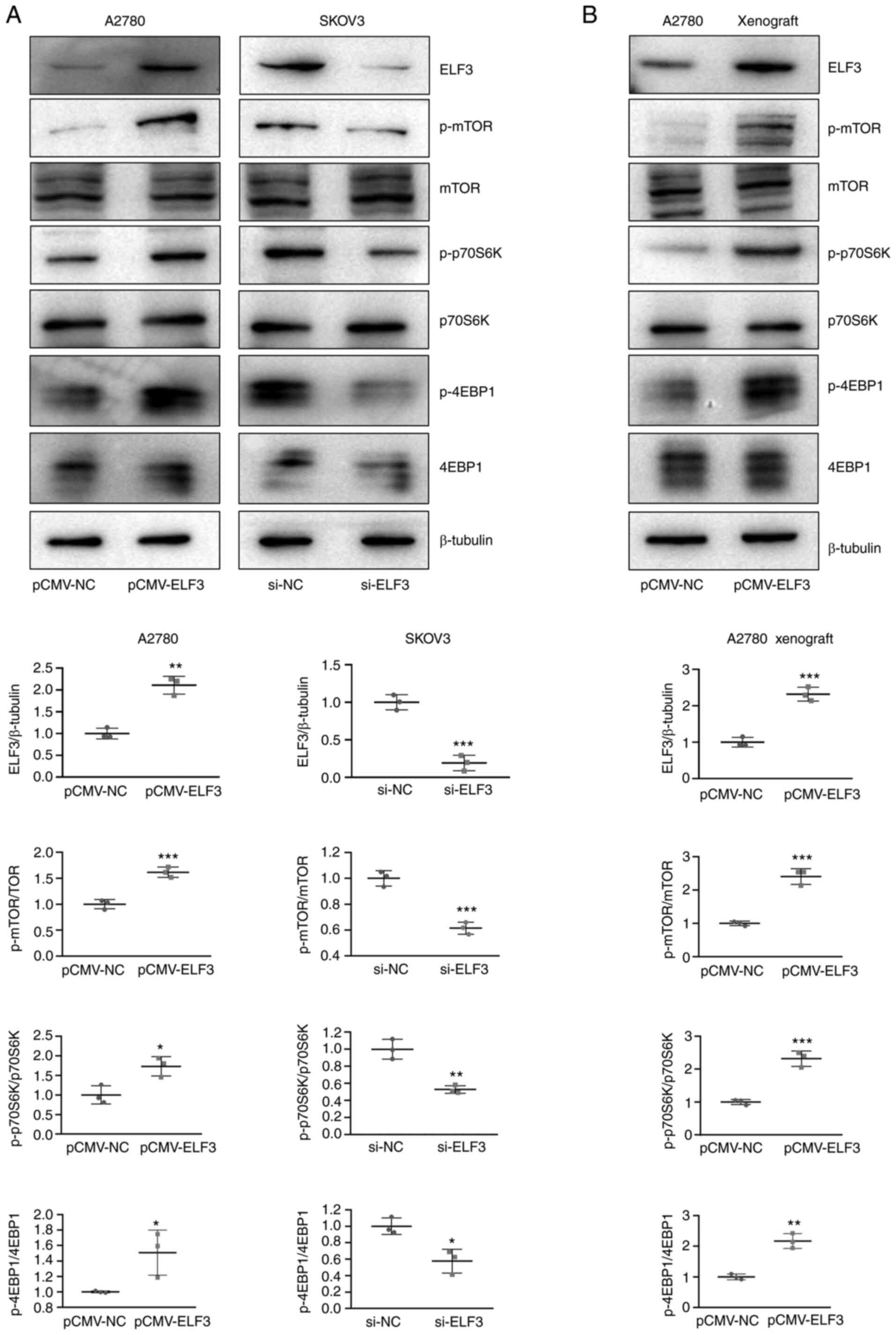
ELF3 activates the mTOR pathway of OC
cells in vitro and in vivo
To further investigate the mechanism by which ELF3
promotes OC proliferation and tumor growth, western blotting was
used to detect alterations in downstream pathway proteins,
following both ELF3 overexpression and knockdown. Western blotting
results showed that the expression of phosphorylated mTOR, P70S6K
and 4EBP1 were increased in ELF3-overexpresssing A2780 cells, and
decreased in ELF3-knockdown SKOV3 cells, compared with their
non-phosphorylated forms (Fig. 6A).
Moreover, downstream mTOR pathway proteins, such as phosphorylated
mTOR, P70S6K and 4EBP1, were also activated in ELF3-overexpresssing
A2780 ×enografts (Fig. 6B). These
results suggested that ELF3 could be activated mTOR pathway.
Collectively, the present study results indicated the promoting
effect of ELF3 on OC, thus providing a potential novel target for
precision OC treatment.
Discussion
ELF3 (also referred to as ERT, ESX, EPR-1 and ESE-1)
is a member of the ETS family (14)
that ELF3 regulates the differentiation of epithelial cells in the
small intestine (15), bronchial
epithelium (16) and urothelium
(17). In previous years, increasing
evidence has revealed that ELF3 plays a role in the regulation of
multiple tumor types. (7,8,18)
However, the effect of ELF3 on tumorigenesis and progression
remains to be elucidated. Mutations in ELF3 have been observed in
gastric cancers, cervical adenocarcinomas, mucinous ovarian
carcinomas and ampullary carcinomas (5,17–19).
Using TCGA, a previous study revealed that mutations in ELF3 were
present in ~6% of both superficial and invasive tumors in bladder
cancer, over half of which were inactivating mutations (20), thus indicating the tumor suppressive
role of ELF3 in bladder cancer. By contrast, increased ELF3
expression promoted cellular proliferation, migration and
invasiveness in hepatocellular carcinoma (8), colorectal cancer (7), prostate cancer (21) and ampullary carcinoma (22).
Ryland et al (5) analyzed somatic mutations in mucinous OC
and found three mutations of ELF3 by exome sequencing. The
mutations of ELF3 in mucinous ovarian cancer were predicted as
deleterious, indicating a suppressive role for ELF3 in mucinous OC.
Yeung et al (6) revealed that
ELF3 was upregulated in long-term OC survivors compared with
short-term survivors, and that ELF3 suppressed OC cell
proliferation by reversing epithelial-mesenchymal transition
(EMT).
In contrast with the findings of a previous study
(6), the results of the present
study revealed that ELF3, a markedly upregulated transcription
factor in high-grade serious OC, was associated with a poor
prognosis, and promoted cellular proliferation in vitro and
in vivo.
According to the present study, genes that were
found to be upregulated in OC (using TCGA) were screened, and due
to its increased expression, ELF3 was selected as a biomarker of
OC. Unlike the unequal grouping in Yeung et al (low ELF3
expression, n=15; high ELF3 expression, n=299) (6), patients with OC in TCGA cohort were my
equally divided into low (n=211) and high (n=212) ELF3 expression
groups in the present study. Kaplan-Meier survival analysis
indicated that high expression of ELF3 was related to the poorer
overall and progression-free survival of patients with OC.
Moreover, increased expression of ELF3 was associated with a higher
cancer stage and a lower patient age, indicating the oncogenic
effect of ELF3 in OC. Subsequent clonogenic, proliferation and
tumor xenograft assays confirmed the role of ELF3 in OC promotion.
mTOR has emerged as a critical factor in human cancer, regulating
protein synthesis, growth, metabolism, aging, regeneration and
autophagy (23,24). ELF3 has previously been reported to
promote EMT (9), as well as PI3K/AKT
and ERK pathway activation (18),
though to the best of our knowledge, the association between ELF3
regulation and the mTOR pathway has not previously been reported.
In the present study, ELF3 was found to activate the mTOR pathway
in OC cells and xenografts by western blotting; however, further
investigation is required to confirm the activation effect of ELF3
on the mTOR pathway in OC.
Cisplatin is a chemotherapeutic agent widely used
for the treatment and management of OC. However, despite an initial
response to treatment, ~70% of patients with OC experience
cisplatin resistance and tumor recurrence (25). A previous study revealed that
chemotherapy resistance was associated with cancer stem cells
(26), the notch receptor/hes family
bHLH transcription factor 1 signaling pathway (27), the hypoxia-inducible factor 1 pathway
(28) and apoptosis (29,30).
However, to the best of our knowledge, the association between ELF3
and OC sensitivity to chemotherapy has not been previously
reported. The present study reported that ELF3 was upregulated in
cisplatin-resistant OC cells, and that ELF3 reduced cisplatin
sensitivity by inhibiting apoptosis. Notably, mTOR signaling
(promoting cisplatin resistance) was reported in non-small cell
lung cancer, OC and hepatocellular carcinoma (31–33),
suggesting that ELF3 reduces cisplatin sensitivity through
activation of the mTOR pathway. However, this speculation requires
further experimental confirmation.
In conclusion, the results of the present study
demonstrated that ELF3 was upregulated in OC, which was associated
with poor clinical characteristics. Increased expression of ELF3
promoted the progression, and reduced the chemotherapeutic
sensitivity, of OC by activating the mTOR pathway, thus providing
novel targets for the treatment of OC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank the Key Laboratory
of Gynecologic Oncology in Universities of Shandong (Jinan, China)
for instrument availability.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GZ and YL were responsible for study conception and
design. SW and RZ acquired the data, which was analyzed and
interpreted by YL and WL. GZ supervised the study. All authors
wrote, reviewed and revised the manuscript. All authors have read
and approved the final manuscript. SW and YL confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
All animal experiments were performed with the
approval of the Shandong University Animal Care and Use
committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Doherty JA, Peres LC, Wang C, Way GP,
Greene CS and Schildkraut JM: Challenges and opportunities in
studying the epidemiology of ovarian cancer subtypes. Curr
Epidemiol Rep. 4:211–220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lheureux S, Gourley C, Vergote I and Oza
AM: Epithelial ovarian cancer. Lancet. 393:1240–1253. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sizemore GM, Pitarresi JR, Balakrishnan S
and Ostrowski MC: The ETS family of oncogenic transcription factors
in solid tumours. Nat Rev Cancer. 17:337–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ryland GL, Hunter SM, Doyle MA, Caramia F,
Li J, Rowley SM, Christie M, Allan PE, Stephens AN, Bowtell DD, et
al: Mutational landscape of mucinous ovarian carcinoma and its
neoplastic precursors. Genome Med. 7:872015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yeung TL, Leung CS, Wong KK,
Gutierrez-Hartmann A, Kwong J, Gershenson DM and Mok SC: ELF3 is a
negative regulator of epithelial-mesenchymal transition in ovarian
cancer cells. Oncotarget. 8:16951–16963. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang JL, Chen ZF, Chen HM, Wang MY, Kong
X, Wang YC, Sun TT, Hong J, Zou W, Xu J and Fang JY: Elf3 drives
β-catenin transactivation and associates with poor prognosis in
colorectal cancer. Cell Death Dis. 5:e12632014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng L, Xu M, Xu J, Wu K, Fang Q, Liang
Y, Zhou S, Cen D, Ji L, Han W and Cai X: ELF3 promotes
epithelial-mesenchymal transition by protecting ZEB1 from
miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell
Death Dis. 9:3872018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res.
45W:W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nagy Á, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sharrocks AD: The ETS-domain transcription
factor family. Nat Rev Mol Cell Biol. 2:827–837. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Flentjar N, Chu PY, Ng AY, Johnstone CN,
Heath JK, Ernst M, Hertzog PJ and Pritchard MA: TGF-betaRII rescues
development of small intestinal epithelial cells in Elf3-deficient
mice. Gastroenterology. 132:1410–1419. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oliver JR, Kushwah R, Wu J, Pan J, Cutz E,
Yeger H, Waddell TK and Hu J: Elf3 plays a role in regulating
bronchiolar epithelial repair kinetics following Clara
cell-specific injury. Lab Invest. 91:1514–1529. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Böck M, Hinley J, Schmitt C, Wahlicht T,
Kramer S and Southgate J: Identification of ELF3 as an early
transcriptional regulator of human urothelium. Dev Biol.
386:321–330. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang H, Yu Z, Huo S, Chen Z, Ou Z, Mai J,
Ding S and Zhang J: Overexpression of ELF3 facilitates cell growth
and metastasis through PI3K/Akt and ERK signaling pathways in
non-small cell lung cancer. Int J Biochem Cell Biol. 94:98–106.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ojesina AI, Lichtenstein L, Freeman SS,
Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio
L, Cibulskis K, Bertelsen B, et al: Landscape of genomic
alterations in cervical carcinomas. Nature. 506:371–375. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo G, Sun X, Chen C, Wu S, Huang P, Li Z,
Dean M, Huang Y, Jia W, Zhou Q, et al: Whole-genome and whole-exome
sequencing of bladder cancer identifies frequent alterations in
genes involved in sister chromatid cohesion and segregation. Nat
Genet. 45:1459–1463. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Longoni N, Sarti M, Albino D, Civenni G,
Malek A, Ortelli E, Pinton S, Mello-Grand M, Ostano P, D'Ambrosio
G, et al: ETS transcription factor ESE1/ELF3 orchestrates a
positive feedback loop that constitutively activates NF-κB and
drives prostate cancer progression. Cancer Res. 73:4533–4547. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yachida S, Wood LD, Suzuki M, Takai E,
Totoki Y, Kato M, Luchini C, Arai Y, Nakamura H, Hama N, et al:
Genomic sequencing identifies ELF3 as a driver of ampullary
carcinoma. Cancer Cell. 29:229–240. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guertin DA and Sabatini DM: Defining the
role of mTOR in cancer. Cancer Cell. 12:9–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Murugan AK: mTOR: Role in cancer,
metastasis and drug resistance. Semin Cancer Biol. 59:92–111. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Agarwal R and Kaye SB: Ovarian cancer:
Strategies for overcoming resistance to chemotherapy. Nat Rev
Cancer. 3:502–516. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li SS, Ma J and Wong AST: Chemoresistance
in ovarian cancer: Exploiting cancer stem cell metabolism. J
Gynecol Oncol. 29:e322018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Islam SS and Aboussekhra A: Sequential
combination of cisplatin with eugenol targets ovarian cancer stem
cells through the Notch-Hes1 signalling pathway. J Exp Clin Cancer
Res. 38:3822019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ai Z, Lu Y, Qiu S and Fan Z: Overcoming
cisplatin resistance of ovarian cancer cells by targeting
HIF-1-regulated cancer metabolism. Cancer Lett. 373:36–44. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Chen W, Jin Y, Xue R, Su J, Mu Z, Li
J and Jiang S: MiR-142-5p enhances cisplatin-induced apoptosis in
ovarian cancer cells by targeting multiple anti-apoptotic genes.
Biochem Pharmacol. 161:98–112. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rada M, Nallanthighal S, Cha J, Ryan K,
Sage J, Eldred C, Ullo M, Orsulic S and Cheon DJ: Inhibitor of
apoptosis proteins (IAPs) mediate collagen type XI alpha 1-driven
cisplatin resistance in ovarian cancer. Oncogene. 37:4809–4820.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gong T, Cui L, Wang H, Wang H and Han N:
Knockdown of KLF5 suppresses hypoxia-induced resistance to
cisplatin in NSCLC cells by regulating HIF-1α-dependent glycolysis
through inactivation of the PI3K/Akt/mTOR pathway. J Transl Med.
16:1642018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wantoch von Rekowski K, König P, Henze S,
Schlesinger M, Zawierucha P, Januchowski R and Bendas G: Insight
into cisplatin-resistance signaling of W1 ovarian cancer cells
emerges mTOR and HSP27 as targets for sensitization strategies. Int
J Mol Sci. 21:92402020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sheng J, Shen L, Sun L, Zhang X, Cui R and
Wang L: Inhibition of PI3K/mTOR increased the sensitivity of
hepatocellular carcinoma cells to cisplatin via interference with
mitochondrial-lysosomal crosstalk. Cell Prolif. 52:e126092019.
View Article : Google Scholar : PubMed/NCBI
|