Introduction
Colorectal cancer (CRC) is the third most common
malignancy worldwide and the main cause of mortality due to tumor
metastasis (1). CRC has a high
mortality rate when detected at advanced stages. However, the use
of stratified treatment options remains limited in standard
clinical practice, which indicates that the curative treatment of
CRC remains intractable (2).
Furthermore, in terms of early diagnosis and successful treatment
of CRC (achieving a high overall survival rate), the 5-year
survival rate can reach ≤90%, whereas this is reduced to 8% in the
case of advanced metastases (3).
Previous evidence showed that multistep processes play a role in
CRC carcinogenesis, which involves a complicated and dynamic
interplay between epigenetics and transcriptomic alterations
(4,5). Therefore, it is essential to understand
the epigenetic mechanisms underlying CRC to identify promising
therapeutic targets.
Previous studies on epigenetics found that nitrogen
6-methyladenosine (m6A) RNA modification serves critical
roles in the regulation of cell fate, proliferation, metabolism and
biogenesis of tumors (6–9). m6A modification is the most
prevalent modification in eukaryotes, and occurs in mammalian mRNAs
(10), long non-coding RNAs
(11) and microRNAs (12). It is involved in numerous
post-transcriptional functions, including mRNA stability, splicing,
transport, localization and translation (13,14).
m6A methylation is a dynamic epigenetic event primarily
modulated by m6A writers, erasers and readers (15). Methyltransferase-like 3 (METTL3; M3)
was identified as an m6A writer that forms a
heterodimeric complex with Wilms' tumor 1-associating protein
(WTAP) (16) and METTL14 (17) to catalyze m6A methylation.
By contrast, fat mass and obesity-associated protein (FTO)
(18) and α-ketoglutarate-dependent
dioxygenase AlkB homolog 5 (ALKBH5) (19) act as m6A erasers and
participate in m6A demethylation. Dysregulation of
m6A methylation may trigger the progression of various
cancer types, including CRC (20).
METTL3 has an oncogenic function, and maintains SRY-box
transcription factor 2 (SOX2) expression through an
m6A-insulin like growth factor 2 mRNA binding protein 2
(IGF2BP2)-dependent mechanism in CRC cells (21), which is upregulated in human CRC and
promotes CRC progression by enhancing MYC expression (22) and epigenetically suppressing yippee
like 5 (23), as well as by
stabilizing cyclin E1 mRNA (24).
Contrarily, METTL3 was reported as a tumor suppressor in CRC
through regulating the p38/ERK signaling pathway (25). These findings reveal that METTL3 acts
as a double-edged sword, which affects the progression of CRC via
diverse mechanisms and targets. Thus, the role of METTL3 and its
specific molecular mechanism in CRC needs to be further
explored.
Snail was found to be a critical transcriptional
factor for epithelial to mesenchymal transition (EMT), which could
promote invasion and proliferation in several cancer types
(26). METTL3 can increase mRNA
stability and translation efficiency to facilitate proliferation
and invasion during EMT in liver cancer (27), while inhibiting Snail results in the
suppression of cell proliferation and motility in CRC (28). However, to the best of our knowledge,
the functional link between m6A methylation and Snail in
CRC progression has not been established to date.
The present study demonstrated that METTL3 is
significantly upregulated in CRC tissues and is associated with
poor survival. METTL3 silencing suppresses CRC proliferation and
invasion in vitro. The present mechanistic studies revealed
that METTL3 promotes the expression of Snail via its m6A
methylase activity. Gain-and-loss of function experiments revealed
that Snail overexpression can counteract the effects of METTL3
knockdown. Overall, the present findings reveal a critical role for
the METTL3/Snail axis in CRC progression, thus representing a
promising therapeutic strategy for CRC treatment.
Materials and methods
Bioinformatics analysis
Clinical data for bioinformatics analysis were
downloaded from the following databases: The Cancer Genome Atlas
(TCGA; http://cancergenome.nih.gov/), Gene
Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) (29) and Oncomine Platform (www.oncomine.org) (30)
analyzed with the R (v3.5.2) and R Bioconductor packages (edgeR;
http://bioconductor.org/packages/release/bioc/html/edgeR.html;
limma; http://bioconductor.org/packages/release/bioc/html/limma.html).
Patients and tumor samples
Tissue specimens, including five paired CRC tissues
and adjacent normal tissues, were obtained from patients with CRC
who received treatment at The Guangdong Second Provincial General
Hospital (Guangzhou, China) between March and July 2018. The
clinical CRC specimens were collected, following ethics approval
from the Institutional Research Ethics Committee of The Guangdong
Second Provincial General Hospital (approval no.
KQ201803005-GSPGH). All subjects were informed of the
investigational nature of the study and provided their written
informed consent. Patients were eligible if they were over 18 years
of age and had no other concurrent tumors, otherwise they were
excluded. Of these patients, three were male (aged 57, 59 and 71
years) and two were female (aged 55 and 63 years). None of the
patients had metastatic tumors. Among them, one patient had stage 1
CRC, two patients had stage 2 CRC and the other two patients had
stage 3 CRC according to the World Health Organization
Classification (31). The tissues
were immediately frozen in dry ice and stored at −80°C until
further use.
Cell culture, transfection and
treatment
The human CRC cell lines HT-29, HCT-116 and SW480;
the normal human colon mucosal epithelial cell lines NCM-460 and
HIEC-6; and the human cell line 293T were obtained from the Cell
Bank of Chinese Academy of Sciences. The HCT-116, SW480 and NCM-460
cell lines were routinely cultured in RPMI-1640 medium with 10% FBS
and 1% penicillin-streptomycin in a humidified incubator at 37°C
with 5% CO2, while the HT-29 cell line was cultured in
McCoy's 5A medium (all from Gibco; Thermo Fisher Scientific, Inc.)
under the aforementioned conditions. The HIEC-6 cell line was
cultured in complete growth medium, which contained Opti-MEM™
Reduced Serum Medium, 4% FBS, 20 mM HEPES, 10 mM GlutaMAX™ and 10
ng/ml Epidermal Growth Factor (all from Gibco; Thermo Fisher
Scientific, Inc.). The 293T cell line was cultured in complete
growth medium, which contained Eagle's Minimum Essential Medium
(Gibco; Thermo Fisher Scientific, Inc.) and 10% FBS.
A total of 1×106 HCT-116 and SW480 cells
were seeded into 6-well plates 24 h prior to transfection, and
2,000 ng plasmid (pcDNA3-vector or pcDNA3-Snail; Guangzhou RioboBio
Co., Ltd.) per well was transfected into the cells using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions.
For the inhibition of METTL3, the HCT-116 and SW480
cells were treated with S-adenosylhomocysteine (100 nM; cat. no.
HY-19528; MedChemExpress) for 48 h then the samples were collected
and analyzed using western blot analysis.
Establishment of stable knockdown cell
lines
A lentivirus-mediated method was used to stably
knock down METTL3 expression in HCT-116 and SW480 cells. The
sequences used, which were synthesized by Guangzhou RiboBio Co.,
Ltd., were as follows: Small hairpin (sh) negative control (NC),
5′-TTCTCCGAACGTGTCACGT-3′; shMETTL3-1, 5′-CGTCAGTATCTTGGGCAAGTT-3′;
shMETTL3-2, 5′-GCACTTGGATCTACGGAATCC-3′; and shMETTL3-3,
5′-GCAAGTATGTTCACTATGAAA-3′. Lentiviruses were packaged by
transfecting the aforementioned shMETTL3 or shNC transfer vector
(pLKO.1), psPAX2 and pMD2.G (all from Guangzhou RiboBio Co., Ltd.)
under a 3:2:1 ratio into 293T cells. The released virus in the
culture medium was collected at 48 and 72 h post-transduction, and
was concentrated by ultracentrifugation at 1.7×105 × g
and 4°C for 2 h.
To construct stable knockdown cells,
1×105 HCT-116 and SW480 cells were transduced with shNC
or shMETTL3 lentivirus at a multiplicity of infection of 10 with 5
µg/ml polybrene (MilliporeSigma; Merck KGaA) at 37°C for 8 h. The
stable expressing cells were screened by treatment with 1 µg/ml
puromycin (MilliporeSigma; Merck KGaA) for 1 month.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
A total of 5×105 HT-29, HCT-116, SW480,
NCM-460 and HIEC-6 cells were collected for RNA extraction using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Total RNA was reverse
transcribed into cDNA using PrimeScript RT Reagent (Takara Bio,
Inc.) according to the manufacturer's instructions. Quantification
of mRNA expression levels was performed using SYBR®
Premix Ex Taq™ Reagent (Takara Bio, Inc.) on a
LightCycler® 480 Instrument II (Roche Applied Science).
The primers were synthesized by Sangon Biotech Co., Ltd., and their
sequences were as follows: METTL3 forward,
5′-CTATCTCCTGGCACTCGCAAGA-3′ and reverse,
5′-GCTTGAACCGTGCAACCACATC-3′; Snail forward,
5′-TGCCCTCAAGATGCACATCCGA-3′ and reverse,
5′-GGGACAGGAGAAGGGCTTCTC-3′; GAPDH forward,
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3′; and 18S forward,
5′-CGGACAGGATTGACAGATTGATAGC-3′ and reverse,
5′-GCGTCCTCCTGGCTGAAGTGG-3′. The expression level was normalized to
the mRNA level of GAPDH and was calculated using the
2−ΔΔCq method (32). The
following thermocycling conditions were used: Initial denaturation
at 95°C for 2 min, followed by 40 cycles at 95°C for 30 sec, 60°C
for 25 sec and 72°C for 25 sec.
Protein isolation and western
blotting
Total protein from the HCT-116 and SW480 cell lines
and tissues was extracted upon lysis with cold RIPA buffer
(Beyotime Institute of Biotechnology) containing protease inhibitor
cocktail (cat. no. P9599; 1:100; Sigma-Aldrich; Merck KGaA). Total
protein concentration was determined by bicinchoninic acid analysis
(Beyotime Institute of Biotechnology). The proteins were then
separated by 10% SDS-PAGE and transferred onto polyvinylidene
difluoride membranes (MilliporeSigma; Merck KGaA). The membranes
were blocked with 5% (w/v) skimmed milk at room temperature for 1
h, and incubated with the following primary antibodies: Anti-METTL3
(cat. no. ab195352; Abcam), anti-Snail (cat. no. 3879S, Cell
Signaling Technology, Inc.) and anti-GAPDH (cat. no. 5174Ss; Cell
Signaling Technology, Inc.) (all at 1,1000) at 4°C overnight,
followed by incubation with a horseradish peroxidase
(HRP)-conjugate secondary antibody (cat. no. SA00001-1; ProteinTech
Group, Inc.; 1:5,000) for 1 h at room temperature. After washing
the membranes with 0.01 M TBS containing 0.1% Tween-20
(Sigma-Aldrich; Merck KGaA) three times, the protein bands were
visualized with an ECL reagent (Tanon Science and Technology Co.,
Ltd.) and were detected using a chemiluminescence system (Bio-Rad
Laboratories, Inc.). To quantify the change in expression levels of
every target, the density of the METTL3 or Snail band in each lane
was determined by densitometric analysis using ImageJ software
(1.47v; National Institutes of Health), and the relative expression
level was normalized to the level of GAPDH.
Immunohistochemistry (IHC)
The CRC and adjacent normal tissue were harvested
and fixed with 4% paraformaldehyde overnight at 4°C then cut into
5-µm thick sections at −20°C. The tissue slides were then
deparaffinized, rehydrated using an alcohol gradient (100, 96 and
70% volume) and subjected to antigen retrieval with sodium citrate
buffer. The tissues sections (5-µm) were then blocked with 5%
normal goat serum (Vector Laboratories, Inc.; Maravai LifeSciences)
with 0.1% Triton X-100 and 3% H2O2 in PBS for
60 min at room temperature, and then incubated with an anti-METTL3
antibody (1:200 dilution; cat. no. ab195352; Abcam) at 4°C
overnight. IHC staining was performed with a HRP conjugate (1:1,000
dilution; cat. no. SA00001-1; ProteinTech Group, Inc.) using DAB
detection. Cell nuclei were counterstained with Hoechst (1:1,000;
cat no. 33342; Invitrogen) for 10 min at room temperature. Images
were obtained with a Nikon E800 light microscope (magnification,
×200; Nikon Corporation) and analyzed with ImageJ software (1.47v;
National Institutes of Health). A non-specific antibody (1:200
dilution; cat. no. ab126820; Abcam) was used as a negative control,
densitometric analysis of the IHC staining signal of METTL3 was
used to analyze any potential significant differences between CRC
tissues and adjacent normal tissues by ImageJ software.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) assay was used to detect the cellular
proliferation of shMETTL3 and shNC cells. The HCT-116 and SW480
cell lines transfected with shM3 and Snail overexpression or NC
vector were seeded into a 96-well plate at a density of 5,000 cells
per well and cultured at 37°C with 5% CO2. CCK-8 reagent
was added to each well at 0, 24, 48 and 72 h, and then incubated at
37°C for 2 h. Next, the absorbance was read at 450 nm using the
PowerWave-X spectrophotometer (BioTek Instruments, Inc.).
Transwell invasion assay
Transwell Matrigel invasion assays were performed
using 24-well Transwell inserts with an 8-µm pore size (Corning,
Inc.). Briefly, the shNC or shM3 of The HCT-116 and SW480 cell
lines transfected with shNC or shM3, or with Snail overexpression
vector or NC were seeded into 24-well culture plates pre-coated
with Matrigel (BD Biosciences) at 37°C for 4 h for evaluating cell
invasion ability. After 24 h, 1×105 cells in 200 µl
culture medium without FBS were plated in the upper chambers, while
700 µl culture medium containing 10% FBS was placed in the
corresponding bottom chamber and incubated for 24 h at 37°C. The
invaded cells were fixed with 4% paraformaldehyde for 15 min and
stained with 0.5% crystal violet solution for 30 min at room
temperature. Images were randomly obtained from different fields of
view, with a Nikon light microscope (magnification, ×200; Nikon
Corporation). The mean number of invaded cells was calculated by
ImageJ software (1.47v; National Institutes of Health) and used to
evaluate the invasion capability of the cells.
Wound healing assay
The HCT-116 and SW480 cell lines transfected with
shNC or shM3 at a density of 5×106 cells per well were
seeded into 6-well plates and grown to 90% confluence in complete
medium prior to scratching. Scratches were made in the cell
monolayer using a Woundmaker™ tool (Essen Bioscience), and
RPMI-1640 medium containing 10% FBS was replaced with medium
containing 1% FBS. Images were captured immediately and after
wounding at the indicated time points (0, 12, 24 and 48 h), and
closure of the wound was monitored using a Nikon light microscope
(magnification, ×200; Nikon Corporation) and analyzed with ImageJ
software (1.47v; National Institutes of Health).
Methylated RNA immunoprecipitation
(MeRIP)
m6A qPCR was performed using Magna MeRIP™
m6A Kit (MilliporeSigma; Merck KGaA) in accordance with
the manufacturer's protocol. Briefly, total RNA was extracted from
shMETTL3- or shNC-transduced cells with TRIzol® reagent
and then treated with DNase R (Qiagen, Inc.). Next, it was randomly
fragmented by treatment with chemical reagents, followed by IP with
10 µg anti-m6A antibody (1:200 dilution; cat. no.
ab151230; Abcam) or mouse IgG, which was linked to Magna ChIP™
Protein A+G Magnetic Beads (cat. no. 16-662; MilliporeSigma). After
washing with IP buffer [10 mmol/l Tris-HCl, 150 mmol/l NaCl and
0.1% (v/v) IGEPAL® CA-630 (cat. no. I8896;
Sigma-Aldrich; Merck KGaA)] three times, the beads were treated
with proteinase K (75 µg/ml; cat. no. P2306; Sigma-Aldrich) for 10
min at 55°C with a vortical shaker. Following phenol extraction and
ethanol precipitation, the IgG and m6A-enriched RNA was
reversely transcribed with random hexamers and the enrichment was
determined using RT-qPCR (33).
RNA stability assay
To measure the half-life of Snail mRNA, shMETTL3 or
shNC cells were seeded at a density of 5×106 cells per
well onto 6-wells plates. The CRC and adjacent normal cells were
extracted from the tissues using sterile surgical instruments in a
biological safety cabinet, then cultured in RPMI-1640 medium
supplemented with 10% FBS. These cells were harvested for total RNA
isolation and subsequent RT-qPCR analysis as aforementioned at 1,
2, 4 and 8 h after the addition of actinomycin D (Act-D;
MilliporeSigma; Merck KGaA) at a final concentration of 5 µg/ml.
The regression slope (k) was used to calculate the half-life of
Snail mature mRNA according to the equation t1/2=ln2/k (34) and 18S was used for normalization.
Statistical analysis
Data are presented as the mean ± standard deviation.
Every experiment was performed independently 3 times. Kaplan-Meier
survival curve was analyzed with the log-rank test and the patients
were divided into low and high expression level group based on the
median expression level (cut-off value, 4.725). Comparisons between
CRC and adjacent normal tissues were analyzed with two-tailed
paired Student's t-test, whereas data obtained from the cell lines
were analyzed by two-tailed unpaired Student's t-test in the case
of comparisons between two groups, or by one-way ANOVA followed by
Bonferroni correction in the case of multiple comparisons.
Statistical analysis was performed with SPSS 19.0 software (IBM
Corp.), and graphical representations were conducted with GraphPad
Prism version 8.0 software (GraphPad Software, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
METTL3 is markedly upregulated in CRC
and is associated with poor prognosis
To study the effects of m6A modification
on the progression and development of CRC, the expression of
m6A-related enzymes, including METTL3, METTL14 and WTAP,
and that of demethyltransferases (FTO and ALKBH5), was evaluated
using TCGA database. The results showed that METTL3 expression was
significantly upregulated in 222 primary CRC tissues compared with
that of 41 normal colonic tissues, while the expression of other
enzymes showed negligible differences (Fig. 1A). The GEPIA database, an online tool
used for analyzing cancer clinical data originated from TCGA
database, showed that METTL3 expression was gradually increased
according to clinical stage classification: Clinical stages 1, 2, 3
and 4 (Fig. 1B). Kaplan-Meier curves
of OS obtained from the GEPIA database revealed that high
expression of METTL3 was associated with less favorable survival
(Fig. 1C). Furthermore, increased
expression of METTL3 was observed in CRC tissues compared with that
in normal tissues using the Oncomine platform (Fig. 1D). Taken together, these
bioinformatics outcomes predict that METTL3 is significantly
upregulated in CRC and was negatively correlated with clinical
stage and OS.
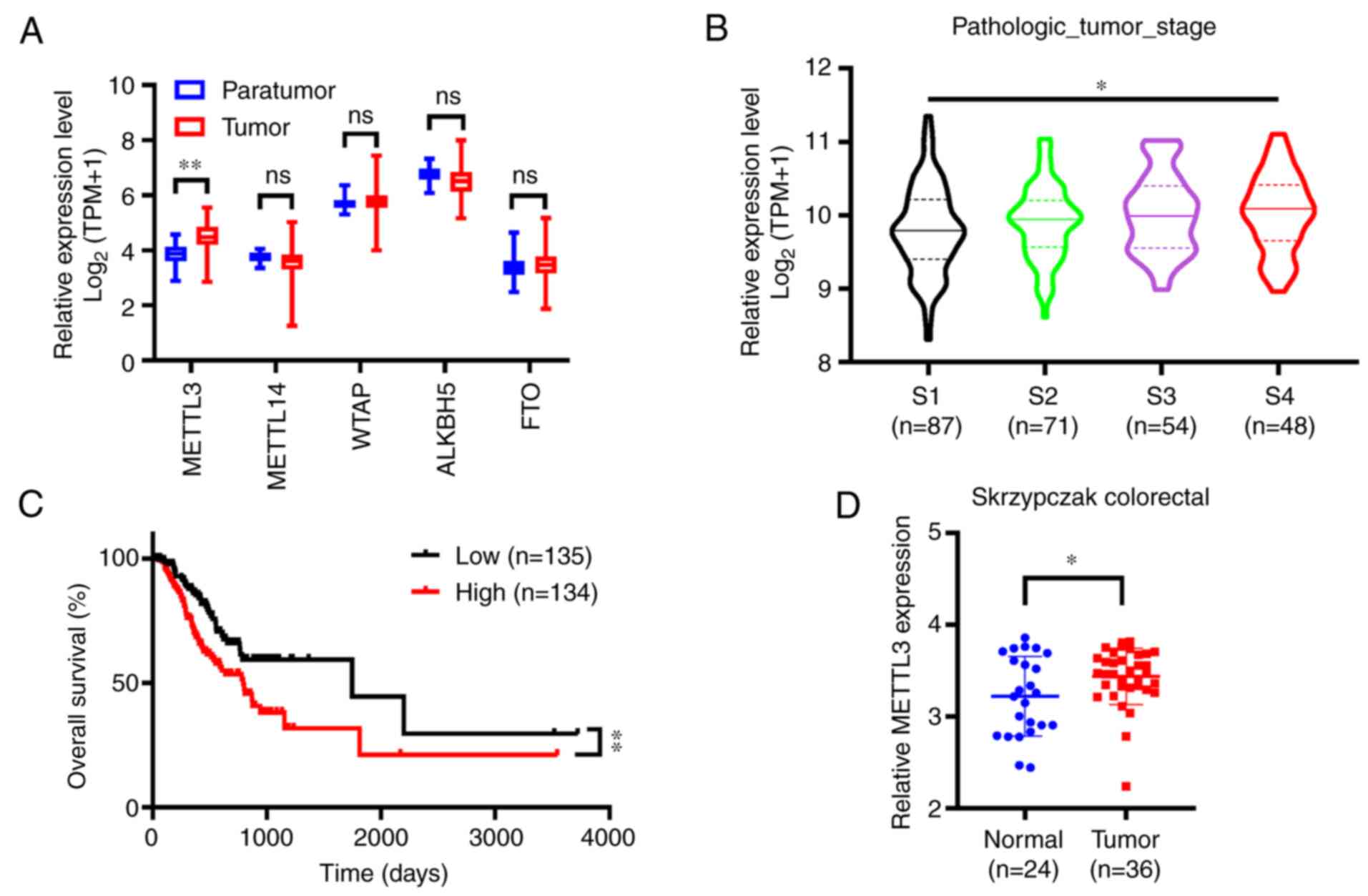 | Figure 1.METTL3 is significantly upregulated
in CRC on TCGA and Oncomine databases. (A) Nitrogen
6-methyladenosine-related enzymes mRNA expression levels in CRC vs.
normal tissues according to TCGA database. (B) METTL3 expression
levels in different clinical stages of CRC exhibited the following
order: Clinical stages 1, 2, 3 and 4, as represented by the violin
plots. (C) The overall survival of patients with CRC who exhibited
high (n=135) vs. low (n=134) levels of METTL3 was plotted by the
Kaplan-Meier method. (D) METTL3 mRNA expression levels in CRC and
normal tissues according to the Oncomine database ‘Skrzypczak
colorectal’. Error bars represent standard deviation. *P<0.05,
**P<0.01. METTL3, methyltransferase-like 3; CRC, colorectal
cancer; TCGA, The Cancer Genome Atlas; TPM, transcript count per
million; ns, not significant; WTAP, Wilms' tumor 1-associating
protein; ALKBH5, α-ketoglutarate-dependent dioxygenase AlkB homolog
5; FTO, fat mass and obesity-associated protein. |
METTL3 expression is markedly
increased in CRC
The expression of METTL3 was examined using two
human colonic epithelial cell lines (HIEC-6 and NCM-460) and in
three CRC cell lines (HT-29, HCT-116 and SW480). RT-qPCR showed
that METTL3 was significantly elevated in CRC cell lines compared
with its expression in normal epithelial cell lines at the mRNA
level (Fig. 2A). The cell lines
exhibiting the most significant differences vs. the controls
(namely, HCT-116 and SW480) were used in subsequent
experiments.
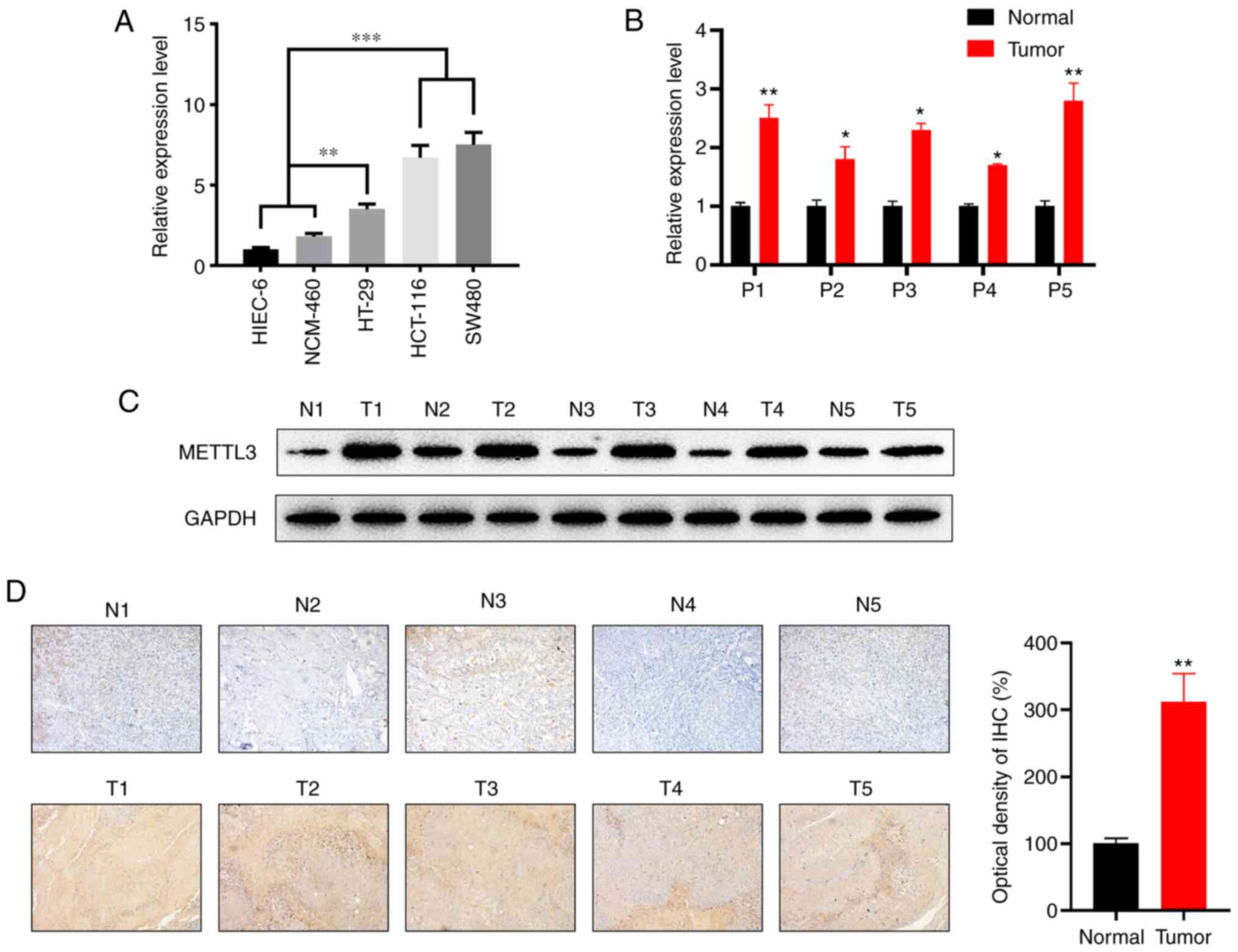 | Figure 2.METTL3 is significantly upregulated
in CRC tissues compared with its expression levels in the
corresponding adjacent normal colon tissues. (A) METTL3 mRNA
expression levels in the human CRC cell lines, HT-29, HCT-116 and
SW480, the normal cell lines NCM-460 and HIEC-6. (B) Relative mRNA
expression levels of METTL3 in CRC tissues compared with those in
the corresponding adjacent normal tissues (n=5). (C) Protein
expression of METTL3 in CRC vs. the corresponding adjacent normal
tissues. GAPDH was used as a loading control. The right panel shows
the quantitative analysis of the western blot results, as analyzed
by paired Student's t-test. (D) The expression of METTL3 in CRC and
adjacent normal tissues was detected by IHC staining
(magnification, ×100; n=5). Error bars represent standard
deviation. *P<0.05, **P<0.01, ***P<0.001. METTL3,
methyltransferase-like 3; CRC, colorectal cancer; P, patient; N,
normal; T, tumor. |
In addition, the mRNA levels of METTL3 were
increased in five CRC tissues compared with those in their
corresponding adjacent normal tissues according to the results of
RT-qPCR (Fig. 2B). Western blotting
further confirmed that METTL3 protein expression was significantly
increased in five CRC tissues relative to that in normal tissues
(Fig. 2C). Furthermore, increased
METTL3-positive staining in primary CRC tissues compared with that
in normal tissues was detected by IHC (Fig. 2D). Overall, these data indicated that
the expression of METTL3 is increased in clinical CRC tissues and
cell lines.
METTL3 facilitates the proliferation
and migration of CRC in vitro
Based on the results of bioinformatics analysis and
the upregulation of METTL3 observed in CRC cell lines and tissues,
it was hypothesized that RNA m6A modification is closely
associated with clinicopathological features, and that the
m6A regulator METTL3 may serve an oncogenic role in CRC.
Therefore, the effects of METTL3 on the proliferation and migration
of CRC cell lines were evaluated.
A scrambled shRNA and three METTL3-silencing shRNAs
were constructed, and their knockdown efficiency was evaluated by
assessing the mRNA expression level of METTL3 after being
transfected into the HCT-116 and SW480 cell lines (Fig. 3A). The results revealed that shM3-3
knockdown was the most efficient shRNA; thus, it was used for
constructing the recombinant lentivirus. Upon transduction of the
lentivirus in wild-type CRC cell lines, RT-qPCR (Fig. 3B) and western blotting (Fig. 3C) confirmed significant knockdown
efficiency of the lentivirus.
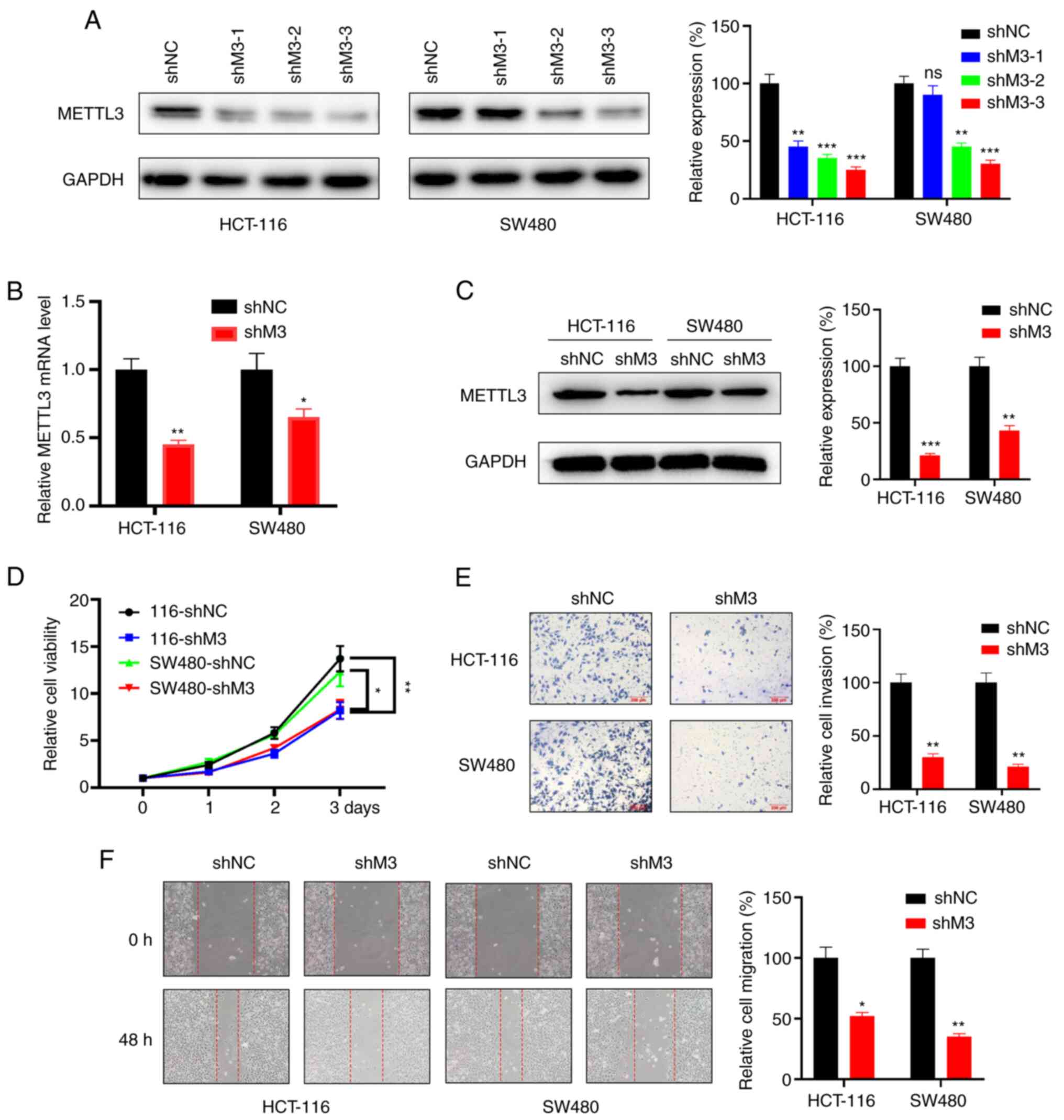 | Figure 3.M3 knockdown represses cell
proliferation, invasion and migration in vitro. (A) Protein
levels (right, quantitative analysis) of M3 in HCT-116 and SW480
cells upon transfection with a scrambled shRNA or with one of three
different M3-silencing shRNAs. (B) mRNA levels of M3 in HCT-116 and
SW480 cells infected with lentivirus carrying the indicated shRNA.
(C) Protein levels of M3 in HCT-116 and SW480 cells infected with
lentivirus carrying the indicated shRNA (right, quantitative
analysis). (D) The proliferation rate of stable scrambled shRNA
(shNC) and stable METTL3 knockdown (shM3) cells was evaluated by
Cell Counting Kit-8 assay in HCT-116 and SW480 cells. (E) Transwell
assay (right, quantitative analysis) was employed to analyze the
difference in the invasion ability of shNC and shM3 cells. (F)
Wound healing assay (magnification, ×200; right, quantitative
analysis) revealed differences in the migration ability of shNC and
shM3 cells. Each experiment was repeated three times independently.
Error bars represent standard deviation. *P<0.05, **P<0.01,
***P<0.001. M3, methyltransferase-like 3; sh, small hairpin RNA;
NC, negative control; ns, not significant. |
Next, stable METTL3 knockdown and corresponding NC
of HCT-116 and SW480 cells were generated to analyze the influence
on cell proliferation of METTL3 depletion. The results revealed
that the proliferation rate of shMETTL3-transduced cells was
significantly decreased compared with that of shNC cells (Fig. 3D). In addition, compared with that of
shNC cells, shM3 cells exhibited a decreased invasion ability
according to the results of Transwell assay (Fig. 3E). Consistently, wound healing assay
showed that the downregulation of METTL3 in CRC cell lines
suppressed cell migration (Fig. 3F).
Collectively, these results indicated that METTL3 promotes the
proliferation, migration and invasion of CRC cell lines.
METTL3 promotes the expression of
Snail in CRC
In a previous study, Snail exhibited oncogenic
roles, and its m6A modification influenced the
progression of liver cancer (24).
The present study hypothesized that METTL3 regulates the expression
of Snail in a m6A-dependent manner, thus contributing to
CRC malignancy. Initially, the m6A modification of Snail
was assessed in shNC and shM3 cells by MeRIP assay, which
identified the presence of m6A modification in shNC
HCT-116 cells, while this enrichment was significantly decreased in
shM3 HCT-116 cells (Fig. 4A).
Consistently, m6A enrichment was increased in shNC SW480
cells compared with that in shMETTL3-infected SW480 cells (Fig. 4B).
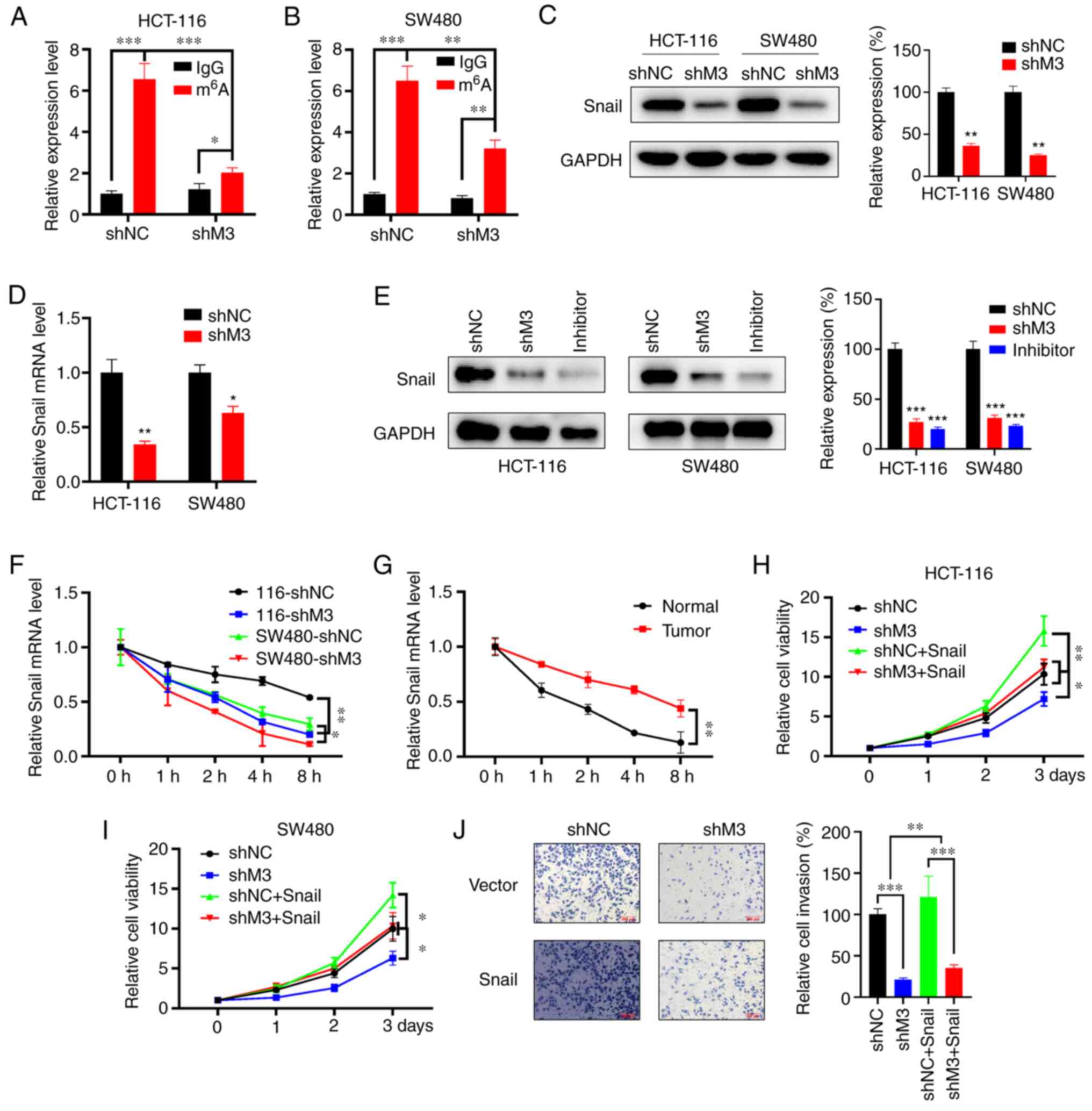 | Figure 4.Snail mediates M3-induced malignancy
in CRC. m6A RIP-qPCR analysis of Snai1 mRNA in shNC- and
shM3-infected (A) HCT-116 and (B) SW480 cells. (C) Snail protein
expression levels (right, quantitative analysis) in stable
with/without M3 knockdown HCT-116 and SW480 cells. (D) Snail mRNA
expression levels in stably infected HCT-116 and SW480 cells
with/without M3 knockdown. (E) Wild-type cells were treated with
S-adenosylhomocysteine at a final concentration of 100 nM for 48 h,
followed by detection of Snail protein expression (right,
quantitative analysis) using western blot analysis. shNC and shM3
cells were included for analysis. (F) Snail mRNA expression levels
in shNC and shM3 cells treated with Act-D (5 µg/ml) for the
indicated time points were evaluated by qPCR. (G) Snail mRNA
expression levels in CRC and adjacent normal tissue cells treated
with Act-D (5 µg/ml) for the indicated time points were evaluated
by qPCR. The proliferation rate of (H) HCT-116 and (I) SW480 cells
transfected with shNC and shM3 and/or overexpressing Snail was
evaluated using a Cell Counting Kit-8 assay at the indicated time
points. (J) shNC or shM3 cells transiently overexpressing Snail or
pcDNA3 vector were subjected to invasion assays for 24 h and
subsequently analyzed with CytoSelect™ 24-well Cell Invasion assay
kits (magnification, ×100; 8 µm; left). The invaded cells were then
quantitatively analyzed (right). All experiments were performed in
triplicate. Error bars represent standard deviation. *P<0.05,
**P<0.01, ***P<0.001. M3, methyltransferase-like 3; CRC,
colorectal cancer; m6A, nitrogen 6-methyladenosine; RIP,
RNA immunoprecipitation; qPCR, quantitative PCR; sh, small hairpin
RNA; NC, negative control; Act-D, actinomycin D. |
Furthermore, western blotting revealed that the
expression of Snail was elevated in shNC cells compared with that
in shM3 cells (Fig. 4C). The mRNA
expression level of Snail was also evaluated in shNC- and
shMETTL3-infected cells, and, similarly, it was observed that the
mRNA level of Snail in shMETTL3-infected cells was downregulated
(Fig. 4D). Inhibiting METTL3 could
decrease Snail expression by addition of an METTL3 inhibitor, akin
to METTL3 knockout (Fig. 4E). To
investigate the potential mechanism responsible for the
downregulation of Snail in shM3 cells, the decay rate of Snail mRNA
in shNC and shM3 cells was determined by adding Act-D at the
indicated time points, and the results showed that the mRNA
stability of Snail was significantly reduced in shMETTL3-infected
cells (Fig. 4F). As expected, the
mRNA stability of Snail in CRC tissues was higher than that in
colon normal tissues (Fig. 4G).
These findings demonstrate that Snail is a direct
downstream effector of METTL3-induced malignancy. To characterize
the potential role of Snail in CRC malignancy, gain- and
loss-of-function experiments were performed by overexpressing Snail
in shM3 and shNC cells. It was observed that Snail overexpression
promoted the proliferation rate of HCT-116 cells and reversed the
effects of METTL3 knockdown in HCT-116 cells (Fig. 4H). Consistently, overexpression of
Snail alleviates the shMETTL3-induced proliferation suppression in
SW480 cells (Fig. 4I). Furthermore,
Snail overexpression rescued the invasive potential of
METTL3-silenced HCT-116 cells (Fig.
4J). Taken together, these results indicate that METTL3
promotes the growth and invasion of CRC through regulating the
level of Snail mRNA in vitro.
Discussion
Recently, m6A modification has gained
attention in the field of epigenomics (35). Previous studies have indicated that
m6A regulators serve important regulatory roles in
diverse biological processes in various human cancer types,
including breast and lung cancer and CRC (36–38).
METTL3, as the most important writer of m6A, is
upregulated in numerous malignancies and can epigenetically
increase gene expression by specifically editing m6A
sites modification, thus eliciting a potential malignant function
(39). Previous studies have
demonstrated that METTL3 is frequently upregulated in CRC, where it
maintains high m6A modification levels (40–42).
Consistently, the present data showed that METTL3 is elevated in
five paired CRC tissues and CRC cell lines. Together with the
previously published results in the literature, the present
findings supported that m6A and elevated METTL3 may
participate in CRC progression. However, the number of clinical
samples is too small in the present study; therefore, additional
paired clinical CRC tissues are required to further support the
conclusions in future studies.
The present study evaluated the effects of METTL3 in
CRC using lentivirus-mediated METTL3 knockdown assays, and the
results showed that the suppression of METTL3 decreased the
invasion, migration and proliferation of the CRC cell lines in
vitro. A previous study demonstrated that METTL3 silencing in
CRC patient-derived organoids and transgenic mouse models exhibited
anticancer effects by inhibiting the translation of glucose
transporter 1 in vivo (43).
Furthermore, knockdown of METTL3 suppressed cell proliferation and
migration in SW620 and HT29 cells (23). These findings indicate that METTL3
serves oncogenic and pivotal roles in CRC carcinogenesis in
vitro and in vivo, which suggests that targeting METTL3
and developing m6A inhibitors may be a novel approach
for CRC therapy.
m6A modification is a complex regulatory
network that participates in various stages of the RNA life cycle
and metabolism, including RNA processing, nuclear output,
regulation of translation and RNA degradation (44). Using m6A-RIP-qPCR and
Act-D assays, the present study demonstrated that METTL3-mediated
m6A enrichment enhances the stability of Snail mRNA
during RNA processing. However, other post-transcriptional
processes were not evaluated and remain to be investigated further.
m6A reader proteins, including IGF2BP1/2/3, eukaryotic
initiation factor 3 and YTH domain family, members 1/2/3, can bind
to an m6A-modified motif indirectly or directly, thus
affecting RNA function (45). A
previous study reported that IGF2BP2 can recognize methylated SOX2
transcripts in the coding sequence regions to prevent SOX2 mRNA
degradation, which contributes to the malignant behavior of CRC
(21). The present findings
demonstrate that m6A can stabilize Snail mature mRNA.
However, further studies are required to characterize the Snail
mRNA m6A editing sites and m6A readers are
involved in their mRNA degradation.
Snail activates EMT signals and pathways as a
transcriptional factor, which can affect cancer progression and the
metastatic potential of tumors (46). The present study confirmed that
overexpression of Snail could increase the proliferation and
migration of the CRC cell lines. Phosphorylation and ubiquitination
of Snail induce protein degradation, which can reverse
tumorigenesis (47). The present
study has showed that METTL3 depletion can repress Snail in an
m6A-dependent manner at the RNA level, indicating that
targeting Snail mRNA m6A sites to induce the suppression
of expression may be a novel method to prevent CRC progression.
The present study used bioinformatics analysis to
screen potential targets, including Snail and METTL3, which were
confirmed in clinical tissues, and explored the mechanisms and
functions of the targets involved in CRC progression. The present
findings provide novel information on the participation of the
m6A epitranscriptome in cancer progression.
In summary, the present study confirmed that METTL3
acts as a critical m6A methyltransferase capable of
facilitating CRC progression, and revealed a novel mechanism by
which METTL3 promotes CRC cell proliferation and invasion via
stabilizing Snail mRNA in an m6A-dependent manner.
Additionally, the present results indicated that the upregulation
of METTL3 may be a major driver of CRC progression in vitro
and in vivo. Overall, these results suggest that targeting
METTL3 and Snail may represent promising therapeutic strategies for
the treatment of CRC.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
Science Foundation of Guangdong Second Provincial General Hospital
(grant no. YQ2015-007).
Availability of data and materials
All data generated and/or analyzed during this study
are included in this published article.
Authors' contributions
Conception and design: JW, GZ and ZY; acquisition of
data: JW, YM and LZ; statistical analysis: JW, GZ and MJ; writing
and revision of the manuscript: JW and ZY. JW and ZY confirmed the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All the study procedures involving human
participants were conducted in accordance with the ethical
standards of the institutional research committee (approval no.
KQ201803005-GSPGH) and patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RG, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
National Health Commission of The People's
Republic of China, . National guidelines for diagnosis and
treatment of colorectal cancer 2020 in China (English version).
Chin J Cancer Res. 32:415–445. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Degiuli M, Reddavid R, Ricceri F, Di
Candido F, Ortenzi M, Elmore U, Belluco C, Rosati R, Guerrieri M
and Spinelli A; Members of the Italian Society of Surgical Oncology
Colorectal Cancer Network (SICO-CCN) Collaborative Group, :
Segmental colonic resection is a safe and effective treatment
option for colon cancer of the splenic flexure: A nationwide
retrospective study of the italian society of surgical
oncology-colorectal cancer network collaborative group. Dis Colon
Rectum. 63:1372–1382. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang R, Cheng AJ, Wang JY and Wang TC:
Close correlation between telomerase expression and adenomatous
polyp progression in multistep colorectal carcinogenesis. Cancer
Res. 58:4052–4054. 1998.PubMed/NCBI
|
|
5
|
Pichler M, Stiegelbauer V,
Vychytilova-Faltejskova P, Ivan C, Ling H, Winter E, Zhang X,
Goblirsch M, Wulf-Goldenberg A, Ohtsuka M, et al: Genome-Wide miRNA
analysis identifies miR-188-3p as a novel prognostic marker and
molecular factor involved in colorectal carcinogenesis. Clin Cancer
Res. 23:1323–1333. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun T, Wu R and Ming L: The role of
m6A RNA methylation in cancer. Biomed Pharmacother.
112:1086132019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma
D, Lv J, Heng J, Ding Y, Xue Y, et al: m6A modulates
haematopoietic stem and progenitor cell specification. Nature.
549:273–276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Di Timoteo G, Dattilo D, Centron-Broco A,
Colantoni A, Guarnacci M, Rossi F, Incarnato D, Oliviero S, Fatica
A, Morlando M and Bozzoni I: Modulation of circRNA metabolism by
m6A modification. Cell Rep. 31:1076412020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen F, Chen Z, Guan T, Zhou Y, Ge L,
Zhang H, Wu Y, Jiang GM, He W, Li J and Wang H:
N6-Methyladenosine regulates mRNA stability and
translation efficiency of KRT7 to promote breast cancer lung
metastasis. Cancer Res. 81:2847–2860. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Y, Yang X, Chen Z, Tian L, Jiang G,
Chen F, Li J, An P, Lu L, Luo N, et al: m6A-induced
lncRNA RP11 triggers the dissemination of colorectal cancer cells
via upregulation of Zeb1. Mol Cancer. 18:872019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Alarcon CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary
microRNAs for processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA
stability. Nature. 505:117–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C:
N6-methyladenosine modulates messenger RNA translation
efficiency. Cell. 161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jia G, Fu Y and He C: Reversible RNA
adenosine methylation in biological regulation. Trends Genet.
29:108–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers
reveals two distinct classes of mRNA methylation at internal and
5′sites. Cell Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou T, Ren Z and Chen C: METTL14 as a
predictor of postoperative survival outcomes of patients with
hepatocellular carcinoma. Nan Fang Yi Ke Da Xue Xue Bao.
40:567–572. 2020.(In Chinese). PubMed/NCBI
|
|
18
|
Mathiyalagan P, Adamiak M, Mayourian J,
Sassi Y, Liang Y, Agarwal N, Jha D, Zhang S, Kohlbrenner E,
Chepurko E, et al: FTO-Dependent N6-methyladenosine
regulates cardiac function during remodeling and repair.
Circulation. 139:518–532. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu K, Mo Y, Li D, Yu Q, Wang L, Lin F,
Kong C, Balelang MF, Zhang A, Chen S, et al:
N6-methyladenosine demethylases Alkbh5/Fto regulate
cerebral ischemia-reperfusion injury. Ther Adv Chronic Dis.
11:20406223209160242020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Cheng X, Wang J, Huang Y, Yuan J
and Guo D: Gene signature and prognostic merit of M6a regulators in
colorectal cancer. Exp Biol Med (Maywood). 245:1344–1354. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN,
Chen ZH, Zeng ZL, Wang F, Zheng J, et al: METTL3 facilitates tumor
progression via an m6A-IGF2BP2-dependent mechanism in
colorectal carcinoma. Mol Cancer. 18:1122019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang DD, Chen ZH, Yu K, Lu JH, Wu QN, Wang
Y, Ju HQ, Xu RH, Liu ZX and Zeng ZL: METTL3 Promotes the
progression of gastric cancer via targeting the MYC Pathway. Front
Oncol. 10:1152020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou D, Tang W, Xu Y, Xu Y, Xu B, Fu S,
Wang Y, Chen F, Chen Y, Han Y and Wang G: METTL3/YTHDF2
m6A axis accelerates colorectal carcinogenesis through
epigenetically suppressing YPEL5. Mol Oncol. Jan 7–2021.doi:
10.1002/1878-0261.12898 (Epub ahead of print). View Article : Google Scholar
|
|
24
|
Zhu W, Si Y, Xu J, Lin Y, Wang JZ, Cao M,
Sun S, Ding Q, Zhu L and Wei JF: Methyltransferase like 3 promotes
colorectal cancer proliferation by stabilizing CCNE1 mRNA in an
m6A-dependent manner. J Cell Mol Med. 24:3521–3533.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng R, Cheng Y, Ye S, Zhang J, Huang R,
Li P, Liu H, Deng Q, Wu X, Lan P and Deng Y: m6A
methyltransferase METTL3 suppresses colorectal cancer proliferation
and migration through p38/ERK pathways. Onco Targets Ther.
12:4391–4402. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu X, Zhao H and Cao Z: The m6A
methyltransferase METTL3 aggravates the progression of
nasopharyngeal carcinoma through inducing EMT by
m6A-modified Snail mRNA. Minerva Med. Jun 5–2020.doi:
10.23736/S0026-4806.20.06653-7 (Epub ahead of print).
|
|
27
|
Lin X, Chai G, Wu Y, Li J, Chen F, Liu J,
Luo G, Tauler J, Du J, Lin S, et al: RNA m(6)A methylation
regulates the epithelial mesenchymal transition of cancer cells and
translation of Snail. Nat Commun. 10:20652019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Wu Z and Hu L: The regulatory
effects of metformin on the [SNAIL/miR-34]:[ZEB/miR-200] system in
the epithelial-mesenchymal transition(EMT) for colorectal
cancer(CRC). Eur J Pharmacol. 834:45–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ahadi M, Sokolova A, Brown I, Chou A and
Gill AJ: The 2019 World Health Organization Classification of
appendiceal, colorectal and anal canal tumours: An update and
critical assessment. Pathology. 53:454–461. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by
m6A-seq. Nature. 485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hentze MW: Determinants and regulation of
cytoplasmic mRNA stability in eukaryotic cells. Biochim Biophys
Acta. 1090:281–292. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang G, Sun Z and Zhang N: Reshaping the
role of m6A modification in cancer transcriptome: A
review. Cancer Cell Int. 20:3532020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu L, Wu D, Ning J, Liu W and Zhang D:
Changes of N6-methyladenosine modulators promote breast
cancer progression. BMC Cancer. 19:3262019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang H, Zhao X and Lu Z: m6A
RNA methylation regulators act as potential prognostic biomarkers
in lung adenocarcinoma. Front Genet. 12:6222332021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tian J, Ying P, Ke J, Zhu Y, Yang Y, Gong
Y, Zou D, Peng X, Yang N, Wang X, et al: ANKLE1
N6-Methyladenosine-related variant is associated with
colorectal cancer risk by maintaining the genomic stability. Int J
Cancer. 146:3281–3293. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huo FC, Zhu ZM, Zhu WT, Du QY, Liang J and
Mou J: METTL3-mediated m6A methylation of SPHK2 promotes
gastric cancer progression by targeting KLF2. Oncogene.
40:2968–2981. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lan H, Liu Y, Liu J, Wang X, Guan Z, Du J
and Jin K: Tumor-associated macrophages promote oxaliplatin
resistance via METTL3-mediated m6A of TRAF5 and
necroptosis in colorectal cancer. Mol Pharm. 18:1026–1037. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen H, Gao S, Liu W, Wong CC, Wu J, Wu J,
Liu D, Gou H, Kang W, Zhai J, et al: RNA
N6-Methyladenosine methyltransferase METTL3 facilitates
colorectal cancer by activating the m6A-GLUT1-mTORC1
axis and is a therapeutic target. Gastroenterology.
160:1284–1300.e16. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Peng W, Li J, Chen R, Gu Q, Yang P, Qian
W, Ji D, Wang Q, Zhang Z, Tang J and Sun Y: Upregulated METTL3
promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK
signaling pathway. J Exp Clin Cancer Res. 38:3932019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X,
Zhang X, Cao Y, Ma D, Zhu X, et al: m6A-dependent
glycolysis enhances colorectal cancer progression. Mol Cancer.
19:722020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Y, Hsu PJ, Chen YS and Yang YG:
Dynamic transcriptomic m6A decoration: Writers, erasers,
readers and functions in RNA metabolism. Cell Res. 28:616–624.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhen D, Wu Y, Zhang Y, Chen K, Song B, Xu
H, Tang Y, Wei Z and Meng J: m6A Reader:
Epitranscriptome target prediction and functional characterization
of N6-methyladenosine (m6A) readers. Front
Cell Dev Biol. 8:7412020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kudo-Saito C, Shirako H, Takeuchi T and
Kawakami Y: Cancer metastasis is accelerated through
immunosuppression during Snail-induced EMT of cancer cells. Cancer
Cell. 15:195–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dominguez D, Montserrat-Sentís B,
Virgós-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J,
Francí C and García de Herreros A: Phosphorylation regulates the
subcellular location and activity of the snail transcriptional
repressor. Mol Cell Biol. 23:5078–5089. 2003. View Article : Google Scholar : PubMed/NCBI
|


















