Introduction
Colorectal cancer (CRC) is a common malignant tumor
in the gastrointestinal tract; its morbidity and fatality rates are
behind only gastric, esophageal and primary liver cancer in
malignant tumors of the digestive system (1,2).
Globally, the incidence and mortality of CRC rank fifth among all
malignant tumors (3). The incidence
of CRC is associated with factors such as age, region and sex. CRC
primarily occurs in middle-aged and elderly people (>40 years
old), and the incidence of CRC in men and women is relatively
similar (4). Previously, the
incidence and mortality rates of CRC have demonstrated a notable
upward trend (5). Numerous studies
have identified KRAS mutations as poor prognostic biomarkers
correlated with poor CRC survival outcomes (6,7). Taking
advantage of synthetic lethal interactions with KRAS mutation may
represent a target for effective therapeutic strategies in patients
with KRAS-mutant CRC (8). Thus,
detection methodology should be used to identify the key prognostic
biomarkers for CRC, specifically KRAS-mutant CRC.
Human coding genes account for <2% of the total
genome. The number of transcripts in the genome is very large; 70%
of the whole human genome is stably transcribed into RNAs, and the
majority of these are non-coding RNAs (9), including microRNAs (miRNAs/miRs) and
long non-coding RNAs (lncRNAs). lncRNAs refer to transcripts
>200 nucleotides in length that do not contain a protein-coding
sequence (10). Long intergenic
non-coding RNAs (lincRNAs) are the largest class of lncRNA
molecules (11). lincRNAs can serve
as transcriptional regulators and influence gene transcription,
acting as decoys to bind proteins or miRNAs (9,12).
Previously, numerous studies have reported that lincRNAs serve
tumor-suppressive or tumor-promoting roles. For instance, lincRNAs
correlated with CRC include CCAT1, CCAT2, CRNDE, HULC and MALAT1
(13,14).
In the past several years, certain studies have
reported crosstalk between lincRNAs and miRNAs during cancer
progression, specifically the hypothesis of competing endogenous
RNAs (ceRNAs) (14,15). The ceRNA hypothesis model states that
all coding and non-coding RNAs sharing common miRNA response
elements may inhibit and indirectly regulate the expression of each
other by competing for miRNA binding sites (16). These models are critical for human
cancer. For instance, CCAT1 epigenetically downregulates c-Myc by
serving as a ceRNA for miR-155, which downregulates c-Myc
expression (16). lincRNAs are
included in the regulatory network of CLDN4 via ceRNA-mediated
miRNA evasion in gastric cancer (17). Thus, ceRNAs comprising lincRNAs,
miRNAs and mRNAs can serve as important prognostic biomarkers in
human cancer.
Targeting KRAS-driven cancer is an effective
strategy that selectively inhibits cancer growth while unharmed
normal cells (18). Thus, it is
necessary to explore and identify key lincRNAs in KRAS-mutant CRC.
However, there appear to be few related previous studies. In the
present study, RNA sequencing (RNA-Seq) and clinical data from The
Cancer Genome Atlas (TCGA) were employed to identify key survival
lincRNAs associated with KRAS mutations in CRC. Furthermore,
RNA-Seq and miRNA-Seq data from TCGA were used to construct ceRNA
models among lincRNAs, miRNAs and mRNAs in CRC. The current study
may provide a new understanding of KRAS-mutated CRC and help to
interpret the mechanisms underlying the functions of these lincRNAs
in CRC.
Materials and methods
Raw data
RNA-Seq, miRNA-Seq, copy number variation (CNV) and
clinical data of CRC (COAD + READ) were downloaded from the TCGA
(https://portal.gdc.cancer.gov). The
RNA-Seq data are presented as fragments per kilobase million
(FPKM). The miRNA-seq data were presented as reads per million
miRNA mapped data.
Dysregulated lincRNA analysis
Dysregulated gene analysis was conducted with R
software version 4.0.3, and the Mann-Whitney U test was performed
to define significantly dysregulated lincRNAs between tumor and
normal samples. To decrease background noise, any lincRNAs with
50th percentile FPKM=0 in tumor or normal samples were eliminated.
Significantly dysregulated lincRNAs were defined as a logarithmic
transformed fold-change (|log2 (FC)|) ≥1 and a false
discovery rate (FDR) ≤0.05. Furthermore, lincRNAs with a mean FPMK
value >1 in tumor samples were considered upregulated, and
lincRNAs with a mean FPMK value >1 in normal samples were
considered downregulated.
Predictive value of lincRNAs for
survival in patients with KRAS mutations
Receiver operating characteristic (ROC) curves were
used to analyze the association between lincRNAs and the survival
status of patients with mutant or wild-type KRAS at 10 and 5 years.
ROC curves were determined to evaluate the sensitivity and
specificity of the expression level of each lincRNA in predicting
mortality in patients (19). Forest
plots were used to demonstrate the result of ROC. An area under the
curve >0.6 and a P<0.05 were the threshold values to indicate
significance.
Identification of key survival-related
lincRNAs with KRAS mutations
Robust likelihood-based survival models were
constructed to identify the key lincRNAs influencing the prognosis
of CRC using rbsurv (https://bioconductor.org/packages/rbsurv) in R
software version 4.0.3. The lincRNAs with the highest frequency
were selected as the final feature lincRNAs. The effect of key
lincRNA expression on patient survival was assessed by the
Kaplan-Meier survival curves and the log-rank test.
Prediction of target lincRNAs and
mRNAs of miRNAs
Target lincRNAs and mRNAs were predicted for miRNAs
using the online tool DIANA (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index).
The cut-off values used to identify a significant correlation were
Pearson correlation coefficient (PCC) of <-0.1 and P<0.05.
Furthermore, a maximal information coefficient (MIC) >0.17 was
set as a cut off for significant correlation (20). PCC and MIC were performed with
Rsoftware version 4.0.3.
Functional enrichment analysis
Functional enrichment analysis was performed with
Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO)
and Reactome under Metascape (http://metascape.org) (21). P<0.05 was set as the cut-toff.
Regulatory network
Regulatory network lincRNAs were directly connected
to miRNAs, and mRNAs were directly connected to the miRNAs,
constituting a regulatory network. The network was constructed and
displayed using Cytoscape 3.7.2 (https://cytoscape.org).
Reverse transcription-quantitative
(RT-q)PCR
A total of 12 pairs of CRC tissues and normal
tissues (normal tissues were 2–3 cm away from cancer tissue) were
obtained from 12 patients (age range, 42–53 years; median age, 48
years; eight men and four women) who underwent radical resection at
The First College of Clinical Medical Science, China Three Gorges
University (Yichang, China) between July 2020 and September 2020.
The patient was diagnosed as CRC by imaging and pathological
examination, and did not receive chemotherapy or radiotherapy
before operation. All samples are stored in −80°C refrigerator for
use. Total RNA was extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA was reverse
transcribed into cDNA using the PrimeScript 1st Strand cDNA
Synthesis Kit Starter kit (Takara Bio, Inc.), according to the
manufacturer's protocol. Relative expression levels of lincRNAs and
mRNAs were quantified using the TB Green® Premix Ex Taq™
II kit (Takara Bio, Inc.), according to the manufacturer's
protocol. Relative expression levels of miRNAs were quantified
using the Mir-X miRNA First-Strand Synthesis kit (Takara Bio,
Inc.), according to the manufacturer's protocol. The primer
sequences are listed in Table SI.
Expression levels of lincRNAs and mRNAs relative to β-actin and
expression of miRNAs relative to U6 snRNA were determined using the
2−ΔΔCq method (22). All
experimental procedures were approved by the Ethics Committee of
The First College of Clinical Medical Science, China Three Gorges
University. Written informed consent was provided by all patients
prior to the study.
Statistical analysis
The Mann-Whitney U test was performed using GraphPad
Prism (GraphPad Software, Inc.) version 9 software and R software
version 4.0.3. Kaplan-Meier survival curves and log-rank tests were
used to evaluate the effect of lincRNA expression on survival.
Receiver operating characteristic (ROC) curves were generated using
GraphPad Prism version 9 software. Data are presented as the mean ±
SEM. P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of significantly
dysregulated lincRNAs in CRC
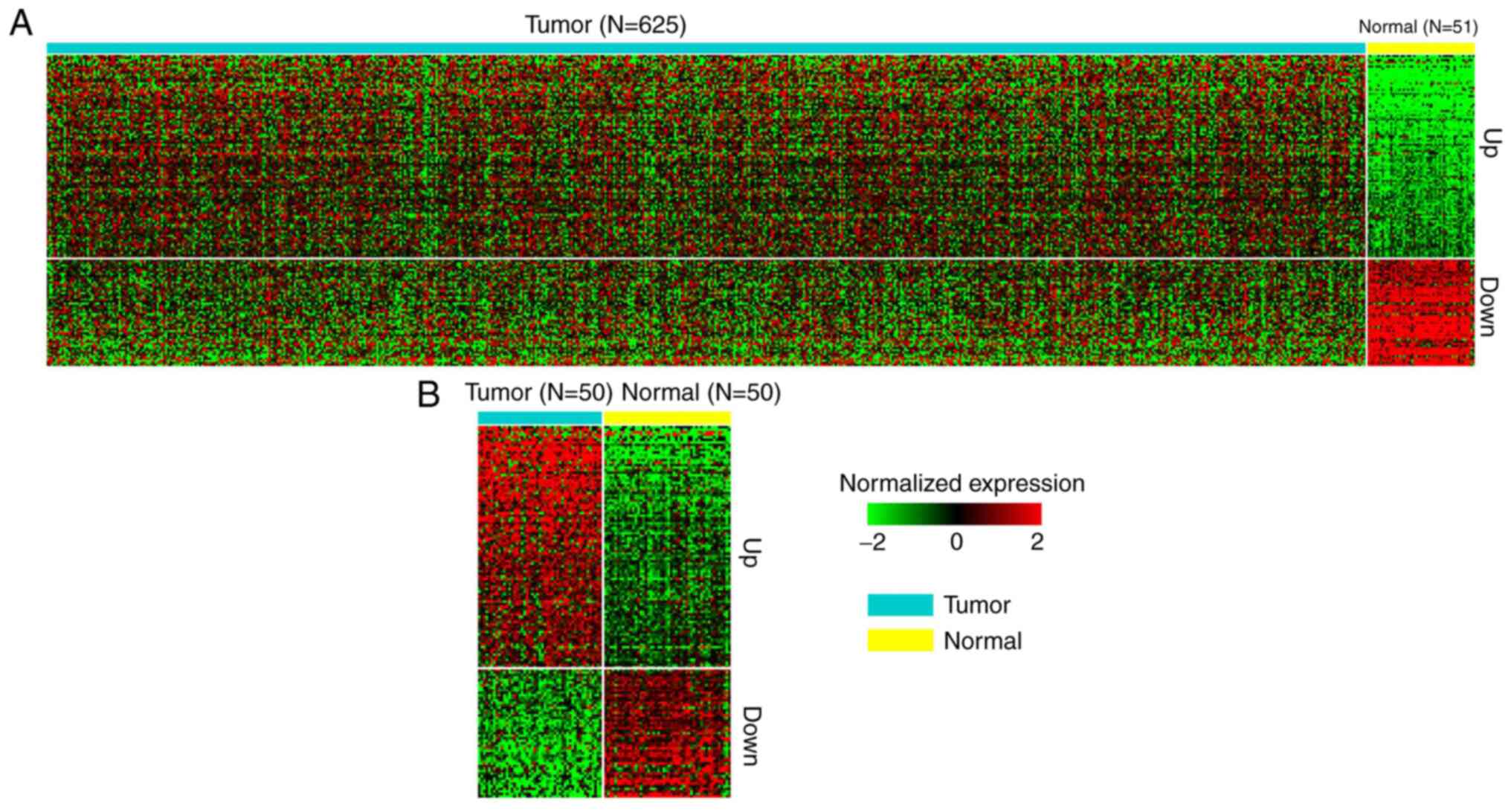
To identify the dysregulated lincRNAs in CRC,
RNA-Seq data from 647 tumor and 51 normal (including 50 pairs) CRC
samples from TCGA datasets were analyzed. Log2(FC) of
lincRNA expression in tumor vs. normal >1 was classified as an
upregulated gene, while <-1 was classified as a downregulated
gene. In total, 7,369 lincRNAs were identified from the datasets.
Subsequently, 96 upregulated lincRNAs and 51 downregulated lincRNAs
were identified. Notably, these lincRNAs were also dysregulated in
50 tumor and adjacent normal samples (Fig. 1). These 147 lincRNAs were
subsequently used to identify significant prognostic
predictions.
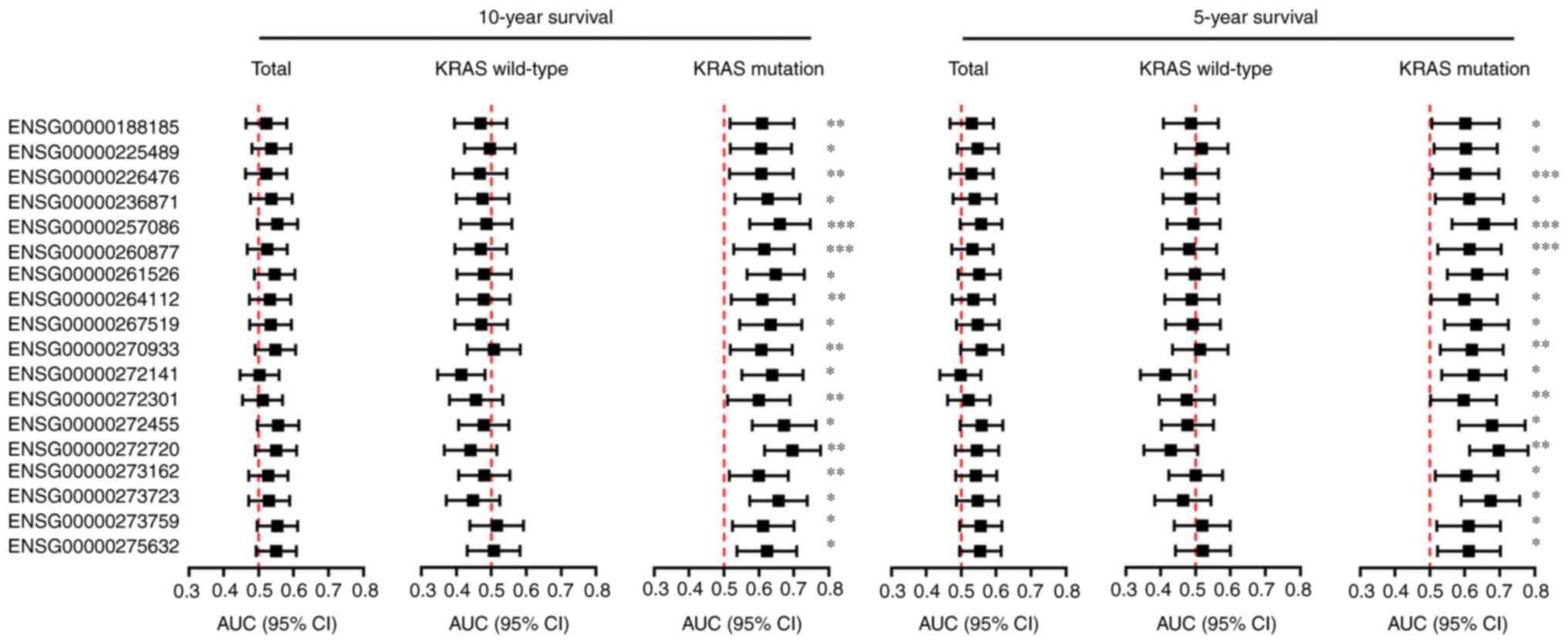
Identification of independent
prognostic lincRNAs with KRAS-mutant CRC
Patient 5- or 10-year survival rates are commonly
used to represent statistical cure rates for those with cancer
(23). Among the 647 tumor samples,
586 were identified as having 10-year survival data, and 545 were
identified as having 5-year survival data. To determine whether the
147 dysregulated lincRNAs influencing the survival of patients with
CRC depended on KRAS mutations, 217 and 198 samples were identified
as having a KRAS mutation at 10 and 5 years, respectively. Using
ROC curve analyses, 18 lincRNAs were identified as independent
prognostic markers in mutant KRAS, rather than wild-type KRAS
clinical samples (Fig. 2). These 18
lincRNAs were used to identify key lincRNAs with KRAS
mutations.
Identification of key prognostic
lincRNAs with KRAS mutations
Random data analysis was performed using robust
likelihood-based modeling 1,000 times. Statistical frequency
analysis of the significantly changed lincRNAs in KRAS-mutant
samples suggested that all the selected lincRNAs had a high
frequency. Two lincRNAs, LINC00265 (Gene Stable ID,
ENSG00000188185) and AL390719.2 (Gene Stable ID, ENSG00000272141),
were identified as key prognostic lincRNAs with KRAS mutations at
both 10 years (Table I) and 5 years
(Table II). LINC00106 was only
significant in the 5-years survival group.
 | Table I.Identification of key survival
lincRNAs with KRAS mutations in 10-year survival. |
Table I.
Identification of key survival
lincRNAs with KRAS mutations in 10-year survival.
| Gene stable ID | Symbols | Nloglik | AIC |
|---|
|
ENSG00000226476 | LINC01748 | 227.81 | 455.62 |
|
ENSG00000188185 | LINC00265 | 224.88 | 451.76a |
|
ENSG00000272141 | AL390719.2 | 221.27 | 446.54a |
|
ENSG00000275632 | AL035461.2 | 221.01 | 448.03 |
|
ENSG00000273759 | AL117379.1 | 220.85 | 449.70 |
|
ENSG00000261526 | AC012615.1 | 219.33 | 450.65 |
|
ENSG00000267519 | AC020916.1 | 219.31 | 448.62 |
|
ENSG00000272455 | AL391244.3 | 217.74 | 451.49 |
|
ENSG00000272301 | AP002360.3 | 217.73 | 449.45 |
|
ENSG00000270933 | AC010719.1 | 217.70 | 453.41 |
|
ENSG00000264112 | AC015813.1 | 217.68 | 455.36 |
|
ENSG00000236871 | LINC00106 | 213.99 | 451.98 |
|
ENSG00000257086 | AP001453.4 | 213.99 | 449.98 |
|
ENSG00000260877 | AP005233.2 | 212.96 | 451.93 |
|
ENSG00000225489 | AL354707.1 | 211.69 | 451.37 |
|
ENSG00000273162 | AL133215.2 | 211.31 | 452.62 |
|
ENSG00000272720 | AL022322.1 | 211.30 | 454.60 |
|
ENSG00000273723 | AL139089.1 | 210.64 | 455.28 |
 | Table II.Identification of key survival
lincRNAs with KRAS mutations in 5-year survival. |
Table II.
Identification of key survival
lincRNAs with KRAS mutations in 5-year survival.
| Gene stable ID | Symbols | Nloglik | AIC |
|---|
|
ENSG00000226476 | LINC01748 | 208.3 | 416.61 |
|
ENSG00000188185 | LINC00265 | 204.93 | 411.85a |
|
ENSG00000236871 | LINC00106 | 201.40 | 406.80a |
|
ENSG00000272141 | AL390719.2 | 199.35 | 404.69a |
|
ENSG00000272720 | AL022322.1 | 198.76 | 405.52 |
|
ENSG00000275632 | AL035461.2 | 198.65 | 407.29 |
|
ENSG00000272301 | AP002360.3 | 198.24 | 408.49 |
|
ENSG00000270933 | AC010719.1 | 197.64 | 411.29 |
|
ENSG00000273759 | AL117379.1 | 197.64 | 409.29 |
|
ENSG00000264112 | AC015813.1 | 197.36 | 412.73 |
|
ENSG00000225489 | AL354707.1 | 196.60 | 413.20 |
|
ENSG00000273723 | AL139089.1 | 196.59 | 415.17 |
|
ENSG00000260877 | AP005233.2 | 192.71 | 409.42 |
|
ENSG00000267519 | AC020916.1 | 192.12 | 410.25 |
|
ENSG00000272455 | AL391244.3 | 191.65 | 411.29 |
|
ENSG00000261526 | AC012615.1 | 191.49 | 412.99 |
|
ENSG00000257086 | AP001453.4 | 188.47 | 408.94 |
|
ENSG00000273162 | AL133215.2 | 188.44 | 410.88 |
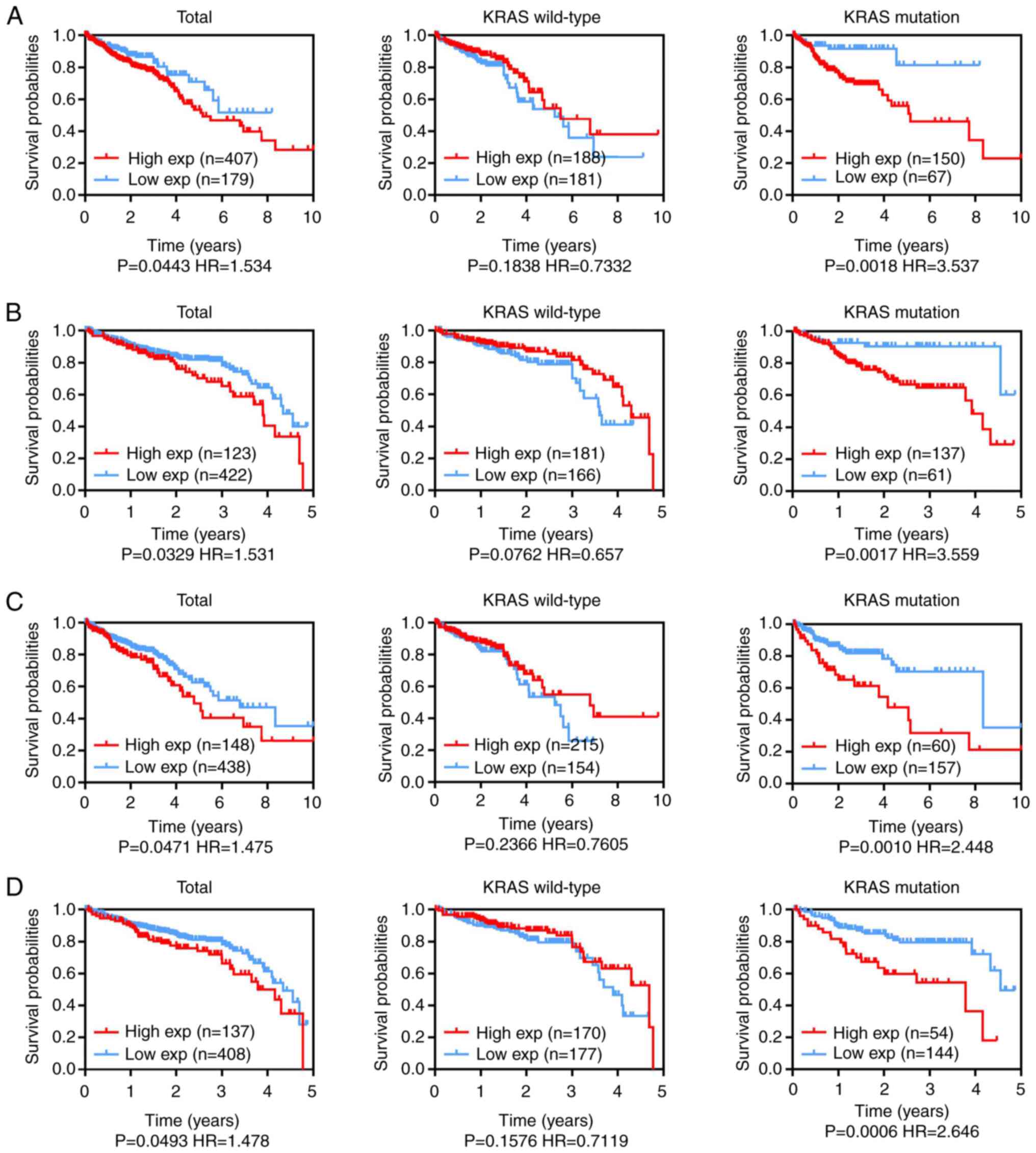
Oncogenicity of key prognostic
lincRNAs with KRAS mutations
To examine whether the expression level of the key
lincRNAs LINC00265 and AL390719.2 was correlated with a less
favorable prognosis in CRC with KRAS mutations, 10- and 5-year
overall survival (OS) rates were analyzed in the CRC TCGA dataset.
To evaluate the clinical significance of the key lincRNAs LINC00265
and AL390719.2 in the survival of patients with CRC, as well as
their associations with the KRAS mutation status, the prognostic
significance of these lincRNAs was determined using TCGA datasets.
The 10- and 5-year OS rates suggested that LINC00265 (Fig. 3A and B) and AL390719.2 (Fig. 3C and D) expression levels were
associated with patient survival in CRC. High LINC00265 (Fig. 3A and B) and AL390719.2 (Fig. 3C and D) expression was significantly
associated with less favorable survival in patients with CRC with
mutant KRAS, but not in those with wild-type KRAS (Fig. 3). Hence, LINC00265 and AL390719.2
upregulation specifically predicts a poor prognosis and represents
an independent prognostic marker in KRAS-mutant CRC.
To investigate the expression of the key lincRNAs
LINC00265 and AL390719.2, the pan-cancer expression levels and CRC
CNV profiles of these lincRNAs were analyzed in TCGA dataset.
LINC00265 and AL390719.2 were identified as being expressed in
cancer, including CRC (Fig. 4A and
B). It was observed that LINC00265 CNV was amplified in 81.54%
of CRC samples (Fig. 4C). LINC00265
expression was significantly higher in CRC with CNV amplification,
and expression showed a positive correlation with CNV amplification
(Fig. 4D). AL390719.2 expression was
also significantly higher in CRC with CNV amplification (Fig. 4E), but CNV amplification was detected
in 32.03% of CRC samples, and AL390719.2 expression was not
associated with CNV amplification (Fig.
4F).
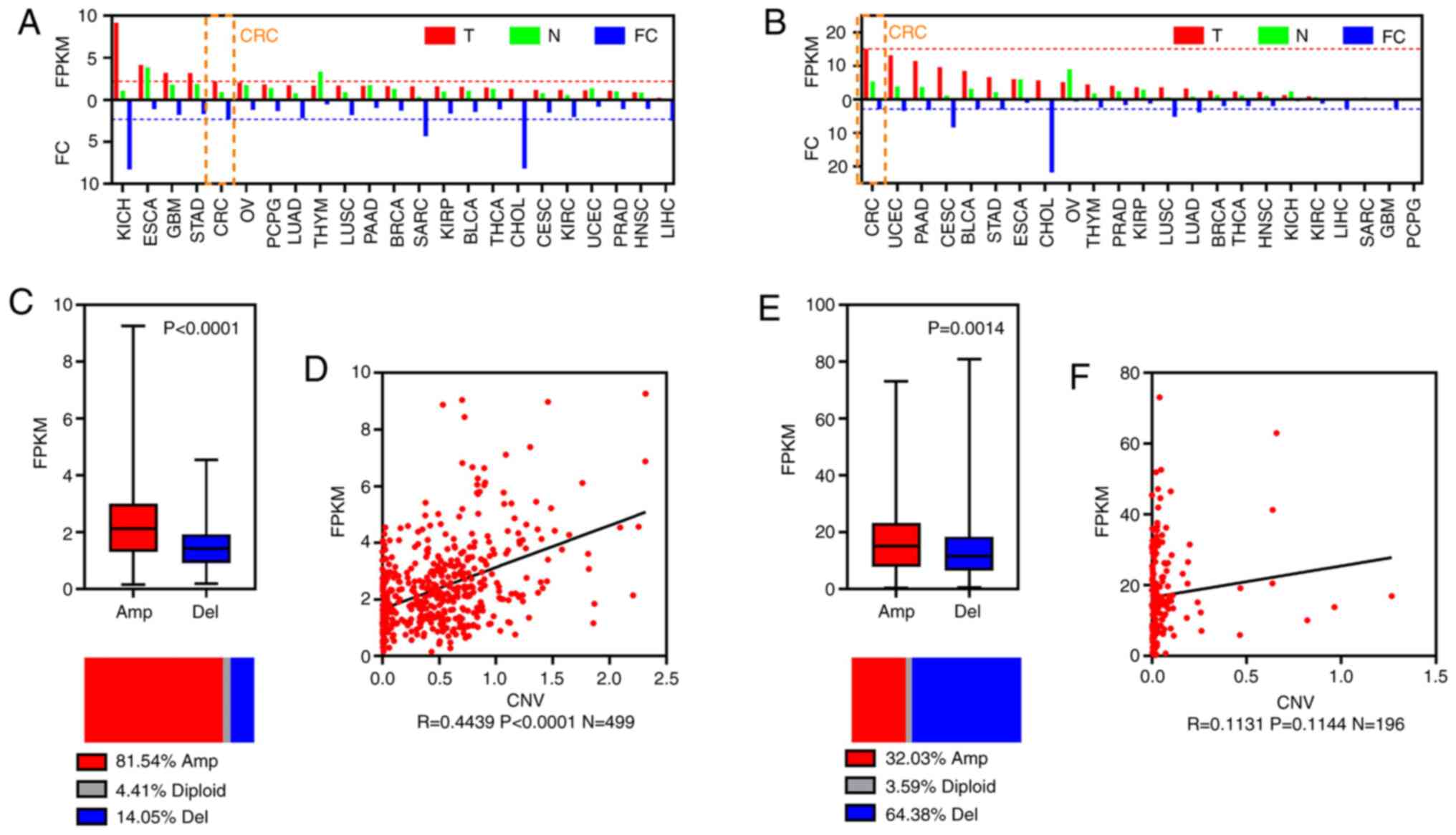 | Figure 4.Oncogenicity of two key prognostic
lincRNAs. Bar plots of FPKM and FC for (A) LINC00265 and (B)
AL390719.2. (C) Boxplot showing FPKM (top) and horizontal slices
showing the percentage group by CNV amplification and deletion of
LINC00265. (D) Scatter diagram showing the correlation between CNV
and FPKM for LINC00265. (E) Boxplot showing FPKM and horizontal
slices showing the percentage group by CNV amplification and
deletion of AL390719.2. (F) Scatter diagram showing the correlation
between CNV and FPKM for AL390719.2. CRC, colorectal cancer;
lincRNA, long intergenic non-coding RNA; FC, fold-change; FKPM,
fragments per kilobase million; CNV, copy number variation; T,
tumor; N, normal; Amp, amplification; Del, deletion; BLCA, bladder
urothelial carcinoma, BRCA, breast invasive carcinoma, CESC,
cervical squamous cell carcinoma and endocervical adenocarcinoma,
CHOL, cholangiocarcinoma, CRC, colorectal cancer, ESCA, esophageal
carcinoma, GBM, glioblastoma multiforme, HNSC, head and neck
squamous cell carcinoma, KICH, kidney chromophobe, KIRC, kidney
renal clear cell carcinoma, KIRP, kidney renal papillary cell
carcinoma, LIHC, liver hepatocellular carcinoma, LUAD, lung
adenocarcinoma, LUSC, lung squamous cell carcinoma, OV, ovarian
serous cystadenocarcinoma, PAAD, pancreatic adenocarcinoma, PCPG,
pheochromocytoma and paraganglioma, PRAD, prostate adenocarcinoma,
SARC, sarcoma, STAD, stomach adenocarcinoma, THCA, thyroid
carcinoma, THYM, thymoma, UCEC, uterine corpus endometrial
carcinoma. |
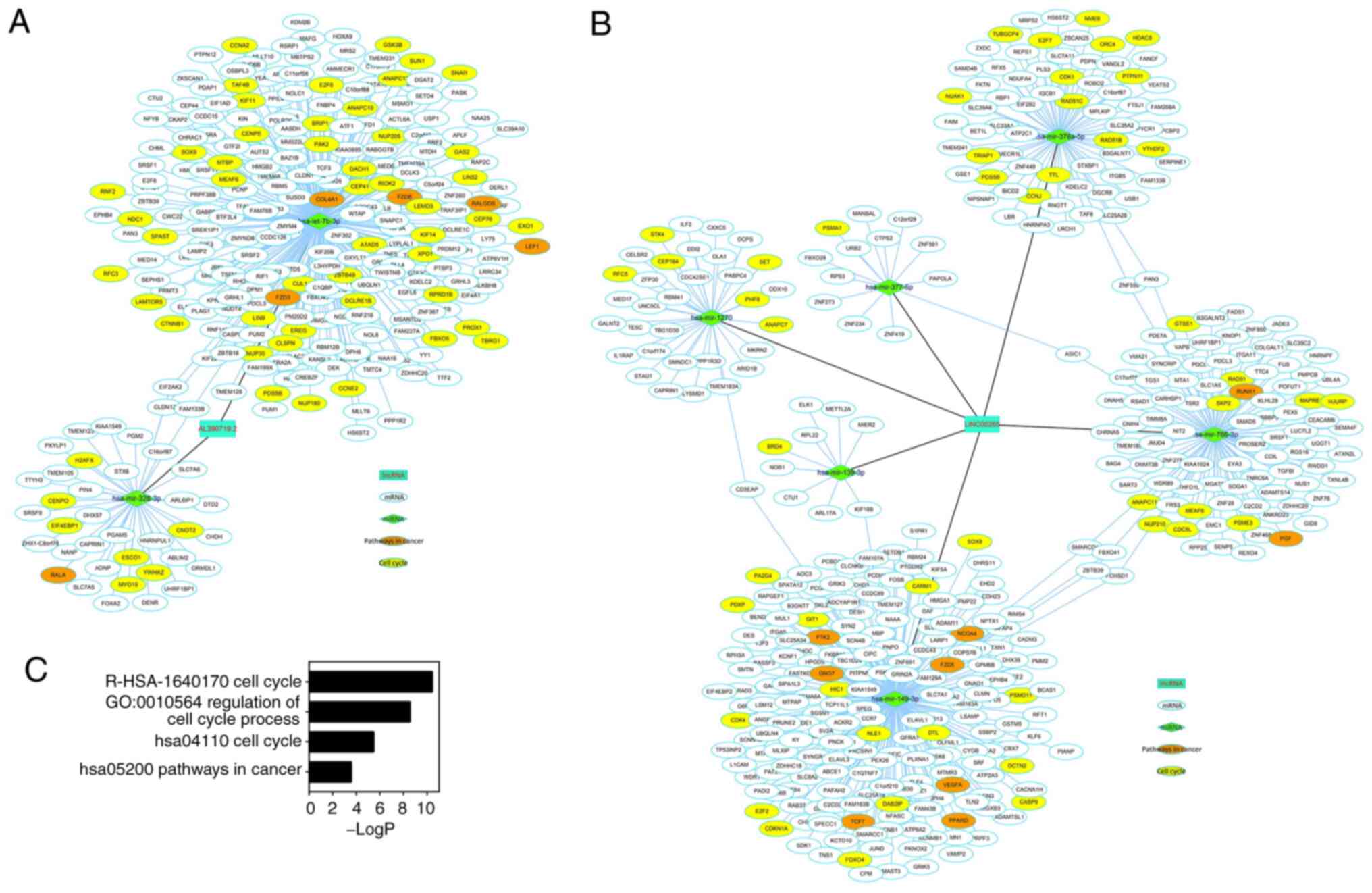
Regulatory modules of key prognostic
lincRNAs with KRAS mutations
miRNA-lincRNA and miRNA-mRNA interactions were
identified using tools from DIANA. PCC and MIC were used to
evaluate the correlation of these interactions. The downregulated
miRNAs were identified between tumor and normal samples with a
log2 (FC) value ≤-1 and FDR <0.05. Finally, 2 miRNAs
and 288 mRNAs connected to AL390719.2 (Fig. 5A) and 6 miRNAs and 415 mRNAs
connected to LINC00265 (Fig. 5B)
were identified. Functional enrichment analysis revealed that the
mRNAs in the regulatory modules may be critical for the cell cycle
in CRC (‘cell cycle’ from Reactome and KEGG, ‘regulation of cell
cycle process’ from GO and ‘pathways in cancer’ from KEGG; Fig. 5C). To validate these findings,
RT-qPCR was used to check lncRNAs, miRNAs and mRNAs (‘hsa05200
pathways in cancer’; orange nodes in Fig. 5 network) in 12 pairs of CRC samples.
RT-qPCR demonstrated that LINC00265, AL390719.2 and 12 genes were
upregulated in tumor samples, while 2 miRNAs were downregulated in
tumor samples (Fig. 6A and C).
Furthermore, RT-qPCR demonstrated that these two miRNAs were
negatively correlated with lncRNAs and mRNAs (Fig. 6B and D).
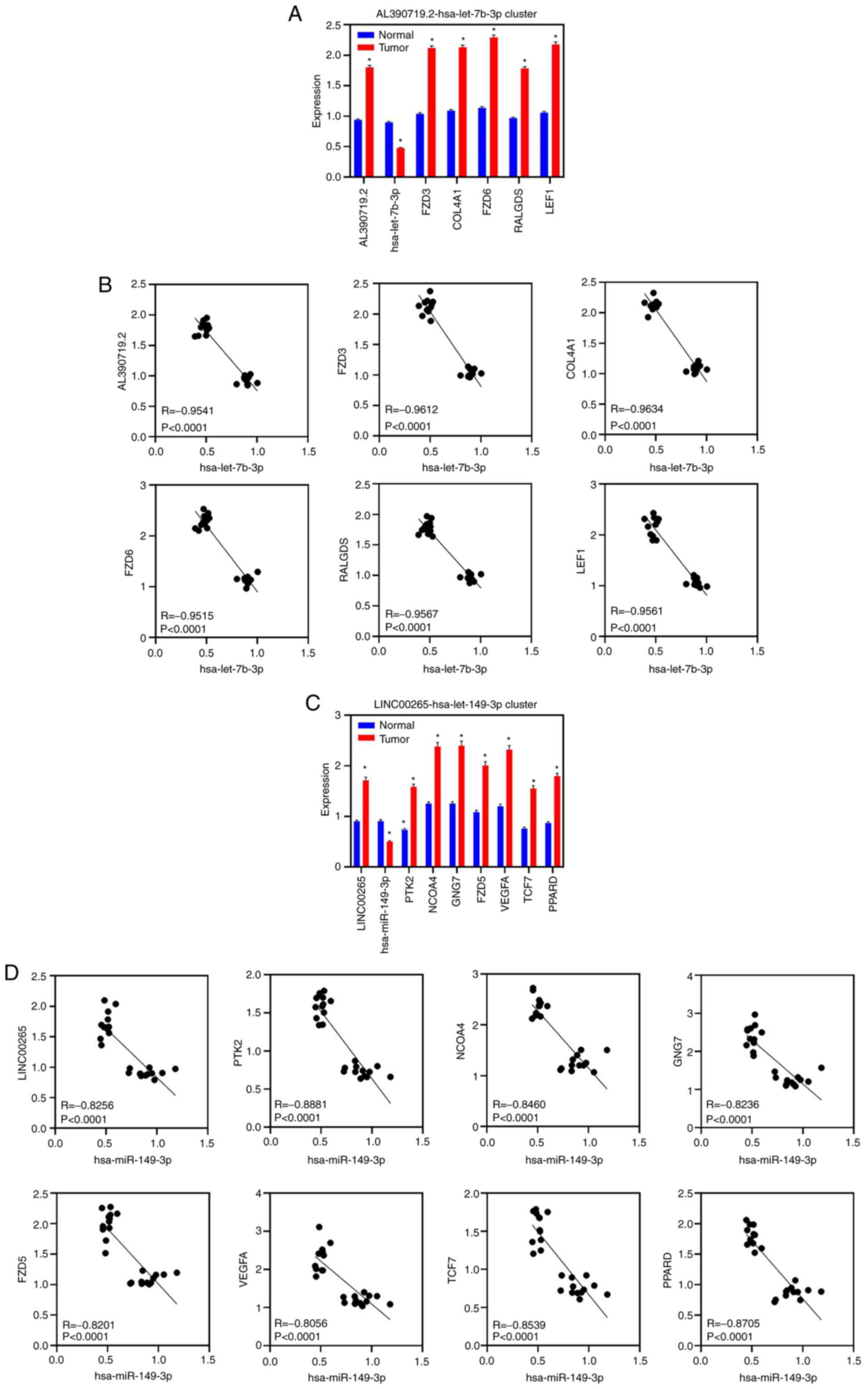 | Figure 6.RT-qPCR validation in CRC samples.
(A) RT-qPCR showing the expression of AL390719.2, hsa-let-7b-3p,
FZD3, COL4A1, FZD6, RALGDS and LEF1 in 12 paired CRC and normal
samples. Data are represented as the mean ± SEM. (B) Scatter
diagram showing the correlation between hsa-let-7b-3p and
AL390719.2, FZD3, COL4A1, FZD6, RALGDS and LEF1 in 12 paired CRC
and normal samples using RT-qPCR. (C) RT-qPCR showing the
expression of LINC00265, hsa-mir-149-3p, PTK2, NCOA4, GNG7, FZD5,
VEGFA, TCF7 and PPARD in 12 paired CRC and normal samples. Data are
presented as the mean ± SEM. (D) Scatter diagram showing the
correlation between hsa-mir-149-3p and LINC00265, PTK2, NCOA4,
GNG7, FZD5, VEGFA, TCF7 and PPARD in 12 paired CRC and normal
samples by RT-qPCR. *P<0.05 (tumor vs. normal) CRC, colorectal
cancer; lincRNA, long intergenic non-coding RNA; RT-qPCR, reverse
transcription-quantitative PCR; miR, microRNA. |
Discussion
CRC is now the third most common malignancy
worldwide. Oncogenic KRAS mutations initiate and sustain CRC
progression. Numerous studies have assessed KRAS mutations
associated with CRC outcomes (24–26). To
provide prognostic lincRNAs to predict the outcomes of patients
with CRC with KRAS mutations, 18 lincRNAs (LINC00265, AL390719.2,
AL035461.2, AL117379.1, AC012615.1, AC020916.1, AL391244.3,
AP002360.3, AC010719.1, AC015813.1, LINC00106, AP001453.4,
AP005233.2, AL354707.1, AL133215.2, AL022322.1, AL139089.1 and
LINC01748) were identified as independent prognostic lincRNAs in
CRC with KRAS mutations. All these lincRNAs are upregulated in
primary CRC tumors, and their increased expression is associated
with a poor prognosis in patients with CRC with KRAS mutations. The
expression levels of these lincRNAs were correlated with 5- and
10-year OS rates in patients with CRC. Furthermore, these 18
lincRNAs were independent prognostic markers in patients with CRC
with mutant KRAS, but not in those with wild-type KRAS. The
aforementioned results suggest that these lincRNAs may serve as
prognostic biomarkers in CRC and correlate with CRC
progression.
Robust likelihood-based survival models were used to
identify two key lincRNAs, LINC00265 and AL390719.2, from 18
lincRNAs. All 18 lincRNAs were first identified as dysregulated and
correlated with poor prognosis with KRAS mutations in human cancer.
LINC00265 was reported to be differentially expressed and revealed
to be a prognostic biomarker in the lung adenocarcinoma (LUAD) TCGA
dataset (27). LINC00265 was also
demonstrated to be upregulated in LUAD samples from TCGA. It was
revealed that LINC00265 expression in CRC tumors was significantly
associated with CNV amplification. Notably, CNV amplification was
reported as an upstream mechanism to increase gene expression
(28,29). The present results indicated that CNV
amplification may cause LINC00265 overexpression in CRC. In
addition, AL390719.2 was shown to be highly expressed in CRC
tumors, but its increased expression was not caused by CNV
amplification. Furthermore, Kaplan-Meier survival curves revealed
that LINC00265 and AL390719.2 expression was associated with 5- and
10-year OS rates. High LINC00265 and AL390719.2 expression was
correlated with less favorable 5- and 10-year OS rates in patients
with mutant KRAS, but not in those with wild-type KRAS. Hence,
LINC00265 and AL390719.2, as key prognostic lincRNAs in KRAS-mutant
CRC, may be used to predict survival for patients with CRC.
To identify the function and molecular mechanism of
LINC00265 and AL390719.2, it was determined whether these two key
lincRNAs, as ceRNAs bound to miRNAs, regulate important genes. It
was revealed that 2 miRNAs and 288 mRNAs were associated with
AL390719.2, and that 6 miRNAs and 415 mRNAs were associated with
LINC00265. Notably, functional enrichment analysis suggested that
mRNAs in the regulatory modules were enriched in cell cycle
biological processes. LINC00265 and AL390719.2 may serve as ceRNAs
to bind these miRNAs and prevent the inhibitory effect of miRNAs on
cell cycle genes, resulting in the upregulation of cell cycle genes
and promoting CRC progression. AL390719.2, as a ceRNA, binds with
hsa-mir-328-3p and hsa-let-7b-3p. LINC00265, as a ceRNA, binds to
hsa-mir-1270, hsa-mir-139-3p, hsa-mir-149-3p, hsa-mir-377-5p,
hsa-mir-378a-5p and hsa-mir-766-3p. These miRNAs were downregulated
in CRC samples from TCGA. In addition, hsa-let-7b-3p,
hsa-mir-328-3p, hsa-miR-139-3p, hsa-mir-149-3p and hsa-miR-378a-5p
have been reported to be downregulated in CRC (30–34).
Certain miRNAs have been reported to impact the cell cycle in human
cancer. For example, hsa-let-7b has been reported to impact the
cell cycle to inhibit prostate cancer cell proliferation in
vitro (35). Moreover,
hsa-mir-149 directly regulates the expression of cyclin-dependent
kinase 4 (CDK4) cell lines, and hsa-mir-149 overexpression results
in G0-G1 arrest and cell death in CRC
(36). Also, CDK1 was the target
gene of hsa-mir-378a-5p, and hsa-mir-378a-5p decreased CDK1
expression in hepatocytes (37).
CDK1 is critical for regulating the G2-M transition
during cell cycle progression (38).
CDK4 controls cell cycle progression via pocket proteins and E2F
transcription factors, and is correlated with cancer development
and progression (39). Therefore,
LINC00265 and AL390719.2 may serve as ceRNAs through competitive
interactions with these miRNAs, resulting in the low expression of
these miRNAs followed by upregulation of cell cycle genes in CRC.
Moreover, RT-qPCR was used to validate LINC00265, AL390719.2 and
related miRNAs and mRNAs in CRC samples.
In conclusion, 18 lincRNAs were identified that were
upregulated and have potential as independent prognostic markers in
CRC with KRAS mutations. From these lincRNAs, LINC00265 and
AL390719.2 were identified as key lincRNAs that serve important
roles in CRC progression, as ceRNAs for the regulation of
lincRNA-miRNA-mRNA networks. The present findings may provide novel
prognostic markers and therapeutic targets for CRC. However, the
investigation of the regulatory models among these genes remains
necessary.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed during the
current study are available in The Cancer Genome Atlas [https://portal.gdc.cancer.gov].
Authors' contributions
JX conceptualized the present study, designed the
research and performed bioinformatics analysis. JX and QYH
performed the experiments. JX and CJG analyzed and interpreted the
data. JX drafted and edited the manuscript, and QYH supervised the
project. JX, QY and CJG confirmed the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First College of Clinical Medical Science, China
Three Gorges University (Yichang, China; approval no.
HEC-KYJJ-2019-056-01). Written informed consent was provided by all
patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nasseri Y and Langenfeld SJ: Imaging for
colorectal cancer. Surg Clin North Am. 97:503–513. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mármol I, Sánchez-de-Diego C, Pradilla
Dieste A, Cerrada E and Rodriguez Yoldi MJ: Colorectal carcinoma: A
general overview and future perspectives in colorectal cancer. Int
J Mol Sci. 18:1972017. View Article : Google Scholar
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Patel SG and Ahnen DJ: Colorectal cancer
in the young. Curr Gastroenterol Rep. 20:152018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Passiglia F, Bronte G, Bazan V, Galvano A,
Vincenzi B and Russo A: Can KRAS and BRAF mutations limit the
benefit of liver resection in metastatic colorectal cancer
patients? A systematic review and meta-analysis. Crit Rev Oncol
Hematol. 99:150–157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Walther A, Johnstone E, Swanton C, Midgley
R, Tomlinson I and Kerr D: Genetic prognostic and predictive
markers in colorectal cancer. Nat Rev Cancer. 9:489–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kaelin WG Jr: The concept of synthetic
lethality in the context of anticancer therapy. Nat Rev Cancer.
5:689–698. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mattick JS and Rinn JL: Discovery and
annotation of long noncoding RNAs. Nat Struct Mol Biol. 22:5–7.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Talyan S, Andrade-Navarro MA and Muro EM:
Identification of transcribed protein coding sequence remnants
within lincRNAs. Nucleic Acids Res. 46:8720–8729. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous rna
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y
and Chen Y: The emerging function and mechanism of ceRNAs in
cancer. Trends Genet. 32:211–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX,
Wu ZH, Chen XW, Gao P, Miao ZF and Wang ZN: Non-coding RNAs
participate in the regulatory network of CLDN4 via ceRNA mediated
miRNA evasion. Nat Commun. 8:2892017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wong CC, Qian Y, Li X, Xu J, Kang W, Tong
JH, To KF, Jin Y, Li W, Chen H, et al: SLC25A22 promotes
proliferation and survival of colorectal cancer cells With KRAS
mutations and xenograft tumor progression in mice via intracellular
synthesis of aspartate. Gastroenterology. 151:945–960.e6. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song Y, Chen QT and He QQ: Identification
of key transcription factors in endometrial cancer by systems
bioinformatics analysis. J Cell Biochem. 120:15443–15454. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reshef DN, Reshef YA, Finucane HK,
Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M and
Sabeti PC: Detecting novel associations in large data sets.
Science. 334:1518–1524. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tripathi S, Pohl MO, Zhou Y,
Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che
J, Mulder LC, et al: Meta- and orthogonal integration of influenza
‘OMICs’ data defines a role for UBR4 in virus budding. Cell Host
Microbe. 18:723–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng Y, Fang YC and Li J: PD-L1
expression levels on tumor cells affect their immunosuppressive
activity. Oncol Lett. 18:5399–5407. 2019.PubMed/NCBI
|
|
23
|
Tai P, Yu E, Cserni G, Vlastos G, Royce M,
Kunkler I and Vinh-Hung V: Minimum follow-up time required for the
estimation of statistical cure of cancer patients: Verification
using data from 42 cancer sites in the SEER database. BMC Cancer.
5:482005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Afrăsânie VA, Marinca MV, Alexa-Stratulat
T, Gafton B, Păduraru M, Adavidoaiei AM, Miron L and Rusu C: KRAS,
NRAS, BRAF, HER2 and microsatellite instability in metastatic
colorectal cancer-practical implications for the clinician. Radiol
Oncol. 53:265–274. 2019. View Article : Google Scholar
|
|
25
|
Cicenas J, Tamosaitis L, Kvederaviciute K,
Tarvydas R, Staniute G, Kalyan K, Meskinyte-Kausiliene E,
Stankevicius V and Valius M: KRAS, NRAS and BRAF mutations in
colorectal cancer and melanoma. Med Oncol. 34:262017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Porru M, Pompili L, Caruso C, Biroccio A
and Leonetti C: Targeting KRAS in metastatic colorectal cancer:
Current strategies and emerging opportunities. J Exp Clin Cancer
Res. 37:572018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z
and Zhang LW: Identification of key long non-coding RNAs as
competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma.
Eur Rev Med Pharmacol Sci. 20:2285–2295. 2016.PubMed/NCBI
|
|
28
|
Liu J, Kruswick A, Dang H, Tran AD, Kwon
SM, Wang XW and Oberdoerffer P: Ubiquitin-specific protease 21
stabilizes BRCA2 to control DNA repair and tumor growth. Nat
Commun. 8:1372017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui K, Liu C, Li X, Zhang Q and Li Y:
Comprehensive characterization of the rRNA metabolism-related genes
in human cancer. Oncogene. 39:786–800. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li X, Li B, Ran P and Wang L:
Identification of ceRNA network based on a RNA-seq shows prognostic
lncRNA biomarkers in human lung adenocarcinoma. Oncol Lett.
16:5697–5708. 2018.PubMed/NCBI
|
|
31
|
Xu XT, Xu Q, Tong JL, Zhu MM, Nie F, Chen
X, Xiao SD and Ran ZH: MicroRNA expression profiling identifies
miR-328 regulates cancer stem cell-like SP cells in colorectal
cancer. Br J Cancer. 106:1320–1330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shen K, Liang Q, Xu K, Cui D, Jiang L, Yin
P, Lu Y, Li Q and Liu J: MiR-139 inhibits invasion and metastasis
of colorectal cancer by targeting the type I insulin-like growth
factor receptor. Biochem Pharmacol. 84:320–330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo H, Hu X, Ge S, Qian G and Zhang J:
Regulation of RAP1B by miR-139 suppresses human colorectal
carcinoma cell proliferation. Int J Biochem Cell Biol.
44:1465–1472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang F, Ma YL, Zhang P, Shen TY, Shi CZ,
Yang YZ, Moyer MP, Zhang HZ, Chen HQ, Liang Y and Qin HL: SP1
mediates the link between methylation of the tumour suppressor
miR-149 and outcome in colorectal cancer. J Pathol. 229:12–24.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu C, Kelnar K, Vlassov AV, Brown D, Wang
J and Tang DG: Distinct microRNA expression profiles in prostate
cancer stem/progenitor cells and tumor-suppressive functions of
let-7. Cancer Res. 72:3393–3404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lulla AR, Slifker MJ, Zhou Y, Lev A,
Einarson MB, Dicker DT and El-Deiry WS: miR-6883 family miRNAs
target CDK4/6 to induce G1 phase cell-cycle arrest in
colon cancer cells. Cancer Res. 77:6902–6913. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang R, Wei M, Yang F, Sheng Y and Ji L:
Diosbulbin B induced G2/M cell cycle arrest in
hepatocytes by miRNA-186-3p and miRNA-378a-5p-mediated the
decreased expression of CDK1. Toxicol Appl Pharmacol. 357:1–9.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Spiller F, Medina-Pritchard B, Abad MA,
Wear MA, Molina O, Earnshaw WC and Jeyaprakash AA: Molecular basis
for Cdk1-regulated timing of Mis18 complex assembly and CENP-A
deposition. EMBO Rep. 18:894–905. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lopez-Mejia IC, Lagarrigue S, Giralt A,
Martinez-Carreres L, Zanou N, Denechaud PD, Castillo-Armengol J,
Chavey C, Orpinell M, Delacuisine B, et al: CDK4 phosphorylates
AMPKα2 to inhibit its activity and repress fatty acid oxidation.
Mol Cell. 68:336–349.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|